Abstract
Simultaneous atomic force microscope (AFM) and sample scanning confocal fluorescence microscope measurements are widely used to obtain mechanistic and structural insights into protein dynamics in live cells. However, the absence of a robust technique to synchronously scan both AFM and confocal microscope piezo stages makes it difficult to visualize force-induced changes in fluorescent protein distribution in cells. To address this challenge, we have built an integrated AFM-confocal fluorescence microscope platform that implements a synchronous scanning method which eliminates image artifacts from piezo motion ramping, produces accurate pixel binning and enables the collection of a scanned image of a sample while applying force to a single point on the sample. As proof of principle, we use this instrument to monitor the redistribution of fluorescent E-cadherin, an essential transmembrane protein, in live cells, upon application of mechanical force.
Keywords: Integrated AFM–fluorescence microscope, AFM-sample scanning confocal microscope, synchronized scanning, simultaneous force-fluorescence measurements, point scanning
Introduction
Integrated force-fluorescence microscopes, which consist of an atomic force microscope (AFM) mounted on an inverted widefield (1, 2) or confocal (3, 4) fluorescence microscope, are widely used to image the topography of cells while simultaneously visualizing the distribution of fluorescently tagged proteins within the cell. While these integrated instruments are useful for locating a point of interest with fluorescence imaging and then probing that point of interest with AFM, the limited optical sectioning capabilities of many widefield setups make it impossible to determine where fluorophores are located along the optical axis. Consequently, these microscopes cannot be used to accurately image a section of interest, such as the apical cell surface. While total internal reflection fluorescence microscopy can be used to image a thin optical slice, the collected fluorescence is typically limited to the region near the cell-substrate interface and as such, the apical surface of a cell cannot be visualized.
AFMs integrated with point scanning confocal microscopes are better suited to image and probe thin optical sections in cells. This integration has previously been achieved by mounting commercial sample-scanning AFMs on confocal microscopes which acquire fluorescence images either by sharing the scanning stage with the AFM (5) or by using a scanning objective (6). Commercial AFMs have also been mounted on confocal microscope platforms that align the laser on an AFM tip using tip/tilt mirrors (7). These integrated microscope designs offer excellent spatial and temporal alignment of the AFM and confocal microscope, which is particularly useful for simultaneous imaging of sample topography and fluorescence intensity (5) or lifetime (8) or for inducing near-field nanophotonic effects using an AFM tip (7-9). Integrated AFM-confocal microscopy setups have also been used to extract quantitative data from indentation of cells (10).
However, since confocal images are acquired either by scanning the sample through a fixed laser position or by scanning the source laser through a stationary area, it is very difficult to acquire a fluorescence image of a sample while simultaneously pressing a single spot on the sample with an AFM tip. Although the sample is stationary in an AFM-confocal microscope that scans the excitation light, moving the beam relative to the input of a powerful objective may cause optical aberrations and the inherent sinusoidal motion of galvo mirrors may result in image distortion. More importantly, a stationary sample cannot be easily stabilized with active feedback, which is essential for monitoring a point of interest over a long period of time and reducing unwanted sample drift (11, 12). In contrast, although image distortion due to a moving laser is reduced in a sample scanning confocal microscopy (SSCM), the AFM tip cannot be used to apply force to a predefined point on a sample during fluorescence image acquisition, unless the AFM tip’s lateral motion is perfectly synchronized with confocal sample scanning. Consequently, previous AFM-confocal microscope setups have not been used to monitor realtime, force-induced, dynamic redistribution of proteins at predefined cellular locations.
To overcome this limitation, we present an integrated AFM-SSCM platform that implements synchronous scanning of both the AFM tip and the sample to obtain fluorescence images while simultaneously applying force to the sample. Our instrument integrates a custom tip-scanning AFM which utilizes a piezo stage capable of long range (100μm) motion in all three spatial axes (13) with a custom SSCM, and acquires images in the presence of force, without any piezo ramp aberrations. We demonstrate the capabilities of our method by directly imaging force-induced redistribution of fluorescently tagged E-cadherin, an essential transmembrane adhesive protein, on the apical cell surface (14, 15).
Methods
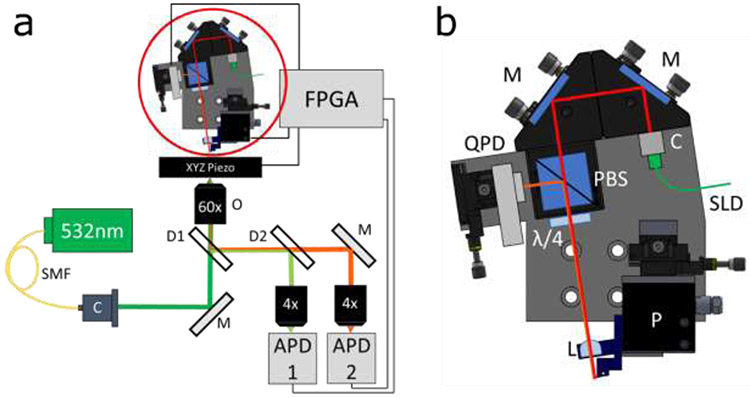
Our instrument (Figure 1a) utilizes a typical sample scanning confocal fluorescence setup (16) with a three-axis piezo sample stage (P-733.3, Physik Instrumente) mounted on a modular microscope frame (RAMM, Applied Scientific Instrumentation). A 532nm laser (OBIS, Coherent) couples into a single mode fiber (SMF). The beam is collimated at the fiber endpoint (C), reflected by a mirror (M) towards a 60x microscope objective (O, Olympus) and focused at the sample mounted on an XYZ piezo stage. The fluorescence and a fraction of the backscattered light are collected through the objective and passed through a dichroic filter (D1, Chroma). A second dichroic (D2, Chroma) splits the backscattered and fluorescent light which are detected using avalanche photodiodes (APD 1 & 2, respectively, Micro Photon Devices). A field programmable gate array (FPGA, National Instruments) is used to command piezo stage position, and acts as a photon counter for the APDs and partially processes the data (with the remaining processing accomplished on the host PC) to form the resulting scan images.
Figure 1: Instrumentation.
(a) Diagram of microscope, SMF- single mode fiber, O- objective, M- mirror, D1/D2- dichroic, C- collimator, FPGA- field programmable gate array, APD1/APD2-avalanche photodiode, (b) Diagram of custom AFM. M- mirror, L- lens, C- collimator, SLD-superluminescent diode, QPD- quadrant photodiode, PBS- polarizing cube beamsplitter, λ/4-quarter waveplate, P- 3 axis piezo.
The AFM is located above the microscope sample (circled in red) and shown in detail in Figure 1b. A fiber-coupled infrared superluminescent diode (SLD, 850nm, QPhotonics) mounts to a collimator (C, Thorlabs) which in turn mounts to two adjustable right-angle mirrors (M, Thorlabs), allowing for fine positioning and angling of the beam onto the AFM cantilever. The beam passes through a polarizing cube beamsplitter (PCB, Thorlabs) and a quarter waveplate (λ/4, Thorlabs) resulting in a circularly polarized collimated beam. This beam is focused through a lens (L, Newport) onto the AFM cantilever. Upon reflection off the cantilever, the polarization handedness flips, resulting in the reflected light being polarized perpendicular to the incoming beam, and as such is reflected by the PCB to the quadrant photodiode (QPD, First Sensor). The AFM chip is mounted at an 8.5° angle on the three-axis piezo stage (P-616, Physik Instrumente) to allow it to probe a surface, and the rest of the optics are mounted to match that angle for perpendicular reflection. The QPD signal is digitized by on board analog to digital converters (ADCs) on the FPGA, which in turn can control the motion of the piezo stage. Each piezo stage is controlled with PI’s E-712.3 controller.
To test our synchronized force-fluorescence microscope we designed a cell pressing assay. Since we wanted to monitor changes in the distribution of cell membrane proteins due to force-induced deformation of a cell, we used Madin-Darby Canine Kidney (MDCK) cells expressing DSRed-tagged E-cadherin. We grew these cells on a piranha (3:1 H2SO4:H2O2) - cleaned glass coverslip which was mounted to our confocal microscope piezo. In order to apply force on these cells without puncturing their membrane, we positioned borosilicate microspheres (diameter < 10μm, Polysciences) at the ends of tipless AFM cantilevers (Arrow TL2, NanoWorld) using a micromanipulator (CellTram, Eppendorf). These microspheres were then sintered to the cantilever (17) in an oven (Isotemp, Fisher Scientific) using a slow heat ramp (room temperature to 500°C at 8°/min, 500°C to 800°C at 5°/min, hold at 800°C for 1 hour). Since the diameters of the microspheres are of a scale comparable to the size of MDCK cells, they do not puncture the cells even with a large applied force. The technique used to synchronously scan the AFM and confocal microscope piezo stage and the results of the cell force-fluorescence application are detailed in the results section.
Results
The AFM and fluorescence microscope piezo stages cover a 2D space by scanning the piezo along one axis and stepping it across the second axis in between scanned lines (Figure 2). Smooth motion along the scan axis was achieved by commanding the piezo to move in a positional “ramp”, that is, to move through the scan line with a constant velocity. To save scanning time and minimize undesired motion, the direction of the ramp was switched with each scan line (Figure 2). Each scan line was distributed across the requested area in such a way that each resulting image pixel represents light collected from the center of that pixel (rather than the edge or corner) yielding a more intuitive sense of the scanned space and a better ability to locate the absolute position of features.
Figure 2: Scanning method and synchronization.
(a) A 2D lateral scan is defined over a 10x10μm area with 10 scan lines. The scan definition is such that when the resulting data is formatted into a 2D image, each pixel value represents light collected from the center of that pixel. The scanning axis (X in this case) also extends before and after the scan area of interest to allow the triggering system which eliminates “warping” at the edges. Note that the range for each trigger extends slightly beyond the scan area, allowing a trigger “hand off’ between scan lines such that the trigger bounce does not signal the start of the next scan line collection. (b) Each piezo (sample and AFM) are programmed to internally record their positions and report the result at the end of the scan. To be sure the scan is not only spatially, but also temporally synced, the 2D scan is separated into each axis and plotted against time, X shown in (c) and Y shown in (d). The traces verify that the two piezo stages are synchronized both spatially and temporally.
While the scan method described above ideally evenly covered the requested 2D space, there was a ramp up and ramp down at either end of the piezo stage travel which warped the image. This undesired effect limited our temporal resolution since it scaled with the time of each scan line, in that over the same scan length a faster scan appeared more warped. To address this problem, we programmatically adjusted a user defined scan line to include scan “padding,” that is, a short distance before and after the requested scan dimensions was built in, and the scan time was adjusted to maintain the requested velocity over the requested scan distance. The padding was chosen to allow enough time and space for the piezo to reach target velocity before reaching the scan line start, and enough time after to reduce velocity to zero before starting the next scan line. The data from these ramp up/ramp down regions were then faithfully excluded from the resulting image. If the piezo stage has a consistent response time, these regions can be excluded by discarding a set amount of data collected over the ramp up/ramp down time and an image can be generated using the remaining data. However, since response time varies between piezo stages, and even between different load weights on the same stage, we relied on the recorded piezo position, using digital position triggers to exclude unwanted ramping data. We set a trigger output to be high only over the user defined scan line (excluding the padding), and only collected detector data when the trigger was high. In this configuration it became necessary to address the problem of digital trigger bouncing (that is rapid changing from high-to-low for a time before stabilizing on its final state), since it breaks image acquisition. Typical “debouncing” solutions eliminate this problem at the cost of time resolution, which would also be detrimental to our images. For this reason we implemented a two trigger scheme, one indicating the start of the scan line and the other indicating the end of the scan line (Fig. 2(a)). This way we used the first switch from each trigger and ignored altogether the bounces that followed.
To faithfully scan the two piezo stages over the same area, synchronized in time and space, the two piezos’ closed-loop motion parameters were tuned such that when the same motion command was sent to both stages, their response time and settling time matched. However, even with their motion tightly coupled, the effort was rendered moot since the scans did not start at the same time for both piezo stages. When the command to start the scan was sent from the host computer (via USB), we saw significant discrepancies in the actual scan start time, resulting in an offset between the two stages for the duration of the scan. To address timing differences, we first synchronized the two controllers’ clocks via cable, ensuring commands received at the same time would execute at the same time. To ensure the commands arrived at the same time, we set the pre-defined scan waveforms to execute when a third digital trigger was set high, sent via FPGA digital output to both piezo controllers.
These features combined allowed an AFM tip to scan synchronously with a feature of interest on the sample to generate a faithful backscatter and fluorescence image. Because this only affects the lateral motion of each stage, the optical Z axis of the AFM is free to move independent of the scan, which will eventually allow force clamp experiments while imaging is taking place. More immediately, this allowed us to image the AFM tip and sample fluorescence simultaneously, while maintaining a user-controlled AFM pressing distance which is useful for gaining mechanistic insight into pliable samples such as cells.
To verify scan synchronization, we glued an AFM cantilever to a glass coverslip which was mounted to the sample piezo stage. We then brought a second AFM cantilever, mounted on the custom AFM apparatus, in close proximity to the coverslip. We commanded both the sample and AFM piezos to start moving as described above and recorded their motion with a camera inserted into the detection arm of the microscope (Supplementary Video). Our data showed that the cantilevers maintained their position relative to one another while scanning through the 2D space, over the laser focus. This visually demonstrated that the AFM and confocal scans begin at the same time and maintain registry. To quantify scan registry, we also recorded the piezos’ positions during the scan shown in Figure 2(b-d). It is important to note these traces show the position measured by each piezo’s capacitive sensor, not the commanded position. Subtracting each AFM piezo position from the corresponding sample stage position within the scan area, showed that the AFM stage lags the sample stage in X by under 12nm (σ < 3nm) and that a small portion of the Y stepping motion (σ < 2nm) was also included in our area of interest (Supplementary Figure S1). This lag can be easily eliminated by optimal tuning of the piezos’ motion response parameters.
With scan synchronization verified, we used our AFM-SSCM to image live cells while simultaneously pressing a point on the cell surface (Figure 3a). We initially roughly aligned a group of cells over the focused laser using manual micrometer actuators on the microscope stage. The objective height was adjusted such that the top of the cells were in focus. We then similarly aligned the AFM cantilever over the cells with actuators on the custom AFM. An initial large synchronized scan was commanded, and a wide view of the cells (via fluorescence) and AFM cantilever with microsphere (via backscattered light) were generated. Next, we selected new scan center points on the fluorescence and backscatter images to redefine the sample and AFM scans, respectively; the result of the following 50x50μm scan is shown in Figure 3b, 3c. Keeping this scan center, we zoomed in on a fluorescent E-cadherin cluster and microsphere by choosing a smaller scan area. We then commanded the AFM piezo to move the cantilever toward the sample until a small deflection was detected from the AFM QPD, indicating the cantilever had made contact with the cell. Next we applied progressively higher forces to the cell by moving the AFM piezo toward the sample, then reduced force by moving away, collecting fluorescence images throughout (50s scan time for each fluorescent image; Figure 3e, 3f). An animated GIF of the fluorescence images are included as Supplementary GIF. As the AFM cantilever moved into the focused waist of the confocal laser beam, it backscattered more light, resulting in a brighter image. At the same time, a ‘dark spot’ formed in the fluorescent cell image, which corresponds in shape and position to the AFM microsphere. We interpret this ‘dark spot’ as E-cadherins on the cell surface being pushed out of the laser focus by the microsphere. Indeed, as the microsphere was lifted from the cell, fluorescence recovery was observed at the pressing site. To estimate the instrumental drift during this experiment, we compared the first and last images from Figure 3e & 3f, and showed that the drift was one pixel or less (pixel size = 100nm), (Supplementary Figure S2).
Figure 3: Imaging force-induced redistribution of cell surface proteins.
To demonstrate the utility of the synchronized force fluorescence platform, we designed an experiment in which we collect fluorescence and backscatter images while applying a varying force on the apical surface of cells expressing DSRed-tagged E-cadherin. (a) Illustration of the experimental setup. The objective focus is set to the apical surface of the cell of interest (cell shown in red, beam waist in green), and the AFM cantilever with microsphere is positioned over the laser focus (or anywhere within the scan area the user chooses). (b) Backscatter and (c) fluorescence image results of a 50x50μm scan from APD 1 & 2, respectively (Figure 1). The triangular shape of the AFM cantilever as well as the silica microsphere sintered to the cantilever (along with a few smaller microspheres) are visible in (b). The fluorescently tagged E-cadherin largely reside at cell-cell junctions, making the edges of the cells bright as seen in (c). (d) Progression of the force applied to the cell by the AFM, and the resulting (e) backscatter and (f) fluorescent images (20x20μm scans). In row (e) we see the microsphere centered in the image, and as it presses into the cell the cantilever becomes brighter as it moves into the laser beam waist. In (f) we see a cluster of fluorescent E-cadherin on the apical surface of a cell. As higher force is applied, these cadherin are pushed out of the beam waist, and we see a dark spot form where the microsphere is pressing into the cell. Upon withdrawal of the AFM tip, we see the fluorescent E-cadherin return to the pressing site as the cell reforms its original shape. These images serve as good confirmation that our synchronized scanning method allows fluorescence imaging while applying force.
Discussion
In this paper, we have demonstrated the design and use of our novel synchronized force-fluorescence confocal scanning microscope. By taking care to ensure the area scan is accurate, repeatable, and synchronized between two piezo stages, we can image the force response of living cells through redistribution of fluorescently tagged membrane proteins. We believe this instrument has great promise in a variety of force-fluorescence applications, mainly in imaging molecules and cells during force application with higher resolution and greater optical axis range than existing methods.
In our ‘proof of concept’ experiments (Figure 3), we applied a very large force (between 1 and 90nN) to effect a clear change in our fluorescence image. While our custom AFM is capable of applying feedback to hold force at a pre-determined set-point, we did not employ this feature while acquiring fluorescence images since the cells continuously relax during AFM pressing (Supplementary Figure S3). We anticipate that correlating cellular force relaxation measurements with fluorescence imaging of membrane protein redistribution can be used in to determine cell and tissue mechanical properties, such as cell viscoelasticity (18, 19). For more delicate experiments, applying lower forces with a controlled force set-point is possible. Our custom AFM has a force resolution of 6pN at an acquisition bandwidth of 1kHz (13) for measurements conducted in air (Supplementary Figure S4). However, for use in an aqueous environment, we include an acrylic block to avoid liquid wicking around the AFM. As a result, our AFM force resolution is limited by the reduction of light intensity falling on the QPD (72pN, Supplementary Figure S4). This limitation can be easily overcome by replacing the superluminescent diode with a more powerful light source and optimizing the AFM mount to further avoid reflection and refraction problems. Of course if the light source is too bright, a fluorescent sample will photobleach, so that balance will need to be struck with further experimentation.
Another issue arises when monitoring the reported force during a 2D scan. Our custom AFM is designed to ideally send the superluminescent diode beam perpendicular to the AFM cantilever, which when aligned correctly should eliminate any change in the beam’s deflection during the cantilever’s lateral scan motion. However our experimental data shows that this alignment is very sensitive, and a small deviation from a perpendicular reflection can cause a change in force signal when no force is being applied during a lateral scan. This is due to the long lever arm of the beam, which provides an advantage for monitoring small deflections, but a disadvantage during scanning. If the beam is not perpendicular to the cantilever and the cantilever moves relative to the beam, we see a resulting shift on the QPD. This artifact can be eliminated in a number of ways, perhaps the easiest being a more precise milling of mounting parts with more robust connections to ensure better angle control. Another option would be to largely replace our AFM optics scheme with a more traditional layout, allowing light source, cantilever, and detector to all mount on the piezo stage and move in unison.
Finally, our instrument can be easily integrated with our previously published fluorescence-compatible active feedback platform (11). With our feedback platform working in conjunction with our custom AFM, we’ll be able to perform single molecule manipulation and force measurement in a repeatable fashion on specifically located fluorescent molecules.
Supplementary Material
Highlights.
Microscope to measure force-induced redistribution of proteins in cells
Acknowledgements:
We thank Dr. Omer Shafraz for assistance with design of the biological assay. Research reported in this publication was supported in part by the National Institute of General Medical Sciences of the National Institutes of Health (R01GM121885).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kellermayer MSZ et al. , Spatially and Temporally Synchronized Atomic Force and Total Internal Reflection Fluorescence Microscopy for Imaging and Manipulating Cells and Biomolecules. Biophys. J 91, 2665–2677 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gumpp H et al. , Triggering Enzymatic Activity with Force. Nano Lett. 9, 3290–3295 (2009). [DOI] [PubMed] [Google Scholar]
- 3.Kassies R et al. , Combined AFM and confocal fluorescence microscope for applications in bio-nanotechnology. Journal of microscopy 217, 109–116 (2005). [DOI] [PubMed] [Google Scholar]
- 4.Meller K, Theiss C, Atomic force microscopy and confocal laser scanning microscopy on the cytoskeleton of permeabilised and embedded cells. Ultramicroscopy 106, 320–325 (2006). [DOI] [PubMed] [Google Scholar]
- 5.Noy A, Huser TR, Combined force and photonic probe microscope with single molecule sensitivity. Rev. Sci. Instrum 74, 1217–1221 (2003). [Google Scholar]
- 6.Schulz O et al. , in Single Molecule Spectroscopy and Imaging III. (International Society for Optics and Photonics, 2010), vol. 7571, pp. 757109. [Google Scholar]
- 7.Gerton JM, Wade LA, Lessard GA, Ma Z, Quake SR, Tip-Enhanced Fluorescence Microscopy at 10 Nanometer Resolution. Phys. Rev. Lett 93, 180801 (2004). [DOI] [PubMed] [Google Scholar]
- 8.Schulz O et al. , Tip induced fluorescence quenching for nanometer optical and topographical resolution. Optical Nanoscopy 2, 1 (2013). [Google Scholar]
- 9.Mangum BD, Mu C, Gerton JM, Resolving single fluorophores within dense ensembles: contrast limits of tip-enhanced fluorescence microscopy. Opt. Express 16, 6183–6193 (2008). [DOI] [PubMed] [Google Scholar]
- 10.Staunton JR, Doss BL, Lindsay S, Ros R, Correlating confocal microscopy and atomic force indentation reveals metastatic cancer cells stiffen during invasion into collagen I matrices. Sci. Rep 6, 19686 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schmidt PD, Reichert BH, Lajoie JG, Sivasankar S, Method for high frequency tracking and sub-nm sample stabilization in single molecule fluorescence microscopy. Scientific Reports 8, 13912 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.King GM, Carter AR, Churnside AB, Eberle LS, Perkins TT, Ultrastable Atomic Force Microscopy: Atomic-Scale Stability and Registration in Ambient Conditions. Nano Letters 9, 1451–1456 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schmidt P, Reichert B, Lajoie J, Sivasankar S, Adaptive atomic force microscope. SPIE BiOS (SPIE, 2020), vol. 11246. [Google Scholar]
- 14.Priest AV, Shafraz O, Sivasankar S, Biophysical basis of cadherin mediated cell-cell adhesion. Exp. Cell Res 358, 10–13 (2017). [DOI] [PubMed] [Google Scholar]
- 15.Priest AV, Koirala R, Sivasankar S, single molecule studies of classical and desmosomal cadherin adhesion. Current opinion in biomedical engineering 12, 43–50 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li H, Yen CF, Sivasankar S, Fluorescence Axial Localization with Nanometer Accuracy and Precision. Nano Lett. 12, 3731–3735 (2012). [DOI] [PubMed] [Google Scholar]
- 17.Sivasankar S, Chu S, Optical bonding using silica nanoparticle sol-gel chemistry. Nano Lett. 7, 3031–3034 (2007). [DOI] [PubMed] [Google Scholar]
- 18.Bausch AR, Ziemann F, Boulbitch AA, Jacobson K, Sackmann E, Local measurements of viscoelastic parameters of adherent cell surfaces by magnetic bead microrheometry. Biophys. J 75, 2038–2049 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rianna C, Radmacher M, Comparison of viscoelastic properties of cancer and normal thyroid cells on different stiffness substrates. European Biophysics Journal 46, 309–324 (2017). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.