Summary
Class 18 myosins are most closely related to conventional class 2 nonmuscle myosins (NM2). Surprisingly, the purified head domains of Drosophila, mouse and human myosin 18A (M18A) lack actin-activated ATPase activity and the ability to translocate actin filaments, arguing that the functions of M18A in vivo do not depend on intrinsic motor activity. M18A has the longest coiled-coil of any myosin outside of the class 2 myosins, suggesting that it might form bipolar filaments similar to conventional myosins. To address this possibility, we expressed and purified full-length mouse M18A using the baculovirus/Sf9 system. M18A did not form large bipolar filaments under any conditions tested. Instead, M18A formed a ~65 nm-long bipolar structure with two heads at each end. Importantly, when NM2 was polymerized in the presence of M18A, the two myosins formed mixed bipolar filaments, as evidenced by cosedimentation, electron microscopy, and single-molecule imaging. Moreover, super-resolution imaging of NM2 and M18A using fluorescently tagged proteins and immunostaining of endogenous proteins showed that NM2 and M18A are present together within individual filaments inside living cells. Together, our in vitro and live-cell imaging data argue strongly that M18A coassembles with NM2 into mixed bipolar filaments. M18A could regulate the biophysical properties of these filaments, and, by virtue of its extra N- and C-terminal domains, determine the localization and/or molecular interactions of the filaments. Given the numerous, fundamental cellular and developmental roles attributed to NM2, our results have far reaching biological implications.
Results and Discussion
The self-assembly of M18Aβ in vitro appears limited to the formation of an antiparallel dimer
Myosins 18Aα and M18β (M18Aα and M18Aβ), two splice variants of the MYO18A gene, have domain architectures similar to that of nonmuscle myosin-2 (NM2), with a myosin motor-like domain, a pair of light chain-binding IQ motifs, and a long, predicted coiled-coil domain ending in a non-helical tailpiece. Additionally, M18Aα and M18Aβ possess extra domains at their N- and C-termini. Specifically, both possess a C-terminal PDZ domain-binding motif, and M18Aα also contains an N-terminal extension containing a KE-rich domain, an ATP-insensitive actin-binding site, and a PDZ domain. Unlike NM2, its closest relative, M18A has no motor activity [1–3]. Despite this, we asked if M18A polymerizes into bipolar filaments like NM2. Electron microscopy (EM) of purified Mus musculus M18Aβ protein at 500 mM KCl revealed a two-headed structure with a long coiled-coil extending from the heads (Figure 1A, upper row), an arrangement similar to NM2 molecules under the same conditions [4]. This structure is hereafter referred to as the “monomer” to maintain the convention established for class-2 myosins. Of note, the coiled-coil tail of M18Aβ has a mean contour length of 97±7 nm (n = 100), significantly shorter than the ~150–160 nm length documented for vertebrate class-2 myosins [5–8]. When the ionic strength was lowered to 100 mM KCl, two M18Aβ molecules associated in an antiparallel configuration, producing a four-headed structure with two heads at each end of the long axis (Figure 1A, lower row; Figure S1A) and a mean contour length of 64±5 nm (n = 100). Based on the dimer length relative to the monomer, along with measured and predicted bend positions in extended monomers (Figure S1D), we interpret this structure as an antiparallel dimer of M18Aβ molecules with folded tails (Figure 1B). This is similar to antiparallel dimers formed by some class-2 myosins in the unphosphorylated, off-state [4, 9, 10]. As with class-2 myosins, the M18Aβ monomer/dimer transition, as assessed by EM (Figure S1B) and analytical centrifugation (Figure S1C), was salt-dependent. Importantly, and in striking contrast to NM2, M18Aβ did not self-assemble into large bipolar filaments at low ionic strength, even following phosphorylation of its regulatory light chains (RLC), or under a range of other conditions, including 1 mM ATP, 1 mM ADP, 2 mM CaCl2, 10 mM MgCl2, or excess EDTA (Figure S1E). Interestingly, analysis of the charge along the coiled-coil demonstrates that the 196-residue charge repeat, important in the formation of NM2 oligomers, is retained by M18A (Figure S1F) [11, 12].
Figure 1. M18A and NM2A form heteropolymers.
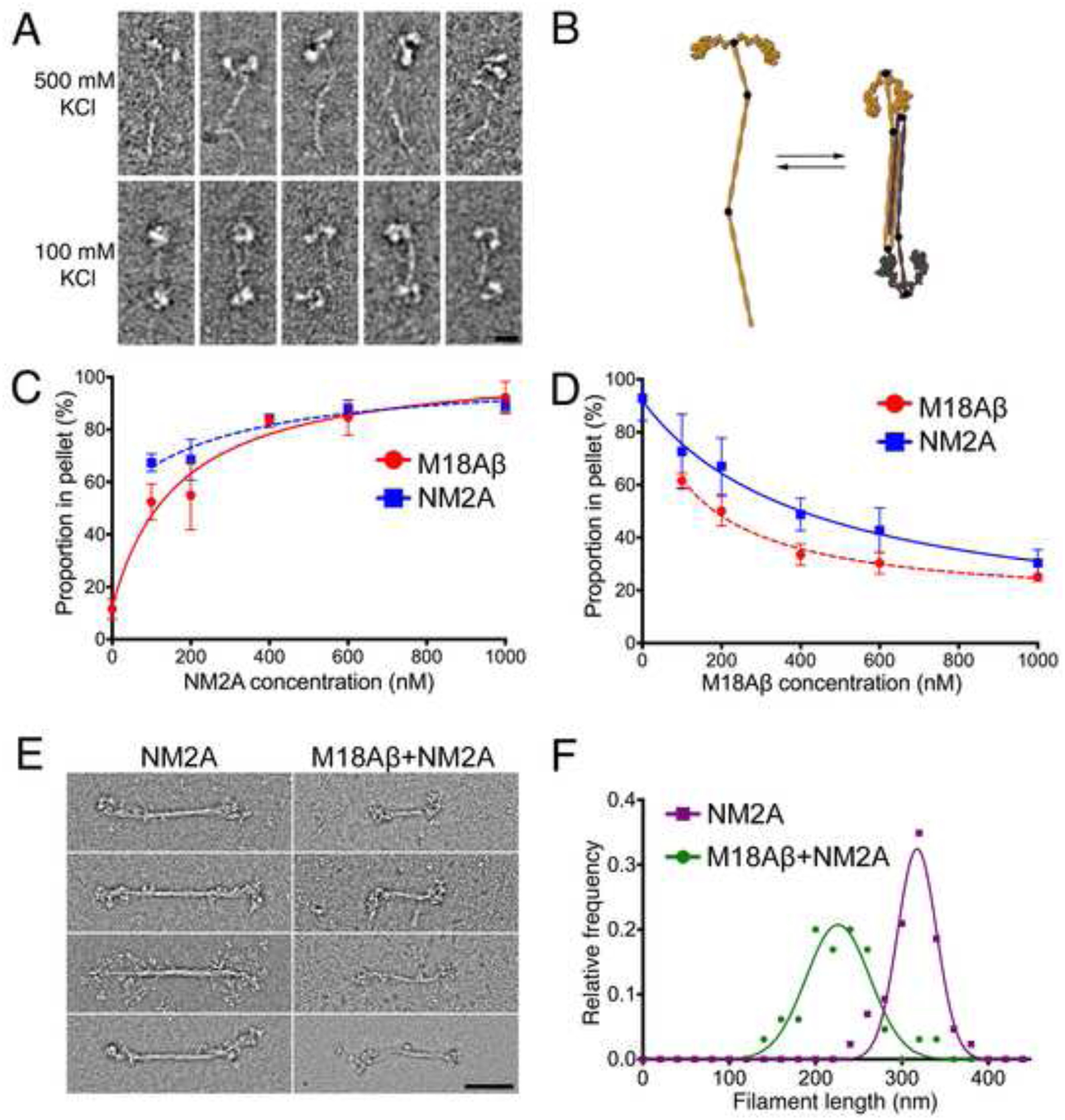
(A) EM images of M18Aβ in high salt buffer (500 mM KCl) (upper row) and low salt buffer (100 mM KCl) (lower row). Scale bar: 20 nm. (B) Model showing proposed antiparallel interaction between two folded M18Aβ molecules. (C) Cosedimentation of a fixed concentration of M18Aβ (200 nM) with a range of NM2A concentrations; error bars denote S.D. (D) Cosedimentation of a fixed concentration of NM2A (200 nM) with a range of M18Aβ concentrations; error bars denote S.D. (E) EM images comparing filaments of pure NM2A (left column) with filaments from an equimolar mixture of M18Aβ and NM2A (150 mM KCl; right column). Scale bar: 100 nm. (F) Length distribution for NM2A filaments alone (purple; n=43) and a 1:1 mixture of M18Aβ and NM2A (green; n=65), measured from electron micrographs.
M18Aβ coassembles with NM2 into mixed bipolar filaments in vitro and regulates the size of these hybrid filaments
Given the similarities between the antiparallel homo-dimers formed by M18Aβ and NM2, as well as the conservation of charge repeats between them, we hypothesized that M18A might be capable of binding NM2 via interactions similar to those used within homo-oligomers of these myosins. We tested this hypothesis using cosedimentation. When analyzed independently in the presence of 150 mM KCl, the majority of NM2A sedimented while the majority of M18Aβ did not (Figure 1C and 1D). In contrast, cosedimentation of a fixed concentration of M18Aβ with increasing concentrations of NM2A led to an increased proportion of M18Aβ in the pellet (Figure 1C), suggesting that M18Aβ interacted with NM2A filaments. Importantly, M18Aβ also cosedimented with increasing concentrations of NM2B or NM2C (Figure S1G), indicating the interaction occurs with all NM2 isoforms. Furthermore, preliminary cosedimentation experiments demonstrated direct interaction between the M18Aα isoform and NM2 tail fragments (data not shown; but see below). Surprisingly, cosedimentation of a fixed concentration of NM2A with increasing concentrations of M18Aβ resulted in a lower proportion of NM2A in the pellet (Figure 1D). Therefore, while M18Aβ and NM2 directly interact, excess M18Aβ appears to interfere with NM2A filament assembly. EM performed on an equimolar mixture of M18Aβ and NM2A (in 150 mM KCl) revealed filaments with a shorter mean length (227±42 nm) than filaments of NM2A alone (314±28 nm) (Figure 1E, 1F). Additionally, the length distribution of copolymers was significantly broader than that of filaments containing only NM2A (Figure 1F). Together, these data are consistent with M18A coassembling with NM2 into mixed bipolar filaments, and with the ability of M18A to influence the size and organization of NM2 filaments.
To further demonstrate M18A/NM2A coassembly in vitro, we performed TIRF microscopy using myosins possessing distinct N-terminal Halotags. AlexaFluor-488 (AF488)-labelled Halotag-NM2A (green) and tetramethylrhodamine(TMR)-labelled Halotag-M18Aβ (red) were mixed in a 1:1 molar ratio and bound to a coverslip (Figure 2A). A variety of fluorescence intensities was observed for individual puncta, consistent with the high standard deviation observed for copolymer length in EM. The brightest puncta (Figure 2A, Spot 1), which likely correspond to filamentous structures, contained both proteins, although their relative amounts varied. Photobleaching traces of these bright puncta (Figure 2C, Spot 1) revealed large numbers of photobleaching events for each myosin, consistent with these puncta corresponding to bipolar filaments assembled from many NM2A and M18Aβ molecules. Some dimmer puncta also appeared to contain both myosins (Figure 2B, Spot 2). Photobleaching analysis of these puncta (Figure 2C, Spot 2) showed two photo-bleaching steps for each color, consistent with the lower limit of hetero-oligomerization being a heterodimer containing one two-headed molecule each of NM2A and M18Aβ. Some of the dimmest puncta appeared to consist of only green or red fluorescence, likely corresponding to individual molecules of each myosin (Figure 2B, Spots 3 and 4). Indeed, these smaller, monochromatic species showed stepwise photobleaching (Figure 2C, Spots 3 and 4) consistent with them being two-headed monomers or four-headed homodimers of the respective myosins. Strong colocalization and large puncta were also observed when polymerization was induced by phosphorylation of RLCs in a mixture of the myosins using MLCK (Figure S2A). Colocalization of the two myosins was absent in high salt buffer, further demonstrating that copolymerization is ionic-strength dependent (Figure S2B). Using the overall intensity of each punctum, along with the average decrease due to quantal photobleaching of single fluorophores in each field, we estimated the number of each myosin per punctum. The distributions of filaments formed in the absence of ATP and formed by RLC phosphorylation were similar, in both cases showing a wide range of ratios of the two myosins (Figure S2C, D). In contrast, puncta in high salt conditions typically consisted of low numbers of the respective myosin (Figure S2E). Together, these data using purified proteins argue strongly that M18A and NM2 coassemble into hybrid filaments in vitro.
Figure 2. TIRF microscopy of M18A-NM2A copolymers.
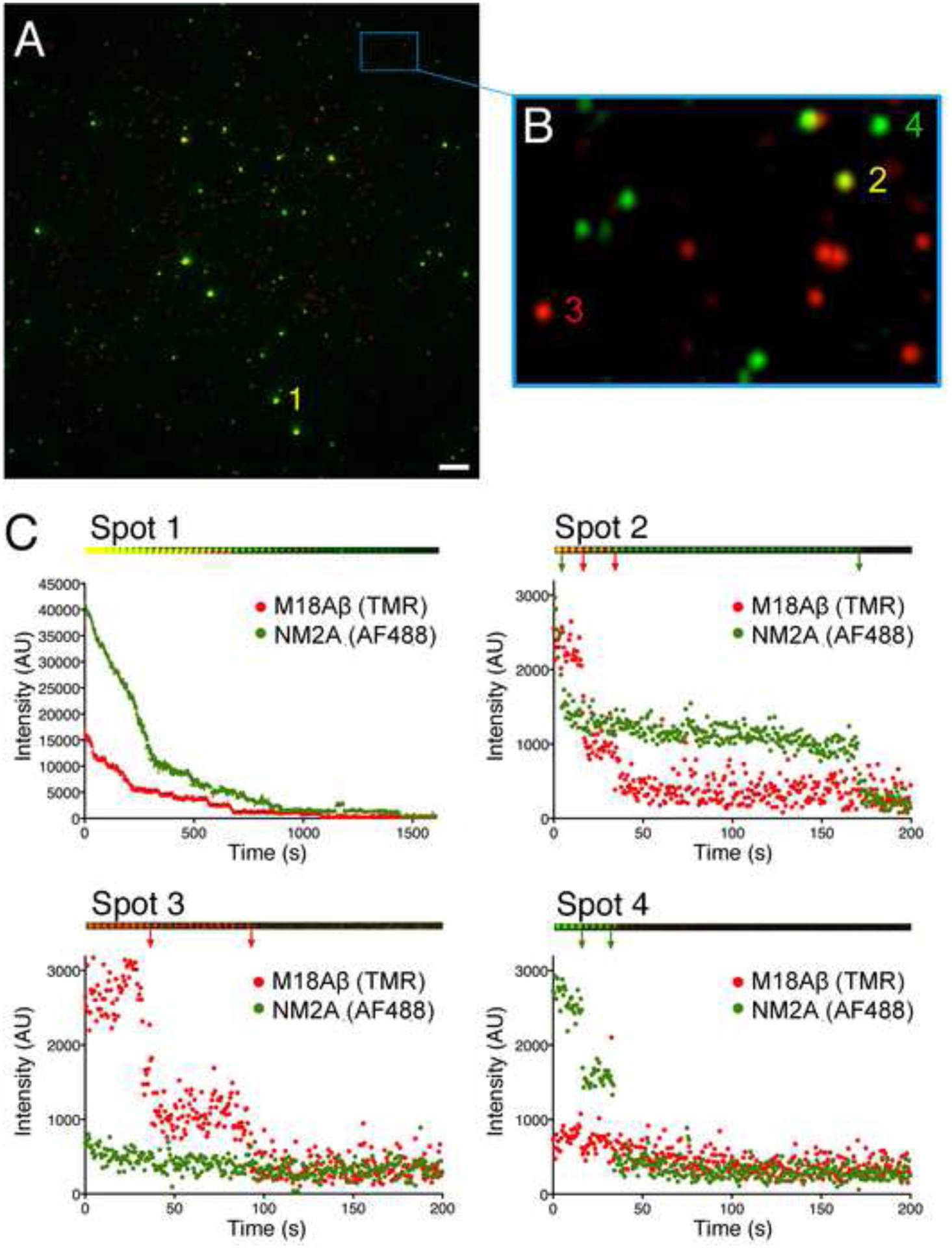
(A) TIRF image showing copolymers of M18Aβ:NM2A (e.g. spot 1). (B) Magnified inset showing low intensity puncta consistent with small M18A:NM2A oligomers (e.g. spot 2) and single molecules of M18A (e.g. spot 3) or NM2A (e.g. spot 4). (C) Photobleaching traces of spots highlighted in (A) and (B). Montages above each plot show images of denoted spots over time. Each image of spot 1 shows the average of a 30 second bin, each image of spots 2, 3 and 4 shows the average of a 4 second bin. Arrows denote photobleaching events.
M18A coassembles with NM2A into mixed bipolar filaments in live cells
In keeping with previous reports [13–15], conventional light microscopy showed that both EGFP-M18Aα and EGFP-M18Aβ colocalize with Apple-NM2A within the lamella of well-spread cells, and that EGFP-M18Aα accumulates with Apple-NM2A under the nucleus (Figure S3A; see Figure S3B and S3C for expression analyses). To determine if this “colocalization” corresponds to coassembly, fluorophore-tagged M18A and NM2 were expressed in multiple cell types and imaged using TIRF structured-illumination microscopy (TIRF-SIM), a super-resolution imaging technique used recently to investigate the isoform composition of individual NM2 filaments [16]. In HeLa cells expressing tdTomato-NM2A and EGFP-M18Aβ, bipolar NM2A filaments were evident in TIRF-accessible regions as two red puncta separated by ~300 nm (Figure 3A). Importantly, in many cases these two red puncta bracketed two green puncta ~125 nm apart (Figure S4A, red plot), corresponding to the heads of EGFP-M18Aβ (Figure 3A and insets A1 and A2; white brackets). This red-green-green-red pattern is consistent with formation of mixed filaments in which EGFP-M18Aβ coassembles via its shorter coiled-coil with tdTomato-NM2A (see cartoon in Figure 3A). Similar results were seen for other M18A and NM2 isoforms in various cell types (Figure S4B). Surprisingly, EGFP-NM2A filaments containing M18A were not obviously shorter than filaments lacking M18A signal (Figure S4A). This discrepancy with the in vitro data almost certainly reflects the low expression of M18A relative to NM2, although there may be additional constraints on filament length in vivo (see Figure S4 for details).
Figure 3. M18A and NM2 coassemble in living cells.
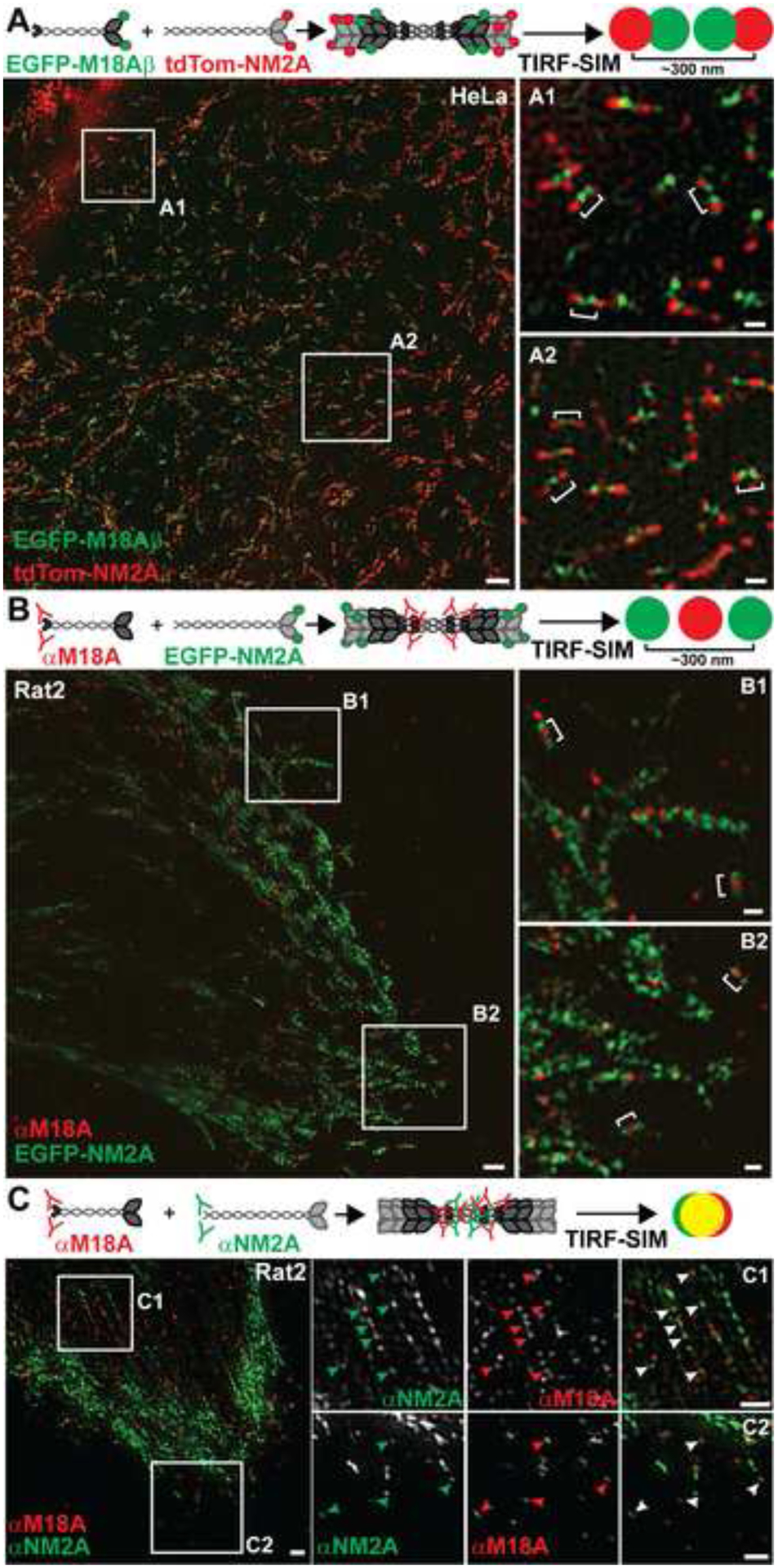
(A) Top: Cartoon depicting coassembly of EGFP-M18Aβ with tdTom-NM2A and the resulting red-green-green-red pattern observed in TIRF-SIM. Bottom: TIRF-SIM image of HeLa cells expressing EGFP-M18Aβ and tdTom-NM2A. White numbered boxes correspond to the magnified insets. White brackets indicate heterotypic filaments of M18A and NM2A. Occasionally, red puncta are bifurcated by a single green punctum. This is probably due to merging of the two green puncta since the distance between M18A heads is close to the axial resolution of TIRF-SIM. Indeed, some slight offset in green and red puncta is normal at this resolution [16]. Scale bars represent 2 μm for the larger image and 300 nm for insets. (B) Top: Cartoon depicting coassembly of EGFP-NM2A with endogenous M18A localized with an antibody to its C-terminus, and the resulting green-red-green pattern observed in TIRF-SIM. Bottom TIRF-SIM image of Rat2 cells expressing EGFP-NM2A and immunostained for M18A (red secondary antibody). White numbered boxes correspond to the magnified insets. White brackets indicate mixed filaments of M18A and NM2A. Scale bars as in (A). (C) Top: Cartoon depicting coassembly of endogenous M18A with endogenous NM2A both localized with antibodies to their respective C-termini, and the yellow puncta with various amounts of closely-associated red and green signals observed in TIRF-SIM. Bottom: Rat2 cells immunostained for endogenous M18A (red secondary antibody) and NM2A (primary antibody directly conjugated to green Alexa FluorAF488) were imaged with TIRF-SIM. Numbered boxes correspond to the row of images on the right, which are presented as gray scale images in the red and green channels and color in the overlay channel. Arrowheads indicate some of the overlapping green and red puncta indicative of mixed filaments. Note that yellow in the overlay channel is only very obvious when the intensities for red and green are approximately equal, which is often not the case when examining the split images. Scale bars represent 1 μm.
To examine the mechanism of M18A and NM2 coassembly in live cells, we expressed a version of M18A with the head and neck regions removed (M18A-tail). If M18A and NM2 coassemble via electrostatic interactions between their coiled-coil domains, then the tail of M18A should be sufficient to create mixed filaments. Consistently, HeLa cells co-expressing EGFP-M18A-tail and tdTomato-NM2A contained numerous red-green-green-red structures indicative of mixed bipolar filaments (Figure S3B). Similar results were seen with mApple-M18A-tail in MEFs expressing EGFP-NM2A (Figure S3B).
To determine if endogenous M18A incorporates into NM2 filaments, we expressed EGFP-NM2A in Rat2 fibroblasts and immunostained with an antibody to the M18A C-terminus (with red secondary antibody). If endogenous M18A coassembles with EGFP-NM2A, TIRF-SIM should reveal two green puncta bifurcated by a single red punctum (Figure 3B cartoon). Indeed, such green-red-green structures were readily apparent (Figure 3B and insets B1 and B2; see white brackets).
Finally, we sought to determine if endogenous M18A coassembles with endogenous NM2 in living cells. Rat2 fibroblasts were immunostained with the C-terminally-directed M18A polyclonal antibody (with red secondary antibody), followed with a C-terminally-directed NM2A polyclonal antibody that had been directly conjugated with green AlexaFluor-488. Examples of coassembly imaged in SIM should yield yellow puncta with various amounts of closely-associated red and green signals, where the intensity of the yellow signal depends on the relative intensities of the individual red and green puncta (Figure 3C cartoon; see also [16]). Such signals are indeed common in stress fibers and lamellae of Rat2 cells, indicating endogenous M18A coassembles with endogenous NM2A (Figure 3C and insets C1 and C2; see arrowheads). Similar results were obtained with endogenous M18A and NM2B (Figure S4C). Of note, not all NM2 filaments appear to contain M18A, and not all M18A appears to partition into NM2 filaments, suggesting theirs is not an obligate interaction. Nonetheless, these results in total argue strongly that the coassembly of M18A and NM2 observed with pure proteins in vitro can be extrapolated to their behavior in live cells.
Conclusions
The surprising finding that class-18 myosins lack motor activity raised many questions regarding their cellular functions. Our demonstration that M18A does not self-assemble into filaments but rather coassembles with NM2 into mixed bipolar filaments offers insights into what M18A might do in cells. One possibility is that M18A regulates the assembly properties of NM2. Consistently, we demonstrated in vitro that M18Aβ reduces the size of NM2A filaments, possibly even preventing their formation when present in great excess. M18A could also regulate the mechanical output of filaments by reducing the number of force generating heads, thereby reducing processivity [17] (Figure 4A). It should be noted, however, that the ratio of M18A to NM2 in U2OS cells is 1:200 [15], arguing that M18A may not be present at sufficiently high levels in vivo to alter NM2 filament size (even in transfected cells; see Figure S4) or negatively regulate NM2 self-assembly. That said, the ratio of M18A to NM2 could be higher in some cell types or subcellular locations, so a role for M18A in regulating NM2 filament size/assembly cannot be ruled out.
Figure 4. Models showing potential roles for M18:NM2 coassembly in vivo.
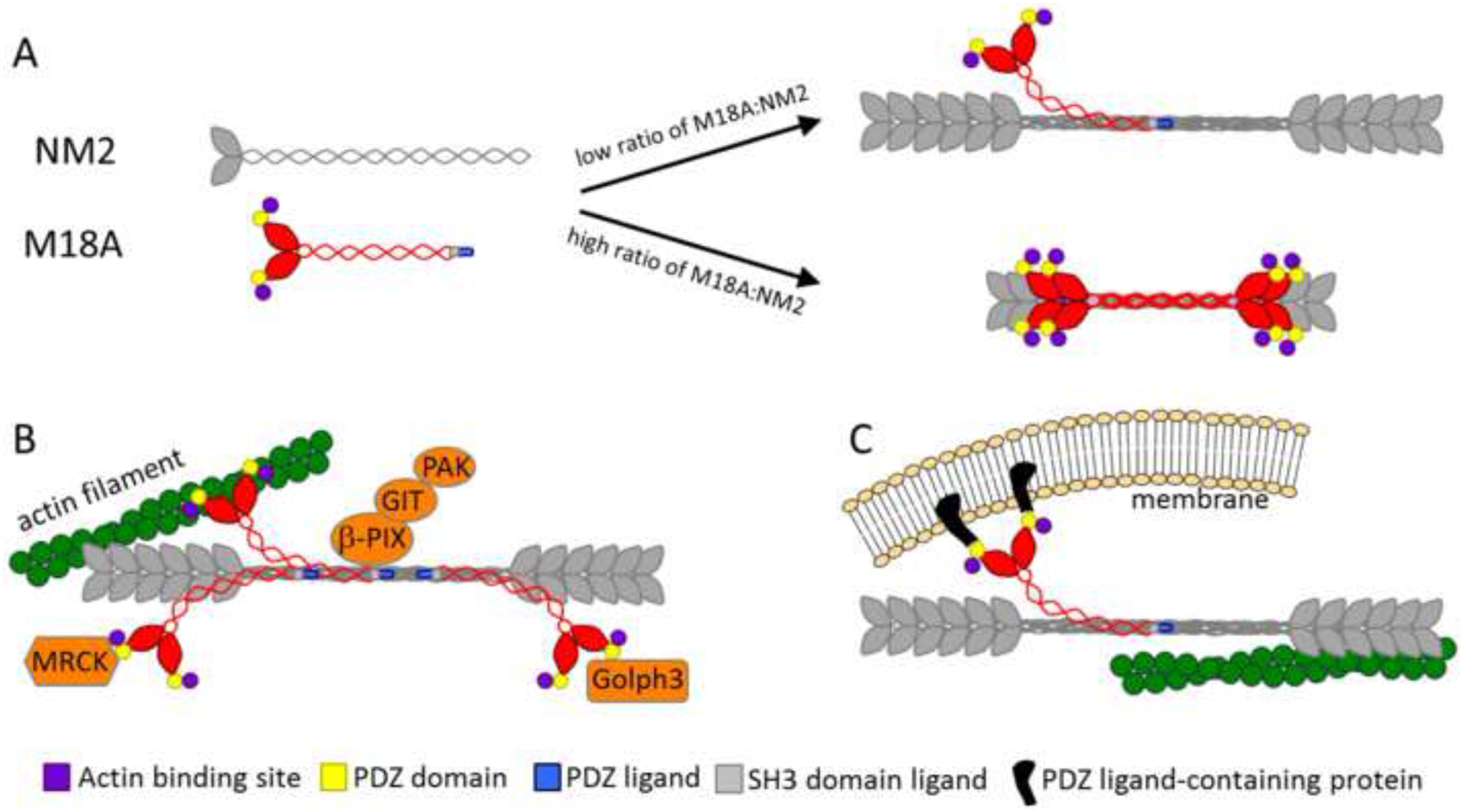
(A) The addition of M18A to NM2 filaments may regulate filament size and mechanochemistry. Based on our in vitro data, the extent to which M18A effects NM2 filament size depends on the relative concentrations of the two myosins, with low ratios of M18A to NM2 (probably the most common situation in vivo) having minimal effects, and with high ratios of M18A to NM2 significantly reducing filament size and likely force output. (B) Addition of M18A, with its protein: protein interaction domains, to NM2 filaments may allow recruitment of specific molecules to hybrid filaments. (C) These protein: protein interaction domains may also serve to attach hybrid filaments to anchored structures against which the filaments can then generate contractile force.
A more likely possibility is that M18A enhances the interaction repertoire of hybrid filaments via the protein:protein interaction sites present within its N- and C-termini. These sites, which include a PDZ domain, PDZ domain ligand, and potential SH3 domain ligands, could recruit specific molecules to filaments (Figure 4B). M18A has been shown to interact with the Rac GEF β–PIX in a complex with PAK2 and GIT1 [14, 18], with a complex containing LRAP35A and the NM2 kinase MRCK [15], and with the Golgi-resident protein GOLPH3 [2, 19]. M18A could also link hybrid filaments to firmly-anchored structures against which hybrid filaments could generate force even when actin filaments are not present on both sides of the bipolar myosin filament (Figure 4C). One such structure could be the PDZ domain-rich postsynaptic density within dendritic spines of neurons, as M18A is highly expressed in neurons, and NM2 is required for normal spine structure/function [20, 21]. Given that M18A protein levels are likely far substoichiometric to NM2 levels, we envision NM2 filaments doped with small amounts of M18A that then drive the above interactions. Importantly, these ATPase-deficient M18A molecules will not interfere with the mechanochemical activities of the NM2 heads in the hybrid filament. In this functional scenario, therefore, the fact that M18A lacks motor activity and does not form filaments on its own makes perfect sense.
Finally, M18A might regulate the formation and/or stability of higher-order acto-NM2 structures like stress fibers [15]. For example, M18Aα might be capable of linking together adjacent stress fibers via its N-terminal nucleotide-insensitive actin binding site. This domain might also serve to stabilize individual stress fibers when levels of active NM2 heads within the hybrid filaments drop. Interactions supported by this and other domains within M18A might also contribute to organizing stress fibers in a sarcomeric-like fashion by recruiting templating molecules, analogous with muscle sarcomeres [22].
The recent demonstration at both the light [16] and EM [23] levels that NM2A, NM2B and NM2C coassemble into heterotypic filaments has expanded our appreciation of NM2 filament diversity. The implications of the results presented here force an even greater reappraisal of this diversity. The ability of cells to mix and match multiple NM2 isoforms with multiple M18A isoforms means filaments with a wide range of enzymatic, mechanical, and interaction properties are possible [24–27]. Importantly, M18 is expressed throughout Metazoa, is widely expressed in vertebrate tissues, and appears essential for viability [28] (our unpublished data). Given this, and the myriad cellular functions supported by NM2 [29, 30], our results have far reaching functional implications. Future studies on the mechanical properties and cellular functions of hybrid M18A:NM2 filaments should yield important new insights into the roles of NM2 in cells and tissues.
Experimental Procedures
Cosedimentation assays
Myosins were mixed in Buffer A (2 mM MgCl2, 0.1 mM EGTA, 10 mM MOPS, pH 7.0) containing 300 mM KCl. Polymerization was initiated by dilution into Buffer A to give a final KCl concentration of 150 mM. Following 10 minute incubation on ice, mixtures were sedimented at 100,000 RPM (RCF ~386,000 × g) for 30 minutes in a Beckman TLA-100 rotor at 4°C. The amount of each myosin in the supernatants and pellets was determined using SDS-PAGE and gel densitometry following staining with Coomassie Blue. Data were plotted using GraphPad Prism software and fit to the one site (total) binding equation.
TIRF microscopy of fluorescently-labeled Halotag proteins
HaloTag M18A and NM2A were labeled with 5-fold molar excess HaloTag TMR Ligand and HaloTag Alexa Fluor 488 Ligand, respectively, overnight on ice. Excess dye was removed using Sephadex G-25 spin columns. Labeled myosins were incubated in Buffer A containing various modifications as indicted in the text. Flow cells (~40μl) were constructed as described previously [17]. Proteins at 5 nM were adsorbed directly onto glass coverslips and the chamber washed with 10-fold excess of Buffer A containing 150 mM KCl and 50 mM DTT. TIRF microscopy was conducted on a Nikon N-STORM microscope operating in TIRF mode using a 100x oil objective (NA 1.49). Data were analyzed using ImageJ. To estimate the number of myosins in individual puncta, the size of an individual photobleaching step was first calculated. The mean of 10 clear photobleaching steps in each field of view (where each step is the average value over 5 seconds pre-step minus average value over 5 seconds post-step), for each channel, was calculated. The total intensity drop in each punctum in the micrograph, following complete photobleaching of the entire field, was then measured and divided by the drop due to single-step photobleaching. A 5×5 pixel square was used for all measurements. Data from 3 separate fields of view in each condition were combined. Particle detection was carried out using the ImageJ ‘Find Maxima’ plugin.
SIM imaging
TIRF-SIM imaging was performed as previously described [16] for all images except Figure 3C, which was collected on a DeltaVision OMX 3D-SIM Imaging System (Applied Precision), as previously described [16].
Supplementary Material
Acknowledgements
The authors thank the Electron Microscopy, Light Microscopy, and Flow Cytometry Core Facilities of the NHLBI for their support, advice and the use of facilities, Michelle Baird and Clare Waterman (NHLBI) for reagents, Bob Adelstein and the Lab of Molecular Cardiology (NHLBI) for mice, and Grzegorz Piszczek (NHLBI Biophysical Core Facility) for help with ultracentrifugation studies.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Guzik-Lendrum S, Heissler SM, Billington N, Takagi Y, Yang Y, Knight PJ, Homsher E, and Sellers JR (2013). Mammalian myosin-18A, a highly divergent myosin. J Biol Chem 288, 9532–9548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Taft MH, Behrmann E, Munske-Weidemann LC, Thiel C, Raunser S, and Manstein DJ (2013). Functional characterization of human myosin-18A and its interaction with F-actin and GOLPH3. J Biol Chem 288, 30029–30041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guzik-Lendrum S, Nagy A, Takagi Y, Houdusse A, and Sellers JR (2011). Drosophila melanogaster myosin-18 represents a highly divergent motor with actin tethering properties. J Biol Chem 286, 21755–21766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Billington N, Wang A, Mao J, Adelstein RS, and Sellers JR (2013). Characterization of three full-length human nonmuscle myosin II paralogs. J Biol Chem 288, 33398–33410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lowey S, Slayter HS, Weeds AG, and Baker H (1969). Substructure of the myosin molecule. I. Subfragments of myosin by enzymic degradation. J Mol Biol 42, 1–29. [DOI] [PubMed] [Google Scholar]
- 6.Huxley HE (1963). Electron Microscope Studies on Structure of Natural and Synthetic Protein Filaments from Striated Muscle. Journal of Molecular Biology 7, 281-&. [DOI] [PubMed] [Google Scholar]
- 7.Niederman R, and Pollard TD (1975). Human platelet myosin. II. In vitro assembly and structure of myosin filaments. J Cell Biol 67, 72–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burgess SA, Yu S, Walker ML, Hawkins RJ, Chalovich JM, and Knight PJ (2007). Structures of smooth muscle myosin and heavy meromyosin in the folded, shutdown state. J Mol Biol 372, 1165–1178. [DOI] [PubMed] [Google Scholar]
- 9.Onishi H, and Wakabayashi T (1984). Electron microscopic studies on myosin molecules from chicken gizzard muscle III. Myosin dimers. Journal of biochemistry 95, 903–905. [DOI] [PubMed] [Google Scholar]
- 10.Trybus KM, and Lowey S (1984). Conformational states of smooth muscle myosin. Effects of light chain phosphorylation and ionic strength. J Biol Chem 259, 8564–8571. [PubMed] [Google Scholar]
- 11.Straussman R, Squire JM, Ben-Ya’acov A, and Ravid S (2005). Skip residues and charge interactions in myosin II coiled-coils: implications for molecular packing. J Mol Biol 353, 613–628. [DOI] [PubMed] [Google Scholar]
- 12.Ricketson D, Johnston CA, and Prehoda KE (2010). Multiple tail domain interactions stabilize nonmuscle myosin II bipolar filaments. Proc Natl Acad Sci U S A 107, 20964–20969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mori K, Furusawa T, Okubo T, Inoue T, Ikawa S, Yanai N, Mori KJ, and Obinata M (2003). Genome structure and differential expression of two isoforms of a novel PDZ-containing myosin (MysPDZ) (Myo18A). Journal of biochemistry 133, 405–413. [DOI] [PubMed] [Google Scholar]
- 14.Hsu RM, Tsai MH, Hsieh YJ, Lyu PC, and Yu JS (2010). Identification of MYO18A as a novel interacting partner of the PAK2/betaPIX/GIT1 complex and its potential function in modulating epithelial cell migration. Molecular biology of the cell 21, 287–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tan I, Yong J, Dong JM, Lim L, and Leung T (2008). A tripartite complex containing MRCK modulates lamellar actomyosin retrograde flow. Cell 135, 123–136. [DOI] [PubMed] [Google Scholar]
- 16.Beach JR, Shao L, Remmert K, Li D, Betzig E, and Hammer JA 3rd (2014). Nonmuscle myosin II isoforms coassemble in living cells. Curr Biol 24, 1160–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nagy A, Takagi Y, Billington N, Sun SA, Hong DK, Homsher E, Wang A, and Sellers JR (2013). Kinetic characterization of nonmuscle Myosin IIb at the single molecule level. J Biol Chem 288, 709–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hsu RM, Hsieh YJ, Yang TH, Chiang YC, Kan CY, Lin YT, Chen JT, and Yu JS (2014). Binding of the extreme carboxyl-terminus of PAK-interacting exchange factor beta (betaPIX) to myosin 18A (MYO18A) is required for epithelial cell migration. Biochim Biophys Acta 1843, 2513–2527. [DOI] [PubMed] [Google Scholar]
- 19.Dippold HC, Ng MM, Farber-Katz SE, Lee SK, Kerr ML, Peterman MC, Sim R, Wiharto PA, Galbraith KA, Madhavarapu S, et al. (2009). GOLPH3 bridges phosphatidylinositol-4- phosphate and actomyosin to stretch and shape the Golgi to promote budding. Cell 139, 337–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ryu J, Liu L, Wong TP, Wu DC, Burette A, Weinberg R, Wang YT, and Sheng M (2006). A critical role for myosin IIb in dendritic spine morphology and synaptic function. Neuron 49, 175–182. [DOI] [PubMed] [Google Scholar]
- 21.Hodges JL, Newell-Litwa K, Asmussen H, Vicente-Manzanares M, and Horwitz AR (2011). Myosin IIb activity and phosphorylation status determines dendritic spine and post-synaptic density morphology. PLoS One 6, e24149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tojkander S, Gateva G, Schevzov G, Hotulainen P, Naumanen P, Martin C, Gunning PW, and Lappalainen P (2011). A molecular pathway for myosin II recruitment to stress fibers. Curr Biol 21, 539–550. [DOI] [PubMed] [Google Scholar]
- 23.Shutova MS, Spessott WA, Giraudo CG, and Svitkina T (2014). Endogenous Species of Mammalian Nonmuscle Myosin IIA and IIB Include Activated Monomers and Heteropolymers. Curr Biol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kovacs M, Wang F, Hu A, Zhang Y, and Sellers JR (2003). Functional divergence of human cytoplasmic myosin II: kinetic characterization of the non-muscle IIA isoform. J Biol Chem 278, 38132–38140. [DOI] [PubMed] [Google Scholar]
- 25.Kim KY, Kawamoto S, Bao J, Sellers JR, and Adelstein RS (2008). The B2 alternatively spliced isoform of nonmuscle myosin II-B lacks actin-activated MgATPase activity and in vitro motility. Biochem Biophys Res Commun 369, 124–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jana SS, Kim KY, Mao J, Kawamoto S, Sellers JR, and Adelstein RS (2009). An alternatively spliced isoform of non-muscle myosin II-C is not regulated by myosin light chain phosphorylation. J Biol Chem 284, 11563–11571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heissler SM, and Manstein DJ (2011). Comparative kinetic and functional characterization of the motor domains of human nonmuscle myosin-2C isoforms. J Biol Chem 286, 21191–21202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bonn BR, Rudolf A, Hornbruch-Freitag C, Daum G, Kuckwa J, Kastl L, Buttgereit D, and Renkawitz-Pohl R (2013). Myosin heavy chain-like localizes at cell contact sites during Drosophila myoblast fusion and interacts in vitro with Rolling pebbles 7. Exp Cell Res 319, 402–416. [DOI] [PubMed] [Google Scholar]
- 29.Vicente-Manzanares M, Ma X, Adelstein RS, and Horwitz AR (2009). Non-muscle myosin II takes centre stage in cell adhesion and migration. Nat Rev Mol Cell Biol 10, 778–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heissler SM, and Manstein DJ (2013). Nonmuscle myosin-2: mix and match. Cell Mol Life Sci 70, 1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


