Abstract
We aimed to describe the SARS-CoV-2 lineages circulating early pandemic among samples with S gene dropout and characterize the receptor-binding domain (RBD) of viral spike protein. Adults and children older than 2 months with signs and symptoms of COVID-19 were prospectively enrolled from May to October in Porto Alegre, Brazil. All participants performed RT-PCR assay, and samples with S gene dropout and cycle threshold < 30 were submitted to high-throughput sequencing (HTS). 484 out of 1,557 participants tested positive for SARS-CoV-2. The S gene dropout was detected in 7.4% (36/484) and a peak was observed in August. The B.1.1.28, B.1.91 and B.1.1.33 lineages were circulating in early pandemic. The RBD novel mutation (Y380Q) was found in one sample occurring simultaneously with C379W and V395A, and the B.1.91 lineage in the spike protein. The Y380Q and C379W may interfere with the binding of neutralizing antibodies (CR3022, EY6A, H014, S304).
Keywords: COVID-19, SARS-CoV-2, RBD, Variants, Novel mutation
1. Introduction
SARS-CoV-2 is a single RNA-stranded virus with high mutation rates. Strategies to mitigate the pandemic include the knowledge of its viral genome and expected mutations. These features could impact disease severity, virus transmission, and vaccine strategies (Anichini et al., 2021; Awadasseid et al., 2021). As the COVID-19 pandemic evolves, there has been concern about the emergence of new SARS-CoV-2 mutations in the receptor binding domain (RBD) from the S region, due to probable effects on both virus transmissibility and the generation of escape mutants from antibodies previously formed to heterologous lineages and vaccines (Vilar and Isom, 2020).
Genetic alterations in the RBD of SARS-CoV-2 may improve the affinity of the virus to binding host cells, possibly increasing transmission rates (Korber et al., 2020; Yurkovetskiy et al., 2020) and making this region a key target for potential therapies and diagnosis (Wrapp et al., 2020). COVID-19 molecular diagnostic tests directed to the S gene use it as one of the RT-PCR multiple target-regions.
2. Objectives
Our aim was to measure the prevalence of the S dropout and characterize the SARS-CoV-2 mutations in the RBD region in a cohort during the early pandemic.
3. Materials and methods
3.1. Participants’ selection
A prospective cohort study enrolled adults and children seeking care at emergency rooms, outpatient clinics, or hospitalized in general wards or intensive care units (ICU) at Hospital Moinhos de Vento and Hospital Restinga e Extremo Sul, in Porto Alegre, Brazil. From May to early October 2020 were included participants presenting signs or symptoms suggestive of COVID-19 (cough, fever, or sore throat). The key exclusion criteria was a negative SARS-CoV-2 RT-PCR result or failure to sample collection. The study was performed in accordance with the Decree 466/12 of the National Health Council (Ministerio da Saude, 2021) and Clinical Practice Guidelines, after approval by the Hospital Moinhos de Vento IRB n° 4.637.933. All participants included in this study provided written informed consent.
3.2. SARS-CoV-2 detection and sequencing
All participants performed qualitative RT-PCR assay (TaqManTM 2019-nCoV Kit v1, catalog number A47532, ThermoFisher Scientific, Pleasanton, California, EUA) to SARS-CoV-2 detection as described elsewhere (Polese-Bonatto et al., 2021). Additionally, S gene dropout samples with cycle threshold less than 30 (Ct < 30.0) were submitted to high-throughput sequencing (HTS) using the Illumina MiSeq. RNA was extracted from naso-oropharyngeal swab samples and the reverse transcription reaction was performed using SuperScript IV reverse transcriptase kit (Thermo Fisher Scientific, Waltham, MA, USA). Libraries were prepared using QIAseq SARS-CoV-2 Primer Panel and QIAseq FX DNA Library UDI kit, according to the manufacturer instructions (Qiagen, Hilden, Germany). The QIAseq SARS-CoV-2 Primer Panel contains a PCR primer set for whole genome amplification of SARS-CoV-2 whose primer sequences were based on the ARTIC network nCov-2019. A pool of all of the normalized libraries was prepared and diluted to a final concentration of 8pM and sequenced on the Illumina MiSeq platform using the MiSeq Reagent kit v3 600 cycles (Illumina). FASTQ reads were imported to Geneious Prime, trimmed (BBDuk 37.25), and mapped against the reference sequence hCoV-19/Wuhan/WIV04/2019 (EPI_ISL_402124) available in EpiCoV database from GISAID (GISAID - Initiative, 2021). Complete genome alignment was performed with the sequences generated. 59 Brazilian SARS-CoV-2 complete genomes and the reference sequence (EPI_ISL_402124) (>29 kb) were retrieved from the GISAID database using Clustal Omega. Maximum Likelihood phylogenetic analysis was applied under the General Time Reversible model allowing for a proportion of invariable sites and substitution rates in Mega X applying 200 replicates and 1000 bootstrap.
3.3. Co-localization of Y380Q with B and T-cell epitopes
3.3.1. B-cell epitopes
Wild type (Y380) and mutated spike protein sequences (Q380) were submitted to Bepipred 1.0 and 2.0 to detect putative humoral epitopes through HMMs and Random forest algorithms (Jespersen et al., 2017; Larsen et al., 2006). To increase sensitivity we set a threshold of -0.2 (Bepipred 1.0) and 0.45 (Bepipred 2.0).
3.3.2. T-cell epitopes
The search in Immune Epitope Database (IEDB) considered T-cells epitopes for SARS-CoV-2 spike protein (region of 10 residues flanking the Y380Q) with 70% similarity in BLAST. Potential binder sequences of representative supertypes MHC-I alleles (HLA-A*01:01, HLA-A*02:01, HLA-A*03:01, HLA-A*24:02, HLA-A*26:01, HLA-B*07:02, HLA-B*08:01, HLA-B*27:05, HLA-B*39:01, HLA-B*40-01, HLA-B*58:01, HLA-B*15:01) were predicted by NetMHCPan-4.1 (O'Donnell et al., 2020; Reynisson et al., 2020).
3.4. Structural modifications and their impact on B and T-cells epitopes recognition
3.4.1. Homology modeling
The wild type SARS-CoV-2 surface glycoprotein sequence (NCBI accession number: YP_009724390) was submitted to BLAST and SwissModel tools. Template crystal candidates were evaluated by the GMQE, QMEAN, Z-score, and residues distribution in the Ramachandran plot, using ERRAT, PROCHECK, PDBsum, ModFold, SwissModel (Benkert et al., 2011; Laskowski et al., 1996; Laskowski et al., 1997; Waterhouse et al., 2018). Protein Data Bank (PDB) 7CWL (3.8Å) was chosen for the approach. Phyre-2 software was employed to homology modeling using the expert mode (one-to-one threading job) for constructing models based on wild protein (P0DTC2) and mutated sequences (Kelley et al., 2015).
3.4.2. Physicochemical properties
Electrostatic potential (EP) was verified through Delphi web server calculations and PIPSA (Richter et al., 2008; Sarkar et al., 2013). Residue exposure characteristics (hydrophobicity and the Accessible Solvent Surface Area - aSAS) were estimated using the Chimera interface (Pettersen et al., 2004).
3.4.3. Antibodies interaction
Crystal complexes of the RBD region with antibodies were recovered from PDB. The LigPlot program was applied to infer protein-antibody interaction sites (Wallace et al., 1995). Hydrogen-bonds (H-bonds) inferences among the RBD domain and antibodies were calculated in the Chimera interface.
3.5. Statistical analysis
Data normality assumptions were verified for continuous variables, and median values and interquartile ranges (IQR) were calculated. Pearson's Chi-square test was used to evaluate proportions between the identified and undetermined results from S gene target, on the epidemiological week; Fisher's exact test was used to compare the frequencies of S dropout considering outpatient and inpatient populations. All analyses were performed in R 3.5.0 statistical software.
4. Results
A total of 1557 participants were screened and 484 were detected positive for SARS-CoV-2 (Supplementary Fig. 1). Of these, 98 (20.2%) subjects were hospitalized and 386 (79.8%) were seen as outpatients only.
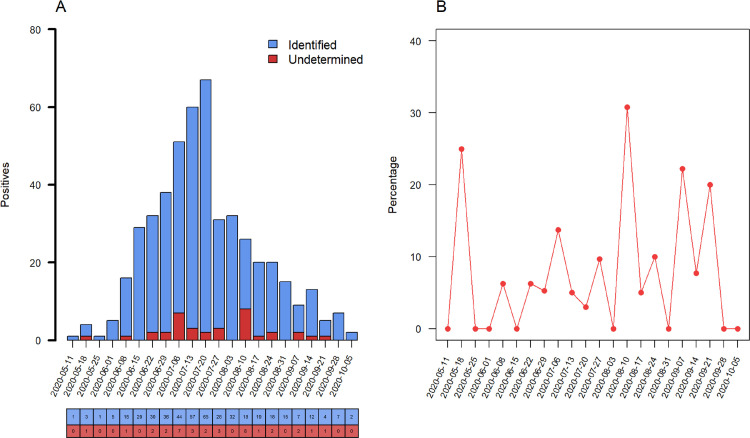
S dropout was characterized as undetermined RT-PCR values for the S gene target, and detected values for ORF1ab and N target probes. We observed a total S dropout of 36 of 484 (7.4%), while ORF1ab and N gene targets showed no dropout (Fig. 1 ). Additionally, an increase in frequency of undetermined results of the S gene target was identified on 10th August week with 8 of 26 dropouts (30.8%, P = 0.007). No differences were observed for other epidemiological weeks. S gene dropout was detected in 12 of 98 hospitalized subjects (12.2%), whereas for outpatients the frequency was 24 of 386 (6.2%) with an OR (95% CI) of 2.10 (0.92-4.57, P = 0.052).
Fig. 1.
Prevalence of S dropout profile from May to October 2020. (A) Number of positive RT-PCR assays, blue columns represent the raw counts of positive assays identified by S gene target and red columns the raw counts of S dropout (characterized as undetected RT-PCR by S, and detected by ORF1ab and N gene targets). (B) Percentage of S dropout over epidemiological week with an increase of undetermined results of the S gene target on the 10th August week, (8/26, 30.8%, P = 0.007). (Color version of figure is available online.)
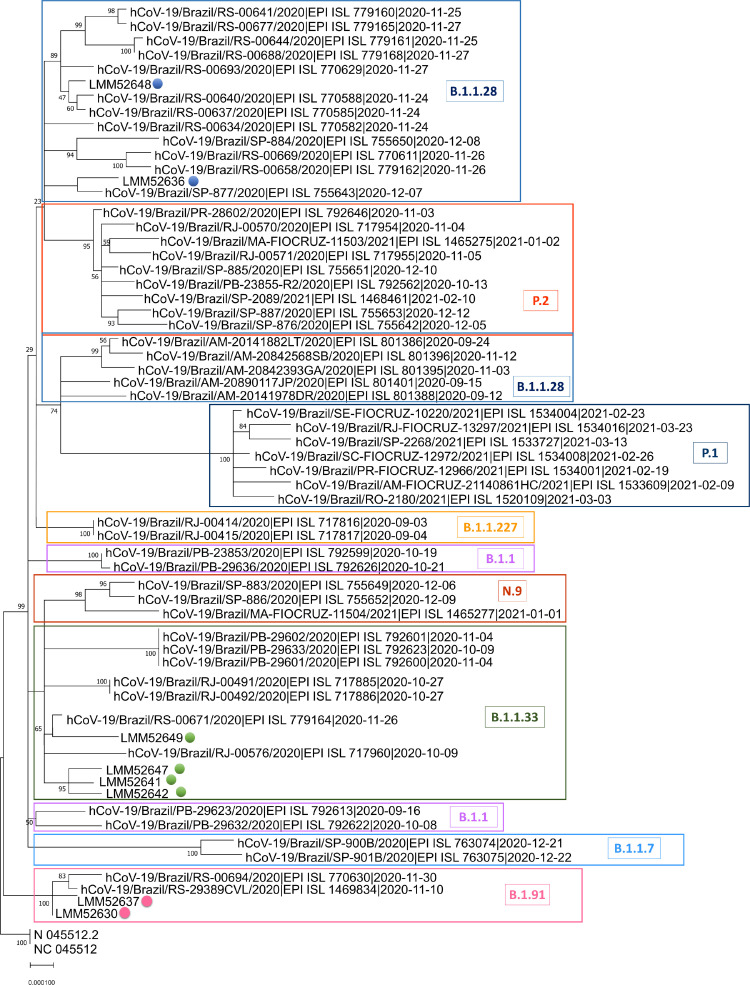
All samples with S gene dropout eligible for sequencing were from adults, and 24 of 36 were submitted to WSG. Eight high-quality SARS-CoV-2 whole-genome sequences were recovered (GISAID accession numbers: EPI_ISL_1799498, EPI_ISL_1799499, EPI_ISL_1799501, EPI_ISL_1799502, EPI_ISL_1823203, EPI_ISL_1799504, EPI_ISL_1799505, EPI_ISL_1799507). The other 16 samples were not analyzed, due to the low quality reads and/or low genome coverage. According to the Pangolin COVID-19 Lineage Assigner tool (cov-lineages/pangolin, 2021) three different lineages were detected: B.1.1.28, B.1.91 and B.1.1.33. Phylogenetic tree corroborated previous results, as shown in Fig. 2 . Spike protein mutations and INDELs were found, including one mutation (Y380Q) that have never been reported (Table 1 ).
Fig. 2.
SARS-CoV-2 complete genome phylogenetic tree. The Maximum Likelihood phylogenetic analysis under General Time Reversible allows a proportion of invariable sites, and the substitution rates were inferred empirically in MEGAX web server applying 200 replicates and 1000 bootstrap.
Table 1.
The consensus and additional mutations, and INDELs observed in the amino acid sequences from eight SARS-CoV-2 infected subjects. Underlined, the novel RBD spike protein mutation.
| ID | Lineage | Consensus mutations of lineages | Additional mutations | INDELS | Sex | Age | Date of sample collection |
|---|---|---|---|---|---|---|---|
| LMM52630 | B.1.91 | D614G, D839Y | C379W, Y380Q, V395A | del:ORF7b:27795:3 | Male | 66.0 | 2020-06-24 |
| LMM52636 | B.1.1.28 | D614G, V1176F | - | - | Male | 68.5 | 2020-07-28 |
| LMM52637 | B.1.91 | G232A, D614G, D839Y, Y1272F | - | - | Male | 71.7 | 2020-07-30 |
| LMM52641 | B.1.1.33 | D614G | T76I, K77N, R78M, F133S | - | Male | 84.6 | 2020-08-25 |
| LMM52642 | B.1.1.33 | D614G | - | del:ORF1ab:686:9 | Female | 62.0 | 2020-08-26 |
| LMM52647 | B.1.1.33 | D614G | F133S | - | Female | 36.1 | 2020-08-10 |
| LMM52648 | B.1.1.28 | D614G, T768N, V1176F | - | - | Male | 39.6 | 2020-08-10 |
| LMM52649 | B.1.1.33 | D614G | - | - | Male | 39.4 | 2020-08-12 |
Median age of the eight sequenced participants was 64.0 years (IQR 39.6-69.3), 75.0% (6/8) were male, and the median days of symptoms onset to inclusion was 7.5 (IQR 2.0-10.0). The most commonly reported symptoms were chills and appetite loss (75.0%), cough, malaise and myalgia (62.5%), and fever (50.0%). Six out of eight participants (75.0%) were hospitalized, 5 (62.5%) required only supplemental oxygen, 3 (37.5%) were admitted to ICU, 2 (25.0%) required mechanical ventilation, and one participant died (1/8, 12.5%). The participant that presented the novel Y380Q mutation was hospitalized for 18 days, including 8 days in ICU, requiring the use of mechanical ventilation.
Wild C379 and Y380 residues of the RBD region were predicted as part of B-cell epitopes in the Bepipred 1.0 and 2.0, even after the amino acid substitutions. NetMHCpan4.1 returns seven binding sequences involving these sites, and two of them were also described as epitopes in the IEDB positive T-cell assays: SASFSTFKCY (for HLA-A*01:01 allele) and KCYGVSPTK (for HLA-A*03:01 allele). The wild sequence (SASFSTFKCY), predicted as a weak binder, turns to a non-binder when mutated (SASFSTFKWQ). The wild strong binder sequence (KCYGVSPTK) turns into a weak binder (KWQGVSPTK).
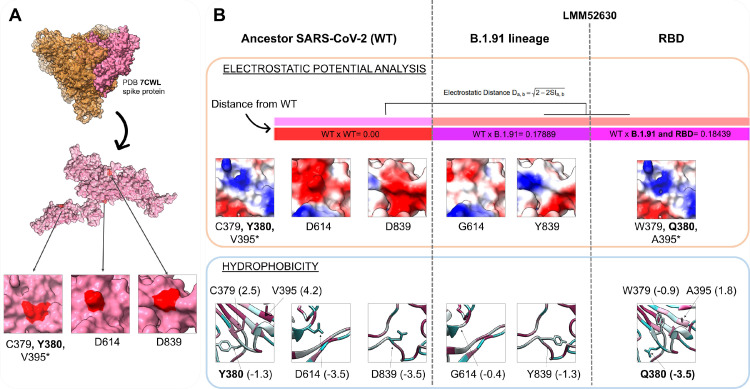
Location of identified mutations in the spike protein is depicted in Fig. 3 A. Structural analysis revealed EP modifications (orange rectangle, Fig. 3B) on the LMM52630 models (which includes the B.1.91 lineage model only, and the B.1.91 lineage associated with the RBD substitutions: C379W, V395A and non-reported Y380Q, found exclusively in this participant) compared to the ancestral monomer sequence model. Greater modifications in electrostatic distance (ED = 0.18) are due to the mutations in the B.1.91 lineage (G614 and Y839). Additionally, mutations in RBD presented similar ED = 0.18, shown in the epogram analysis (color bars in Fig. 3B). These results can be even more evident examining the surface distribution charges: the D614 and D839 wild residues are negatively charged (red pattern, Fig. 3B) and this pattern is disrupted in G614 and Y839 mutated residues; while the surface EP distribution and models conformation show more discreet modifications for the RBD region variants.
Fig. 3.
Physicochemical modifications of mutations in the B.1.91 lineage and RBD region (C379W, Y380Q, V395A). (A) The PDB 7CWL crystal with the model generated for the spike protein monomer is detached in pink. Variants' positions at the tridimensional structure surface are highlighted in red. The neighbor residues C379 and Y380 are depicted together, while the V395* is buried and not perceptible at the surface. Both regions (D614 and D839) of the B.1.91 lineage are in exposure regions of the spike protein surface. (B) The SARS-COV-2 ancestor lineage (represented by C379, Y380, V395, D614 and D839 residues) was compared to the G614 and Y839 residues of the B.1.91 variant of concern (VOC), and to the mutations presented by the LMM52630 subject (namely: G614 and Y839 of B.1.91 lineage, in addition to W379, Q380 and A395 from RBD residues) considering the electrostatic potential (EP) (orange rectangle) and hydrophobicity (blue rectangle) properties. The epogram (color bars) presents the electrostatic distances (ED) of the mutated models compared to the wild type (ancestor). Both models (B.1.91 lineage and RBD mutations) are similar with ED = 0.18, and diverging from the ancestor. Below of the epogram, are shown the divergences on electrostatic surface distribution, considering the wild and mutated residues. The color scale is represented by a variation from red (more electronegative residues, -5) to blue (more electropositive, +5) passing through neutral (white, 0). The hydrophobicity scale varies from more hydrophobic residues (represented in magenta, positive values) to more hydrophilic (in blue, negative values). PDB = Protein Data Bank; RBD = Receptor binding domain; WT = Wild type. (Color version of figure is available online.)
Amino acid substitutions revealed alterations in hydrophobicity, either by changing the direction of this property (from hydrophobic to hydrophilic and vice versa) or even its intensity. In a general way, substitutions observed in the B.1.91 lineage turn their regions to more hydrophobic, while the RBD mutations to more hydrophilic, denoted by negative and positive values (blue rectangle, Fig. 3B) considering hydrophilic and hydrophobic patterns, respectively. Substitutions from wild residues D614 (-3.5) and D839 (-3.5) to G614 (-0.4) and Y839 (-1.3) incremented the hydrophobicity in B.1.91 lineage, while the substitution of Y380 (-1.3) and V395 (4.2) to Q380 (-3.5) and A395 (1.8) reduced this property. The C379W mutation changed the direction from highly hydrophobic (2.5) to hydrophilic (-0.9). Altered residues in the RBD region, especially the C379W and Y380Q mutations, are located close to each other and likely gain strength, thus providing an overall shift to hydrophilic profile. The changing potential of these two close mutations induced to the buried A395 a more hydrophilic profile when compared to the ancestor (from 4.2 to 1.8).
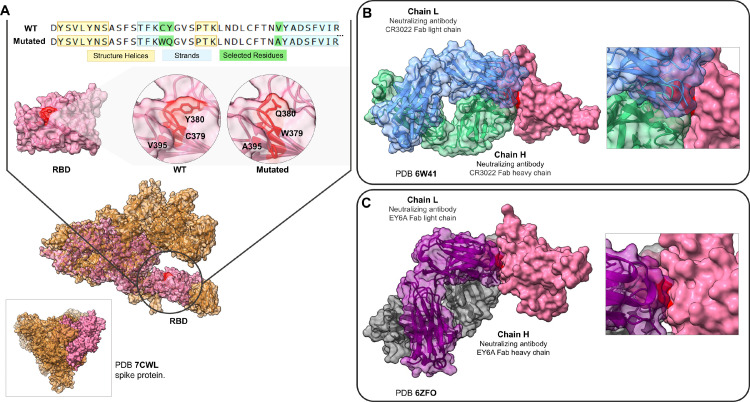
A structural investigation of more than 20 crystals of viral spike protein from PDB, revealed that the mutations C379W, Y380Q and V395A are in a contact area complexed with antibodies in the RBD region. And four crystals that presented Fab antibodies (fragment antigen-binding) are in contact with 379 and 380 RBD residues. Fig. 4 B exhibits the CR3022 human antibody complexed with the RBD region in contact with mutation sites. We observed the same when evaluating the S2H97, EY6A and S304 antibodies. H-bonds are important non-covalent interaction forces which can assist in protein residue bindings, especially in stabilizing antibody-antigen interactions (Miyanabe et al., 2018). Mutated Q380 residue leads to an H-bond disruption observed previously between the wild Y380 with the S99 residue of the CR3022 neutralizing antibody. While the W379 alteration disables the previous H-bond between the wild C379 with T94 residue of the EY6A neutralizing antibody. Mutated W379 also affects the H-bond neighboring of the G381 with Y92 residue of EY6A antibody. RBD-antibody interaction sites are shown in Supplementary Table 1.
Fig. 4.
RBD mutation regions of the LMM52630 subject in contact with neutralizing antibodies. (A) The comparison of the mutated sequences with the wild type. Below, the structural model with the selected residues, depicted (red color) for wild and mutated RBD region. The model of the monomer was obtained from the 7CWL PDB structure. (B) The contact region among the RBD (pink), the Fab CR3022 neutralizing antibody (light and heavy chains, represented in blue and green colors, respectively) and the residues C379 and Y380 (red). (C) The same RBD region contacting the neutralizing antibody EY6A (the light chain in purple and heavy in grey). RBD = Receptor binding domain; WT = Wild type. (Color version of figure is available online.)
5. Discussion
Our results suggest that SARS-CoV-2 S gene dropouts were present in the community as early as in the beginning of the pandemic in Southern Brazil. Further, a new mutation (Y380Q) was identified, and with C379W modify important properties in the RBD region, which may interfere with the binding of the CR3022, EY6A, H014, S304 neutralizing antibodies (NAbs).
S gene dropout has been found strongly associated with a six-nucleotide deletion resulting in the loss of two amino acids: H69 and V70. Despite the HTS, we could not directly link the S dropout to specific viral mutations. This result may be due to the small sample size submitted to sequencing. Though, we found the presence of three different lineages B.1.1.28, B.1.91 and B.1.1.33. Additionally, the Y380Q mutation was identified in the S gene, and thus could be associated with the natural history of virus evolution.
It is well known that modifications in the EP and hydrophobicity distribution may interfere in protein-protein interaction. Interface regions are usually composed of residues presenting opposite charges and hydrophobic pairs, where small changes on these properties, in important functional sites, may impact canonical interactions (Yan et al., 2008). NAbs are fundamental elements of the immune system against viral infections (Dong et al., 2020), and the H-bonds of wild C379 and Y380 residues with the S304, CR3022, S2H97 and EY6A NAbs reinforce the importance of these regions. Therefore, the H-bond disruptions observed in W379 and Q380 substitutions plus the alteration in hydrophobicity disfavor the RBD-antibodies interactions. As the C379 residue is part of one (1/4) disulfide bonds in the RBD region (C379-C432) its disruption could generate instability since it contributes to β sheet conformation maintenance (Lan et al., 2020).
A previous study reported that the C379 and Y380 residues are part of an epitope for H014 antibody, which could sterically compete with the ACE2 host molecule for the RBD interaction (Verkhivker and Di, 2021). It also reported an overlap among the binding epitopes for the H014 and CR3022 antibodies. The CR3022 monoclonal antibody neutralizes the RBD region of SARS-CoV-2, disrupts the prefusion spike conformation, and also competes sterically with ACE2 (Huo et al., 2020) without physically blocking it (Yu et al., 2020). The recently described S2H97 is a potential NAb (Starr et al., 2021) that exhibits a notable tight binding even with divergent RBD regions from other Sarbecoviruses. Other SARS-CoV-2 NAbs that bind to the RBD were also described as non-overlapping with the ACE2 binding site (Barnes et al., 2020).
This study has some limitations. The small sample size in which HTS was feasible may limit any conclusion about the clinical severity related to the mutations found. Moreover, individuals were enrolled in a single city in Southern Brazil, which may limit the generality of our findings. Nonetheless, despite such limitations, a novel mutation (Y380Q) in the RBD region of SARS-CoV-2 spike protein was described. Analysis based on crystal structures reinforces the importance of the Y380 and C379 residues in the NAbs binding, and thus mutations in these regions may affect the interaction effectiveness between the NAbs and SARS-CoV-2 protein, as inferred by computational analysis.
Our findings indicate that SARS-CoV-2 variants were circulating quite early in the community. A possible role of the new described mutation with clinical severity can be speculated, but further studies are needed to confirm this hypothesis. Studies assessing mutations and their relation to prognosis are necessary, and also to evaluate vaccine effectiveness in a challenging scenery that is continuously changing.
COVIDa study group
Adriane Isabel Rohden, Ana Paula dos Santos, Camila Dietrich, Caroline Cabral Robinson, Catia Moreira Guterres, Débora Vacaro Fogazzi, Denise Arakaki-Sanchez, Fernanda Lutz Tolves, Fernando Rovedder Boita, Francieli Fontana Sutile Tardetti Fantinato, Gisele Alcina Nader Bastos, Jaina da Costa Pereira, Maicon Falavigna, Maristênia Machado Araújo, Patricia Bartholomay Oliveira, Regis Goulart Rosa, Shirlei Villanova Ribeiro, Thainá Dias Luft, Tiago Fazolo.
Acknowledgments
We thank the Scientific Committee of the Research Support Nucleus (Núcleo de Apoio à Pesquisa, NAP) of Hospital Moinhos de Vento for technical-scientific consultancy. We thank the inclusion personal, laboratory team, and site staff from Hospital Moinhos de Vento and from Hospital Restinga e Extremo Sul. Aline Andrea da Cunha, Joao Ronaldo Mafalda Krauser, Paulo Sergio Kroeff Schmitz, Sidiclei Machado Carvalho, Fabio Jose Rockenbach, Thaís Pacheco Oliveira, Cláudia Josiel Oliveira do Coito, Kelly Viegas Antunes, Marcelo da Silva Ferreira, Rafael Garcia Trindade, Thayna Silva Lino, Tiago da Silva Silvano, Adriana da Silva Silveira, Alceu Kuckoski, Ana Paula da Silva Lopes, Andreia Escobar da Costa, Clarice Cardoso Machado, Erica Vieira da Silva, Evelin Inácia da Silva, Luciana Rodrigues Ribeiro, Marcely Mayr da Costa, Morgana Thais Carollo Fernandes, Rafael da Silva Cassafuz.
Funding
This work was supported by the Brazilian Ministry of Health, through the Institutional Development Program of the Brazilian National Health System (PROADI-SUS) in collaboration with Hospital Moinhos de Vento.
Declaration of competing interest
The authors report no conflicts of interest relevant to this article.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.diagmicrobio.2022.115636.
Appendix. Supplementary materials
References
- Anichini G, Terrosi C, Gori Savellini G, Gandolfo C, Franchi F, Cusi MG. Neutralizing antibody response of vaccines to SARS-CoV-2 variants. Vaccines. 2021;9:517. doi: 10.3390/vaccines9050517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awadasseid A, Wu Y, Tanaka Y, Zhang W. SARS-CoV-2 variants evolved during the early stage of the pandemic and effects of mutations on adaptation in Wuhan populations. Int J Biol Sci. 2021;17:97–106. doi: 10.7150/ijbs.47827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes CO, Jette CA, Abernathy ME, Dam K-MA, Esswein SR, Gristick HB, et al. SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies. Nature. 2020;588:682–687. doi: 10.1038/s41586-020-2852-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benkert P, Biasini M, Schwede T. Toward the estimation of the absolute quality of individual protein structure models. Bioinformatics. 2011;27:343–350. doi: 10.1093/bioinformatics/btq662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- cov-lineages/pangolin. CoV-lineages; 2021.
- Dong Y, Dai T, Wei Y, Zhang L, Zheng M, Zhou F. A systematic review of SARS-CoV-2 vaccine candidates. Sig Transduct Target Ther. 2020;5:1–14. doi: 10.1038/s41392-020-00352-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GISAID - Initiative n.d. Available at: https://www.gisaid.org/. (accessed July 13, 2021).
- Huo J, Zhao Y, Ren J, Zhou D, Duyvesteyn HME, Ginn HM, et al. Neutralization of SARS-CoV-2 by Destruction of the Prefusion Spike. Cell Host & Microbe. 2020;28:445–454.e6. doi: 10.1016/j.chom.2020.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jespersen MC, Peters B, Nielsen M, Marcatili P. BepiPred-2.0: improving sequence-based B-cell epitope prediction using conformational epitopes. Nucleic Acids Research. 2017;45:W24–W29. doi: 10.1093/nar/gkx346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJE. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc. 2015;10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korber B, Fischer WM, Gnanakaran S, Yoon H, Theiler J, Abfalterer W, et al. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;182:812–827.e19. doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan J, Ge J, Yu J, Shan S, Zhou H, Fan S, et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- Larsen JEP, Lund O, Nielsen M. Improved method for predicting linear B-cell epitopes. Immunome Res. 2006;2:2. doi: 10.1186/1745-7580-2-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski RA, Hutchinson EG, Michie AD, Wallace AC, Jones ML, Thornton JM. PDBsum: a Web-based database of summaries and analyses of all PDB structures. Trends Biochem Sci. 1997;22:488–490. doi: 10.1016/s0968-0004(97)01140-7. [DOI] [PubMed] [Google Scholar]
- Laskowski RA, Rullmannn JA, MacArthur MW, Kaptein R, Thornton JM. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J Biomol NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- Ministerio da Saude n.d. Available at: https://bvsms.saude.gov.br/bvs/saudelegis/cns/2013/res0466_12_12_2012.html. (accessed July 13, 2021).
- Miyanabe K, Akiba H, Kuroda D, Nakakido M, Kusano-Arai O, Iwanari H, et al. Intramolecular H-bonds govern the recognition of a flexible peptide by an antibody. J Biochem. 2018;164:65–76. doi: 10.1093/jb/mvy032. [DOI] [PubMed] [Google Scholar]
- O'Donnell TJ, Rubinsteyn A, Laserson U. MHCflurry 2.0: Improved Pan-Allele Prediction of MHC Class I-Presented Peptides by Incorporating Antigen Processing. Cell Systems. 2020;11:42–48.e7. doi: 10.1016/j.cels.2020.06.010. [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, et al. UCSF Chimera–a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Polese-Bonatto M, Sartor ITS, Varela FH, Gianinni GLT, Azevedo TR, Kern LB, et al. Children have similar RT-PCR cycle threshold for SARS-CoV-2 in comparison with adults. MedRxiv. 2021;2021 doi: 10.1101/2021.04.20.21255059. [DOI] [Google Scholar]
- Reynisson B, Alvarez B, Paul S, Peters B, Nielsen M. NetMHCpan-4.1 and NetMHCIIpan-4.0: improved predictions of MHC antigen presentation by concurrent motif deconvolution and integration of MS MHC eluted ligand data. Nucleic Acids Res. 2020;48:W449–W454. doi: 10.1093/nar/gkaa379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter S, Wenzel A, Stein M, Gabdoulline RR, Wade RC. webPIPSA: a web server for the comparison of protein interaction properties. Nucleic Acids Res. 2008;36:W276–W280. doi: 10.1093/nar/gkn181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar S, Witham S, Zhang J, Zhenirovskyy M, Rocchia W, Alexov E. DelPhi web server: a comprehensive online suite for electrostatic calculations of biological macromolecules and their complexes. Commun Comput Phys. 2013;13:269–284. doi: 10.4208/cicp.300611.201011s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr TN, Czudnochowski N, Zatta F, Park Y-J, Liu Z, Addetia A, et al. Antibodies to the SARS-CoV-2 receptor-binding domain that maximize breadth and resistance to viral escape. BioRxiv. 2021 doi: 10.1101/2021.04.06.438709. [DOI] [Google Scholar]
- Verkhivker GM, Di Paola L. Integrated biophysical modeling of the SARS-CoV-2 spike protein binding and allosteric interactions with antibodies. J Phys Chem B. 2021;125:4596–4619. doi: 10.1021/acs.jpcb.1c00395. [DOI] [PubMed] [Google Scholar]
- Vilar S, Isom DG. One Year of SARS-CoV-2: how much has the virus changed? BioRxiv. 2020 doi: 10.1101/2020.12.16.423071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace AC, Laskowski RA, Thornton JM. LIGPLOT: a program to generate schematic diagrams of protein-ligand interactions. Protein Eng. 1995;8:127–134. doi: 10.1093/protein/8.2.127. [DOI] [PubMed] [Google Scholar]
- Waterhouse A, Bertoni M, Bienert S, Studer G, Tauriello G, Gumienny R, et al. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46:W296–W303. doi: 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrapp D, Wang N, Corbett KS, Goldsmith JA, Hsieh C-L, Abiona O, et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan C, Wu F, Jernigan RL, Dobbs D, Honavar V. Characterization of protein–protein interfaces. Protein J. 2008;27:59–70. doi: 10.1007/s10930-007-9108-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F, Xiang R, Deng X, Wang L, Yu Z, Tian S, et al. Receptor-binding domain-specific human neutralizing monoclonal antibodies against SARS-CoV and SARS-CoV-2. Sig Transduct Target Ther. 2020;5:1–12. doi: 10.1038/s41392-020-00318-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yurkovetskiy L, Wang X, Pascal KE, Tomkins-Tinch C, Nyalile TP, Wang Y, et al. Structural and functional analysis of the D614G SARS-CoV-2 spike protein variant. Cell. 2020;183:739–751.e8. doi: 10.1016/j.cell.2020.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.