Abstract
Objective
This study analyzed physician treating behavior through the use of a multiplex gastrointestinal polymerase chain reaction (GI PCR) test compared with usual testing in emergency department (ED) patients with suspected acute infectious diarrhea to assess differences in antibiotic management.
Methods
A prospective, single‐center, randomized control trial was designed to investigate antibiotic use in ED patients with moderate to severe suspected infectious diarrhea, comparing those who received GI PCR to those who received usual testing. ED patients with signs of dehydration, inflammation, or persistent symptoms were randomized to either the experimental arm (GI PCR) or the control arm (usual testing or no testing).
Results
A total of 74 patients met study criteria and were randomized to either the experimental GI PCR arm (n = 38) or to the control arm (n = 36). Participants in the GI PCR arm received antibiotics in 87% of bacterial or protozoal diarrheal infections (13/15) whereas those in the control arm received antibiotics in 46% of bacterial or protozoal infections (6/13) (P value 0.042) with 2‐proportion difference 0.41 (95% confidence interval 0.07 and 0.68).
Conclusions
ED use of multiplex GI PCR led to an increase in antibiotic use for bacterial and protozoal causes of infectious diarrhea compared to usual testing. This increase in antibiotics appears to be appropriate given patients’ moderate to severe symptoms and a definitive identification of a likely bacterial or protozoal cause of symptoms. Results should be interpreted with caution because of the small sample size.
Keywords: Antibiotic Stewardship, diagnostic testing, emergency medicine, infectious diarrhea, PCR testing, Point‐of care testing
1. INTRODUCTION
1.1. Background
Acute infectious diarrhea occurs commonly in the United States and leads to ≈500,000 hospitalizations and 5000 deaths annually. 1 There are many potential infectious etiologies including viral, bacterial, and protozoal classes but the causative microbe is usually not identified. 2 Traditional tests, such as stool cultures, have a low sensitivity and a long processing time and are not useful in the ED for real‐time treatment decisions. In settings outside the ED, analysis of stool samples with polymerase chain reaction (PCR) tests have allowed for an increased detection of gastrointestinal (GI) pathogens and a decrease in inappropriate antibiotic use.
1.2. Importance
ED clinicians must balance the well‐established risk of overtreatment with unnecessary antibiotics in cases of viral diarrhea with the fact that appropriate antibiotic usage can reduce the duration and severity of symptoms and mitigate risk of future spread in cases of bacterial diarrhea. 3 , 4 , 5 Because of concerns about overtreatment of viral diarrhea, current guidelines do not recommend empiric antibiotic therapy for infectious diarrhea except for traveler's diarrhea or when a clinically plausible organism is identified. 6 With the introduction of multiplex GI PCRs, diarrheal sources can be identified in about an hour thus allowing testing and treatment during a single visit. It is unknown how the use of a multiplex PCR test changes ED management for patients with suspected acute infectious diarrhea.
1.3. Goals of this investigation
In this single‐center randomized control study, we aimed to compare how appropriate antibiotic use differed for ED patients with acute infectious diarrhea who received a GI PCR test in the ED versus those that received usual diagnostic testing.
2. METHODS
2.1. Study design and setting
The design of this study is a single‐center randomized controlled trial. ED patients were screened for enrollment from November 17, 2018 until March 15, 2020 when the study was stopped early because of the COVID‐19 pandemic. The study was conducted at a single university hospital ED (George Washington University Hospital, Washington, DC), which treats a diverse patient population and has an annual census of approximately 80,000 visits per year. Institutional review board approval was obtained and the trial was registered on clinicaltrials.gov (NCT03809117). This trial is also in accordance with Consolidated Standards of Reporting Trials reporting guidelines.
2.2. Selection of participants
Research assistants (RAs) prescreened all adult ED patients for a complaint of diarrhea of at least moderate severity during mostly daytime hours but specific times varied throughout study. RA coverage was approximately 80 hours per week, 7 days per week. To be eligible, a patient qualified for the study if they had (1) 3 or more loose stools in the past 24 hours plus 1 of the following markers of at least moderate severity: (2a) persistent symptoms (more than 7 days); (2b) evidence of dehydration defined as need for intravenous fluids or clinician judgment assessed by direct questioning to clinician by RA after standard clinical assessment of the general appearance and alertness of the patient including the pulse, the blood pressure, the presence or absence of postural hypotension, the mucous membranes and tears, presence of sunken eyes, skin turgor, capillary refill, or jugular venous pressure; and (2c) signs of inflammation including fever, blood in stool, cramping rectal pain, or clinician judgment. Patients were excluded for the following reasons: no stool sample provided; diarrheal symptoms have lasted longer than 14 days; diarrhea likely due to a non‐infectious cause such as Crohn's disease, radiation colitis, irritable bowel syndrome, or celiac disease; informed consent could not be obtained; no reliable contact information for follow‐up was available; cause of current episode diarrhea was likely from previously confirmed Clostridium difficile; clinician stated that the patient needed to receive GI PCR for clinical care and thus could not be randomized; patient recently already recently received GI PCR before screening; patient was discharged/eloped prior to enrollment; and patient was incarcerated at time of screening. After inclusion and exclusion criteria were assessed, the RA confirmed with the ED clinician that the diarrhea was “presumed to be infectious” before enrollment.
The Bottom Line.
This single‐center randomized controlled trial of 74 emergency department patients with moderate to severe diarrheal illness found that a 1‐hour gastrointestinal polymerase chain reaction test increased the use of antibiotics for bacterial and protozoal infections by 40% compared to usual treatment
2.3. Interventions
If a patient met study criteria and consented to enroll in the trial, they were randomized to either the experimental arm with the GI PCR or to the control arm with usual testing or no testing. Participants were randomized in a 1:1 ratio via the computerized randomization feature embedded within the REDCap data management software. Initially, randomization occurred after consent and eligibility was established. However, owing to the fact that many patients qualified for the study but were unable to provide a timely stool sample, we made an in‐study modification to the protocol to change the time of randomization until after the stool sample was provided for patients who qualified.
In the GI PCR arm, the collected stool was analyzed in a Clinical Laboratory Improvement Amendments‐certified hospital lab by trained staff using a multiplex PCR assay that tested for a panel of potentially causative agents. Only liquid stool could be analyzed. The GI PCR test used in this study was the Biofire FilmArray GI Panel, a Food and Drug Administration (FDA)‐cleared diagnostic test reported to have more than 95% specificity for a variety of microbial agents. 7 The test has the ability to detect 7 bacterial species (Aeromonas, Campylobacter, C. difficile, Plesiomonas shigelloides, Salmonella, Vibrio, and Yersinia enterocolitica), 6 diarrheagenic Escherichia coli species (enteroaggregative, enteropathogenic, enterotoxigenic, enteroinvasive, Shiga‐like toxin‐producing, and E. coli O157), 4 parasites (Cryptosporidium, Cyclospora cayetanensis, Entamoeba histolytica, and Giardia lamblia), and 5 viruses (adenovirus, astrovirus, norovirus, rotavirus A, and sapovirus). Each panel contains an internal nucleic acid extraction control and a PCR control. The test runs were considered valid if the run was completed normally and internal controls were passed. Results of this test were available in about 1 hour per run per specimen and were reported in the electronic health record (EHR) while the patient was still in the ED. In the control arm, clinicians were asked not to order the GI PCR on initial stool sample but received no restrictions on other test ordering. For these patients, the stool sample was frozen in a negative 80‐degree freezer to be analyzed at the conclusion of the study to ensure that both arms were balanced by type and class of pathogen. All other ED care was per physician discretion in both groups including the ordering of intravenous fluids, radiographic testing, antibiotic administration, medication prescription, and hospital admission.
2.4. Outcomes
The primary outcome was administration of antibiotics for bacteria or protozoal diarrhea either in the ED or as a prescription at the time of discharge. Antibiotic use was generally considered appropriate because of identification of a treatable and plausible cause of illness and because patients represented a group of ED patients in whom diarrhea was combined with inflammation, dehydration, or prolonged symptoms. Rationale for “appropriate” antibiotic use is based on extrapolation of guidelines from Infectious Diseases Society of America and American College of Gastroenterology. 6 , 8 In these guidelines, patients with non‐viral moderate severity diarrhea, treatment is recommended except in cases of Shiga‐toxin producing E. coli or E. coli O157 in which case it is unknown if antibiotics harm or benefit the patient. 9 , 10 People with fever or bloody diarrhea should be evaluated for enteropathogens for which antimicrobial agents may confer clinical benefit, including Salmonella enterica subspecies, Shigella, and Campylobacter. 6
2.5. Measurements
Although patients were enrolled prospectively, the primary outcome was generally obtained via a chart review using a structured data sheet with trained abstractors per established methods. 11 At the point of enrollment, abstractors who were blinded to group assignment collected participant data from the EHR using a standardized data collection tool including demographic information, history of present illness, recent travel, employment history, current medications, past medical history, triage vital signs, type and amount of intravenous hydration given, serum chemistry lab results, complete blood count, ED medications, prescription medications, imaging procedures, hospital disposition, and symptomatology at Day 2, Day 7, and Day 30. Follow‐up telephone contacts were conducted for all participants and were attempted at postrandomization Days 2, 7, and 30. Follow‐up attempts were made at least 4 times with calls at different times before the participant was considered lost to follow‐up.
2.6. Analysis
Mann‐Whitney U test was used to compare the group differences for the continuous variables. A chi‐square test or Fisher's exact test was performed to test the group differences for categorical variables. For the purposes of our primary outcome, antibiotic usage, Farrington‐Manning score was used to calculate the confidence interval (CI) for the proportion difference between the experimental and control groups, and for each individual arm, exact Clopper‐Pearson CIs were calculated. 12 , 13 Unless otherwise specified, P values reported are nominal P values and are based on the usual chi‐square statistic (or Fisher's exact test in the case of infrequent outcomes) for discrete variables or the Wilcoxon Rank Sum test for continuous variables. In planning this study, in order to detect a 20% increase in appropriate antibiotics (defined previously), we anticipated 88 patients in each group to detect a difference of 80% versus 60% with 95% confidence and 80% power. This estimation of native disease incidence was based on data collected in preliminary studies at this site. 14 The sample size was adjusted to take into consideration the potential for participant losses. A logistic regression model was created to determine if there were clinical or historical factors associated with bacterial or protozoal infection versus viral. Based on symptoms and signs identified a priori by the investigative team as relevant to the diagnosis, these clinical or historical factors were chosen to be included in the full logistic model: "How many episodes of diarrhea have you had?"+ "When did this episode of diarrhea start?"+ "Nausea" + "Vomiting"+ "Abdominal Pain" + "Fever" + "Blood in Stool"+ "Recent travel" + "Currently using any prescription antibiotics?" + "Triage Pulse (bpm)" + "Triage sbp" + "SpO2 (%)" + "Temperature (Fahrenheit)" + "Respiratory Rate." Model selection was conducted using stepwise selection based on Akaike Information Criterion. Analysis was performed using SAS 9.4 (SAS Institute, Cary, NC).
3. RESULTS
3.1. Characteristics of study subjects
There were 663 screenings that led to 87 (13%) randomizations and 74 participants who received study allocation (Figure 1). For many eligible patients, a stool sample was not provided. Baseline characteristics, microbes identified, and ED management are summarized by treatment group. A total of 74 patients provided diarrheal stool samples and were randomized to either the experimental GI PCR arm or to the control arm consisting of no testing or usual testing. There were no statistically significant differences between treatment groups in characteristics at baseline except for heart rate and systolic blood pressure (Table 1).
FIGURE 1.
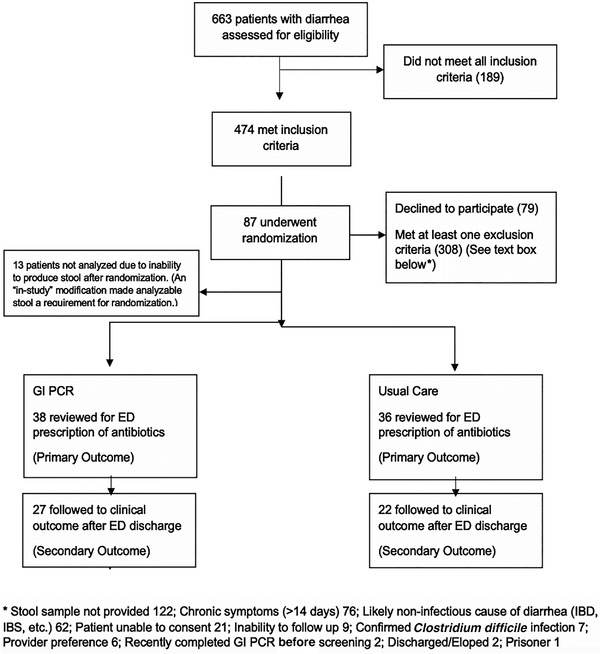
CONSORT diagram. Abbreviations: CONSORT, Consolidated Standards of Reporting Trials; ED, emergency department; GI PCR, gastrointestinal polymerase chain reaction; IBD, inflammatory bowel disease; IBS, irritable bowel syndrome
TABLE 1.
Summary of participants
| GI PCR (n = 38) | Control (n = 36) | P value | |
|---|---|---|---|
| Baseline characteristics | |||
| Median age at screening (year.) [IQR] | 34 [26.00, 51.25] | 33 [25.75, 52.25] | 0.84 |
| Female (%) | 23 (60.5) | 19 (52.8) | 0.66 |
| Black (%) | 21 (61.8) | 21 (63.6) | 1 |
| White (%) | 10 (29.4) | 9 (27.3) | |
| Asian, Native American, other, unknown (%) | 3 (8.8) | 3 (9.1) | |
| Hispanic or Latino (%) | 5 (13.9) | 7 (21.2) | 0.63 |
| Not Hispanic or Latino (%) | 31 (86.1) | 26 (78.8) | |
| 3 or more loose stools in the past 24 hours (%) | 38 (100.0) | 36 (100.0) | NA |
| Symptoms greater than 24 hours (%) | 28 (73.7) | 28 (77.8) | 0.89 |
| Dehydration (%) | 25 (65.8) | 19 (52.8) | 0.37 |
| Vomiting (%) | 26 (68.4) | 21 (58.3) | 0.51 |
| Abdominal pain (%) | 32 (84.2) | 32 (88.9) | 0.80 |
| Fever (%) | 14 (36.8) | 13 (36.1) | 1 |
| Symptoms lasting more than 7 days (%) | 1 (2.6) | 2 (5.6) | 0.61 |
| Heart rate at triage (bpm, median [IQR]) | 95.50 [87.00, 106.00] | 87.00 [76.75, 97.25] | 0. 03 |
| Systolic blood pressure (mmHg, median [IQR]) | 124.50 [117.25, 132.75] | 132.00 [122.50, 145.50] | 0. 04 |
| Temperature (F, median [IQR]) | 98.30 [98.10, 98.60] | 98.40 [98.10, 98.80] | 0. 85 |
| Recent travel (%) | 9 (23.7) | 13 (36.1) | 0. 36 |
| Prescription antibiotics (%) | 3 (7.9) | 2 (5.6) | 1 |
Abbreviations: GI PCR, gastrointestinal polymerase chain reaction; IQR, interquartile range.
3.2. Main results
Participants in the GI PCR arm received antibiotics in 87% of bacterial or protozoal diarrheal infections (13/15) whereas those in the control arm received antibiotics in 46% of bacterial or protozoal infections (6/13) (P value 0.042) (Table 2). The 2‐proportion difference was 0.41 with 95% CI of 0.07 and 0.68. The intensity of the ED work‐up is evident in the high rates of intravenous rehydration therapy and computed tomography scans (Table 2). In addition, the median ED length of stay was 8 hours. Symptom improvement was tracked in both groups and reported from Day 2, Day 7, and Day 30 (Table 3). Finally, as part of our model section process, we identified 2 clinical predictors that were positively associated with bacterial versus viral causes of diarrhea in a logistic regression model: fever and symptoms duration longer than 2 days; plus, we identified 1 symptom that was negatively associated with bacterial causes: the presence of vomiting (Table 4).
TABLE 2.
Main results by group allocation
| Microbe Identified by GI PCR | GI PCR (n = 38) | Control (n = 36) | P value |
|---|---|---|---|
| Campylobacter (%) | 4 (10.5) | 3 (8.3) | 1 |
| Clostridium difficile toxin A/B (%) | 1 (2.7) | 2 (5.7) | 0.61 |
| Plesiomonas shigelloides (%) | 0 | 0 | NA |
| Salmonella (%) | 1 (2.6) | 2 (5.6) | 0.61 |
| Vibrio, non‐cholerae (%) | 1 (2.6) | 0 (0.0) | 1 |
| Vibrio cholerae (%) | 1 (2.6) | 0 (0.0) | 1 |
| Yersinia enterocolitica (%) | 0 | 0 | NA |
| Enteroaggregative Escherichia coli (EAEC) (%) | 2 (5.3) | 5 (13.9) | 0.26 |
| Enteropathogenic E. coli (EPEC) (%) | 3 (7.9) | 6 (16.7) | 0.30 |
| Enterotoxigenic E. coli (ETEC) (%) | 2 (5.3) | 1 (2.8) | 1 |
| Shiga‐like toxin‐producing E. coli (STEC) stx1/stx2 | 0 | 0 | NA |
| Shigella/enteroinvasive E. coli (EIEC) (%) * | 5 (13.2) | 0 (0.0) | 0.06 |
| Cryptosporidium (%) | 0 (0.0) | 2 (5.6) | 0.23 |
| Cyclospora cayetanensis (%) | 0 | 0 | NA |
| Entamoeba histolytics (%) | 0 | 0 | NA |
| Giardia lamblia (%) | 1 (2.7) | 0 (0.0) | 1 |
| Adenovirus 40/41 (%) | 0 (0.0) | 2 (5.6) | 0.23 |
| Astrovirus (%) | 0 (0.0) | 1 (2.8) | 0.49 |
| Norovirus GI/GII (%) | 13 (34.2) | 7 (19.4) | 0.19 |
| Rotavirus (%) | 2 (5.3) | 2 (5.6) | 1 |
| Sapovirus (%) | 1 (2.6) | 1 (2.8) | 1 |
| Summary of microbial class detected | |||
| Bacteria only (%) | 11 (28.9) | 9 (25.0) | 0.91 |
| Both bacteria and virus (%) | 3 (7.9) | 2 (5.6) | |
| Parasite (%) | 1 (2.6) | 2 (5.6) | |
| Virus only (%) | 12 (31.6) | 10 (27.8) | |
| No microbe detected (%) | 11 (28.9) | 13 (36.1) | |
| Emergency department therapy | |||
| Antibiotics administered in ED (%) | 10 (26.3) | 3 (6.8) | 0.07 |
| Antibiotics prescribed for diarrhea (%) | 13 (34.2) | 8 (22.2) | 0.38 |
| IV rehydration therapy (%) | 31 (81.6) | 25 (69.4) | 0.44 |
| Antidiarrheal medication prescribed (%) | 4 (10.5) | 2 (5.7) | 0.68 |
| Admitted to hospital (%) | 7(18.4) | 3 (8.3) | 0.31 |
| Computed tomography ordered (%) | 8 (21.1) | 8 (22.2) | 1 |
| Length of stay (hours, median [IQR]) | 7.68 [6.43, 10.67] | 8.15 [5.90,11.23] | 0.80 |
| Summary antibiotics for diarrhea | |||
|
Bacterial or protozoal infection (exact fraction, proportion [95% CI]) |
13/15 0.87, (0.62. 0.96) |
6/13 0.46, (0.23, 0.71) |
0.04 |
| Viral infection (exact fraction) | 1/12 | 1/10 | 1 |
| None detected (exact fraction) | 3/11 | 2/13 | 0.63 |
Abbreviations: CI, confidence interval; GI PCR, gastrointestinal polymerase chain reaction; IQR, interquartile range; IV, intravenous.
TABLE 3.
Follow‐up symptoms
| GI PCR, n (%)N = 27 | Control, n (%)N = 22 | P value | |
|---|---|---|---|
| Day 2: Do you still have the same symptoms as when you came to the emergency department? (n, % “yes”) | 12 (44.4) | 11 (50.0) | 0.92 |
| Day 7: Do you still have the same symptoms as when you came to the emergency department? (n, % “yes”) | 2 (8.7) | 4 (20.0) | 0.39 |
| Day 30: Do you still have the same symptoms as when you came to the emergency department? (n, % “yes”) | 2 (6.7) | 4 (14.8) | 0.41 |
TABLE 4.
Clinical predictors of bacterial or protozoal infection versus viral infection
| Effect | OR | 95% CI | P value | |
|---|---|---|---|---|
| When did this episode of diarrhea start? (2 days or greater) vs (Less than 24 hours ago) | 16.62 | 1.36 | 202.71 | 0.03 |
| Vomiting (Yes) vs (No) | 0.002 | <0.001 | 0.19 | 0.007 |
| Fever (Yes) vs (No) | 11.28 | 1.03 | 124.17 | 0.047 |
| Triage pulse (bpm) | 0.90 | 0.80 | 1.00 | 0.05 |
| Triage SBP (mmHg) | 0.92 | 0.83 | 1.01 | 0.07 |
| SpO2 (%) | 0.55 | 0.23 | 1.29 | 0.17 |
| Temperature (Fahrenheit) | 8.64 | 0.69 | 107.47 | 0.09 |
Abbreviations: CI, confidence interval; OR, odds ratio; SBP, systolic blood pressure.
4. LIMITATIONS
The findings of this study have to be seen in light of some limitations. First, this was a single‐center study and the results may not be generalizable to other sites. Second, the small sample size meant that only 23 patients were identified as having a bacterial or protozoal infection; as this study was underpowered, the observed effect of the intervention may be overemphasized. The study team planned for a greater sample size; however, because of the COVID‐19 pandemic, we ended the study prematurely. It is important to emphasize that the study was terminated early because the pandemic and not because of a stopping rule or an interim analysis that might limit the risk of bias.
An additional limitation is that by the time the study started, the GI PCR was available to clinicians as a test outside of the study. Although we discouraged this practice, 6 patients who might have been eligible for the study were not enrolled because the clinician felt that a GI PCR was required for patient care, thus introducing a selection bias into our study. Furthermore, interpreting the results of this study is complicated by several factors that may suggest alternate understandings of our findings. First, patients who were not administered or prescribed antibiotics in the ED were recorded in this study as not receiving antibiotic treatment during their disease course; however, patients may have been prescribed antibiotics during subsequent health care visits for the same complaint. This discrepancy could falsely inflate the difference between the rate of antibiotic administration in the GI PCR arm versus the control one.
5. DISCUSSION
In this single‐center randomized control trial, ED patients with acute infectious diarrhea caused by bacteria or protozoa were more likely to be treated with antibiotics if they received a GI PCR test than those who received usual testing or no testing. A GI PCR testing strategy increased the rate of antibiotic prescriptions for a bacterial or protozoal infection by 40% compared to the usual practice control group. To our knowledge, this is the first randomized controlled trial on the topic in the ED setting but our findings are similar to prior studies in other settings. In the primary care setting, a GI PCR strategy showed an increased detection of GI pathogens when using compared to routine testing (28.3% vs 8.3%) for patients with presumed infectious diarrhea. 15 In an inpatient study of 699 patients with acute watery diarrhea, GI PCR led to a decrease in inappropriate antibiotic use from 42.9% to 25.8% compared to traditional diagnostic modalities. 16 Furthermore, the more rapid test results led to a reduced time in isolation for hospitalized patients and a reduced total number of antibiotic days. 17 In addition, a rapid diagnostic strategy led to 1‐day reduction in time to initiation of targeted treatment versus empiric treatment in hospitalized patients with diarrhea (1 day vs 2 days) associated with use of rapid GI PCR panel. 18 Finally, patients with likely infectious diarrhea who were tested with the GI PCR were more likely to receive antibiotics for bacterial infection as part of a retrospective chart review. 19
Ideally, rapid testing panels increase a clinician's ability to identify treatable pathogens at the point of care and may improve treatment and infection control. 19 , 20 One implication of this study is that it demonstrates that microbiological testing may be necessary for physicians to distinguish bacterial from viral causes of acute infectious diarrhea. Prior literature shows that neither bloody nor persistent diarrhea were associated with positive stool culture. 21 A logistic regression analysis performed as part of secondary outcomes for this study identified 3 factors as predictive of bacterial or protozoal source of infectious diarrhea. Despite the limitations of a small sample size, the 3 predictive factors identified were (1) duration longer than 2 days, (2) fever and, (3) coexisting vomiting (negative predictor). The identification of these predictive factors raises the possibility that a clinical decision rule might be created to augment microbiological testing and should be compared to GI PCR testing.
Before the pandemic, the biggest challenge to enrollment was that eligible patients were often unable to provide a timely stool sample in the ED. We made the presumption that if a patient was unable to provide a stool sample after several hours in the ED, they most likely belonged to a more benign cohort and were more likely to represent a benign cohort. In the future, a rectal swab to collect samples might be a promising collection strategy if approved by FDA and manufacturer.
A test‐and‐treat strategy for ED diarrhea could challenge current guidelines that recommend diarrheal testing only on the basis of history of travel and to withhold antibiotics for community acquired diarrhea owing to the likelihood of viral etiology such as norovirus, rotavirus, and adenovirus. 8 A test‐and‐treat strategy has been shown feasible with other ED GI complaints and could also be feasible with infectious diarrhea. 22 One potential criticism of using GI PCR in the ED is that testing will lead to overuse of antibiotics and patients with mild disease will be unnecessarily treated for a condition that is typically self‐limited. As a counterargument, patients who are appropriately treated for diarrhea are known to get better sooner and be less likely to spread to other people. Even 1 less day of profuse diarrhea may represent a significant improvement in a patient‐centered outcome such as missed school or workdays. Further studies addressing the possibility of combining GI PCR and physician education on antibiotic use in acute diarrheal illness may be necessary to determine whether the use of GI PCR is efficacious in decreasing the amount of inappropriate antibiotics in the ED. Another option is to combine treatment options with shared decision making to better elicit patient's values and priorities.
This study illustrates how physicians must carefully balance the competing risks of overtreatment and undertreatment for a given condition. Antibiotic treatment confers a risk of adverse drug events such as acute hypersensitivity reaction, C. difficile infection, acute kidney injury, or cardiotoxicity. 23 One study even asserts that up to 19.3% of all ED visits for adverse drug reactions were due to antibiotic‐associated adverse events. 24 Future research should include a quality‐improvement style study comparing physician behavior and patient outcomes before and after the introduction of these PCR‐based diagnostic tests.
The ED setting draws patients for a variety of reasons including the desire to get a rapid diagnostic test that may not be available in other settings. Technology such as GI PCR that allows rapid identification of specific infection provides actionable information and minimizes the need for empiric treatment of disease. The public health implications of rapid microbiological testing are well known to physicians and the lay‐public as a result of the COVID‐19 pandemic. 25 Rapid testing may also be important for surveillance of GI diseases in community outbreaks and scenarios such as restaurants, schools, and cruise ships. In certain cases, rapid diagnosis may lead to earlier public health interventions.
In summary, this randomized clinical trial showed that ED patients with moderately severe acute infectious diarrhea were more likely to receive antibiotics for treatment of bacterial diarrhea if they were tested with a multiplex PCR test than if they received usual diagnostic testing. Although the study was limited by early termination, it shows how early detection changes physician management. If a clinical benefit of antibiotic antidiarrheal therapy for bacterial infectious diarrhea is established, testing with GI PCR challenges current guidelines that recommend antibiotic antidiarrheal treatment only on the basis of history of travel. Future studies will need to address how GI PCR affects overall cost, risks of antibiotic side effects, and risks of antibiotic overuse and resistance.
CONFLICTS OF INTEREST
None.
AUTHOR CONTRIBUTIONS
Andrew C. Meltzer conceived the study, designed the trial, and obtained research funding. Andrew C. Meltzer and Nataly Montano Vargas supervised the conduct of the trial and data collection. Andrew C. Meltzer and Nataly Montano Vargas undertook recruitment of patients and managed the data, including quality control. Yihe Huang and Yan Ma provided statistical advice on study design and analyzed the data; Andrew C. Meltzer and Nataly Montano Vargas oversaw the data collection and data analysis. Andrew C. Meltzer, Sophia Newton, Joel Lange, Nicole C. Hall, Nataly Montano Vargas, Yihe Huang, Seamus Moran, and Yan Ma drafted the manuscript, and all authors contributed substantially to its revision. Andrew C. Meltzer takes responsibility for the paper as a whole.
Biography
Andrew C. Meltzer, MD, MS, is an Associate Professor of Emergency Medicine at George Washington University in Washington, DC, whose research focuses on the use of novel bedside diagnostics and therapeutics to identify and treat patients with emergency conditions with a focus on gastrointestinal emergencies.

Meltzer AC, Newton S, Lange J, et al. A randomized control trial of a multiplex gastrointestinal PCR panel versus usual testing to assess antibiotics use for patients with infectious diarrhea in the emergency department. JACEP Open. 2022;3:e12616. 10.1002/emp2.12616
Funding and support: This research study was funded by BioFire Diagnostics, LLC. This grant was given to Andrew C. Meltzer, MD, as an investigator‐initiated project. Manufacturer had no contribution to the writing or reviewing of the manuscript.
Supervising Editor: Grant Lipman, MD
ClinicalTrials.gov Identifier: NCT03809117
REFERENCES
- 1. Roy SL, Scallan E, Beach MJ. The rate of acute gastrointestinal illness in developed countries. J Water Health. 2006;4(S2):31‐69. [DOI] [PubMed] [Google Scholar]
- 2. Scallan E, Hoekstra RM, Angulo FJ, et al. Foodborne illness acquired in the united states—major pathogens. Emerg Infect Dis. 2011;17(1):7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Guerrant RL, Van Gilder T, Steiner TS, et al. Practice guidelines for the management of infectious diarrhea. Clin Infect Dis. 2001;32(3):331‐351. [DOI] [PubMed] [Google Scholar]
- 4. Dryden MS, Gabb RJ, Wright SK. Empirical treatment of severe acute community‐acquired gastroenteritis with ciproftoxacin. Clin Infect Dis. 1996;22(6):1019‐1025. [DOI] [PubMed] [Google Scholar]
- 5. Goodman LJ, Trenholme GM, Kaplan RL, et al. Empiric antimicrobial therapy of domestically acquired acute diarrhea in urban adults. Arch Intern Med. 1990;150(3):541‐546. [PubMed] [Google Scholar]
- 6. Shane AL, Mody RK, Crump JA, et al. 2017 infectious diseases society of america clinical practice guidelines for the diagnosis and management of infectious diarrhea. Clin Infect Dis. 2017;65(12):e45‐e80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Buss SN, Leber A, Chapin K, et al. Multicenter evaluation of the biofire filmarray gastrointestinal panel for etiologic diagnosis of infectious gastroenteritis. J Clin Microbiol. 2015;53(3):915‐925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Riddle MS, DuPont HL, Connor BA. ACG clinical guideline: diagnosis, treatment, and prevention of acute diarrheal infections in adults. Am J Gastroenterol. 2016;111(5):602‐622. [DOI] [PubMed] [Google Scholar]
- 9. Panos GZ, Betsi GI, Falagas ME. Systematic review: are antibiotics detrimental or beneficial for the treatment of patients with escherichia coli O157: h7 infection?. Aliment Pharmacol Ther. 2006;24(5):731‐742. [DOI] [PubMed] [Google Scholar]
- 10. Kakoullis L, Papachristodoulou E, Chra P, et al. Shiga toxin‐induced haemolytic uraemic syndrome and the role of antibiotics: a global overview. J Infect. 2019;79(2):75‐94. [DOI] [PubMed] [Google Scholar]
- 11. Kaji AH, Schriger D, Green S. Looking through the retrospectoscope: reducing bias in emergency medicine chart review studies. Ann Emerg Med. 2014;64:292‐298. [DOI] [PubMed] [Google Scholar]
- 12. Fleiss JL, Levin B, Paik MC. Statistical Methods for Rates and Proportions. Third Edition. John Wiley & Sons; 2003. [Google Scholar]
- 13. Newcombe RG. Two‐sided confidence intervals for the single proportion: comparison of seven methods. Stat Med. 1998;17(8):857‐872. [DOI] [PubMed] [Google Scholar]
- 14. Li A, Tran S, Wang H, LeSaux M, Ma Y, Meltzer AC. Multiplex polymerase chain reaction test to diagnose infectious diarrhea in the emergency department. Am J Emerg Med. 2019;37(7):1368‐1370. Epub 2018 Dec 12. PMID: 30587396. [DOI] [PubMed] [Google Scholar]
- 15. Khare R, Espy MJ, Cebelinski E, et al. Comparative evaluation of two commercial multiplex panels for detection of gastrointestinal pathogens by use of clinical stool specimens. J Clin Microbiol. 2014;52(10):3667‐3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Keske Ş, Zabun B, Aksoy K, et al. Rapid molecular detection of gastrointestinal pathogens and its role in antimicrobial stewardship. J Clin Microbiol. 2018;56(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Machiels JD, Cremers AJ, van Bergen‐Verkuyten MC, et al. Impact of the biofire filmarray gastrointestinal panel on patient care and infection control. PLoS One. 2020;15(2):e0228596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Torres‐Miranda D, Akselrod H, Karsner R, et al. Use of biofire filmarray gastrointestinal PCR panel associated with reductions in antibiotic use, time to optimal antibiotics, and length of stay. BMC Gastroenterol. 2020;20(1):1‐7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Li A, Tran S, Wang H, et al. Multiplex polymerase chain reaction test to diagnose infectious diarrhea in the emergency department. Am J Emerg Med. 2019;37(7):1368‐1370. [DOI] [PubMed] [Google Scholar]
- 20. Rand KH, Tremblay EE, Hoidal M, et al. Multiplex gastrointestinal pathogen panels: implications for infection control. Diagn Microbiol Infect Dis. 2015;82(2):154‐157. [DOI] [PubMed] [Google Scholar]
- 21. Chan SSW, Ng KC, Lam PK, et al. Predictors of positive stool culture in adult patients with acute infectious diarrhea. J Emerg Med. 2002;23(2):125‐130. [DOI] [PubMed] [Google Scholar]
- 22. Meltzer AC, Winter LE, Kulie P, et al. Treating gastritis, peptic ulcer disease, and *‐dyspepsia in the emergency department: the feasibility and patient‐reported outcomes of testing and treating for helicobacter pylori infection. Ann Emerg Med. 2015;66(2):131‐139. [DOI] [PubMed] [Google Scholar]
- 23. Tamma PD, Avdic E, Li DX, et al. Association of adverse events with antibiotic use in hospitalized patients. JAMA Intern Med. 2017;177(9):1308‐1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Shehab N, Patel PR, Srinivasan A, et al. Emergency department visits for antibiotic‐associated adverse events. Clin Infect Dis. 2008;47(6):735‐743. [DOI] [PubMed] [Google Scholar]
- 25. Mina MJ, Peto TE, García‐Fiñana M, Semple MG, Buchan IE. Clarifying the evidence on SARS‐CoV‐2 antigen rapid tests in public health responses to COVID‐19. Lancet. 2021;397(10283):1425‐1427. [DOI] [PMC free article] [PubMed] [Google Scholar]


