Abstract
Background
The survival of glioblastoma patients is poor. Median survival after diagnosis is 15 months, despite treatment involving surgical resection, radiotherapy, and/or temozolomide chemotherapy. Identification of novel targets and stratification strategies of glioblastoma patients to improve patient survival is urgently needed. Whole-genome sequencing (WGS) is the most comprehensive means to identify such DNA-level targets. We report a unique set of WGS samples along with comprehensive analyses of the glioblastoma genome and potential clinical impact of WGS.
Methods
Our cohort consisted of 42 glioblastoma tumor tissue and matched whole-blood samples, which were whole-genome sequenced as part of the CPCT-02 study. Somatic single-nucleotide variants, small insertions/deletions, multi-nucleotide variants, copy-number alterations (CNAs), and structural variants were analyzed. These aberrations were harnessed to investigate driver genes, enrichments in CNAs, mutational signatures, fusion genes, and potential targeted therapies.
Results
Tumor mutational burden (TMB) was similar to other WGS efforts (1–342 mutations per megabase pair). Mutational analysis in low TMB samples showed that the age-related CpG demethylation signature was dominant, while hyper- and ultramutated tumors had additional defective DNA mismatch repair signatures and showed microsatellite instability in their genomes. We detected chromothripsis in 24% of our cohort, recurrently on chromosomes 1 and 12. Recurrent noncoding regions only resulted in TERT promoter variants. Finally, we found biomarkers and potentially druggable changes in all but one of our tumor samples.
Conclusions
With high-quality WGS data and comprehensive methods, we identified the landscape of driver gene events and druggable targets in glioblastoma patients.
Keywords: genomics, glioblastoma, precision medicine, whole-genome sequencing
Key Points.
Mutational signatures varied greatly in low and hyper- or ultramutated tumors.
Chromothripsis was found in 24% of our patients, repeatedly on chromosomes 1 and 12.
We found biomarkers and potentially druggable targets in nearly all our tumor samples.
Importance of the Study.
We characterized the genomes of 42 glioblastoma samples. By utilizing whole-genome sequencing data, identification of structural variants, copy-number alterations, mutational signatures, catastrophic events, and microsatellite instability is much more accurate than based on panel-based or whole-exome data. We compared our results with previously published glioblastoma studies (panel-based, whole-exome sequencing, and whole-genome sequencing) and showed the heterogeneity through concordant (mutational burden) and discordant (chromothripsis, prevalence of fusion genes) findings. By linking our molecular data with current clinical practice and trials, we show researchers and neuro-oncologists the frequencies, biomarkers, and potential targeted treatment options for glioblastoma patients.
Glioblastoma is the most common and aggressive malignant primary brain tumor in adults. Patients have a median survival of fewer than 15 months after diagnosis, and standard treatment consists primarily of surgical resection and subsequent radiation therapy with concurrent and adjuvant temozolomide chemotherapy.1–3 The short survival is due to the aggressiveness of the tumor, its infiltrative nature and therefore cannot be resected completely, and its refractoriness to treatments, which may be due to intratumoral heterogeneity of glioblastoma.4
Research of the genomic landscape of glioblastomas has primarily focused on changes in the coding regions of genes. These studies revealed mutations in common cancer markers such as TP53 and PTEN, and glioblastoma-associated hallmarks as TERT promoter, IDH1 and PDGFRA mutations, MGMT promoter methylation, EGFR amplification and mutations, and homozygous deletion of the CDKN2A/CDKN2B locus.5 However, as only a fraction of the entire genome is protein coding, genetic changes outside the coding genome require further investigation in relation to driving glioblastoma. By assessing a patient’s entire cancer genome, tumor mutational burden (TMB) can be quantified more precisely compared to panel- and whole-exome sequencing (WES)-based data, optimizing cutoffs for hypermutation and potential immunotherapy susceptibility.6 Analyzing noncoding regions may provide new insights into tumor biology and may point toward new directions for diagnosis and treatments.7 Moreover, detection of structural variants (SVs), copy-number alterations (CNAs), microsatellite instability (MSI), and common mutational patterns are more uniformly covered and more accurate using whole-genome sequencing (WGS) compared to the commonly used exome and panel sequencing technologies. Whole-genome analyses can lead to the identification of novel treatment targets, which may ultimately improve the standard treatment modalities for glioblastoma patients.
We therefore investigated WGS data from 42 glioblastoma tumor tissue samples that were sequenced as part of the CPCT-02 study.8 Here, we show the genomic characterization of the CPCT-02 glioblastoma cohort, compare our data to other WGS datasets, and discuss which genetic characteristics are potential targets for current or novel treatments. In the end, we remodel our genetic insights into clinically relevant hypotheses which could be applied in precision medicine strategies to increase survival at an individual patient level.
Methods
Center for Personalized Medicine: Patient cohort, study procedures, sample collection
Details about the study protocol, sampling, and sequencing have recently been discussed by Priestley et al.8 and van Dessel et al.9 This study and its protocol were approved by the medical ethical committee (METC) of the University Medical Center Utrecht, as well as the Netherlands Cancer Institute. In accordance with the Declaration of Helsinki, patients with glioblastoma provided their written informed consent for participation in the CPCT-02 (NCT01855477) clinical study before procedures. Core needle biopsies from the tumor lesion and peripheral blood samples were collected, along with clinical data in the same manner across Dutch hospitals.
Hartwig Medical Foundation: Whole genome sequencing and preprocessing
Tumor and whole-blood pairs were whole-genome sequenced on a HiSeqX system generating 2 × 150 base read pairs using standard settings (Illumina) at the Hartwig Medical Foundation central sequencing center. Patients included in this study were whole-genome sequenced between June 5, 2017 and September 14, 2018. Preprocessing was performed as described by Priestley et al.,8 as well as tumor profiling of 32 of these samples. Briefly, read pairs were mapped to the human reference genome (GRCh37) using BWA-mem,10 and subsequent variant calling was applied (see below). Next, several quality control or correction steps were performed systematically. SV calling was performed using the Genome Rearrangement IDentification Software Suite (GRIDSS).11,12 Computational ploidy estimation and copy-number (CN) analysis were performed using the custom pipeline PURPLE (PURity & PLoidy Estimator),8,12 estimating tumor purity and CN profile by combining B-allele frequency, read depth, and SVs. Gene fusion events were called using LINX12 (v1.11). MSI status was determined by assessing small insertions/deletions (InDels) per million bases in repeat regions with the tool MSISeq13 (MSI with a score >4).
Variant annotation and filtering.
—Somatic variants were determined using Strelka and provided by the Hartwig Medical Foundation as part of the data request. Somatic Variant Call Format files were annotated based on GRCh37 with HUGO gene symbols, HGVS notations, gnomAD14 frequencies, and COSMIC15 (9 733 455 entries) and dbSNP16 (686 600 501 entries) identifiers, using VEP17 (database release 95, merged cache) with setting ”—per_gene”. Exclusively somatic single-nucleotide variants (SNVs), small InDels, multi-nucleotide variants (MNVs) with ≥3 alternative read observations, and passing variant caller quality control were included in the analyses. Furthermore, population variants were removed to prevent germline leakage, based on the gnomAD database (v2.0.2): gnomAD exome (ALL) allele frequency ≥0.001 and gnomAD genome (ALL) ≥0.005. Variants specific for the Dutch CPCT cohort were removed based on a panel of normals from 1762 representative normal blood HMF samples. The most deleterious mutation was used to annotate the overlapping gene for each sample.
TMB calculation.
—The number of mutations per megabase pair was calculated as the amount of somatic genome-wide SNVs, MNVs, and InDels divided by the amount of callable nucleotides in the human reference genome (GRCh37) FASTA file:
Ploidy and copy-number analysis.
—Broad and focal somatic CN alterations were identified by GISTIC2.0 (v2.0.23), using the following parameters: genegistic 1, gcm extreme, maxseg 4000, broad 1, brlen 0.98, conf 0.95, rx 0, cap 3, saveseg 0, armpeel 1, smallmem 0, res 0.01, ta 0.1, td 0.1, savedata 0, savegene 1, and qvt 0.1. Distinction between shallow and deep CN events per region was based on thresholding performed by GISTIC2.0.18 The alterations are assigned a score that takes both the amplitude and the frequency of its occurrence across samples into account (G-score). Thresholding divides into 5 CN categories, 2 for deletions (−2 = deep, possibly homozygous loss, −1 = shallow, possibly heterozygous loss), 1 for diploid (0 = diploid), and 2 for amplifications (1 = few additional copies, often broad gain, 2 = more copies, often focal gain). Annotation of GISTIC2.0 peaks was performed as follows: (1) Wide peaks were annotated with all overlapping canonical UCSC genes within its limits. (2) Focal peaks were annotated based on overlapping genomic coordinates, using custom R scripts and UCSC gene annotations.
SV analysis.
—SVs were imported using custom R scripts, overlapping genes on at least one breakpoint, using GRCh37 genomic coordinates. SVs with an upstream or downstream tumor allele frequency (TAF) below 0.1 as determined by PURPLE and GRIDSS were discarded along with SVs covering all exons of a gene. In the case of both (multiple) mutations and/or SVs in the same gene, these were annotated as “multiple mutations.”
Fusion gene analysis.
—WGS-based TSV files were imported using R and overlapped with the 3 pillars of ChimerDB19; deep sequencing data (ChimerSeq), text mining of PubMed publications (ChimerPub), with extensive manual annotations (ChimerKB). Events that were not present in any pillar of ChimerDB and intragene fusions were filtered out. RNA-Seq-based fusion genes detected with Isofox (https://github.com/hartwigmedical/hmftools/tree/master/isofox) were imported using R and overlapped with the fusion events detected in the DNA sequencing.
Somatic driver genes analysis.
—We utilized the dN/dS model20 (192 Poisson rate parameters; under the full trinucleotide model) to identify genes undergoing mutational selection with the R package dndscv20 (v0.0.0.9). Both the substitution model and InDel model were used and were corrected for sequence composition, gene length, and mutational signatures. These models test the ratio between nonsynonymous (missense, nonsense, and essential splice site) and background (synonymous) mutations. To identify genes that drive selection, a q value of <0.05 (both including and excluding the InDel model) was used.
Mutational signatures analysis.
—Mutational signatures analysis was performed using the MutationalPatterns R package21 (v1.10.0). The mutational signatures based on single base substitutions (N = 90 v3 signatures), as established by Alexandrov et al.,22 were downloaded from COSMIC. SNVs were categorized according to their respective trinucleotide context (GRCh37) into a mutational spectrum matrix Mij (where i represents 1:96 trinucleotide contexts and j represents the number of 1:42 samples) and subsequently, a constrained linear combination of the 90 mutational signatures was constructed per sample using nonnegative least squares regression implemented in the R package pracma23 (v2.2.9).
Catastrophic mutational processes.
—Detection of chromothripsis (CT)-like events and localized hypermutation (kataegis) were performed as described in van Dessel et al.9 CT, also known as chromosomal shattering, followed by seemingly random re-ligation, was detected using Shatterseek (v0.4) with default settings. The following definition of CT was applied: (1) total number of intrachromosomal SVs involved in the event ≥25; (2) maximum number of oscillating CN segments (2 CN states) ≥7 or maximum number of oscillating CN segments (3 CN states) ≥14; (3) total size of CT event ≥20 Mb; (4) satisfying the test of equal distribution of SV types (P > .05); and (5) satisfying the test of nonrandom SV distribution within the cluster region or chromosome (P ≤ .05). Kataegis was detected using a piecewise constant fit method on SNVs for each chromosome separately. The resultant segments with a mean intermutational distance of ≤2000 bp and at least 5 SNVs were taken into account. Samples with more than 200 events were ignored, as these show global hypermutation instead of local.
Recurrent somatic variants.
—All filtered somatic mutations occurring in at least 4 samples were selected as recurrent. Noncoding constraint mutations (NCCMs; classified as “related to key GBM genes” and “select genes outside the key GBM genes”) retrieved from Sakthikumar et al.24 were compared to all variants by overlapping genomic ranges with the IRanges R package.25
Actionable targets.
—Iclusion (https://iclusion.com) data, which connect specific or gene-level aberrations to clinical cancer studies, while also integrating clinical interpretations from Precision Oncology Knowledge Base (OncoKB),26 Clinical Interpretation of Variants in Cancer (CIViC),27 and Cancer Genome Interpreter (CGI),28 were provided by Hartwig Medical foundation. These data were imported into R, attaching a label based on the actionability for each sample and gene. On-Label indicates treatment registered by federal authorities for glioblastoma, whereas Off-Label indicates a registration for other tumor types. The Off-Label data were filtered for cancer types fitting gliomas and solid tumors, excluding entries for chemotherapy (for its general applicability). All targets and biomarkers were overlapped with filtered molecular data to verify the presence and assessed by our neurooncological experts.
Hartwig Medical Foundation: RNA Sequencing
RNA was isolated from biopsy using the QIAsymphony RNA Kit (Qiagen) for tissue and quantified on the Qubit. A total of 50–100 ng of RNA was used as input for KAPA RNA HyperPrep Kit with RiboErase (Human/Mouse/Rat) library preparation (Roche) on an automated liquid handling platform (Beckman Coulter). RNA was fragmented (high temperature in the presence of magnesium) to a targeted fragment length of 300 bp. Barcoded libraries were sequenced as pools on either a NextSeq 500 (V2.5 reagents) generating 2 × 75 base read pairs or on a NovaSeq 6000 generating 2 × 150 base read pairs using standard settings (Illumina). BCL output from the sequencing platform was converted to FASTQ using Illumina’s bcl2fastq tool (versions 2.17–2.20) using default parameters. RNA-Seq data were aligned to GRCh37 using STAR29 to unsorted BAMs with chimeric reads included. Gene and transcript counts were generated and used for subsequent fusion detection using Isofox (https://github.com/hartwigmedical/hmftools/tree/master/isofox).
Results
Genomic Overview
We started off by characterizing the genomes of the 42 glioblastoma samples in our cohort. We detected 1 246 206 SNVs, 218 519 InDels, and 2224 MNVs in the coding regions of the genome (Table 1; Supplementary Figure 1). The types of SNV are unequally distributed (Supplementary Figure 1B). Transitions are, for instance, more frequent than transversions (Supplementary Figure 1E). All these small genetic aberrations lead to 2 dominant types of variants at protein level: (1) missense variants (54%) but also (2) synonymous variants (26%), which have no effect on the function of the protein (Supplementary Figure 1G). With a mean tumor purity of 70% (Supplementary Figure 1C), we were able to assess, accurately, the genome ploidy (Supplementary Figure 1D), which is mostly diploid and the number and types of SVs (1408 deletions, 1099 translocations, 1200 tandem duplications, 599 insertions, 1456 inversions; Supplementary Figure 1F). Next, we compared our results to an independent glioblastoma set from the Pan-Cancer Analysis of Whole Genomes (PCAWG) consortium30 (N = 39) (Table 1). Our findings are in concordance with PCAWG; however, we detected 966 176 more noncoding variants than the PCAWG dataset in our CPCT-02 dataset. Of these noncoding variants, 99% were detected in a single, ultramutated sample.
Table 1.
Median Mutational Frequencies of the CPCT-02 Glioblastoma Cohort (N = 42) and PCAWG Cohort (N = 39)
| Median [Q1–Q3] | CPCT-02 | PCAWG | P |
|---|---|---|---|
| Coding variants | 63.5 [58–82.5] | 61 [51.5–80.5] | 0.41 |
| SNVs | 60.5 [54–78.5] | 59 [48–76] | 0.5 |
| InDels | 4 [2–6.75] | 2 [1–3.5] | 0.0047 |
| MNVs | 0 [0–0.75] | NA | NA |
| Noncoding variants | 7957 [7016–8749] | 7322 [5590–8959] | 0.15 |
| SVs | 98 [69.75–180.25] | 100 [77.25–146] | 0.87 |
SNVs, single-nucleotide variants; MNVs, multi-nucleotide variants; SVs, structural variants. Quartiles 1 and 3 are indicated in brackets and the Mann–Whitney U test was applied as a statistical test.
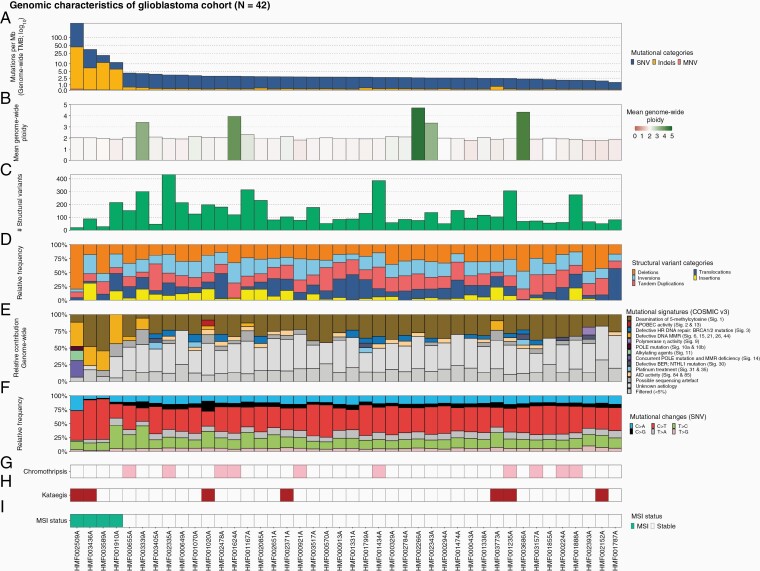
As a natural measure of mutational load, we determined TMB (number of mutations per megabase pair) and performed mutational signature calling. Ninety percent (N = 38) of tumors showed a TMB below 5.0 (Figure 1A), with a mean of 2.6. Three tumors had a TMB exceeding 10.0 (hypermutated) and one tumor exceeded 100 (ultramutated).31 Mutational signatures of tumors with low TMB included clock-like and age-related signature 1, whereas tumors with high TMB presented additional defective DNA mismatch repair (MMR) signatures (Figure 1E).32 The ultramutated sample was the only sample with a contribution from the alkylating agent resembling mutational signature 11. All high-TMB samples also showed MSI profiles, with genomes enriched for InDels in satellite regions.33 The genome-wide ploidy (Figure 1B) is generally diploid, but 5 samples showed whole-genome duplication events, even though many chromosomal arms are altered in copy number.
Figure 1.
Overview of genomic characteristics of CPCT-02 glioblastoma whole-genome sequencing cohort (N = 42). Track A shows the tumor mutational burden (mutations per Mb; blue for single-nucleotide variants [SNVs], yellow for insertions/deletions [Indels], and red for multi-nucleotide variants [MNVs]). Track B confers the mean genome-wide ploidy for each sample. Tracks C and D illustrate the abundance of structural variants and the relative frequency of the type of variants. Tracks E and F show the relative mutational signature contribution (COSMIC signatures v3) and the relative frequency of mutational changes at base level. Tracks G and H display which samples exhibit predicted catastrophic events, namely chromothripsis and kataegis. Finally, track I shows predicted microsatellite instability profiles of the sample. The figure is ordered descendingly by tumor mutational burden on the x-axis.
Copy-Number Alterations
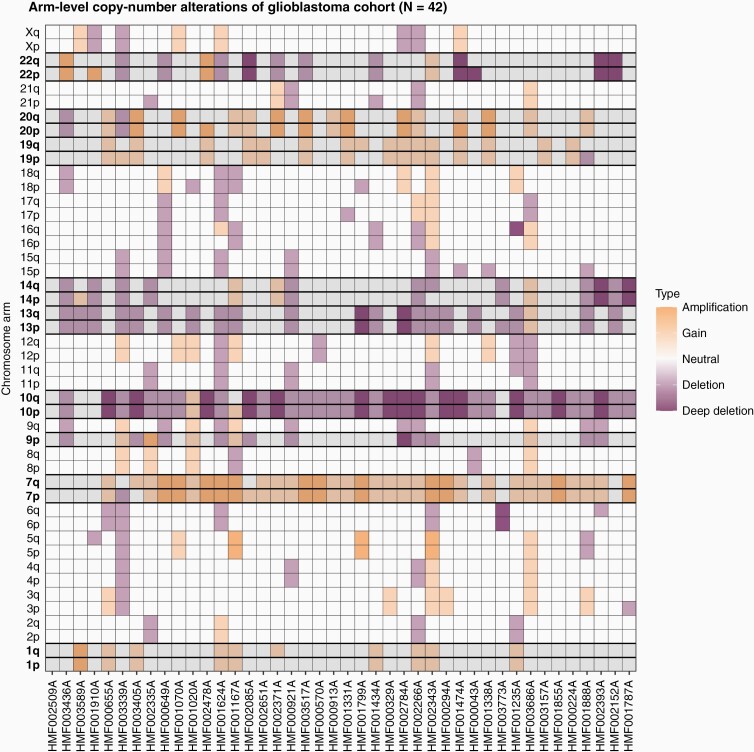
We detailed both arm-level (Figure 2) and focal CNAs (Figure 3). Amplification of chromosome 7 (79%) and deletion of chromosome 10 (86%) were most prevalent in our cohort. Loss of 9p occurred in 26%, making it the only chromosome to have statistically significant alterations limited to the small arm. Eight chromosomes (1, 7, 10, 13, 14, 19, 20, and 22) were significantly altered (q value < 0.05) in copy number on both arms.
Figure 2.
Arm-level copy-number alterations and significantly altered (q value < 0.05) chromosome arms according to GISTIC2.0 in the CPCT-02 glioblastoma whole-genome sequencing cohort (N = 42). Each column is a glioblastoma sample (ordered descendingly by tumor mutational burden), with the chromosome arm listed on the y-axis. Increase in copy number is displayed in the grid cells in yellow and decrease in purple, white squares show no change in copy number. Highlighted rows with bold chromosome-arm names indicate statistically significant arm-level changes.
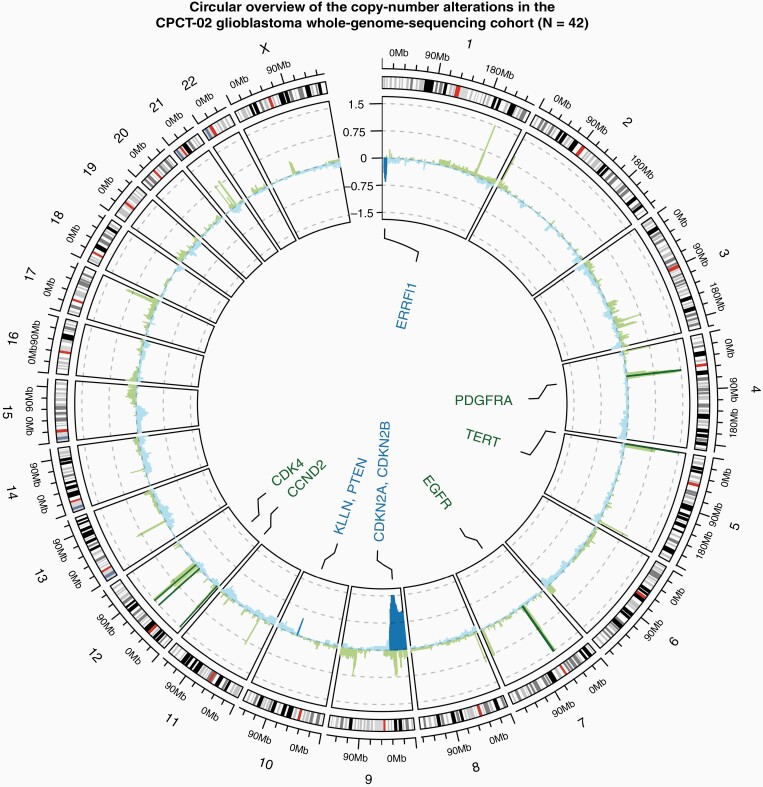
Figure 3.
Circular overview of the copy-number alterations in the CPCT-02 glioblastoma whole-genome sequencing cohort (N = 42). The outer ring shows the chromosomal ideogram, followed by a cohort-wide GISTIC2.0 G-score track with large peaks rounded to 1.5 and −1.5. Negative copy numbers on the y-axis (blue) indicate deletions, with positive (green) indicating amplifications. The darker color is indicative of passing the statistical q-value threshold of 0.05. Known cancer driver genes overlapping copy-number peaks found to be significant by GISTIC2.0 are labeled in the center of the circle, utilizing the same color scheme as the G-score track.
Structural Variants
The median number of SVs per sample was 98 (ranging from 20 to 433) and was similar to that of the PCAWG glioblastoma set (Table 1). The absolute number of the different types of SVs per sample and the associated genomic widths are depicted in Supplementary Figure 2. While the absolute number of SVs differed from sample to sample, the relative frequency was relatively narrow with quartile 1 (Q1) from 9.67 to Q3 of 12.6. Deletions showed the highest interquartile range, as well as the highest mean (28.2%) and maximum (80%) relative to the sample. Insertions were detected least, with a mean of 7.9% and a maximum of 30.7% relative to the sample. CT occurred in 10 patients (24%, Figure 1G)34 and kataegis in 7 patients (16%, Figure 1H). While CT in other cancer types typically shatters entire chromosomes or chromosome arms, events tend to affect smaller regions on single chromosomes, as previously reported in glioblastoma.30 CT of chromosomes 1 and 12 were the most common events in our cohort (Supplementary Figure 3, Supplementary Datafile 1). We detected no CT on chromosomes 7 and 9, whereas previous studies linked this to a higher incidence of EGFR, MDM2 and/or CDK4 (hyper-)amplifications, and CDKN2A homozygous deletions. All these amplifications/deletions were independent of CT events in our cohort based on χ 2 tests (alpha ≤ 0.05). Five samples contained an FGFR3–TACC3 fusion and there was one instance of a CCDC6–RET fusion (Supplementary Figure 4). With the access to 2 RNA-Seq samples, we could confirm actual fusion transcripts of both fusions in their respective samples; one with a FGFR3–TACC3 and one with a CCDC6–RET fusion. This 100% concordance was identified with a minimum of 38 fragment counts supporting the fusion transcripts in RNA-Seq data. The incidence of the FGFR3–TACC3 fusion (11.9%) is higher (P = .00435; Fisher’s exact test) than previous reports, which ranged 1.2%–3.1%.7,35
Noncoding Variants
WGS is well suited to study both coding and noncoding variants. We therefore scrutinized recurrent variants in our cohort and found 2 reliable variants in the TERT promoter (C228T in 19 samples and C250T in 10 samples). Sakthikumar et al.24 pose that NCCMs could play an essential role in glioblastoma, finding enrichments for these variants in their cohort. They split these NCCMs between “related to key glioblastoma genes” (354 variants in 60 genes) and “other protein-coding genes” (591 variants in 43 genes). We also assessed which of their posited NCCMs were present in our cohort, resulting in just 2 occurrences: one in SULT1B1 and one in MET.
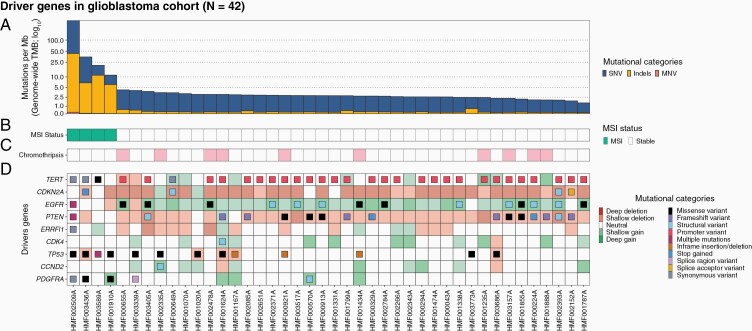
Driver Gene Analysis
Next, we screened for enrichments (q value < 0.05) of coding mutations and CNAs, which presented 9 well-known driver genes of glioblastomas (Figure 4D). The most noticeable drivers were CDKN2A (homozygous deletion in 69%), TERT (promoter variants in 67%), EGFR (amplified in 42.9% and enriched in mutations), and PTEN (deletions in 9.5% and enriched in mutations). Most EGFR-amplified samples also contained SVs or missense variants within the gene. Furthermore, we detected deletions in the ERBB receptor feedback inhibitor 1 encoding gene ERRFI1 (homozygous deletions, N = 2), amplifications of CDK4 (14.3%), PDGFRA (11.9%), and CCND2 (9.5%), and disruptive TP53 mutations. This last gene was the only driver exclusively significantly enriched based on small mutations. Mutation status of genes commonly related to glioblastoma (such as IDH1/IDH2) not passing our threshold value of significance (P-adjusted < .05) is depicted in Supplementary Figure 5.
Figure 4.
Overview of coding mutations and copy-number alterations in driver genes (determined by dN/dS and GISTIC2.0) in the CPCT-02 glioblastoma whole-genome sequencing cohort (N = 42). Track A shows the tumor mutational burden (mutations per Mb; blue for single-nucleotide variants [SNVs], yellow for insertions/deletions [Indels], and red for multi-nucleotide variants [MNVs]). Tracks B and C show predicted microsatellite instability (MSI) status of the sample and chromothripsis status. The oncoplot in track D shows mutations (filled center) and copy-number alterations (grid cell background). The figure is ordered descendingly by tumor mutational burden on the x-axis.
Potential Drug Targets
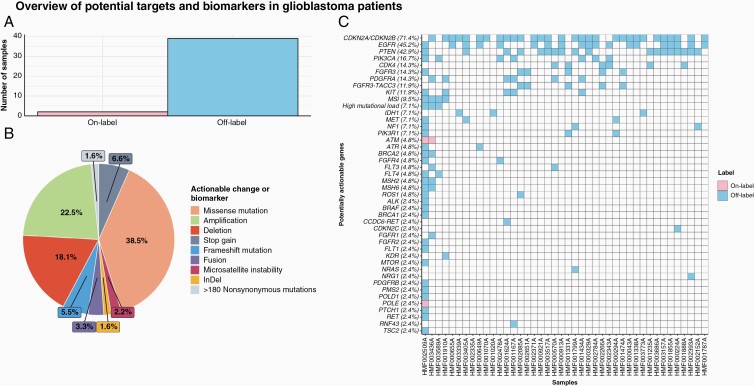
Finally, we assessed potential targets detectable through WGS by utilizing databases with clinical trial data and drug-target combinations in cancer patients, such as iClusion data. We found biomarkers and potentially targetable events in all but one of our 42 samples (Figure 5A and C), with a median of 3 targetable genes per sample. Results of clinical trials have been published for some of these (eg, EGFR36). The type of targetable change or biomarker is represented in Figure 5B, with the specific change and treatment listed in Supplementary Datafile 1.
Figure 5.
Overview of DNA-based biomarkers and potential treatment options in 42 whole-genome sequenced glioblastoma samples (CPCT-02). (A) Highest level of potential treatment options available based on genomic characteristics. On-Label indicates treatment registered by federal authorities for glioblastoma, whereas Off-Label indicates a registration for other tumor types. (B) The distribution of actionable target types in percentages. (C) Potentially actionable alterations at gene level with every column portraying a sample, ordered descendingly by tumor mutational burden on the x-axis. A tile is colored pink in case of an On-Label treatment and blue for Off-Label.
Discussion
By investigating driver gene events, CNAs, we found that our cohort contained most of the prevalent glioblastoma hallmarks, such as TERT promoter mutations, EGFR amplification, and CDKN2A deletion. These findings are in line with other glioblastoma datasets, as all events that drive mutational selection in our set of glioblastoma tumors have been previously described. Noncoding variants presented the largest difference in mutational frequencies between the CPCT-02 study and PCAWG.30 Since we found just 2 noncoding variants out of 945 Sakthikumar et al.24 previously reported in glioblastoma, these noncoding variants were not of relevance in our cohort. Also in line with previous reports for glioblastoma, CT (24% of our cohort) generally occurs in smaller regions than in other cancer types. Contrary to the findings of Ah-Pine et al.,34 no CT occurred on chromosome 7 or 9 and most commonly detected on chromosomes 1 and 12 in our cohort. On chromosome 12, MDM2 and CDK4 amplification were independent of CT events. It should be noted that not only different platforms were used (WGS vs SNP arrays), but also different definitions of CT. While it is encouraging to see the hallmarks represented in our dataset, it does raise the question if the differences in CT are purely due to disparity in the definition. However, this can likely be answered when the field settles on a gold-standard CT definition in general and in glioblastoma specifically.
We investigated mutational signatures, which greatly differed between low- and high-TMB samples. This is akin to the (panel-based) data presented by Touat et al.,37 with 2 types of high-TMB tumors (treatment-induced and de novo). One patient pretreated with temozolomide showed signature 11 (resembling the mutational pattern of alkylating agents) in its mutational landscape. The larger contribution of defective DNA MMR signatures in the 4 high-TMB samples aligns with MSI, which is a marker of this phenomenon.31–33 Touat et al. suggested a high-TMB cutoff of 17.0 based on the MSK-IMPACT panels. Based on MSI status prediction and the larger contribution of defective DNA MMR signatures in our cohort, 4 samples fit the high-TMB mold in our cohort, passing a WGS cutoff of 10 somatic mutations per Mb. Additionally, we have one genomically distinct sample with 342 mutations per Mb, which was categorized as ultramutated.31 This ultramutated sample shows a similar mutational signature pattern as the treatment-induced hypermutated samples Touat et al. present, while the other 3 high TMB match the de novo samples. The fact that the genomic MSI status overlaps perfectly with our high-TMB samples suggests that this might be an alternative means of determining TMB status in glioblastoma.33
We presented an overview of potential targets and biomarkers based on genomic characteristics of our cohort where 41 out of 42 samples showed at least one targetable gene alteration. In an effort to remain comprehensive, we did not exclude any targets that have not shown promise in clinical trials in the past. The fact that there is a median of 3 potentially targetable gene alterations for these samples provides optimism for the improvement on the standard treatment modalities for glioblastoma patients at a personal level. Since this list consists of targets (genes or alterations) linked with drugs and is not glioblastoma-specific, there is no guarantee that these therapeutics have the capacity to cross the blood–brain barrier.38 For instance, EGFR has been posited as a prime target for glioblastoma patients, due to the amplification of the gene and protein overexpression. However, its promise is as of yet unfulfilled, with many hypotheses for the reasons behind previously unsuccessful trials.36 Drug delivery across the blood–brain barrier seems to be the most problematic issue, but intratumoral adaptations at a pharmacodynamic level may also constitute resistance to therapy. The negative impact of these limitations needs to be considered before the observed genetic alterations are targeted clinically. For example, CDKN2A and CDKN2B were frequently deleted in our cohort. Clearly, a homozygously deleted gene cannot be targeted, but alternate means of reactivating the pathway can prove useful. While not applicable for most patients, our 5 samples with the FGFR3–TACC3 fusion events show a higher incidence (11.9%) than the previously reported 1%–3.1%. This oncogenic fusion might facilitate targeted treatment with FGFR3 kinase inhibition.39 Our other reported fusion (CCDC6–RET) is rather prevalent in thyroid and lung cancers and shows selective sensitivity to RET inhibitors in clinical trials with those cancer types.35,40 We confirmed fusion gene detection data based on RNA-Seq from 2 of the 5 samples we reported. As multiple potential targetable genes are present in these samples, in-depth genomic knowledge by WGS will improve clinical study design and treatment allocation with targeted therapies in glioblastoma patients.
In conclusion, we have described the genomic landscape by means of the TMB, driver gene events, and mutational signatures in a cohort of 42 glioblastoma patients included in CPCT-02. Several factors (such as mutational frequencies) coincide with other datasets, while others do not (CT locations, noncoding mutations). Finally, we have alluded to several potential treatment options and biomarkers for 41 out of 42 patients, including within ongoing clinical trials, based on molecular traits. This reveals new avenues for clinical trials and can improve patient management through potential personalized medicine in glioblastoma patients.
Supplementary Material
Acknowledgments
This publication and the underlying study have been made possible partly on the basis of the data that Hartwig Medical Foundation (HMF) and the Center of Personalised Cancer Treatment (CPCT) have made available to the study.
Funding
No funding was received for this work.
Conflict of interest statement. The authors declare that they have no competing interests related to this study.
Authorship statement. P.F., H.J.G.v.d.W., and F.Y.F.d.V. contributed equally to this work; conceptualization: F.Y.F.d.V., H.J.G.v.d.W., and P.F.; formal analysis: W.S.v.d.G.; supervision: P.F., H.J.G.v.d.W., and F.Y.F.d.V.; writing—original draft: W.S.v.d.G., P.F., H.J.G.v.d.W., and F.Y.F.d.V.; writing—review and editing: Y.H., K.D., P.A.R., S.B., and R.H.J.M.
Data availability
The data that support the findings of this study are available from Hartwig Medical Foundation, which were used under data request number DR-093 for the current study. Both WGS and clinical data are freely available for academic use from the Hartwig Medical Foundation through standardized procedures and request forms can be found at https://www.hartwigmedicalfoundation.nl. All tools and scripts used for processing the WGS data are available at https://github.com/hartwigmedical/ and/or can be provided by authors upon request.
References
- 1. Ostrom QT, Gittleman H, Truitt G, Boscia A, Kruchko C, Barnholtz-Sloan JS. CBTRUS statistical report: primary brain and other central nervous system tumors diagnosed in the United States in 2011–2015. Neuro Oncol. 2018;20(suppl 4):iv1–iv86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Stupp R, Mason WP, van den Bent MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352(10):987–996. [DOI] [PubMed] [Google Scholar]
- 3. Akgül S, Patch A-M, D’Souza RCJ, et al. Intratumoural heterogeneity underlies distinct therapy responses and treatment resistance in glioblastoma. Cancers (Basel). 2019;11(2):190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Draaisma K, Chatzipli A, Taphoorn M, et al. Molecular evolution of IDH wild-type glioblastomas treated with standard of care affects survival and design of precision medicine trials: a report from the EORTC 1542 study. J Clin Oncol. 2020;38(1):81–99. [DOI] [PubMed] [Google Scholar]
- 5. Ceccarelli M, Barthel FP, Malta TM, et al. ; TCGA Research Network . Molecular profiling reveals biologically discrete subsets and pathways of progression in diffuse glioma. Cell. 2016;164(3):550–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mankor JM, Paats MS, Groenendijk FH, et al. ; CPCT Consortium . Impact of panel design and cut-off on tumour mutational burden assessment in metastatic solid tumour samples. Br J Cancer. 2020;122(7):953–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Brennan CW, Verhaak RG, McKenna A, et al. ; TCGA Research Network . The somatic genomic landscape of glioblastoma. Cell. 2013;155(2):462–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Priestley P, Baber J, Lolkema MP, et al. Pan-cancer whole-genome analyses of metastatic solid tumours. Nature. 2019;575(7781):210–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. van Dessel LF, van Riet J, Smits M, et al. The genomic landscape of metastatic castration-resistant prostate cancers reveals multiple distinct genotypes with potential clinical impact. Nat Commun. 2019;10(1):5251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Vasimuddin M, Misra S, Li H, Aluru S. Efficient architecture-aware accel- eration of BWA-MEM for multicore systems. In: 2019 IEEE International Paralleland Distributed Processing Symposium (IPDPS), Rio de Janeiro, Brazil. 2019:314–324. doi: 10.1109/IPDPS.2019.00041 [DOI] [Google Scholar]
- 11. Cameron DL, Schröder J, Penington JS, et al. GRIDSS: sensitive and specific genomic rearrangement detection using positional de Bruijn graph assembly. Genome Res. 2017;27(12):2050–2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cameron DL, Baber J, Shale C, et al. GRIDSS, PURPLE, LINX: unscrambling the tumor genome via integrated analysis of structural variation and copy number. bioRxiv. 2019. doi: 10.1101/781013, preprint: not peer reviewed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Huang MN, McPherson JR, Cutcutache I, Teh BT, Tan P, Rozen SG. MSIseq: software for assessing microsatellite instability from catalogs of somatic mutations. Sci Rep. 2015;5:13321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Karczewski KJ, Francioli LC, Tiao G, et al. ; Genome Aggregation Database Consortium . The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581(7809):434–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tate JG, Bamford S, Jubb HC, et al. COSMIC: the catalogue of somatic mutations in cancer. Nucleic Acids Res. 2019;47(D1):D941–D947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Smigielski EM, Sirotkin K, Ward M, Sherry ST. dbSNP: a database of single nucleotide polymorphisms. Nucleic Acids Res. 2000;28(1):352–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. McLaren W, Gil L, Hunt SE, et al. The Ensembl variant effect predictor. Genome Biol. 2016;17(1):122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mermel CH, Schumacher SE, Hill B, Meyerson ML, Beroukhim R, Getz G. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 2011;12(4):R41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jang YE, Jang I, Kim S, et al. ChimerDB 4.0: an updated and expanded database of fusion genes. Nucleic Acids Res. 2020;48(D1):D817–D824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Martincorena I, Raine KM, Gerstung M, et al. Universal patterns of selection in cancer and somatic tissues. Cell. 2017;171(5):1029–1041.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Blokzijl F, Janssen R, van Boxtel R, Cuppen E. Mutational patterns: comprehensive genome-wide analysis of mutational processes. Genome Med. 2018;10(1):33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Alexandrov LB, Kim J, Haradhvala NJ, et al. The repertoire of mutational signatures in human cancer. Nature. 2020;578(7793):94–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Pracma. https://cran.r-project.org/web/packages/pracma/index.html. Accessed 11 December, 2020.
- 24. Sakthikumar S, Roy A, Haseeb L, et al. Whole-genome sequencing of glioblastoma reveals enrichment of non-coding constraint mutations in known and novel genes. Genome Biol. 2020;21(1):127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lawrence M, Huber W, Pagès H, et al. Software for computing and annotating genomic ranges. PLoS Comput Biol. 2013;9(8):e1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chakravarty D, Gao J, Phillips S, et al. OncoKB: a precision oncology knowledge base. JCO Precis Oncol. 2017;1(1):1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Griffith M, Spies NC, Krysiak K, et al. CIViC is a community knowledge base for expert crowdsourcing the clinical interpretation of variants in cancer. Nat Genet. 2017;49(2):170–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tamborero D, Rubio-Perez C, Deu-Pons J, et al. Cancer Genome Interpreter annotates the biological and clinical relevance of tumor alterations. Genome Med. 2018;10(1):25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Dobin A, Davis CA, Schlesinger F, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Campbell PJ, Getz G, Korbel JO, et al. Pan-cancer analysis of whole genomes. Nature. 2020;578(7793):82–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Barresi V, Simbolo M, Mafficini A, et al. Ultra-mutation in IDH wild-type glioblastomas of patients younger than 55 years is associated with defective mismatch repair, microsatellite instability, and giant cell enrichment. Cancers. 2019;11(9):1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Martinez R, Schackert HK, Plaschke J, Baretton G, Appelt H, Schackert G. Molecular mechanisms associated with chromosomal and microsatellite instability in sporadic glioblastoma multiforme. Oncology. 2004;66(5):395–403. [DOI] [PubMed] [Google Scholar]
- 33. Leelatian N, Hong CS, Bindra RS. The role of mismatch repair in glioblastoma multiforme treatment response and resistance. Neurosurg Clin N Am. 2021;32(2):171–180. [DOI] [PubMed] [Google Scholar]
- 34. Ah-Pine F, Casas D, Menei P, Boisselier B, Garcion E, Rousseau A. RNA-sequencing of IDH-wild-type glioblastoma with chromothripsis identifies novel gene fusions with potential oncogenic properties. Transl Oncol. 2021;14(1):100884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Singh D, Chan JM, Zoppoli P, et al. Transforming fusions of FGFR and TACC genes in human glioblastoma. Science. 2012;337(6099):1231–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Westphal M, Maire CL, Lamszus K. EGFR as a target for glioblastoma treatment: an unfulfilled promise. CNS Drugs. 2017;31(9):723–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Touat M, Li YY, Boynton AN, et al. Mechanisms and therapeutic implications of hypermutation in gliomas. Nature. 2020;580(7804):517–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sarkaria JN, Hu LS, Parney IF, et al. Is the blood-brain barrier really disrupted in all glioblastomas? A critical assessment of existing clinical data. Neuro Oncol. 2018;20(2):184–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Guagnano V, Kauffmann A, Wöhrle S, et al. FGFR genetic alterations predict for sensitivity to NVP-BGJ398, a selective pan-FGFR inhibitor. Cancer Discov. 2012;2(12):1118–1133. [DOI] [PubMed] [Google Scholar]
- 40. Woo HY, Na K, Yoo J, et al. Glioblastomas harboring gene fusions detected by next-generation sequencing. Brain Tumor Pathol. 2020;37(4):136–144. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from Hartwig Medical Foundation, which were used under data request number DR-093 for the current study. Both WGS and clinical data are freely available for academic use from the Hartwig Medical Foundation through standardized procedures and request forms can be found at https://www.hartwigmedicalfoundation.nl. All tools and scripts used for processing the WGS data are available at https://github.com/hartwigmedical/ and/or can be provided by authors upon request.