Abstract
Primordial germ cells (PGCs) are specified by diverse mechanisms in early development. In some animals, PGCs are specified via inheritance of maternal determinants, while in others, in a process thought to represent the ancestral mode, PGC fate is induced by cell interactions. Although the terminal factors expressed in specified germ cells are widely conserved, the mechanisms by which these factors are regulated can be widely diverse. Here we show that a post-translational mechanism of germ cell specification is conserved between two echinoderm species thought to employ divergent germ line segregation strategies. Sea urchins segregate their germ line early by an inherited mechanism. The DEAD-box RNA - helicase Vasa, a conserved germline factor, becomes enriched in the PGCs by degradation in future somatic cells by the E3-ubiquitin-ligase Gustavus (Gustafson et al., 2011). This post-translational activity occurs early in development, substantially prior to gastrulation. Here we test this process in germ cell specification of sea star embryos, which tive signaling mechanisms after gastrulation for PGC fate determination. We find that Vasa-GFP protein becomes restricted to the PGCs in the sea star even though the injected mRNA is present throughout the embryo. Gustavus depletion, however, results in uniform accumulation of the protein. These data demonstrate that Gustavus-mediated Vasa turnover in somatic cells is conserved between species with otherwise divergent PGC specification mechanisms. Since Gustavus was originally identified in Drosophila melanogaster to have similar functions in Vasa regulation (Kugler et al., 2010), we conclude that this node of Vasa regulation in PGC formation is ancestral and evolutionarily transposable from the ancestral, induced PGC specification program to an inherited PGC specification mechanism.
Keywords: Echinoderm, Vasa, Gustavus, CRISPR Cas9, primordial germ cell, sea urchin, sea star
Introduction
A critical step in animal development is the segregation of primordial germ cells (PGCs), which will ultimately become the stem cells that produce eggs and sperm in the adult. PGC fate is induced by cell interactions during embryogenesis in mice, salamanders, crickets and sea stars (Donoughe et al., 2014; Extavour and Akam, 2003; Fresques and Wessel, 2018). In other animals, like flies, worms, zebrafish and sea urchins, PGC fate is specified by inheritance of maternal factors early in development (Extavour and Akam, 2003; Voronina et al., 2008; Wessel et al., 2014a). Sea stars and sea urchins are convenient systems to compare mechanisms of PGCs specification as they use inductive (sea stars) and inherited (sea urchins) mechanisms, yet both belong to the echinoderm clade. In sea urchins, the primordial germ cells are specified early in development and are derived from the small micromeres which form at the 5th cleavage following two asymmetric cell divisions. These cells inherit maternally stored information in the egg and early embryo to direct germ cell fate. This process is analogous to how the PGCs are specified in e.g. Drosophila, and Zebrafish, (Seydoux and Braun, 2006; Strome and Updike, 2015). In the sea urchin Strongylocentrotus purpuratus, maternally-supplied and ubiquitous Vasa is inherited by the PGCs, but is eliminated from somatic cells post-translationally (Gustafson et al., 2011; Voronina et al., 2008). The E3 ligase Gustavus regulates this post-translational process, which is recapitulated with expression of exogenous Vasa-GFP by mRNA injection into eggs (Gustafson et al., 2011; Zazueta-Novoa and Wessel, 2014).
Embryos of the sea star instead rely on cell interactions to specify their putative PGCs. Germ cell factors are present broadly in early development of the sea star, but following gastrulation, the factors become restricted into progressively smaller cohorts of presumptive germ cells (Fresques et al., 2014; Wessel et al., 2014b). These cells then form a pouch-like structure on left side of the gut called the posterior enterocoel (PE) (Inoue, 1992). Although the lineage of PE cells has not been traced throughout development to conclude definitively their PGC nature, we hypothesize they are, or contribute to, PGCs based on selective expression of a variety of germ cell factors presumably important for making functional germ cells (Fresques et al., 2014). Cell interactions are essential for this process in the sea star Patiria miniata, and Nodal is one such pathway that restricts the putative PGCs (hereby named PGCs in the text for simplicity) to the future germ line on the left side of the endoderm (Fresques and Wessel, 2018).
Thus, the mechanisms of germ-cell factor regulation appear distinct between the sea urchin, using inherited information, and in the sea star, relying more on cell - signaling pathways. Here we investigated how the enrichment of germ cell factors is controlled in the sea star embryo. We anticipated that exogenous mRNAs encoding Vasa- and Nanos-GFP introduced into the sea star egg would behave distinctly from the localization mechanism seen in the sea urchin. This appears not to be the case.
Materials and Methods
Embryo and larva cultures
Patiria miniata adult animals were shipped obtained from Pete Halmay (peterhalmay@gmail.com) and Marinus Scientific (info@marinusscientific.com) and kept in a sea water aquarium at 15°C. Gonads were surgically obtained by a tiny cut between the arms on the oral side, and oocytes, embryos and larvae cultured at 16°C as described (Foltz et al., 2004). Oocytes were microinjected, then treated with 1-methyladenine (Acros Organics) at a final concentration of 10 μM to induce meiotic resumption. Eggs were fertilized by adding sperm to the culture at a 1:1,000,000 dilution.
Plasmid constructs and synthetic RNA
Patiria miniata Vasa and Nanos were identified using previously published ovary transcriptomes (Cameron et al., 2009; Kudtarkar and Cameron, 2017; Reich et al., 2015) and the genomic resources at echinobase.org. Genes were amplified from first strand cDNA reverse transcribed from total ovary mRNA and were then cloned into pCS2+8 as c-terminal GFP or mCherry fusions (Gökirmak et al., 2012). The GFP-stop construct was a N-terminal GFP followed by a stop codon and cloned into pCS2+8 vector. For expression of constructs in embryos and larvae, plasmids were linearized with NotI to yield a linear template DNA. Synthetic mRNA was transcribed in vitro with the mMessage mMachine SP6 and the polyadenylation kit (Life Technologies), then precipitated using ammonium acetate solution. Synthetic mRNA for vasa, nanos or GFP-stop were injected at a final concentration of 500 ng/μl.
Perturbation experiments with MO injection and CRISPR Cas9
A morpholino antisense oligonucleotide (MASO, GeneTools, Philomath, OR). complementary to the Pm-gustavus transcript 5′ UTR was used at a final concentration of 500 μM to block protein translation (MASO sequence is 5’-AAGGCACTACCGCTCCGAAGATC-3’). A scrambled MASO sequence was used as control. Three Cas9 guide RNAs (gRNAs; 150 ng/μl of each gRNA) were used to target the Gus gene. gRNAs were mixed with 750 ng/μl of Cas9 mRNA while control larvae were injected with Cas9 mRNA without gRNAs. gRNAs were purchased from Synthego and the oligonucleotide sequences areas follows: PmGus_18: ATGGGGCAGAAACTGTCCGG; PmGus_133: CACTGGGGGCATGTCCAGCA; PmGus_262: AGAGCACGGACTGCATTCGG. Prophase arrested oocytes were injected with approximately 20 picoliters of injection solutions in nuclease free water.
Image and statistical analyses
Images of fixed or live larvae were taken on a Nikon Yokogawa W1 spinning disk microscope with 40x silicon oil objective. Images were processed and analyzed in Fiji (ImageJ). Videos were made with the 3D plug-in on the Nikon Elements software. For image analysis, on each image, we measured both the PE (posterior enterocoel) and the stomach 4 to 6 ROIs (Regions of Interest) of Vasa-GFP intensity and subtracted the background. We then calculated the Vasa-GFP ratio by dividing the signal in the PE by the signal in the stomach: a value >1 indicates that the signal intensity is greater in the PE than in the stomach, a value <1 indicates that the signal is more intense in the stomach than the PE, and a value =1 means that the signal intensity is equal in the PE and the stomach. Student’s t-test was performed to assess statistical significance, values of p<0.001 are represented as ****. Graphs and statistical analyses were performed using Prism (GraphPad Software). Figures were constructed using Adobe Illustrator.
In situ hybridization
Larvae were fixed in 4% paraformaldehyde and probed for the expression of mCherry RNA by fluorescence in situ hybridization (FISH) as described in (Perillo et al., 2021). The mCherry probe primers used were Fw:5’AAGGGCGAGGAGGATAACAT3’ and Rv: 5’CTTCAGCTTCAGCCTCTGCT3’. The P. miniate Nanos probe template was synthesized using primers Fw:5’ AGTGACCGGACGACATTCTC3’ and Rv: 5’CGTCTGACTGGACTGGGTTT3’; and the vasa probe was synthesized as reported in Fresques 2014
Results
Vasa mRNA injected into the egg results in Vasa protein enriched selectively in the PGCs.
To identify how Vasa protein is regulated during sea star development, we cloned the complete Vasa open reading frame fused to GFP. Following injection into the oocyte and fertilization, the mRNA became uniformly distributed throughout the developing embryo. Prior to formation of the PE (posterior enterocoel), Vasa-GFP protein accumulated in all cells of the embryo in a perinuclear subcellular localization (Figure 1A). However, when the PE formed at the early larval stage, the exogenously encoded protein became enriched in cells of the PE (i.e. the future PGC-like cells) and was depleted in the somatic cells (Figure 1B,C,D). Thus, when the PE is formed as a distinct structure, a transition occured wherein Vasa protein is eliminated in somatic cells, but retained selectively in the PE, as well as some cells within the anterior pouches (AP, two hollow tubes of mesodermal origin that extend parallel to the esophagus) (Figure 1B, arrow). Further, we found that the Vasa protein is always enriched perinuclearly in cells of the PE (Figure 1 D and I). To determine if other germ line associated factors also accumulate in the PE post-translationally, we tested the exogenous expression of Nanos-GFP in the same manner. In contrast to Vasa-GFP, exogenous Nanos was equally distributed throughout the embryo and did not become enriched in the PE in larvae (Figure 1E–G) suggesting that endogenous Nanos enrichment in the PE is not controlled post-translationally (Fresques et al., 2014). Indeed, endogenous Nanos transcripts are specifically expressed in the vegetal region of the gastrula and subsequently in the PE, suggesting instead some transcriptional control mechanisms (Fresques et al., 2014; Fresques and Wessel, 2018). We quantitated the differential signal intensities for Vasa-GFP, Nanos-GFP and a construct containing GFP alone as a control, and compared the intensity of the fluorescent signals for each construct in the PE relative to the stomach, the tissue that lies adjacent to the PE. We found that while exogenous Vasa protein is enriched in the PE compared to the stomach, the ratio of both Nanos and of GFP control constructs in the PE and the stomach were equal (Figure 1 H–K). We concluded that Vasa protein is selectively restricted to the PE through unique post-translational mechanisms, while Nanos-GFP, and GFP alone, do not show any such cell type enrichment.
Figure 1. The PE selectively retains Vasa-GFP, but not Nanos-GFP nor GFP alone.
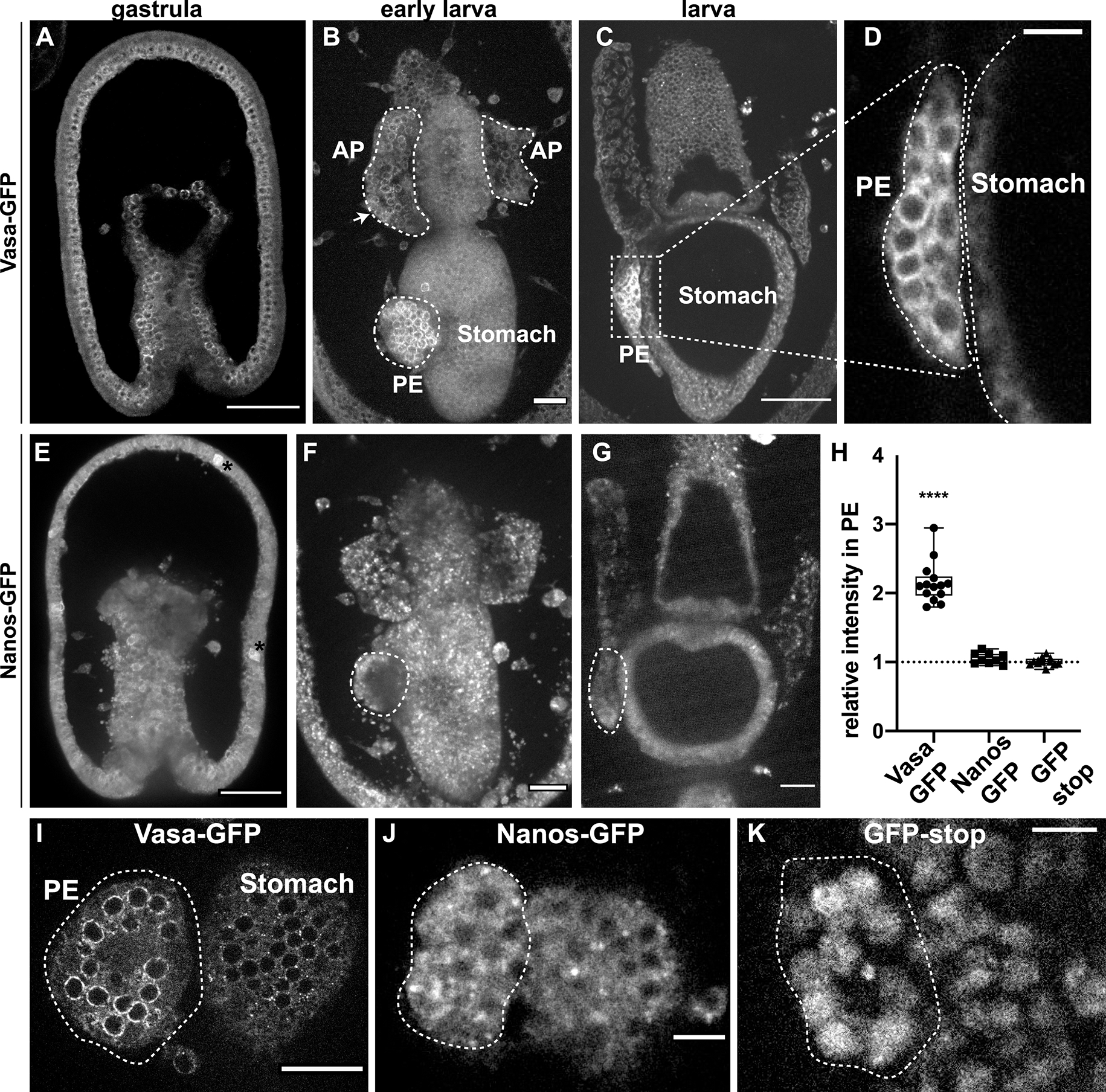
A-D) GFP fluorescence from an exogenous full-length Vasa-GFP mRNA seen from gastrula to late larva. The microinjected mRNA is translated by the embryos and the fluorescent signal corresponds to the location of the Vasa protein. Note that the Vasa protein is equally distributed in all cells in gastrulae (A) but becomes restricted to the PE when this tissue forms in larvae (B,C). Arrow in B shows Vasa protein in the AP. D). Magnification of the PE showing Vasa localization is perinuclear. E-G) Nanos-GFP signal from gastrula to larva shows that Nanos protein does not accumulate in a specific region and it is uniformly distributed throughout the embryo. Asterisk in E shows autofluorescence of mesenchyme cells embedded in the ectoderm. H) Quantification of the microinjected exogenous Vasa, Nanos or GFP-stop fluorescence in the PE relative to the stomach shows that only Vasa protein is enriched in the PE. For t-test **** significance the p-value is < 0,0001. I-K) Subcellular localization of the Vasa, Nanos and GFP-stop proteins in the PE (left) compared to the stomach (right). The stomach has been chosen to copare the fluorescence signals because it is the tissue immediately adjacent to the PE for a stringent imaging comparison. Dotted white lines highlight the PE. Scale bar 50μm (A, E), 20μm (B,C,F, G, I) and 10μm (D,J,K). PE = posterior enterocoel; AP = anterior pouch
Exogenously encoded Vasa protein is stable selectively in the PE
Is the exogenously-encoded Vasa-GFP protein enrichment in the PE due to selective retention of the transcripts to the PGCs of the PE? Selective transcript retention is seen in animals where the germ cell factors are inherited, as it is in the sea urchin (Gustafson and Wessel, 2010; Oulhen and Wessel, 2013). To test this mechanism, we injected Vasa-mCherry mRNA, cultured embryos to the larval stage when the Vasa protein selectively accumulates in the PE (Figure 2A) and then tested for the presence of the exogenous Vasa-mch RNA by in situ RNA hybridization. We found that the injected Vasa-mch transcripts were expressed throughout the larva and showed no enrichment for the PE or any other tissue (Figure 2B). As a control, the mCherry probe showed no signal in uninjected embryos (Figure 2C), while endogenous vasa mRNA was detected selectively in the PE and in few cells of the anterior pouches (Figure D). These findings suggest that the Vasa protein is either selectively translated in the PE or is selectively degraded in all other tissues
Figure 2. Exogenous Vasa mRNA is not retained selectively anywhere in the embryo.
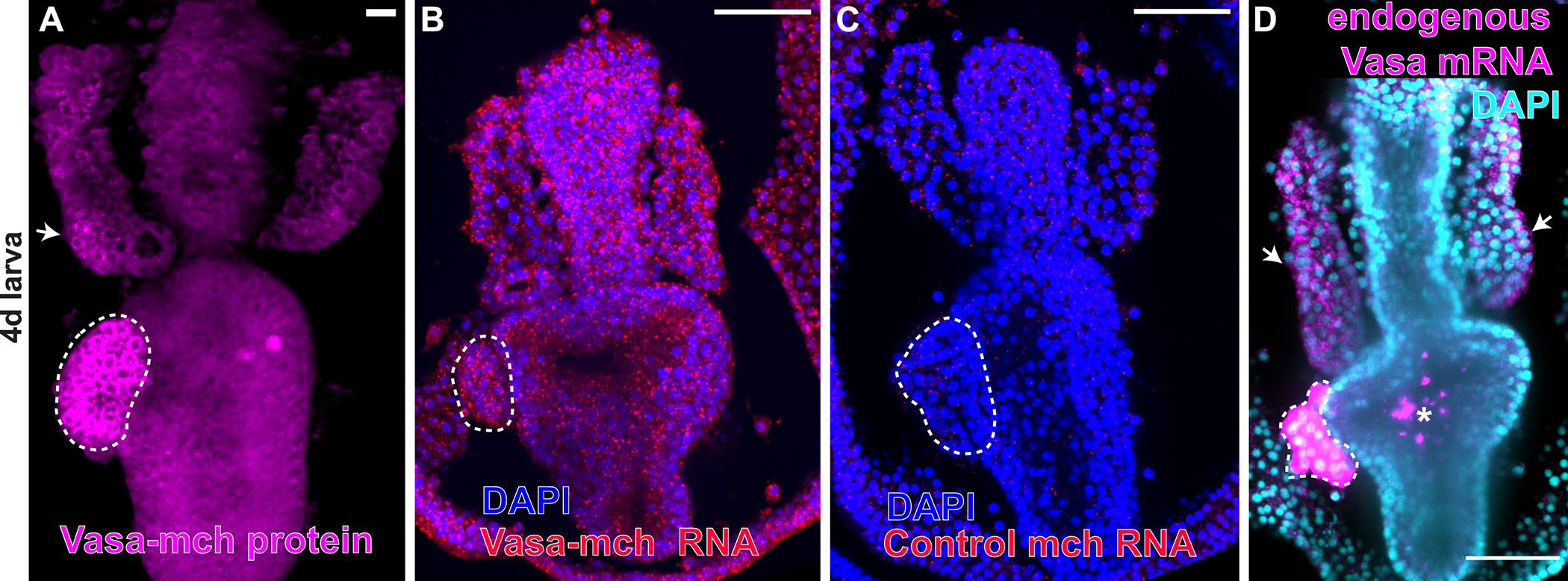
A) Exogenous Vasa-mCherry protein is enriched in the PE and in some cells of the anterior pouches (AP), white arrow. B) Fluorescent in situ hybridization of larvae expressing Vasa-mch using the mCherry probe detects Vasa-mCherry mRNA in every cell of the larva. C) A probe for mcherry used as a negative control shows no endogenous mcherry gene activity in wild type larvae. D) Detection of endogenous vasa mRNA in wild type larvae using a vasa antisense probe showing that vasa transcripts are expressed in the PE and in some cells of the AP (white arrows). Asterisk indicates non-specific fluorescence in the stomach of fed larvae. Scale bars = 50 μm. Dotted white lines highlight the PE. All figures show larvae 90–96h old.
Vasa protein is selectively degraded in somatic cells by a Gustavus-dependent mechanism
The E3 ubiquitin ligase Gustavus (Gus) has been reported to target Vasa to the proteasome in a variety of animals (Gustafson et al., 2011; Kugler et al., 2010; Sellars et al., 2015; Zhang et al., 2011). In the sea star, Gus is expressed in all tissues except the PE and the anterior pouches (Fresques et al., 2014), suggesting that this protein could be responsible for Vasa protein degradation outside of the PE. To test if Vasa is a target of Gustavus, we injected sea star oocytes with a Gustavus-specific morpholino antisense nucleotides (GusMO) and the resulting Vasa-GFP protein accumulation was visualized in live embryos. In gastrulae, Vasa-GFP showed uniform perinuclear localization of all cells in the embryo for both control and morphant embryos (Figure 3A, B). In larvae, the Gustavus morphants revealed a distinct phenotype with disorganized anterior pouches and PE (Figure 3C, D,E, F ). In 5 day larvae, the morphants developed a more severe phenotype wherein Vasa protein was abundant throughout the larva, the PE fragmented into smaller pieces that remained around the stomach (Figure 3H, white arrowheads), and two additional anterior pouches (AP) grew posteriorly (video 1 and 2; Figure 3G,H).
Figure 3. Gustavus is required for clearing Vasa protein in somatic tissues.
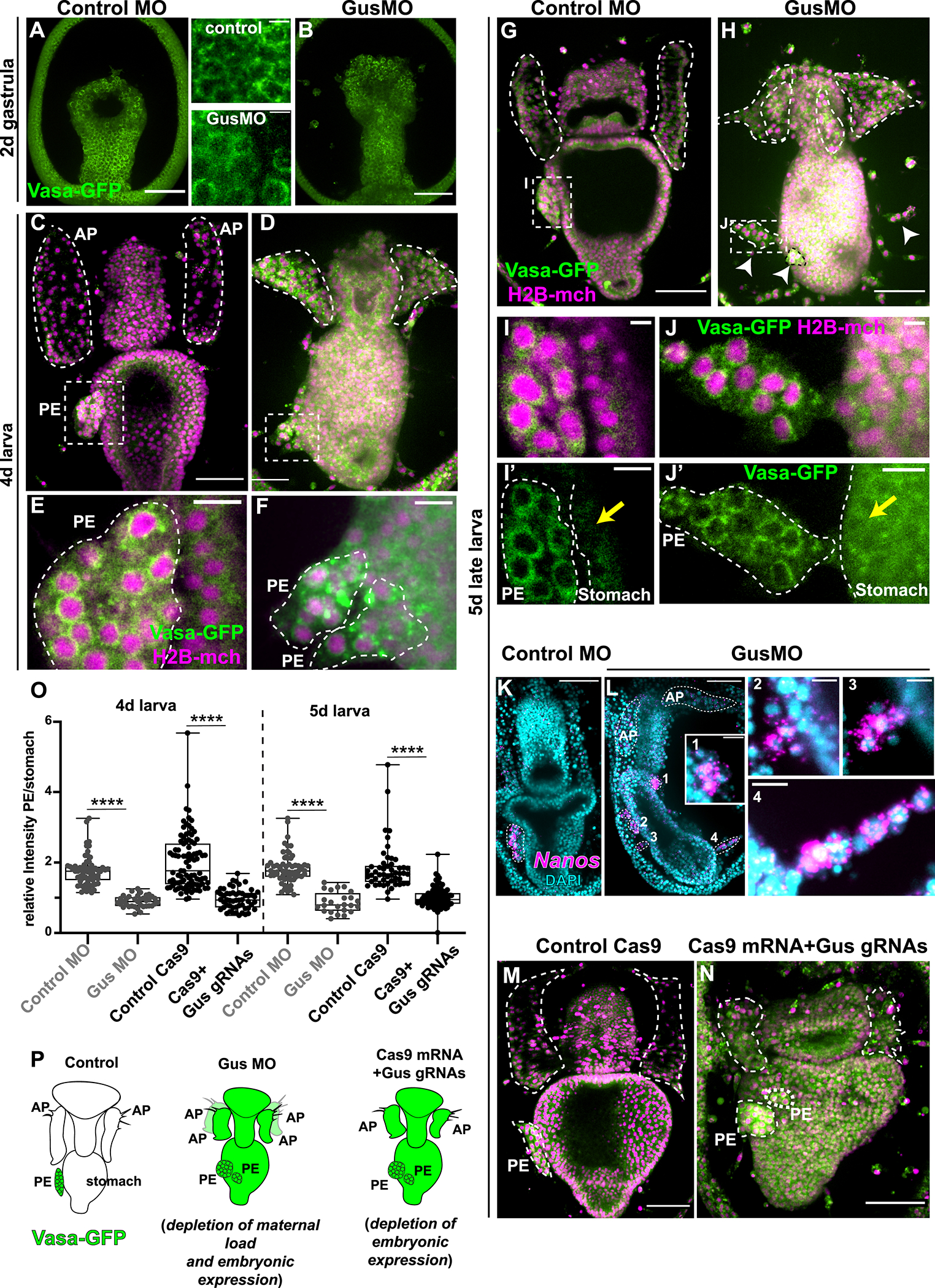
A,B) In gastrulae, Vasa-GFP protein is enriched perinuclearly in every cell-type in both controls and Gus morphants (see also inserts). C,D, E, F) 4 day larvae showing that in Gus morphants Vasa protein accumulates also in extra-PE tissues and the PE is fragmented G, H). At 5 days, controls have one PE on the left side of the stomach while Gus morphants have fragmented PEs (white arrowheads). I, J) In both controls and Gus-morphants, Vasa protein is localized perinuclearly in cells of the PE. Note that Vasa protein has accumulated in cells of the stomach of the Gus morphants only (yellow arrows). (For the GusMO phenotype at 5 days see also video 1 and 2. K, L) Fluorescent in situ hybridization showing that nanos transcripts are enriched in the PE in controls (K) and in the fragmented PEs in Gus morphants (L). Panels 1, 2, 3 and 4 are magnifications of the fragmented PEs from L that are enriched in nanos transcripts. AP and PEs are highlighted by white dotted lines. M) Controls (injected with Cas9 mRNA) and N) Gus knockout larvae (injected with Cas9 mRNA and 3 sgRNAs targeting Gus DNA) showing that preventing embryonic Gus expression causes fragmented PEs, vasa protein accumulation in all larval tissues and impaired APs. O) Quantification of Vasa protein enrichment in the PE with respect to the stomach in control MO and GusMO and in Cas9 control and Cas9 mRNA+ Gus sgRNAs in 4 and 5 day larvae. The ratio is close to 1 if the Vasa signal intensity is similar in the PE and the stomach, while it is >1 if the Vasa signal is stronger in the PE respect to the stomach. Student’s t-test gives p value <0.001 (****). P) Cartoon summarizing the main results. Exogenous vasa protein is shown in green. Vasa protein is degraded in the somatic tissue by a Gustavus-mediated mechanism so that Vasa is enriched in the PE only. Lack of Gustavus (using both morpholino and CRISPR-Cas9 approaches) results in ectopic Vasa protein expression in all somatic tissues and in the fragmented PEs. Note that when Gus is maternally depleted by blocking the overall protein translation there are 4 anterior pouches, while specifically blocking embryonic Gus expression results in smaller, but not supernumerary pouches. PE=posterior enterocoele (presumptive PGCs); AP= anterior pouches. Scale bar is 50μm in A,B,C,D,G,H,K,L, M,N and 5μm in I and J and 10μm in E,F, I’, J’, L1,L2,L3,L4.
In the Gus morphant larvae, we found an increase of Vasa protein in the stomach and throughout the embryo compared to control larvae (Figure 3 I,J yellow arrows), while the Vasa-GFP in the PE remained the same (Figure 3E, F, I, J). Moreover, in extra PE tissues the Vasa-GFP accumulated in the cytoplasm instead of perinuclearly in Gus morphants (Figure 3 I, J, I’, J’) suggesting that the protein is translated but not targeted to the correct cellular compartment. Since the extra PEs showed Vasa perinuclear localization, like in the control PE, we hypothesized that these were PE fragments or dispersed cell clusters from the PE. To further test this hypothesis, we investigated whether transcripts for germ line factors were expressed in the fragmented PEs of Gus morphants. Since these larvae expressed ectopic Vasa-GFP (and therefore vasa transcripts would be detected everywhere in the larva, as shown in Figure 2B), we focused on nanos transcripts as a marker for PE cells. We found that Nanos is enriched in the PE of control larvae as well as in the many PE fragments that appeared in the Gus morphants (Figure K, L for whole larvae and panels 1,2,3,4 for magnifications of PEs seen in GusMO). Moreover, as stomach cells did not display Vasa perinuclear localization, we propose that these extra PE structures that expressed Nanos were presumptive PE cells that did not associate with the majority PE cluster, rather than cells budding from the stomach.
To further test this model, and the specificity of the Gus morpholino, we used a CRISPR/Cas9 approach using guide RNAs targeting the Gus locus to generate a Gus-knockout. We quantified the resulting signal intensity of the Vasa-GFP in the PE versus the stomach and found that as in the Gus morphants, Gus knockouts displayed a significant increase in Vasa-GFP protein outside of the PE (Figure 3O). We further found that Gus knockout larvae also showed fragmented PEs, while the anterior pouches were smaller and disorganized but did not develop the two extra APs (M, N). Collectively, our results blocking Gus expression with two complementary approaches indicates that Gustavus regulates Vasa protein turnover in the somatic tissues exclusive of the PE and AP.
Discussion
We have found through comparative studies that two organisms representing extremes in germ line determination strategy, sea urchins and sea stars, use a similar mode of post-translational regulation for Vasa. It is striking that even though the site and the timing of deployment of its function in development is distinct, the node of posttranslational regulation selectively for Vasa is the same. Gustavus (Gus) was originally identified in Drosophila and was shown to be involved in regulation of Vasa in the embryo posterior, and is present in the PGCs, potentially as a protection mechanism for Vasa there (Kugler et al., 2010). Gus may play both a somatic degradation mechanism, and a PGC protective mechanism for Vasa also in the sea urchin (Gustafson et al., 2011). In the mouse, Gus orthologs are present in the granulosa cells of the ovary, immediately adjacent to the developing oocytes, perhaps to repress any germ line commitment from those somatic cells (Xing et al., 2006). Now seen from results here, we conclude that the Gus in the sea star P. miniata, functions to degrade Vasa in somatic cells, thereby restricting Vasa location to the PGCs early in their specification. We surmise from these various contexts that the Gus-Vasa interaction is ancestral to the diverse mechanisms of germ line specification, but the timing and effect of this interaction varies through evolution.
Another revealing finding was that two complementary approaches to block Gus expression gave similar results. Using a morpholino approach we depleted Gus transcripts that were maternally loaded into the embryo, in addition to the newly transcribed Gus mRNA. On the other hand, by editing the Gus gene using a CRISPR Cas9 approach we blocked the embryonic Gus expression but not the maternally loaded Gus transcripts that are enriched in the oocyte (Fresques et al., 2014). The outcome of both approaches was an upregulation of Vasa protein in extra-PE tissues and the lack of coalescence in cells of the PE (Figure 3 D, H, L, N, O and cartoon in Figure 3P). The only noticeable difference between the two approaches was that while the depletion of maternal and embryonic Gus resulted in four anterior pouches instead of two, depletion of embryonic Gus only resulted in two smaller impaired anterior pouches. A similar phenotype of radial anterior pouches was observed in larvae depleted of Nodal (Fresques 2018). We hypothesize that the radialized phenotype observed with morpholino knockdown is likely due to more complete depletion of the Gus maternal load, which would disrupt the Nodal pathway early in embryogenesis, prior to the transcription of Gus-null embryonic transcript accumulation.
The similarities in the Gus-Vasa utilization between the sea star and the sea urchin are significant and comparing these closely related taxa makes important distinctions. Since the two embryos specify their germ line distinctly, we anticipated dramatic differences in implementation. The relationship of the two gene products were found, however, to be shared, with any apparent differences more a feature of developmental differences; in timing of expression, in phenotype of the Gus-morphants, the selectivity of Vasa in the somatic cells, and of selectivity for Vasa in the germ line. A significant difference in the results of the sea urchin and the sea star however, is still a result of an unexplained mechanism. Gus knockdown in the sea urchin S. purpuratus, resulted in no Vasa accumulation anywhere, whereas expression of a Gus lacking its SOC-box resulted in accumulation of significant levels of Vasa throughout the embryo. We interpret this phenotype as Gus in the sea urchin has a protective and/or a degradative function, depending on the cell type within which is working. This is similar to what is concluded in Drosophila, in which Gustavus was originally identified (Kugler et al., 2010). In the sea star, however, Gus knockdown or knockout resulted in a dramatic increase in Vasa protein throughout the embryo, and perhaps by virtue of the absence of Gus mRNA in the PE, a protective role of Gus for Vasa is not seen (summarized in Figure 3P). Perhaps the difference between the species balances on the difference in PGC specification mechanisms, the sea urchin utilizing early specification by inherited factors in the egg and early embryo – as in Drosophila - whereas the sea star uses a late, inductive mechanism, as in the mouse. It will be important to test this distinction in other animals that use an inductive PGC mechanism and that have Gus and Vasa expression coincidently in the embryos.
Supplementary Material
Highlight.
The RNA helicase Vasa is regulated spatially by post-translational mechanisms in the sea star embryo
Ubiquitous Vasa-GFP mRNA yields selective protein accumulation in the presumptive germ line.
Clearance of Vasa from somatic cells in the embryo depends on the E3-ligase, Gustavus.
Sea urchin and sea stars use distinct mechanisms of specification for primordial germ cells (PGC), but with a similar node in the regulation of Vasa
Acknowledgments
The authors are grateful for the support for this work from the Eunice Kennedy Shriver National Institute of Child Health and Human Development (K99HD099315) to S.Z.S and the National Institutes of Health grant 1R35GM140897 to GMW.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Cameron RA, Samanta M, Yuan A, He D, Davidson E, 2009. SpBase: the sea urchin genome database and web site. Nucleic acids research 37, D750–D754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donoughe S, Nakamura T, Ewen-Campen B, Green DA 2nd, Henderson L, Extavour CG, 2014. BMP signaling is required for the generation of primordial germ cells in an insect. Proceedings of the National Academy of Sciences of the United States of America 111, 4133–4138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Extavour CG, Akam M, 2003. Mechanisms of germ cell specification across the metazoans: epigenesis and preformation. Development 130, 5869–5884. [DOI] [PubMed] [Google Scholar]
- Foltz K, Adams N, Runft L, 2004. Echinoderm eggs and embryos: procurement and culture, in: Ettensohn C, Wessel G, Wray G (Eds.), Methods Cell Biology Academic Press, pp. 39–73. [DOI] [PubMed] [Google Scholar]
- Fresques T, Zazueta-Novoa V, Reich A, Wessel GM, 2014. Selective accumulation of germ-line associated gene products in early development of the sea star and distinct differences from germ-line development in the sea urchin. Developmental dynamics : an official publication of the American Association of Anatomists 243, 568–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fresques TM, Wessel GM, 2018. Nodal induces sequential restriction of germ cell factors during primordial germ cell specification. Development 145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gökirmak T, Campanale JP, Shipp LE, Moy GW, Tao H, Hamdoun A, 2012. Localization and substrate selectivity of sea urchin multidrug (MDR) efflux transporters. The Journal of biological chemistry 287, 43876–43883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafson EA, Wessel GM, 2010. Exogenous RNA is selectively retained in the small micromeres during sea urchin embryogenesis. Mol Reprod Dev 77, 836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafson EA, Yajima M, Juliano CE, Wessel GM, 2011. Post-translational regulation by gustavus contributes to selective Vasa protein accumulation in multipotent cells during embryogenesis. Developmental biology 349, 440–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue C, Kiyomoto M, Shirai H, 1992. Germ cell differentiation in starfish: The posterior enterocoel as the origin of germ cells in Asterina pectinifera. Development Growth & Differentiation 34, 413–418. [DOI] [PubMed] [Google Scholar]
- Kudtarkar P, Cameron RA, 2017. Echinobase: an expanding resource for echinoderm genomic information. Database : the journal of biological databases and curation 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kugler JM, Woo JS, Oh BH, Lasko P, 2010. Regulation of Drosophila vasa in vivo through paralogous cullin-RING E3 ligase specificity receptors. Molecular and cellular biology 30, 1769–1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oulhen N, Wessel GM, 2013. Retention of exogenous mRNAs selectively in the germ cells of the sea urchin requires only a 5’-cap and a 3’-UTR. Mol Reprod Dev 80, 561–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perillo M, Paganos P, Spurrell M, Arnone MI, Wessel GM, 2021. Methodology for Whole Mount and Fluorescent RNA In Situ Hybridization in Echinoderms: Single, Double, and Beyond. Methods in molecular biology 2219, 195–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich A, Dunn C, Akasaka K, Wessel G, 2015. Phylogenomic analyses of Echinodermata support the sister groups of Asterozoa and Echinozoa. PloS one 10, e0119627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellars MJ, Trewin C, McWilliam SM, Glaves RS, Hertzler PL, 2015. Transcriptome profiles of Penaeus (Marsupenaeus) japonicus animal and vegetal half-embryos: identification of sex determination, germ line, mesoderm, and other developmental genes. Mar Biotechnol (NY) 17, 252–265. [DOI] [PubMed] [Google Scholar]
- Seydoux G, Braun RE, 2006. Pathway to totipotency: lessons from germ cells. Cell 127, 891–904. [DOI] [PubMed] [Google Scholar]
- Strome S, Updike D, 2015. Specifying and protecting germ cell fate. Nature reviews. Molecular cell biology 16, 406–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voronina E, Lopez M, Juliano CE, Gustafson E, Song JL, Extavour C, George S, Oliveri P, McClay D, Wessel G, 2008. Vasa protein expression is restricted to the small micromeres of the sea urchin, but is inducible in other lineages early in development. Developmental biology 314, 276–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessel GM, Brayboy L, Fresques T, Gustafson EA, Oulhen N, Ramos I, Reich A, Swartz SZ, Yajima M, Zazueta V, 2014a. The biology of the germ line in echinoderms. Mol Reprod Dev 81, 679–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessel GM, Fresques T, Kiyomoto M, Yajima M, Zazueta V, 2014b. Origin and development of the germ line in sea stars. Genesis 52, 367–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing Y, Gosden R, Lasko P, Clarke H, 2006. Murine homologues of the Drosophila gustavus gene are expressed in ovarian granulosa cells. Reproduction 131, 905–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zazueta-Novoa V, Wessel GM, 2014. Protein degradation machinery is present broadly during early development in the sea urchin. Gene Expr Patterns 15, 135–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Chen L, Qin J, Zhao W, Wu P, Yu N, Ma L, 2011. cDNA cloning and expression analysis of gustavus gene in the oriental river prawn Macrobrachium nipponense. PloS one 6, e17170. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


