Abstract
Assisted oocyte activation (AOA) has been proposed as an effective technique to overcome the problem of impaired fertilization after intracytoplasmic sperm injection (ICSI) but the safety of AOA remains a concern. We aimed to investigate if AOA induces imprinting effects on embryos. We used 13 cleavage embryos, nine blastocysts, and eight placentas from 15 patients. The subjects were divided into six groups by tissue type and with or without AOA. The methylation levels of imprinted genes (H19, paternally expressed gene [PEG3] and small nuclear ribonucleoprotein polypeptide N [SNRPN]) were tested by pyrosequencing. We observed different methylation levels among cleavage embryos. The variability was much more remarkable between cleavage embryos than blastocysts and placenta tissues. The methylation levels were especially higher in SNRPN and lower in the H19 gene in AOA embryos than those without AOA. No significant difference was found either among blastocysts or among placenta tissues regardless of AOA. The methylation levels of the three genes in blastocysts were very similar to those in the placenta. Compared to conventional ICSI, AOA changed imprinting methylation rates at H19 and SNRPN in cleavage embryos but not in the blastocyst stage and placenta. We recommend that blastocyst transfer should be considered for patients undergoing AOA during in vitro fertilization.
Keywords: assisted oocyte activation, intracytoplasmic sperm injection, ionophore A23187, imprinted genes, methylation
1. Introduction
The introduction and implementation of intracytoplasmic sperm injection (ICSI) have become the most successful micromanipulation procedure for treating male infertility. However, although the fertilization rate of ICSI is 70–80% [1,2], total fertilization failure still occurs in 1–3% of ICSI cycles and can reoccur in subsequent cycles [3,4,5]. Thus, although total fertilization failure after ICSI is a rare event, it may occur in the presence of a presumptively normal spermatozoon. Moreover, low fertilization (<30%) can be observed in repeated ICSI cycles for some patients [6].
After appropriate counseling, the combination of ICSI with assisted oocyte activation (AOA) is often recommended for couples dealing with total or nearly total fertilization failure after ICSI. At present, several chemical, mechanical or physical stimuli are applied to promote oocyte activation during a subsequent ICSI cycle to overcome this failed fertilization [7]. Previous studies have reported an increase in fertilization rates and utilization of cleavage stage embryos with AOA [8]. The AOA protocol is usually based on Ca2+ ionophores [9,10,11,12], strontium [13,14], a modified ICSI technique [15,16], or electric pulses [17,18]. Among these protocols, Ca2+ ionophore A23187 treatment has been widely applied in human oocyte activation [3].
During the physiological process of fertilization, the oocyte is activated by phospholipase C zeta, a sperm-borne factor [19,20,21], which induces the production of inositol-triphosphate in the ooplasm and releases calcium from the endoplasmic reticulum in an oscillatory mode [3]. Sperm-induced Ca2+ oscillations stimulate mitochondrial respiration and, in turn, the resulting adenosine triphosphate production is required to maintain sperm-triggered calcium waves. Nevertheless, during the AOA process, the oocyte activation with A23187 induces Ca2+ elevation in the form of a single transient, which is not followed by further Ca2+ oscillations. Beyond that, the action of A23187 can release calcium in an uncontrolled fashion from all intracellular stores, including those that would not normally be involved in the activation process. Therefore, because of the nonphysiological effect of A23187, the safety of AOA in the process of assisted reproductive technology (ART) should be carefully monitored.
The potential of calcium ionophores to support oocyte activation and achieve acceptable fertilization rates has been already tested in mice [22,23]. Moreover, retrospective studies also analyzed their oocyte activation and proposed its benefit, which resulted in healthy babies [6,10,24,25]. Yet, these studies have mainly focused on investigating the effectiveness of AOA in reproductive medicine, while few studies reported on the safety of this approach. In fact, ionophores, including A23187, exert many effects on cell homeostasis that might have a long-term effect on gene expression, some of which might be a threat and possible risk for epigenetics [26,27,28].
In this study, we evaluated the effects of AOA on imprinted genes during ART treatment. Paternal imprinted maternally expressed transcript (H19), paternally expressed gene (PEG3), and small nuclear ribonucleoprotein polypeptide N (SNRPN), which are well-studied imprinted genes, were selected for analysis. Abnormal methylation of the SNRPN gene has been reported in imprinting syndromes that with an increased prevalence in children conceived using ART [29]. H19 is a paternally methylated imprinted gene; its alterations have been described in placentas from ART pregnancy [30]. The loss of PEG3 imprinted methylation has been observed in mouse blastocysts derived from ART [31]. By using the donated embryos and placenta tissue, we compared the methylation status of differentially methylated regions (DMRs) of these three key imprinted genes between patients with and without AOA.
2. Methods
2.1. Ethical approval
The study was approved by the Ethical Committee of Peking University People’s Hospital (approval number 2011-67). All the patients delivered a healthy baby after ART treatment. Surplus embryos and placenta tissues were donated for research with written consent (Table A1).
2.2. IVF-ET treatment protocols and artificial oocyte activation
The women who were offered AOA had at least one total or nearly total fertilization failure after ICSI in previous cycles, and one couple with globozoospermia was offered AOA on half oocytes and conventional ICSI on the remaining half oocytes. The patients who underwent conventional ICSI treatment (nonassisted oocyte activation [NOA]) were used as controls; the two groups were matched by age.
The individual stimulation protocols for in vitro fertilization & embryo transfer (IVF-ET) were determined according to the age of the patient and the ovarian reserve status, including the antral follicle count, basal levels of follicle-stimulating hormone (FSH), luteinizing hormone, and estradiol (E2). Most women underwent the long luteal downregulation protocol. Briefly, 1.25 mg of gonadotropin-releasing hormone agonist (GnRHa, Diphereline®, Beaufour-Ipsen Pharmaceuticals Ltd., Paris, France) was injected on menstrual day 21. The initial gonadotropin dose was based on the physician’s discretion but always contained an amount of rFSH, supplemented with at least one ampoule (75 IU) of human menopausal gonadotropin. For the flare-up agonist stimulation, a dose of rFSH along with a fixed dose of GnRHa (0.1 mg/day, triptorelin, Ferring, Saint-Prex, Switzerland) was administered beginning on menstrual day 2. Human chorionic gonadotropin (HCG, 10,000 IU, Lizhu Ltd., Guangdong, China) was administered when at least two follicles were 18 mm in diameter. Oocytes were retrieved by transvaginal ultrasound-guided follicular aspiration 36 h later. Oocytes were fertilized using ICSI.
For artificial oocyte activation, 30 min after ICSI, oocytes were incubated in a culture medium containing 10 mM calcium ionophore A23187 (Sigma) for 10 min at 37°C and 6% CO2. The oocytes were then extensively washed and placed in a culture medium (G-1; Vitrolife) in the incubator under 6% CO2, 5% O2, and 89% N2.
The fertilization results (two pronuclei, 2PN) were assessed 16–20 h after insemination. High-quality transferred or frozen embryos were defined as embryos developed from normally fertilized eggs, with no more than 20% fragmentation, no multinucleation, and 7–8 blastomeres, 72 h after egg retrieval. Blastocysts scored as Gardner’s classification were transferred or frozen if they reached at least third-stage expansion with A or B for inner cell mass (ICM) or trophectoderm. One or two embryos per patient were transferred on the third or fifth day after oocyte retrieval. Surplus embryos were frozen by vitrification procedure (KITAZATO). After the patients agreed and decided to donate, frozen embryos were thawed, and embryo quality was evaluated. Next, samples were frozen in nitrogen for DNA methylation analyses. Donated placenta tissue was collected within 30 min after delivery and frozen in nitrogen.
2.3. Pyrosequencing
Genomic DNA from a single embryo or placenta was isolated with the DNeasy Blood and Tissue Kit (Qiagen). Subsequent bisulfite conversion was performed using an EpiTect Bisulfite Kit (Qiagen), following the manufacturer’s guidelines. The primers for a polymerase chain reaction and pyrosequencing (Table 1) were designed using PyroMark Assay Design 2.0 software (Qiagen). Polymerase chain reaction amplification for H19, SNRPN, and PEG3 was performed with an initial denaturation step at 95°C for 2 min, 36 cycles at 95°C for 15 s, primer-specific annealing temperature for 15 s, and 72°C for 15 s; a final extension step was completed at 72°C for 7 min. The amplification reaction was carried out in a final volume of 50 µL, containing 2 µL of DNA, 12.5 µL of ready-mix (KAPA 2 G Robust HS ReadyMix), 1 µL of each primer (50 pM/µL) and 8.5 µL H2O. Pyrosequencing of the PCR fragments was performed on a Pyro-Mark Q96 ID pyrosequencing system (Qiagen). Pyro Q-CpG software (Qiagen) was used for data analysis.
Table 1.
Primers used for H19, PG3, and SNRPN methylation analysis
| Imprinted gene | Primer sequence | Amplicon length (base pair) | Number of CpGs site |
|---|---|---|---|
| H19 | Forward | 253 | 6 |
| AGGGTTTTTGGTAGGTATAGAG | |||
| Reverse | |||
| CCTATTCCCAAATAACCCC | |||
| Sequencing | |||
| GTGGAATAGGAAGTGGT | |||
| PEG3 | Forward | 154 | 4 |
| GGTGTAGAAGTTTGGGTAGTT | |||
| Reverse | |||
| ACTCACCTCACCTCAATACTAC | |||
| Sequencing | |||
| GTTTATTTTGGGTTGGT | |||
| SNRPN | Forward | 220 | 7 |
| GGGAGGGAGTTGGGATTTTTGTA | |||
| Reverse | |||
| AAACCACCCACACAACTAACCTTAC | |||
| Sequencing | |||
| GGAGTTGGGATTTTTGTAT |
2.4. Statistical analyses
Statistical analyses were performed with SPSS version 17.0. For each imprinted gene, the difference in DNA methylation between embryos was assessed by a one-way ANOVA test followed by Turkey multiple comparison tests. If the data sets were not normally distributed, the nonparametric Mann–Whitney U test was used to assess between-group differences. A P-value of <0.05 was considered statistically significant.
3. Results
In order to evaluate the possible impact of AOA on epigenetics, we quantified DNA methylation of three imprinted genes (PEG3, SNPRN, and H19) using pyrosequencing on cleavage embryos, blastocysts, and placenta. A total of 13 cleavage embryos, nine blastocysts, and eight placentas were included in this study. As highlighted in Table 2, four cleavage embryos and four blastocysts were derived from AOA, and nine cleavage embryos and five blastocysts from NOA (conventional ICSI). All these embryos came from the same cycle, in which the patients had one or two healthy babies after treatment. In addition to these embryos, placentas were also collected from eight patients; three underwent AOA and five underwent NOA. There were no significant differences between the two groups with regard to women’s age, body mass index, treatment protocol, oocytes, and high-quality embryos (Table A2).
Table 2.
Summary of different groups for DNA methylation analysis
| Group | Tissue type | Number | Method of fertilization | AOA | Live birth (patient who donated) |
|---|---|---|---|---|---|
| AOA-C | D3 cleavage embryo | 4 | ICSI | Yes | Yes |
| NOA-C | D3 cleavage embryo | 9 | ICSI | No | Yes |
| AOA-B | Blastocyst | 4 | ICSI | Yes | Yes |
| NOA-B | Blastocyst | 5 | ICSI | No | Yes |
| AOA-P | Placenta | 3 | ICSI | Yes | Yes |
| NOA-P | Placenta | 5 | ICSI | No | Yes |
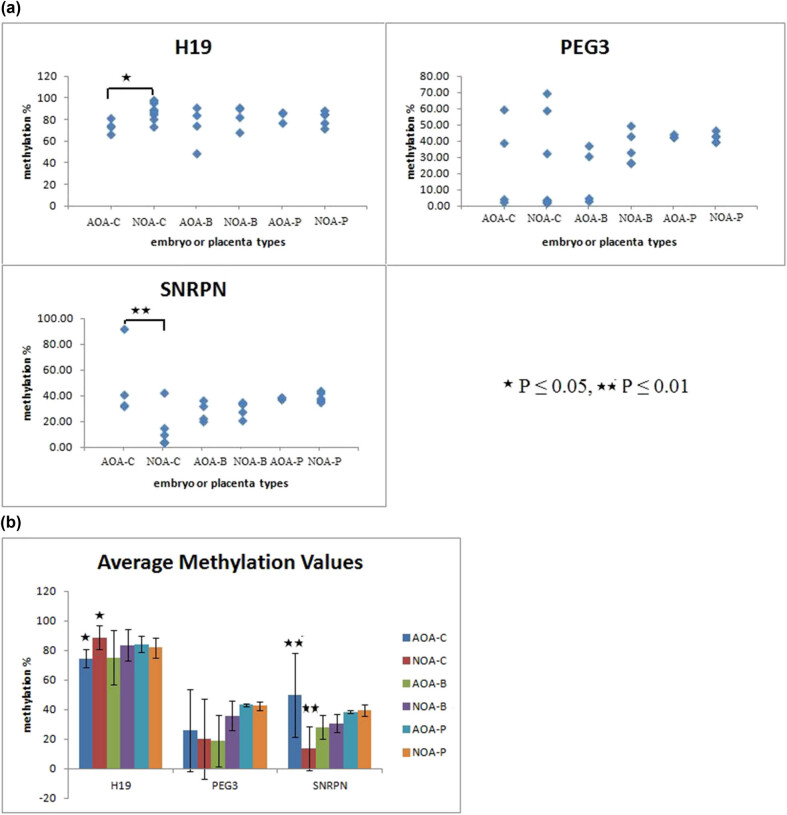
The average methylation levels of H19, PEG3, and SNRPN for each embryo or placenta are shown in Figure 1. First, we compared the methylation levels in cleavage embryos (AOA-C versus NOA-C), blastocysts (AOA-B versus NOA-B), and placentas (AOA-P verse NOA-P) between groups. The greatest range in methylation values occurred in the groups AOA-C and NOA-C, especially for PEG3 (2.5–59.25% in the AOA-C group and 2.0–59% in the NOA-C group) and SNRPN (32.18–92% in the AOA-C group and 4.14–42.57% in NOA-C group); for H19 and SNRPN, the difference in DNA methylation between AOA-C and NOA-C groups was significant (P-value ≤0.05 for H19 and P-value ≤0.001 for SNRPN). For the gene PEG3, no significant difference was observed between AOA-C and NOA-C groups. More importantly, in the blastocysts and placentas the variance of three genes was no difference between two groups.
Figure 1.
Pyrosequencing analysis of DNA methylation at three imprinted genes for different types of embryos or placentas. (a) The global methylation percentage in the methylated allele of H19, PEG3, and SNRPN for different embryos or placentas. Each dot corresponds to an embryo or a placenta. (b) Average methylation values plotted with standard deviation values for a different group. For genes H19 and SNRPN, the difference between AOA-C and NOA-C was significant (P ≤ 0.05 and P ≤ 0.001).
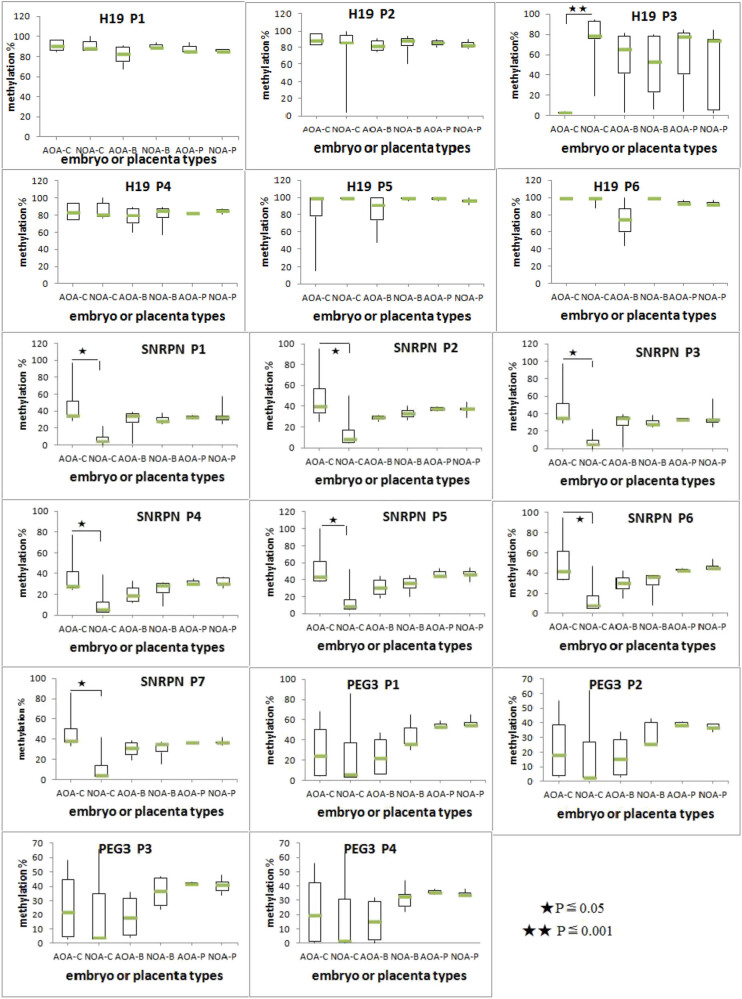
The methylation profiles of individual imprinted alleles for H19, SNRPN, and PEG3 are shown in Figure 2. The number of DMR-associated CpG dinucleotides analyzed for each gene is as follows: H19 (n = 6), SNRPN (n = 7), and PEG3 (n = 4). The DNA methylation status of each individual CpG was then determined by pyrosequencing. For the gene SNRPN, the comparison between AOA-C and NOA-C groups revealed a significant difference; the methylation levels of all the 7 CpG sites were significantly higher in cleavage embryos of AOA than those of NOA (P < 0.05). For the gene H19, methylation values on the third site were significantly different between the cleavage embryos, which were lower in the AOA-C group than in the NOA group (3.45 ± 0.47% versus 76.33 ± 22.69%, P < 0.05). For PEG3, although there was a great deviation on cleavage embryos, no significant difference was observed between AOA-C and NOA-C groups (P > 0.05). Notably, for all these CpG sites of three imprinted genes, the methylation levels showed no significant difference in blastocysts with or without AOA (AOA-B versus NOA-B, P > 0.05). Similar data were obtained when comparing the methylation variance between placenta in the two groups (AOA-P versus NON-P).
Figure 2.
The methylation profile of methylated alleles of H19, SNRPR, and PEG3 for a different group. Boxplot diagrams present the methylation percentage of all CpG sites across H19 (H19 P1-P6), SNRPR (SNRPN P1-7), and (PEG3 P1-P4) in six groups (AOA-C, NOA-C, AOA-B, NOA-B, AOA-P, and NOA-P). The box represents the interquartile range, which contains 50% of values. The whiskers are lines that extend from the box to the highest and lowest levels. A line across the box indicates the median value for each group. Statistical significance values are as follows: *P ≤ 0.05 and **P ≤ 0.001.
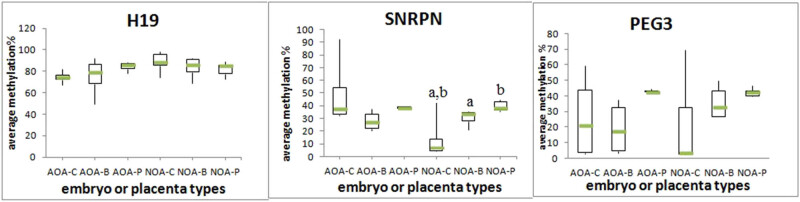
The development trend of the three imprinted genes from cleavage embryos, the blastocyst to the placenta is shown in Figure 3. For the three genes, the relative methylation level had a similar kinetic trend from the blastocyst stage to the placenta, both in the AOA group and NOA group. The methylation level in the blastocyst and placenta was very close in both groups. When comparing among AOA-C, AOA-B, and AOA-P or comparing among NOA-C, NOA-B, and NOA-P, there was no significant variance for the methylation levels of genes H19 and PEG3 (P > 0.05). The results were different for the gene SNRPN. Comparison across the groups without activation revealed that a significant difference existed between NOA-C and NOA-B and NOA-C and NOA-P. The methylation level of SNRPN in cleavage embryos was much lower than those in blastocysts and placentas. However, there were no significant differences in any of the performed comparisons across the group with activation.
Figure 3.
Boxplot diagrams showing the average methylation percentage of H19, SNRPR, and PEG3 in six groups (AOA-C, AOA-B, AOA-P, NOA-C, NOA-B, and NOA-P). Comparison results between groups without activation and between groups with activation were separately conducted. Boxplots with the same superscripts are significantly different (P value <0.05).
4. Discussion
AOA is an effective method for avoiding total fertilization failure. It can repair defective activation and improve ICSI outcomes. Clinical trials suggested that birth outcomes and health for children from artificial oocyte activation techniques are comparable to those children conceived by conventional ICSI [32] and that the developmental outcomes of children 3–10 years born after AOA are within expected ranges [25]. Yet, the number of live births after AOA is still low. Nonetheless, the sole concept of using live birth as the end-point for successful IVF is somewhat controversial [33]. Although the use of calcimycin or ionomycin, which can increase membrane permeability to extracellular Ca2+, had not been linked with any deleterious effects and did not cause chromosomal abnormalities [34,35], their safety and potential long-term effects during embryogenesis are still unknown. Recently, Chen et al. showed that a high concentration of ionomycin increased DNA damage and decreased mouse blastocyst formation [36]. This study indicated that the improper application of AOA may have adverse effects on pre-implantation embryo development.
Biochemical processes during artificial oocyte activation are well-investigated [37]; yet, most data come from preclinical studies. Because gametogenesis and embryogenesis exhibit considerable species differences, particularly between humans and rodents, experimental findings in animal models cannot directly and accurately reflect the real situation in humans. Knowing that ionophores can affect cell homeostasis and, in turn, have long-term effects on gene expression, concerns have been raised regarding the interference of AOA in the epigenetic quality of the oocyte and embryo. Sill, evidence reporting epigenetic effects of AOA on humans is limited. ARTs and infertility may also be associated with epigenetic disorders such as the disruption of genomic imprinting [38,39,40]. Thus, further investigation is needed to investigate whether oocyte activation may cause epigenetic modifications as assumed for in vitro operation or culture media.
In many studies, imprinted genes were used as a model for studying ART-induced epigenetic changes in oocytes [41,42,43,44,45,46]. In this study, surplus embryos donated by patients who underwent ART were used to investigate the impact of oocyte activation on the methylation of imprinted genes. Our data suggested that the methylation levels for all three genes (H19, PEG3, and SNRPN) were altered in embryos obtained by AOA [31]. The methylation levels of H19 and SNRPN in cleavage embryos obtained by activation were significantly different from embryos obtained without activation. The methylation level of SNRPN was much higher, and the methylation level of H19 was much lower in a group with activation, while no difference was observed for PEG3. Based on these results, it was not possible to assess which level was exactly right as we did not have the information on the epigenetic variation for embryos from fertile couples. Yet, these data suggested that AOA might impact certain imprinted genes compared to traditional ICSI. Our results support the hypothesis that the combination of ARTs could induce more epimutations in the embryos as was previously suggested [40,47].
Apart from being accurately established during gametogenesis, genome-wide changes in DNA methylation may occur during the pre-implantation period. The dramatic DNA demethylation occurs from fertilization and the two-cell stage human embryo and reaches the lowest DNA methylation at the blastocyst’s ICM. Greater global demethylation and DNA remethylation changes make the embryo more susceptible to disturbances [46,48,49]. In this study, most of the obvious changes were observed in cleavage embryos, suggesting that cleavage embryos were more vulnerable to AOA. We also noticed that the methylation values of imprinted genes were more stable in the blastocyst and placenta either from the activation group or from traditional ICSI. Also, the methylation level in the blastocyst was very close to that in the placenta. Thus, it seems that the influence of AOA on imprinted genes tends to be stable after the embryos develop to blastocyst. Because the blastocyst in this study could not be the same one that developed from cleavage embryos involved in the study, it was not easy to ascertain whether the process of self-adjustment to the influence of AOA during the embryo development does exist. However, based on the similar methylation states of blastocysts and placentas in either the AOA group or NOA group, it seems that if the embryo survived and developed to a blastocyst, the impact of AOA on future generations might be reduced to a minimum.
An increased incidence of imprinting disorder after ART has been described in humans. For example, ART-associated Angelman syndrome associated with hypomethylation at the SNRPN imprinting control region had been previously reported [50,51,52]. Previous studies in mice showed that ARTs might result in significantly lower global methylation and a higher number of abnormal alleles for maternal SNRPN in embryos when compared with embryos developed in vivo. Interestingly, our results showed that the methylation level of SNRPN was significantly higher in embryos from AOA, complicating the study of the individual effect of oocyte activation. It seemed that the imprinted gene SNRPN was a very sensitive epigenetic mark to survey the effects of ARTs on epigenetics. We also noticed that some individual embryos presented the lowest methylation levels for all three genes. Whether this might be related to their reduced developmental competence should be further investigated.
This study has some limitations. First, the number of samples was relatively small. Our results only elucidated the epigenetic effect of AOA on some specific embryos donated by patients. Second, we only focused on the three well-selected imprinted genes; more imprinted genes should be examined in future studies. Third, the embryos and placentas came from different couples. Because the imprinted genes can show considerable methylation variation among normal individuals, further experiments need to be performed by increasing the sample size, and the epigenetic alteration of baby born from AOA should be specified. Although we found that AOA had some effect on imprinted genes, especially for SNRPN, the functional consequences of methylation changes on the imprinted gene remain to be elucidated. Before that, AOA still needs to be considered as experimental [6,25]; its application requires thorough consultation with the patient and should only be done if correct indications are present.
Abbreviations
- AOA
assisted oocyte activation
- ARTs
assisted reproductive technologies
- DMRs
differentially methylated regions
- E2
estradiol
- FSH
follicle stimulating hormone
- HMG
human menopausal gonadotropin
- ICSI
intracytoplasmic sperm injection
- IP3
inositol-triphosphate
- LH
luteinizing hormone
- NOA
nonassisted oocyte activation
- PEG3
paternally expressed gene
- SNRPN
small nuclear ribonucleoprotein polypeptide N
Acknowledgements
The authors are grateful to the patients who donated their embryos or placenta tissue and participate in this study.
Appendix
Table A1.
Summary of patients for donation of embryos and placenta
| Group | Patients | Age | Indication for IVF | History of total or nearly total fertilzation failure after ICSI | Tissue type | Number | Live birth |
|---|---|---|---|---|---|---|---|
| AOA-C | 1 | 30 | Nonobstructive-azoospermia | 1 | D3 cleavage embryo | 2 | A healthy female baby |
| AOA-P | Placenta | 1 | |||||
| AOA-C | 2 | 20 | Nonobstructive-azoospermia | 1 | D3 cleavage embryo | 2 | A healthy female and a male baby |
| AOA-B | Blastocyst | 1 | |||||
| AOA-B | 3 | 32 | Globozospermia | No | Blastocyst | 2 | A healthy male baby |
| AOA-B | 4 | 30 | Nonobstructive-azoospermia | 2 | Blastocyst | 1 | A healthy female baby |
| AOA-P | 5 | 36 | Nonobstructive-azoospermia | 3 | Placenta | 1 | A healthy male baby |
| AOA-P | 6 | 31 | Severe oligoasthenospermia | 2 | Placenta | 1 | A healthy male baby |
| NOA-C | 1 | 34 | Severe oligozoospermia | No | D3 cleavage embryo | 4 | A healthy male baby |
| NOA-C | 2 | 34 | Severe teratozoospermia | No | D3 cleavage embryo | 5 | Two healthy female babies |
| NOA-B | 3 | 31 | Severe teratozoospermia | No | Blastocyst | 2 | A healthy female baby and a male baby |
| NOA-B | 4 | 32 | Globozospermia | No | Blastocyst | 2 | A healthy male baby |
| NOA-P | 5 | 28 | Obstructive-azoospermia | No | Blastocyst | 1 | A healthy male baby |
| NOA-P | 6 | 32 | Oligozoospermia | No | Placenta | 1 | A healthy female baby |
| NOA-P | 7 | 31 | Severe oligozoospermia | No | Placenta | 1 | A healthy female baby |
| NOA-P | 8 | 28 | Severe oligozoospermia | No | Placenta | 1 | A healthy male baby |
| NOA-P | 9 | 30 | Obstructive-azoospermia | No | Placenta | 1 | A healthy female baby |
| NOA-P | 10 | 33 | Nonobstructive-azoospermia | No | Placenta | 1 | A healthy male baby |
Table A2.
Characteristics and Ovarian hyperstimulation parameters of AOA and conventional ICSI undergoing IVF-ET
| Characteristics | Controls (n = 9) | Cases (n = 6) | p |
|---|---|---|---|
| Age (year) | 31.30 ± 2.16 | 29.83 ± 5.31 | 0.445 |
| BMI (kg/m2) | 22.00 ± 1.41 | 22.09 ± 2.10 | 0.917 |
| Fasting glucose | 4.97 ± 0.45 | 5.06 ± 0.46 | 0.678 |
| Fasting insulin | 7.68 ± 1.07 | 8.00 ± 0.82 | 0.543 |
| Insulin resistance | 1.70 ± 0.32 | 1.79 ± 0.14 | 0.442 |
| Primary infertility | 9 (100%) | 6 (100%) | 1 |
| Treatment protocol | 0.287 | ||
| I | 5 (55.56%) | 5 (83.33%) | |
| II | 3 (33.33%) | 0 (0) | |
| III | 1 (11.11%) | 1 (16.67%) | |
| Basal FSH (IU/L) | 7.27 ± 0.70 | 7.78 ± 1.68 | 0.403 |
| Basal LH (IU/L) | 4.84 ± 0.53 | 4.73 ± 1.26 | 0.815 |
| Basal E2 (pg/L) | 38.30 ± 6.96 | 47.33 ± 10.91 | 0.06 |
| Antral follicle count (n) | 12.80 ± 0.92 | 12.17 ± 5.38 | 0.716 |
| Days of stimulation | 9.90 ± 1.79 | 11.33 ± 2.16 | 0.173 |
| Total dose of gonadotropins (U) | 2212.50 ± 710.85 | 2875.00 ± 1144.00 | 0.171 |
| Number of follicles >13 mm | 16.50 ± 9.52 | 15.67 ± 7.50 | 0.858 |
| Number of oocytes retrieved | 17.80 ± 9.51 | 18.83 ± 10.79 | 0.941 |
| Number of MII oocytes (n) | 13.90 ± 4.33 | 13.67 ± 8.12 | 0.796 |
| Number of 2PN (n) | 9.90 ± 5.30 | 9.17 ± 5.38 | 0.856 |
| Number of high-quality embryos | 4.50 ± 2.65 | 3.40 ± 1.95 | 0.494 |
Footnotes
Funding information: This work was supported by the National Natural Science Foundation of China (No. 82071715), and Capital Clinical Medical Application and Development Funds (No. Z111107058811028). The funders had no role in study design, collection, analysis, and interpretation of data, writing of the report, and decision to submit the article for publication.
Author contributions: The data management and analysis were all completed by Dr Rong Liang, Dr Fang Fang, Dr Sen Li, Dr Xi Chen, and Dr Xiaohong Zhang under the guidance of Dr Qun Lu. Manuscript preparation and editing were completed by Dr Rong Liang and Dr Fang Fang. The authors were all involved in project design and literature search. All authors read and approved the final manuscript.
Conflict of interest: None.
Data availability statement: All data generated or analyzed during this study are included in this published article and its supplementary information files.
References
- [1].Palermo GD, Neri QV, Takeuchi T, Rosenwaks Z. ICSI: where we have been and where we are going. Semin Reprod Med. 2009;27(2):191–201. [DOI] [PubMed]; Palermo GD, Neri QV, Takeuchi T, Rosenwaks Z. ICSI: where we have been and where we are going. Semin Reprod Med. 2009;27(2):191–201. doi: 10.1055/s-0029-1202309. [DOI] [PubMed] [Google Scholar]
- [2].Neri QV, Lee B, Rosenwaks Z, Machaca K, Palermo GD. Understanding fertilization through intracytoplasmic sperm injection (ICSI). Cell Calcium. 2014;55(1):24–37. [DOI] [PMC free article] [PubMed]; Neri QV, Lee B, Rosenwaks Z, Machaca K, Palermo GD. Understanding fertilization through intracytoplasmic sperm injection (ICSI) Cell Calcium. 2014;55(1):24–37. doi: 10.1016/j.ceca.2013.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Kashir J, Heindryckx B, Jones C, De Sutter P, Parrington J, Coward K. Oocyte activation, phospholipase C zeta and human infertility. Hum Reprod Update. 2010;16(6):690–703. [DOI] [PubMed]; Kashir J, Heindryckx B, Jones C, De Sutter P, Parrington J, Coward K. Oocyte activation, phospholipase C zeta and human infertility. Hum Reprod Update. 2010;16(6):690–703. doi: 10.1093/humupd/dmq018. [DOI] [PubMed] [Google Scholar]
- [4].Flaherty SP, Payne D, Matthews CD. Fertilization failures and abnormal fertilization after intracytoplasmic sperm injection. Hum Reprod. 1998;13(Suppl 1):155–64. [DOI] [PubMed]; Flaherty SP, Payne D, Matthews CD. Fertilization failures and abnormal fertilization after intracytoplasmic sperm injection. Hum Reprod. 1998;13(Suppl 1):155–64. doi: 10.1093/humrep/13.suppl_1.155. [DOI] [PubMed] [Google Scholar]
- [5].Esfandiari N, Javed MH, Gotlieb L, Casper RF. Complete failed fertilization after intracytoplasmic sperm injection–analysis of 10 years’ data. Int J Fertil Womens Med. 2005;50(4):187–92. [PubMed]; Esfandiari N, Javed MH, Gotlieb L, Casper RF. Complete failed fertilization after intracytoplasmic sperm injection–analysis of 10 years’ data. Int J Fertil Womens Med. 2005;50(4):187–92. [PubMed] [Google Scholar]
- [6].Montag M, Köster M, van der Ven K, Bohlen U, van der Ven H. The benefit of artificial oocyte activation is dependent on the fertilization rate in a previous treatment cycle. Reprod Biomed Online. 2012;24(5):521–6. [DOI] [PubMed]; Montag M, Köster M, van der Ven K, Bohlen U, van der Ven H. The benefit of artificial oocyte activation is dependent on the fertilization rate in a previous treatment cycle. Reprod Biomed Online. 2012;24(5):521–6. doi: 10.1016/j.rbmo.2012.02.002. [DOI] [PubMed] [Google Scholar]
- [7].Vanden Meerschaut F, Nikiforaki D, Heindryckx B, De Sutter P. Assisted oocyte activation following ICSI fertilization failure. Reprod Biomed Online. 2014;28(5):560–71. [DOI] [PubMed]; Vanden Meerschaut F, Nikiforaki D, Heindryckx B, De Sutter P. Assisted oocyte activation following ICSI fertilization failure. Reprod Biomed Online. 2014;28(5):560–71. doi: 10.1016/j.rbmo.2014.01.008. [DOI] [PubMed] [Google Scholar]
- [8].Sfontouris IA, Nastri CO, Lima ML, Tahmasbpourmarzouni E, Raine-Fenning N, Martins WP. Artificial oocyte activation to improve reproductive outcomes in women with previous fertilization failure: a systematic review and meta-analysis of RCTs. Hum Reprod. 2015;30(8):1831–41. [DOI] [PubMed]; Sfontouris IA, Nastri CO, Lima ML, Tahmasbpourmarzouni E, Raine-Fenning N, Martins WP. Artificial oocyte activation to improve reproductive outcomes in women with previous fertilization failure: a systematic review and meta-analysis of RCTs. Hum Reprod. 2015;30(8):1831–41. doi: 10.1093/humrep/dev136. [DOI] [PubMed] [Google Scholar]
- [9].Heindryckx B, De Gheselle S, Gerris J, Dhont MDe, Sutter P. Efficiency of assisted oocyte activation as a solution for failed intracytoplasmic sperm injection. Reprod Biomed Online. 2008;17(5):662–8. [DOI] [PubMed]; Heindryckx B, De Gheselle S, Gerris J, Dhont MDe, Sutter P. Efficiency of assisted oocyte activation as a solution for failed intracytoplasmic sperm injection. Reprod Biomed Online. 2008;17(5):662–8. doi: 10.1016/s1472-6483(10)60313-6. [DOI] [PubMed] [Google Scholar]
- [10].Ebner T, Köster M, Shebl O, Moser M, Van der Ven H, Tews G, et al. Application of a ready-to-use calcium ionophore increases rates of fertilization and pregnancy in severe male factor infertility. Fertil Steril. 2012;98(6):1432–7. [DOI] [PubMed]; Ebner T, Köster M, Shebl O, Moser M, Van der Ven H, Tews G. et al. Application of a ready-to-use calcium ionophore increases rates of fertilization and pregnancy in severe male factor infertility. Fertil Steril. 2012;98(6):1432–7. doi: 10.1016/j.fertnstert.2012.07.1134. [DOI] [PubMed] [Google Scholar]
- [11].Nasr-Esfahani MH, Deemeh MR, Tavalaee M. Artificial oocyte activation and intracytoplasmic sperm injection. Fertil Steril. 2010;94(2):520–6. [DOI] [PubMed]; Nasr-Esfahani MH, Deemeh MR, Tavalaee M. Artificial oocyte activation and intracytoplasmic sperm injection. Fertil Steril. 2010;94(2):520–6. doi: 10.1016/j.fertnstert.2009.03.061. [DOI] [PubMed] [Google Scholar]
- [12].Vanden Meerschaut F, Nikiforaki D, De Gheselle S, Dullaerts V, Van den Abbeel E, Gerris J, et al. Assisted oocyte activation is not beneficial for all patients with a suspected oocyte-related activation deficiency. Hum Reprod. 2012;27(7):1977–84. [DOI] [PubMed]; Vanden Meerschaut F, Nikiforaki D, De Gheselle S, Dullaerts V, Van den Abbeel E, Gerris J. et al. Assisted oocyte activation is not beneficial for all patients with a suspected oocyte-related activation deficiency. Hum Reprod. 2012;27(7):1977–84. doi: 10.1093/humrep/des097. [DOI] [PubMed] [Google Scholar]
- [13].Yanagida K, Morozumi K, Katayose H, Hayashi S, Sato A. Successful pregnancy after ICSI with strontium oocyte activation in low rates of fertilization. Reprod Biomed Online. 2006;13(6):801–6. [DOI] [PubMed]; Yanagida K, Morozumi K, Katayose H, Hayashi S, Sato A. Successful pregnancy after ICSI with strontium oocyte activation in low rates of fertilization. Reprod Biomed Online. 2006;13(6):801–6. doi: 10.1016/s1472-6483(10)61027-9. [DOI] [PubMed] [Google Scholar]
- [14].Kyono K, Kumagai S, Nishinaka C, Nakajo Y, Uto H, Toya M, et al. Birth and follow-up of babies born following ICSI using SrCl2 oocyte activation. Reprod Biomed Online. 2008;17(1):53–8. [DOI] [PubMed]; Kyono K, Kumagai S, Nishinaka C, Nakajo Y, Uto H, Toya M. et al. Birth and follow-up of babies born following ICSI using SrCl2 oocyte activation. Reprod Biomed Online. 2008;17(1):53–8. doi: 10.1016/s1472-6483(10)60293-3. [DOI] [PubMed] [Google Scholar]
- [15].Tesarik J, Rienzi L, Ubaldi F, Mendoza C, Greco E. Use of a modified intracytoplasmic sperm injection technique to overcome sperm-borne and oocyte-borne oocyte activation failures. Fertil Steril. 2002;78(3):619–24. [DOI] [PubMed]; Tesarik J, Rienzi L, Ubaldi F, Mendoza C, Greco E. Use of a modified intracytoplasmic sperm injection technique to overcome sperm-borne and oocyte-borne oocyte activation failures. Fertil Steril. 2002;78(3):619–24. doi: 10.1016/s0015-0282(02)03291-0. [DOI] [PubMed] [Google Scholar]
- [16].Ebner T, Moser M, Sommergruber M, Jesacher K, Tews G. Complete oocyte activation failure after ICSI can be overcome by a modified injection technique. Hum Reprod. 2004;19(8):1837–41. [DOI] [PubMed]; Ebner T, Moser M, Sommergruber M, Jesacher K, Tews G. Complete oocyte activation failure after ICSI can be overcome by a modified injection technique. Hum Reprod. 2004;19(8):1837–41. doi: 10.1093/humrep/deh325. [DOI] [PubMed] [Google Scholar]
- [17].Zhang J, Wang CW, Blaszcyzk A, Grifo JA, Ozil J, Haberman E, et al. Electrical activation and in vitro development of human oocytes that fail to fertilize after intracytoplasmic sperm injection. Fertil Steril. 1999;72(3):509–12. [DOI] [PubMed]; Zhang J, Wang CW, Blaszcyzk A, Grifo JA, Ozil J, Haberman E. et al. Electrical activation and in vitro development of human oocytes that fail to fertilize after intracytoplasmic sperm injection. Fertil Steril. 1999;72(3):509–12. doi: 10.1016/s0015-0282(99)00264-2. [DOI] [PubMed] [Google Scholar]
- [18].Yanagida K, Katayose H, Yazawa H, Kimura Y, Sato A, Yanagimachi H, et al. Successful fertilization and pregnancy following ICSI and electrical oocyte activation. Hum Reprod. 1999;14(5):1307–11. [DOI] [PubMed]; Yanagida K, Katayose H, Yazawa H, Kimura Y, Sato A, Yanagimachi H. et al. Successful fertilization and pregnancy following ICSI and electrical oocyte activation. Hum Reprod. 1999;14(5):1307–11. doi: 10.1093/humrep/14.5.1307. [DOI] [PubMed] [Google Scholar]
- [19].Saunders CM, Larman MG, Parrington J, Cox LJ, Royse J, Blayney LM, et al. PLC zeta: a sperm-specific trigger of Ca(2+) oscillations in eggs and embryo development. Development. 2002;129(15):3533–44. [DOI] [PubMed]; Saunders CM, Larman MG, Parrington J, Cox LJ, Royse J, Blayney LM. et al. PLC zeta: a sperm-specific trigger of Ca(2+) oscillations in eggs and embryo development. Development. 2002;129(15):3533–44. doi: 10.1242/dev.129.15.3533. [DOI] [PubMed] [Google Scholar]
- [20].Swann K, Larman MG, Saunders CM, Lai FA. The cytosolic sperm factor that triggers Ca2+ oscillations and egg activation in mammals is a novel phospholipase C: PLCzeta. Reproduction. 2004;127(4):431–9. [DOI] [PubMed]; Swann K, Larman MG, Saunders CM, Lai FA. The cytosolic sperm factor that triggers Ca2+ oscillations and egg activation in mammals is a novel phospholipase C: PLCzeta. Reproduction. 2004;127(4):431–9. doi: 10.1530/rep.1.00169. [DOI] [PubMed] [Google Scholar]
- [21].Anifandis G, Messini CI, Dafopoulos K, Daponte A, Messinis IE. Sperm contributions to oocyte activation: more that meets the eye. J Assist Reprod Genet. 2016;33(3):313–6. [DOI] [PMC free article] [PubMed]; Anifandis G, Messini CI, Dafopoulos K, Daponte A, Messinis IE. Sperm contributions to oocyte activation: more that meets the eye. J Assist Reprod Genet. 2016;33(3):313–6. doi: 10.1007/s10815-016-0653-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Heytens E, Soleimani R, Lierman S, De Meester S, Gerris J, Dhont M, et al. Effect of ionomycin on oocyte activation and embryo development in mouse. Reprod Biomed Online. 2008;17(6):764–71. [DOI] [PubMed]; Heytens E, Soleimani R, Lierman S, De Meester S, Gerris J, Dhont M. et al. Effect of ionomycin on oocyte activation and embryo development in mouse. Reprod Biomed Online. 2008;17(6):764–71. doi: 10.1016/s1472-6483(10)60403-8. [DOI] [PubMed] [Google Scholar]
- [23].Heytens E, Schmitt-John T, Moser JM, Jensen NM, Soleimani R, Young C, et al. Reduced fertilization after ICSI and abnormal phospholipase C zeta presence in spermatozoa from the wobbler mouse. Reprod Biomed Online. 2010;21(6):742–9. [DOI] [PubMed]; Heytens E, Schmitt-John T, Moser JM, Jensen NM, Soleimani R, Young C. et al. Reduced fertilization after ICSI and abnormal phospholipase C zeta presence in spermatozoa from the wobbler mouse. Reprod Biomed Online. 2010;21(6):742–9. doi: 10.1016/j.rbmo.2010.07.006. [DOI] [PubMed] [Google Scholar]
- [24].Heindryckx B, Van der Elst J, De Sutter P, Dhont M. Treatment option for sperm- or oocyte-related fertilization failure: assisted oocyte activation following diagnostic heterologous ICSI. Hum Reprod. 2005;20(8):2237–41. [DOI] [PubMed]; Heindryckx B, Van der Elst J, De Sutter P, Dhont M. Treatment option for sperm- or oocyte-related fertilization failure: assisted oocyte activation following diagnostic heterologous ICSI. Hum Reprod. 2005;20(8):2237–41. doi: 10.1093/humrep/dei029. [DOI] [PubMed] [Google Scholar]
- [25].Vanden Meerschaut F, D’Haeseleer E, Gysels H, Thienpont Y, Dewitte G, Heindryckx B, et al. Neonatal and neurodevelopmental outcome of children aged 3–10 years born following assisted oocyte activation. Reprod Biomed Online. 2014;28(1):54–63. [DOI] [PubMed]; Vanden Meerschaut F, D’Haeseleer E, Gysels H, Thienpont Y, Dewitte G, Heindryckx B. et al. Neonatal and neurodevelopmental outcome of children aged 3–10 years born following assisted oocyte activation. Reprod Biomed Online. 2014;28(1):54–63. doi: 10.1016/j.rbmo.2013.07.013. [DOI] [PubMed] [Google Scholar]
- [26].Ducibella T, Fissore R. The roles of Ca2+, downstream protein kinases, and oscillatory signaling in regulating fertilization and the activation of development. Dev Biol. 2008;315(2):257–79. [DOI] [PMC free article] [PubMed]; Ducibella T, Fissore R. The roles of Ca2+, downstream protein kinases, and oscillatory signaling in regulating fertilization and the activation of development. Dev Biol. 2008;315(2):257–79. doi: 10.1016/j.ydbio.2007.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Ebner T, Montag M. Artificial oocyte activation: evidence for clinical readiness. Reprod Biomed Online. 2016;32(3):271–3. [DOI] [PubMed]; Ebner T, Montag M. Artificial oocyte activation: evidence for clinical readiness. Reprod Biomed Online. 2016;32(3):271–3. doi: 10.1016/j.rbmo.2015.12.004. [DOI] [PubMed] [Google Scholar]
- [28].Anifandis G, Michopoulos A, Daponte A, Chatzimeletiou K, Simopoulou M, Messini CI, et al. Artificial oocyte activation: physiological, pathophysiological and ethical aspects. Syst Biol Reprod Med. 2019;65(1):3–11. [DOI] [PubMed]; Anifandis G, Michopoulos A, Daponte A, Chatzimeletiou K, Simopoulou M, Messini CI. et al. Artificial oocyte activation: physiological, pathophysiological and ethical aspects. Syst Biol Reprod Med. 2019;65(1):3–11. doi: 10.1080/19396368.2018.1516000. [DOI] [PubMed] [Google Scholar]
- [29].van Montfoort AP, Hanssen LL, de Sutter P, Viville S, Geraedts JP, de Boer P. Assisted reproduction treatment and epigenetic inheritance. Hum Reprod Update. 2012;18(2):171–97. [DOI] [PMC free article] [PubMed]; van Montfoort AP, Hanssen LL, de Sutter P, Viville S, Geraedts JP, de Boer P. Assisted reproduction treatment and epigenetic inheritance. Hum Reprod Update. 2012;18(2):171–97. doi: 10.1093/humupd/dmr047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Sakian S, Louie K, Wong EC, Havelock J, Kashyap S, Rowe T, et al. Altered gene expression of H19 and IGF2 in placentas from ART pregnancies. Placenta. 2015;36(10):1100–5. [DOI] [PubMed]; Sakian S, Louie K, Wong EC, Havelock J, Kashyap S, Rowe T. et al. Altered gene expression of H19 and IGF2 in placentas from ART pregnancies. Placenta. 2015;36(10):1100–5. doi: 10.1016/j.placenta.2015.08.008. [DOI] [PubMed] [Google Scholar]
- [31].Market-Velker BA, Zhang L, Magri LS, Bonvissuto AC, Mann MR. Dual effects of superovulation: loss of maternal and paternal imprinted methylation in a dose-dependent manner. Hum Mol Genet. 2010;19(1):36–51. [DOI] [PubMed]; Market-Velker BA, Zhang L, Magri LS, Bonvissuto AC, Mann MR. Dual effects of superovulation: loss of maternal and paternal imprinted methylation in a dose-dependent manner. Hum Mol Genet. 2010;19(1):36–51. doi: 10.1093/hmg/ddp465. [DOI] [PubMed] [Google Scholar]
- [32].Lu Q, Chen X, Shen H, Zhang X, Li Y, Liang R, et al. No genetic alterations in infants from intracytoplasmic sperm injection in combination with artificial oocyte activation: a pilot study. Chin Med J (Engl). 2014;127(2):383–5. [PubMed]; Lu Q, Chen X, Shen H, Zhang X, Li Y, Liang R. et al. No genetic alterations in infants from intracytoplasmic sperm injection in combination with artificial oocyte activation: a pilot study. Chin Med J (Engl) 2014;127(2):383–5. [PubMed] [Google Scholar]
- [33].Santella L, Dale B. Assisted yes, but where do we draw the line? Reprod Biomed Online. 2015;31(4):476–8. [DOI] [PubMed]; Santella L, Dale B. Assisted yes, but where do we draw the line? Reprod Biomed Online. 2015;31(4):476–8. doi: 10.1016/j.rbmo.2015.06.013. [DOI] [PubMed] [Google Scholar]
- [34].Deemeh MR, Tavalaee M, Nasr-Esfahani MH. Health of children born through artificial oocyte activation: a pilot study. Reprod Sci. 2015;22(3):322–8. [DOI] [PubMed]; Deemeh MR, Tavalaee M, Nasr-Esfahani MH. Health of children born through artificial oocyte activation: a pilot study. Reprod Sci. 2015;22(3):322–8. doi: 10.1177/1933719114542017. [DOI] [PubMed] [Google Scholar]
- [35].Xu Z, Yao G, Niu W, Fan H, Ma X, Shi S, et al. Calcium ionophore (A23187) rescues the activation of unfertilized oocytes after intracytoplasmic sperm injection and chromosome analysis of blastocyst after activation. Front Endocrinol (Lausanne). 2021;12:692082. [DOI] [PMC free article] [PubMed]; Xu Z, Yao G, Niu W, Fan H, Ma X, Shi S. et al. Calcium ionophore (A23187) rescues the activation of unfertilized oocytes after intracytoplasmic sperm injection and chromosome analysis of blastocyst after activation. Front Endocrinol (Lausanne) 2021;12:692082. doi: 10.3389/fendo.2021.692082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Chen C, Sun T, Yin M, Yan Z, Yu W, Long H, et al. Ionomycin-induced mouse oocyte activation can disrupt preimplantation embryo development through increased reactive oxygen species reaction and DNA damage. Mol Hum Reprod. 2020;26(10):773–83. [DOI] [PubMed]; Chen C, Sun T, Yin M, Yan Z, Yu W, Long H. et al. Ionomycin-induced mouse oocyte activation can disrupt preimplantation embryo development through increased reactive oxygen species reaction and DNA damage. Mol Hum Reprod. 2020;26(10):773–83. doi: 10.1093/molehr/gaaa056. [DOI] [PubMed] [Google Scholar]
- [37].Vanden Meerschaut F, Nikiforaki D, De Roo C, Lierman S, Qian C, Schmitt-John T, et al. Comparison of pre- and post-implantation development following the application of three artificial activating stimuli in a mouse model with round-headed sperm cells deficient for oocyte activation. Hum Reprod. 2013;28(5):1190–8. [DOI] [PubMed]; Vanden Meerschaut F, Nikiforaki D, De Roo C, Lierman S, Qian C, Schmitt-John T. et al. Comparison of pre- and post-implantation development following the application of three artificial activating stimuli in a mouse model with round-headed sperm cells deficient for oocyte activation. Hum Reprod. 2013;28(5):1190–8. doi: 10.1093/humrep/det038. [DOI] [PubMed] [Google Scholar]
- [38].Huntriss J, Picton HM. Epigenetic consequences of assisted reproduction and infertility on the human preimplantation embryo. Hum Fertil (Camb). 2008;11(2):85–94. [DOI] [PubMed]; Huntriss J, Picton HM. Epigenetic consequences of assisted reproduction and infertility on the human preimplantation embryo. Hum Fertil (Camb) 2008;11(2):85–94. doi: 10.1080/14647270802116250. [DOI] [PubMed] [Google Scholar]
- [39].Manipalviratn S, DeCherney A, Segars J. Imprinting disorders and assisted reproductive technology. Fertil Steril. 2009;91(2):305–15. [DOI] [PMC free article] [PubMed]; Manipalviratn S, DeCherney A, Segars J. Imprinting disorders and assisted reproductive technology. Fertil Steril. 2009;91(2):305–15. doi: 10.1016/j.fertnstert.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Simopoulou M, Sfakianoudis K, Rapani A, Giannelou P, Anifandis G, Bolaris S, et al. Considerations regarding embryo culture conditions: from media to epigenetics. Vivo. 2018;32(3):451–60. [DOI] [PMC free article] [PubMed]; Simopoulou M, Sfakianoudis K, Rapani A, Giannelou P, Anifandis G, Bolaris S. et al. Considerations regarding embryo culture conditions: from media to epigenetics. Vivo. 2018;32(3):451–60. doi: 10.21873/invivo.11261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Denomme MM, Mann MR. Genomic imprints as a model for the analysis of epigenetic stability during assisted reproductive technologies. Reproduction. 2012;144(4):393–409. [DOI] [PubMed]; Denomme MM, Mann MR. Genomic imprints as a model for the analysis of epigenetic stability during assisted reproductive technologies. Reproduction. 2012;144(4):393–409. doi: 10.1530/REP-12-0237. [DOI] [PubMed] [Google Scholar]
- [42].Trapphoff T, El Hajj N, Zechner U, Haaf T, Eichenlaub-Ritter U. DNA integrity, growth pattern, spindle formation, chromosomal constitution and imprinting patterns of mouse oocytes from vitrified pre-antral follicles. Hum Reprod. 2010;25(12):3025–42. [DOI] [PubMed]; Trapphoff T, El Hajj N, Zechner U, Haaf T, Eichenlaub-Ritter U. DNA integrity, growth pattern, spindle formation, chromosomal constitution and imprinting patterns of mouse oocytes from vitrified pre-antral follicles. Hum Reprod. 2010;25(12):3025–42. doi: 10.1093/humrep/deq278. [DOI] [PubMed] [Google Scholar]
- [43].Heinzmann J, Hansmann T, Herrmann D, Wrenzycki C, Zechner U, Haaf T, et al. Epigenetic profile of developmentally important genes in bovine oocytes. Mol Reprod Dev. 2011;78(3):188–201. [DOI] [PubMed]; Heinzmann J, Hansmann T, Herrmann D, Wrenzycki C, Zechner U, Haaf T. et al. Epigenetic profile of developmentally important genes in bovine oocytes. Mol Reprod Dev. 2011;78(3):188–201. doi: 10.1002/mrd.21281. [DOI] [PubMed] [Google Scholar]
- [44].Kuhtz J, Romero S, De Vos M, Smitz J, Haaf T, Anckaert E. Human in vitro oocyte maturation is not associated with increased imprinting error rates at LIT1, SNRPN, PEG3 and GTL2. Hum Reprod. 2014;29(9):1995–2005. [DOI] [PubMed]; Kuhtz J, Romero S, De Vos M, Smitz J, Haaf T, Anckaert E. Human in vitro oocyte maturation is not associated with increased imprinting error rates at LIT1, SNRPN, PEG3 and GTL2. Hum Reprod. 2014;29(9):1995–2005. doi: 10.1093/humrep/deu155. [DOI] [PubMed] [Google Scholar]
- [45].Huntriss J, Woodfine K, Huddleston JE, Murrell A, Rutherford AJ, Elder K, et al. Quantitative analysis of DNA methylation of imprinted genes in single human blastocysts by pyrosequencing. Fertil Steril. 2011;95(8):2564–7. [DOI] [PubMed]; Huntriss J, Woodfine K, Huddleston JE, Murrell A, Rutherford AJ, Elder K. et al. Quantitative analysis of DNA methylation of imprinted genes in single human blastocysts by pyrosequencing. Fertil Steril. 2011;95(8):2564–7. doi: 10.1016/j.fertnstert.2011.04.035. [DOI] [PubMed] [Google Scholar]
- [46].Anifandis G, Messini CI, Dafopoulos K, Messinis IE. Genes and conditions controlling mammalian pre- and post-implantation embryo development. Curr Genomic. 2015;16(1):32–46. [DOI] [PMC free article] [PubMed]; Anifandis G, Messini CI, Dafopoulos K, Messinis IE. Genes and conditions controlling mammalian pre- and post-implantation embryo development. Curr Genomic. 2015;16(1):32–46. doi: 10.2174/1389202916666141224205025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Ludwig M, Katalinic A, Gross S, Sutcliffe A, Varon R, Horsthemke B. Increased prevalence of imprinting defects in patients with Angelman syndrome born to subfertile couples. J Med Genet. 2005;42(4):289–91. [DOI] [PMC free article] [PubMed]; Ludwig M, Katalinic A, Gross S, Sutcliffe A, Varon R, Horsthemke B. Increased prevalence of imprinting defects in patients with Angelman syndrome born to subfertile couples. J Med Genet. 2005;42(4):289–91. doi: 10.1136/jmg.2004.026930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Guo H, Zhu P, Yan L, Li R, Hu B, Lian Y, et al. The DNA methylation landscape of human early embryos. Nature. 2014;511(7511):606–10. [DOI] [PubMed]; Guo H, Zhu P, Yan L, Li R, Hu B, Lian Y. et al. The DNA methylation landscape of human early embryos. Nature. 2014;511(7511):606–10. doi: 10.1038/nature13544. [DOI] [PubMed] [Google Scholar]
- [49].Zhu P, Guo H, Ren Y, Hou Y, Dong J, Li R, et al. Single-cell DNA methylome sequencing of human preimplantation embryos. Nat Genet. 2018;50(1):12–9. [DOI] [PubMed]; Zhu P, Guo H, Ren Y, Hou Y, Dong J, Li R. et al. Single-cell DNA methylome sequencing of human preimplantation embryos. Nat Genet. 2018;50(1):12–9. doi: 10.1038/s41588-017-0007-6. [DOI] [PubMed] [Google Scholar]
- [50].Ørstavik KH, Eiklid K, van der Hagen CB, Spetalen S, Kierulf K, Skjeldal O, et al. Another case of imprinting defect in a girl with Angelman syndrome who was conceived by intracytoplasmic semen injection. Am J Hum Genet. 2003;72(1):218–9. [DOI] [PMC free article] [PubMed]; Ørstavik KH, Eiklid K, van der Hagen CB, Spetalen S, Kierulf K, Skjeldal O. et al. Another case of imprinting defect in a girl with Angelman syndrome who was conceived by intracytoplasmic semen injection. Am J Hum Genet. 2003;72(1):218–9. doi: 10.1086/346030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Sutcliffe AG, Peters CJ, Bowdin S, Temple K, Reardon W, Wilson L, et al. Assisted reproductive therapies and imprinting disorders–a preliminary British survey. Hum Reprod. 2006;21(4):1009–11. [DOI] [PubMed]; Sutcliffe AG, Peters CJ, Bowdin S, Temple K, Reardon W, Wilson L. et al. Assisted reproductive therapies and imprinting disorders–a preliminary British survey. Hum Reprod. 2006;21(4):1009–11. doi: 10.1093/humrep/dei405. [DOI] [PubMed] [Google Scholar]
- [52].Cox GF, Bürger J, Lip V, Mau UA, Sperling K, Wu BL, et al. Intracytoplasmic sperm injection may increase the risk of imprinting defects. Am J Hum Genet. 2002;71(1):162–4. [DOI] [PMC free article] [PubMed]; Cox GF, Bürger J, Lip V, Mau UA, Sperling K, Wu BL. et al. Intracytoplasmic sperm injection may increase the risk of imprinting defects. Am J Hum Genet. 2002;71(1):162–4. doi: 10.1086/341096. [DOI] [PMC free article] [PubMed] [Google Scholar]