Abstract
During 2014, highly pathogenic (HP) influenza A viruses (IAVs) of the A/Goose/Guangdong/1/1996 lineage (GsGD-HP-H5), originating from Asia, were detected in domestic poultry and wild birds in Canada and the US. These clade 2.3.4.4 GsGD-HP-H5 viruses included reassortants possessing North American lineage gene segments; were detected in wild birds in the Pacific, Central, and Mississippi flyways; and caused the largest HP IAV outbreak in poultry in US history. To determine if an antibody response indicative of previous infection with clade 2.3.4.4 GsGD-HP-H5 IAV could be detected in North American wild waterfowl sampled before, during, and after the 2014–15 outbreak, sera from 2,793 geese and 3,715 ducks were tested by blocking enzyme-linked immunosorbent assay and hemagglutination inhibition (HI) tests using both clade 2.3.4.4 GsGD-HP-H5 and North American lineage low pathogenic (LP) H5 IAV antigens. We detected an antibody response meeting a comparative titer-based criteria (HI titer observed with 2.3.4.4 GsGD-HP-H5 antigens exceeded the titer observed for LP H5 antigen by two or more dilutions) for previous infection with clade 2.3.4.4 GsGD-HP-H5 IAV in only five birds, one Blue-winged Teal (Spatula discors) sampled during the outbreak and three Mallards (Anas platyrhynchos) and one Canada Goose (Branta canadensis) sampled during the post-outbreak period. These serologic results are consistent with the spatiotemporal extent of the outbreak in wild birds in North America during 2014 and 2015 and limited exposure of waterfowl to GsGD-HP-H5 IAV, particularly in the central and eastern US.
Keywords: Clade 2.3.4.4, ducks, geese, hemagglutination inhibition, H5, influenza A virus, North America, serology
INTRODUCTION
In November 2014, a highly pathogenic (HP) H5N2 influenza A virus (IAV) of the A/Goose/Guangdong/1/1996 (GsGD-HP-H5) lineage was detected in domestic turkeys in Canada (Pasick et al. 2015). Subsequently, this virus, a progenitor HP H5N8 IAV, and additional reassortant viruses containing North American lineage IAV gene segments were detected in wild birds and domestic poultry in the US (Lee et al. 2016). Most of the isolates from wild birds originated from ducks, geese, and raptors (Ip et al. 2015; Bevins et al. 2016; Ramey et al. 2017). Among poultry, backyard and commercial chicken and turkey flocks were infected, with the most extensive losses occurring in Minnesota, Iowa, and surrounding Midwestern states (US Department of Agriculture 2015). The H5N2 subtype was the predominant GsGD-HP-H5 IAV isolated from both domestic poultry and wild birds during 2014–15 (Saito et al. 2015).
The clade 2.3.4.4 GsGD-HP-H5 IAV, which was first detected in Southeast Asia in 2014, probably entered North America with migratory wild birds via Alaska and was further dispersed by wild birds within the continent (Lee et al. 2015; Ramey et al. 2016). From November 2014 to June 2015, various subtypes of clade 2.3.4.4 GsGD-HP-H5 IAV were detected from numerous waterfowl species sampled in the Pacific, Central, and Mississippi flyways (Bevins et al. 2016; Krauss et al. 2016). However, from June 2015 to present in North America, there have been no cases of clade 2.3.4.4 GsGD-HP-H5 IAV infection in domestic poultry and few detections from wild waterfowl. After June 2015, there have been only four detections from wild waterfowl, all from individual Mallards (Anas platyrhynchos), including birds sampled during July 2015 in Utah, November 2015 in Oregon, August 2016 in Alaska, and December 2016 in Montana (Lee et al. 2017; US Department of Agriculture 2017). While these results and subsequent sequencing of the August 2016 sample provide evidence that clade 2.3.4.4 GsGD-HP-H5 IAV persisted in North America for more than a year after these viruses were eradicated from poultry, there is no recent evidence that HP viruses continue to circulate among North American wild bird populations (Krauss et al. 2016).
Serologic approaches to IAV surveillance have been successfully used as supportive tools in epidemiologic studies of low pathogenic (LP) IAV in wild birds (Brown et al. 2010). Most of these applications have utilized tests for influenza A conserved antigens such as nucleoproteins (NPs) that provide no information related to the presence of antibodies to specific IAV subtypes or strains. There have been several attempts to serologically identify birds that have been naturally infected with GsGD-HP-H5 lineage viruses. Gilbert et al. (2014) tested wild bird sera collected in Mongolia, the Netherlands, and Norway by hemagglutination inhibition (HI) against a panel of clade 0, 1, 2.1, 2.2, and 2.3 GsGD-HP-H5 antigens. Serum samples from wild birds were collected from areas in Mongolia that had experienced eight outbreaks of GsGD-HP-H5 lineage viruses representing clades 2.2 and 2.3.2.1 (Gilbert et al. 2012), while those from Europe were collected from areas where HP IAV outbreaks were rare or absent. Based on the HI titer differences observed with a panel of H5 antigens, a bias in HI antibody titers suggestive of previous exposure to the GsGD-HP-H5 viruses was detected with the Mongolian samples, but not the European serum samples (Gilbert et al. 2014). More recently, combined virologic and serologic (HI and microneutralization [MNt]) approaches were used in Europe related to surveillance for clade 2.3.4.4 HP H5N8 viruses (Poen et al. 2016, 2018; Hill et al. 2019). In those studies, HI antibodies suggestive of clade 2.3.4.4 GsGD-HP-H5 IAV exposure were detected in Mute Swan (Cygnus olor), Lesser White-fronted Geese (Anser erythropus), Egyptian Goose (Alopo-chen aegyptiaca), Mallard, Eurasian Wigeon (Mareca penelope), Black-headed Gull (Chroicocephalus ridibundus), and Common Coot (Fulica atra). These results were consistent with the wild bird species that were positive for virus isolation of HP H5N8 from Russia, the Netherlands, and Sweden (Poen et al. 2016).
The introduction and subsequent outbreak of clade 2.3.4.4 GsGD-HP-H5 IAV in North America during 2014–15 and the wild bird surveillance efforts that accompanied this outbreak provided an opportunity to further evaluate the utility of serologic-based surveillance to detect evidence of previous infection with these viruses. The objective of this study was to determine if antibody responses suggestive of previous infection with clade 2.3.4.4 GsGD-HP-H5 IAV could be detected in North American waterfowl sampled before (prior to 1 November 2014), during 1 November 2014–30 May 2015), and after (1 June 2015–31 January 2016) the 2014–15 outbreak. Results provided information that facilitates further evaluation of the spatiotemporal extent of the outbreak in North America and information on the potential utility of subtype-specific serology as a complementary surveillance tool related to HP IAV outbreaks that involve both wild and domestic birds.
MATERIALS AND METHODS
Wild birds sampled
Serum samples tested in this study were collected by personnel from the Minnesota Department of Natural Resources, West Virginia Division of Natural Resources, Pennsylvania Game Commission, Georgia Department of Natural Resources, US Department of Agriculture (USDA) Wildlife Services, US Geological Survey (USGS) Alaska Science Center, USGS Patuxent Wildlife Research Center, USGS Western Ecological Research Center, University of California–Davis, the Ohio State University, and University of Georgia. Capture methods included rocket netting, swim-in traps, and driving of flightless molting birds into capture pens. Samples were also collected from hunter-harvested birds. Blood samples from live birds were collected via jugular, brachial, or tarsal veins. All collections were made in compliance with the specific agency’s or institution’s animal care and use policy, and, when applicable, committee approval and all appropriate state and federal wildlife collection permits were obtained.
Antigen preparation
Virus antigens used for antibody testing included 1) wild type (wt) North America H5 low pathogenicity avian influenza (LPAI) A/Blue-winged Teal/Texas/AI12–4150/2012 (H5N2) (LP-wtBWT/TX) and A/Mallard/Minnesota/AI11–3933/2011 (H5N1) (LP-wtmallard/MN) viruses; 2) inactivated GsGD-H5 HPAI A/Gyrfalcon/Washington/41088–6/2014 (H5N8) (HP-GYR/WA) and GsGD A/Turkey/Minnesota/9845/2014 (H5N2) (HP-TKY/MN) viruses, kindly provided by the National Veterinary Services Laboratory (NVSL), USDA, Ames, Iowa; and 3) viruses carrying hemagglutinin (HA) and neuraminidase (NA) from selected virus strains with the internal genes of the laboratory-adapted A/Puerto Rico/08/1934 (H1N1) strain (PR/08). Recombinant viruses were rescued by standard reverse genetics (rg) techniques (Perez et al. 2017) using HA/NA pairs from A/Chicken/Netherlands/EMC-3/2014 (ΔH5N8) (HP-rgCHK/Netherlands, provided by Erasmus Medical Center, Rotterdam, the Netherlands), A/Snow Goose/Missouri/CC15–84a/2015 (DH5N2) (HP-rgSnow/MO, provided by St. Jude Children’s Research Hospital, Memphis, Tennessee), and North America H5 LPAI A/Blue-winged Teal/Texas/AI12–4150/2012 (H5N2) (LP-rgBWT/TX) viruses.
Antibody testing
All serum samples were initially screened using a commercial blocking enzyme-linked immunosorbent assay (bELISA; IDEXX AI MultiS-Screen AB test, IDEXX Laboratories, Westbrook, Maine, USA). Samples were classified based on serum/negative (s/n) optical density ratios as positive (s/n≤0.5), suspect (0.5>s/n<0.7), or negative (s/n>0.7). The s/n≤0.5 value represents the positive threshold recommended by the manufacturer. Classification of samples as suspect was based on results for experimentally and naturally infected waterfowl that suggest such samples may also represent previously IAV-exposed birds (Brown et al. 2010; Tolf et al. 2013; Shriner et al. 2016). All samples testing positive were further tested by HI. In order to validate the use of the bELISA as a screening test, limited numbers of bELISA suspect and negative samples also were tested by HI.
Serum samples were tested for antibodies to H5 IAV using a standard HI procedure. The testing protocol, with minor exceptions, paralleled the testing strategy used in Europe as described by Poen et al. (2016). All samples were treated at a 1:4 dilution with receptor-destroying enzyme (Denka Seiken Co., Ltd., Tokyo, Japan). Sera (25 μL) were initially screened by HI using turkey red blood cells at a serum dilution of 1:16 against LP-rgBWT/TX and HP-GYR/WA antigens at four HA units each. Samples demonstrating nonspecific hemagglutination of turkey red blood cells were not tested further and were not included in the HI data summaries. Samples testing HI positive at the 1:16 screen for either antigen were tested a second time at dilutions from 1:16 to 1:512 against LP-rgBWT/TX, HP-GYR/WA, and HP-TKY/MN antigens.
To validate HI as a means of differentiating between antibodies to clade 2.3.4.4 GsGD-HP-H5 and North American lineage LP H5 IAV, additional antigens that included clade 2.3.4.4 HP-TKY/MN as well as North American lineage LPAI antigens LP-rgBWT/TX, LP-wtBWT/TX, and LP-wtmallard/MN were tested against control and field sera. The serum panel included antisera against HP-GYR/WA and clade 2.3.4.4 GsGD-HP-H5 A/northern pintail/Washington/40964/2014 (H5N2) produced in chickens provided by NVSL and the Southeast Poultry Research Laboratory (SEPRL), Agricultural Research Service, USDA; sera from experimental infections of mallards with HP-GYR/WA provided by SEPRL or North American LP A/mallard/Minnesota/355779/2000 (H5N2) IAV; and field serum samples collected in 2012 from Mute Swans in Michigan (provided by Wildlife Services, USDA) and Snow Geese (Anser caerulescens) sampled in Texas, prior to the 2014–15 detection of clade 2.3.4.4 GsGD-HP-H5 viruses in North America.
In order to investigate the potential utility of MNt to differentiate between antibodies to clade 2.3.4.4 GsGD-HP-H5 viruses and North American LP H5 IAV, LP-rgBWT/TX, HP-rgCHK/Netherlands, and HP-rgSnow/MO were used. Testing was limited to serum samples with HI titers for the clade 2.3.4.4 GsGD-HP-H5 antigens that exceeded titers observed with LP-rgBWT/TX by at least a single dilution. The MNt was conducted as previously described using Madin-Darby canine kidney cells and 100 median tissue culture infective doses of antigen (Wong et al. 2016).
Data analysis
A sample was regarded as having an antibody titer biased to clade 2.3.4.4 GsGD-HP-H5 IAV and indicative of previous exposure if the HI titer observed with HP-GYR/WA or HP-TKY/MN exceeded the titer observed for LP-rgBWT/TX by two or more dilutions. Samples with a one dilution bias were regarded as suggestive of clade 2.3.4.4 GsGD-HP-H5 IAV exposure. The same criteria were used for MNt with LP-rgBWT/TX antibody titer results compared with titers obtained with the HP-rgCHK/Netherlands and HP-rgSnow/MO antigens.
RESULTS
HI validation
Comparisons of antibody titers for clade 2.3.4.4 GsGD-HP-H5 and North American LP IAV antigens for control sera (n=3) and sera from experimentally infected Mallards (n=7) correctly identified exposures to either clade 2.3.4.4 GsGD-HP-H5 or North American lineage LP H5 IAV (Table 1). Comparative antibody titers (≤2 dilution difference) also correctly identified North American lineage LP H5 IAV exposure in 11 of 14 Snow Geese and Mute Swans sampled 2 years prior to the detection of clade 2.3.4.4 GsGD-HP-H5 IAV in North America (Table 1). Antibody titers in the remaining three samples were too low to discriminate. Although there was good agreement among results derived using different clade 2.3.4.4 GsGD-HP-H5 and North American lineage LP IAV, the most consistent results were observed with HP-GYR/WA and LP-rgBWT/TX. Consequently, these viruses were selected as antigens for subsequent HI screening.
Table 1.
Validation of hemagglutination inhibition (HI) testing protocol using clade 2.3.4.4 goose Guangdong lineage highly pathogenic H5 influenza A virus (GsGD-HP-H5 IAV) and North American lineage low pathogenic (LP) H5 influenza A virus (IAV) antigens tested against control antisera, sera from birds experimentally infected with GsGD-HP-H5 IAV and LP IAV, and field samples from wild birds taken prior to detection of clade 2.3.4.4 GsGD-HP-H5 IAV in North America. Twofold higher HI antibody titers to either clade 2.3.4.4 GsGD-HP-H5 IAV or North American lineage LP H5 IAV are in bold.
| Clade 2.3.4.4 GsGD-HP-H5 IAV antigensa |
North American lineage LP H5 IAV antigensb |
|||||
|---|---|---|---|---|---|---|
| Serum source | HP-Turkey MN | HP-GYR/WA | LP-rgBWT/TX | LP-wtBWT/TX | LP-Mallard/MN | |
|
| ||||||
| A/GYR/WA (H5N8) (SEPRL) | Control antiserac | 64 | 128 | <16 | <16 | <16 |
| A/GYR/WA (H5N8) (NVSL) | ≥1,024 | ≥1,024 | 64 | <16 | <16 | |
| A/Northern pintail/WA (H5N2) (SEPRL) | 256 | 256 | <16 | <16 | <16 | |
| A/GYR/WA (H5N8) Mallard 1 (SEPRL) | Experimentally infected Mallards (Anas platyrhynchos)d | 16 | 128 | <16 | <16 | <16 |
| A/GYR/WA (H5N8) Mallard 2 (SEPRL) | 128 | 128 | <16 | <16 | <16 | |
| A/Mallard/MN/355779 (H5N2) Mallard 1 | <16 | <16 | 128 | 128 | 128 | |
| A/Mallard/MN/355779 (H5N2) Mallard 2 | <16 | <16 | 128 | 64 | 64 | |
| A/Mallard/MN/355779 (H5N2) Mallard 3 | <16 | <16 | 32 | 16 | 16 | |
| A/Mallard/MN/355779 (H5N2) Mallard 4 | <16 | <16 | 512 | 128 | 128 | |
| A/Mallard/MN/355779 (H5N2) Mallard 5 | <16 | <16 | 32 | 16 | 16 | |
| Snow Goose 1 | Pre-outbreak field samplese | <16 | 32 | 256 | Not tested | 64 |
| Snow Goose 2 | <16 | <16 | 128 | Not tested | 32 | |
| Snow Goose 3 | <16 | <16 | 128 | Not tested | 32 | |
| Snow Goose 4 | <16 | <16 | 128 | Not tested | 64 | |
| Snow Goose 5 | <16 | <16 | 16 | Not tested | 16 | |
| Snow Goose 6 | <16 | <16 | 32 | Not tested | 16 | |
| Mute Swan 1 | <16 | <16 | 64 | Not tested | 64 | |
| Mute Swan 2 | <16 | <16 | 128 | Not tested | 64 | |
| Mute Swan 3 | <16 | <16 | 32 | Not tested | 16 | |
| Mute Swan 4 | <16 | <16 | 64 | Not tested | <16 | |
| Mute Swan 5 | <16 | <16 | 16 | Not tested | <16 | |
| Mute Swan 6 | <16 | <16 | 16 | Not tested | <16 | |
| Mute Swan 7 | <16 | <16 | 512 | Not tested | 128 | |
| Mute Swan 8 | <16 | <16 | 256 | Not tested | 128 | |
Clade 2.3.4.4 GsGD-HP-H5 IAV antigens included: HP-Turkey/MN = A/Turkey/Minnesota/9845/2014 (H5N2) and HP-GYR/WA = A/Gyrfalcon/Washington/2014 (H5N8).
North American lineage LP H5 IAV antigens included: LP-rgBWT/TX = reverse genetics-A/Blue-winged Teal/AI12-4150/Texas/2012 (ΔH5N2), LP-wtBWT/TX = A/Blue-winged Teal/Texas/AI12-4150/2012 (H5N2), and LP-Mallard/MN = A/Mallard/Minnesota/AI11-3933/2011 (H5N1).
Control antisera (chicken) to HP-A/GYR/WA and A/Northern pintail/WA = A/Northern Pintail/Washington/40964/2014 (H5N2). Sera provided by Southeast Poultry Research Laboratory (SEPRL), Agricultural Research Service, US Department of Agriculture and the National Veterinary Services Laboratories (NVSL), Veterinary Services, US Department of Agriculture.
Sera from mallards experimentally infected with A/GYR/WA and A/Mallard/Minnesota/355779/2000 (H5N2).
Sera from Snow Geese (Anser caerulescens) sampled in Texas during 2012 and Mute Swans (Cygnus olor) sampled in Michigan by Wildlife Services, US Department of Agriculture personnel during 2012.
Waterfowl samples
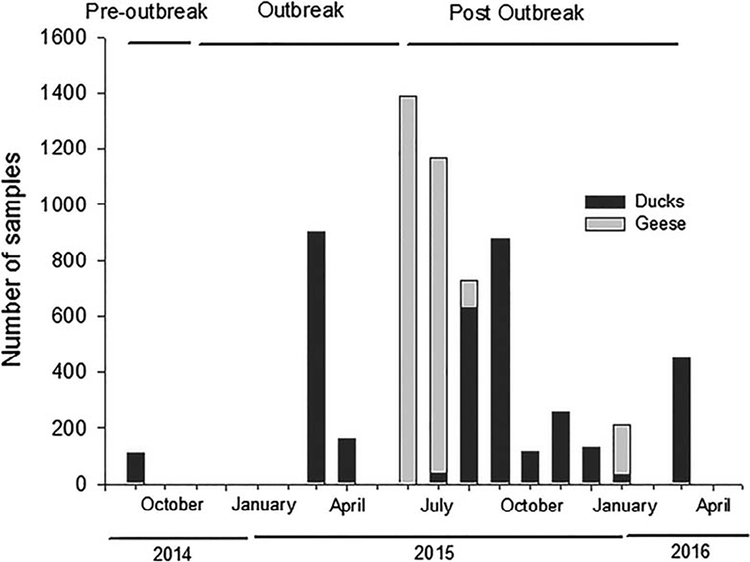
Serum samples were obtained from 2,793 hatch year and adult geese representing three species from seven states (Supplementary Material Table 1). All geese were sampled during the post-outbreak period from 1 June 2015 to 30 January 2016 (Fig. 1). Samples from geese were distributed among the Pacific (n=305), Central/Mississippi (n=1426), and Atlantic (n=1062) flyways. Serum samples were obtained from 3,715 ducks representing 12 species from nine states (Supplementary Material Table 2). Ducks were sampled from March 2014 to March 2016 during the pre-outbreak (n=111), outbreak (n=968), and post-outbreak (n=2636) periods (Fig. 1). Samples were distributed among the Pacific (n=528), Central/Mississippi (n=2205), and Atlantic (n=982) flyways.
Figure 1.
Temporal distribution of serum samples from ducks and geese in relation to the clade 2.3.4.4 goose Guangdong lineage highly pathogenic H5 influenza A virus outbreak in North America. Pre-outbreak (before 1 November 2014), outbreak (1 November 2014–1 June 2015), and postoutbreak (after 1 June 2015) periods are identified.
bELISA, HI, and MNt
Of the 2,793 geese tested, 734 tested positive for antibodies to the IAV NP as determined by bELISA. A higher proportion of antibody-positive samples originated from after hatch year (AHY; 37.3%, 703/1,886, 95% confidence limit [CL] 0.351–0.395) compared to hatch year (HY; 3.4%, 31/907, 95% CL 0.023–0.048) birds (Supplementary Material Table 1). This same relationship was observed with ducks; 1,934 of 3,715 ducks tested were NP antibody–positive by bELISA, and a higher proportion of bELISA-positive samples originated from AHY (57.4%, 1,361/2,370, CL 0.553–0.592) as compared to HY ducks (42.2%, 559/1,327, CL 0.397–0.450; Supplementary Material Table 2).
Although some samples were excluded from HI testing as a result of nonspecific binding to turkey red blood cells, sample losses were not excessive; for geese and ducks, HI results were recorded for 91.3% (1,623/1,776) and 87.1% (2,038/2,340) of samples tested, respectively. To evaluate the use of bELISA as a screening test for subsequent HI testing, all birds collected from Minnesota, Pennsylvania, and West Virginia regardless of bELISA status were tested by HI. With geese (all samples were from Canada Goose [Branta canadensis]), HI results were obtained from negative, suspect, and bELISA-positive samples with 0.4% (3/760), 7.3% (12/165), and 19.3% (75/389) testing positive for one or more H5 IAV antigens, respectively. Overall, 86% of the total HI-positive samples from geese also tested positive on the bELISA screen, and 97% tested either bELISA-positive or suspect. Similar results were observed with ducks, where HI tests results obtained from 295 negative, 150 suspect, and 523 bELISA-positive sera samples yielded 1.0% (3/295), 8.6% (13/150), and 15.3% (80/523) H5 HI–positive results, respectively. Overall, 83% of the total HI-positive samples from ducks also tested positive on the bELISA screen; 97% of these HI-positive samples were classified as positive or suspect on the bELISA screen.
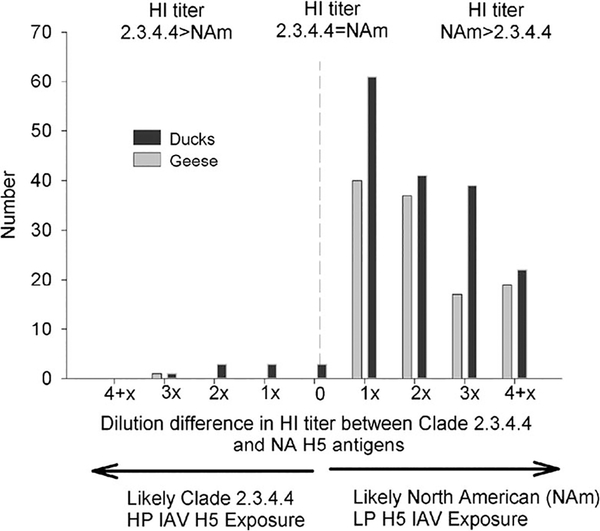
Antibodies to H5 IAV as determined by HI were detected in Canada Goose, Snow Goose, Cackling Goose (Branta hutchinsii), Mallard, Blue-winged Teal (Spatula discors), Northern Pintail (Anas acuta), American Black Duck (Anas rubripes), Green-winged Teal (Anas crecca), Northern Shoveler (Spatula clypeata), White-winged Scoter (Melanitta fusca), Long-tailed Duck (Clangula hymalis), Lesser Scaup (Aythya affinis), and Greater Scaup (Aythya marila; Supplementary Material Tables 1, 2). The distribution of comparative antibody titers to North American lineage LP H5 and GsGD-HP-H5 IAV antigens for goose and duck sera demonstrated a strong bias toward North American lineage antigens (Fig. 2). Comparative HI titers indicative of clade 2.3.4.4 GsGD-HP-H5 IAV exposure (≥2 dilution difference) were restricted to sera obtained from a single Canada Goose, three Mallards, and one Blue-winged Teal (Table 2). Three of these birds were sampled in Minnesota less than 4 mo after the end of the outbreak. A smaller bias toward clade 2.3.4.4 GsGD-HP-H5 IAV suggestive of exposure (a one-dilution HI titer difference) was identified in three samples from Mallards: two from Minnesota and one from Alaska sampled in the summer and fall of 2015 (results not shown).
Figure 2.
Hemagglutination inhibition (HI) titer bias among sera samples obtained from North American waterfowl to clade 2.3.4.4 goose Guangdong lineage highly pathogenic (HP) H5 or North American lineage low pathogenic (LP) H5 influenza A viruses (IAV) antigens.
Table 2.
Hemagglutination inhibition (HI) antibody titers for wild birds with an antibody profile suggestive of previous infection with clade 2.3.4.4 goose Guangdong lineage highly pathogenic (HP) H5 influenza A virus (IAV).
| HI antibody titerc |
|||||||
|---|---|---|---|---|---|---|---|
| Species | Location | Sample date | Agea | bELISAb | LP-rg BWT/TX | HP-GYR/WA | HP-Turkey/MN |
|
| |||||||
| Canada Goose (Branta canadensis) | LeSueur County, Minnesota | June 2015 | AHY | 0.484 | <16 | 64 | 32 |
| Mallard (Anas platyrhynchos) | Crawford County, Pennsylvania | September 2015 | AHY | 0.073 | <16 | 64 | 64 |
| Mallard | Carver County, Minnesota | August 2015 | HY | 0.257 | 32 | 128 | 128 |
| Mallard | Marshall County, Minnesota | September 2015 | HY | 0.103 | <16 | 32 | 32 |
| Blue-winged Teal (Anas discors) | Chambers County, Texas | March 2015 | AHY | 0.303 | <16 | 32 | 16 |
HY = hatch year; AHY = after hatch year.
Antibodies to IAV nucleoprotein as determined by IDEXX Laboratories commercial blocking enzyme-linked immunosorbent assay (bELISA) avian influenza (AI) MultiS-Screen antibody test kit.
HI titers against three IAV antigens: LP-rgBWT/TX = low pathogenic (LP) reverse genetics-A/Blue-winged Teal/AI12 4150/Texas/2012 (H5N2), HP-GYR/WA = HP A/Gyrfalcon/Washington/41088-6/2014 (H5N8), and HP-Turkey/MN = HP A/Turkey/Minnesota/9845/2014 (H5N2). Bolded antibody titers represent ≥2 dilution bias to clade 2.3.4.4 antigens.
Although comparative antibody titers observed with MNt correctly identified previous exposure to North American lineage or clade 2.3.4.4 GsGD-HP-H5 IAV in control sera and experimentally infected Mallards, agreement with the clade 2.3.4.4 GsGD-HP-H5 HI comparative bias indicative of previous exposure (two dilutions higher than the LP-rgBWT/TX titers) and weaker bias suggestive of prior exposure (one dilution higher antibody compared to LP-rgBWT/TX) was limited to field serum samples from one Canada Goose and two Mallards from Minnesota. The 2.3.4.4 GsGD-HP-H5 IAV antibody titer from the Canada Goose observed with MNt exceeded the LP-rgBWT/TX antibody titer by a single dilution (Table 3).
Table 3.
Microtiter neutralization (MN) results for wild birds with hemagglutination inhibiting (HI) antibody titers twofold and onefold higher to clade 2.3.4.4 goose Guangdong lineage highly pathogenic (HP) H5 (GsGD-HP-H5) influenza A virus (IAV) compared to North American lineage low pathogenic (LP) H5 IAV. Also included are wild bird samples that tested HI positive only for North American lineage H5 IAV, experimental, and control sera. Titers with a ≥twofold difference between clade 2.3.4.4 GsGD-HP-H5 and North American lineage LP H5 IAV antigens are in bold.
| HI antibody titera |
MN antibody titerb |
||||||
|---|---|---|---|---|---|---|---|
| Species | Location | LP-rg BWT/TX | HP GYR/WA | HP Turkey/MN | LP-rg BWT/TX | HP-rg CHK/Neth | HP-rg SNOW/MO |
|
| |||||||
| Canada Goose (Branta canadensis)c | LeSueur County, Minnesota | <16 | 64 | 32 | <20 | <20 | 20 |
| Mallard (Anas platyrhynchos)c | Crawford County, Pennsylvania | <16 | 64 | 64 | 40 | 20 | 20 |
| Mallardc | Carver County, Minnesota | 32 | 128 | 128 | 40 | 160 | 80 |
| Mallardc | Marshall County, Minnesota | <16 | 32 | 32 | <20 | 40 | 20 |
| Blue-winged Teal (Anas discors)c | Chambers County, Texas | <16 | 32 | 16 | 40 | <20 | <20 |
| Mallardc | Yukon-Koyukuk, Alaska | <16 | 16 | 16 | 20 | <20 | <20 |
| Mallardc | Marshall County, Minnesota | 32 | 64 | 64 | 160 | 160 | 80 |
| Mallardc | Roseau County, Minnesota | <16 | <16 | 16 | 160 | 40 | 20 |
| Canada Goosed | Anoka County, Minnesota | 256 | 64 | 64 | 320 | 40 | 20 |
| Canada Goosed | Anoka County, Minnesota | 128 | <16 | <16 | 160 | <20 | <20 |
| Mallardd | Carver County, Minnesota | 64 | <16 | <16 | 320 | 20 | 20 |
| Mallardd | Carver County, Minnesota | 64 | <16 | <16 | <20 | <20 | <20 |
| Mallard 6e | Experimental | 128 | <16 | <16 | 80 | <20 | <20 |
| Mallard 7e | Experimental | 128 | <16 | <16 | 160 | <20 | <20 |
| Chickenf | NVSL antisera | 64 | ≥1,024 | ≥1,024 | <20 | 640 | 640 |
HI antigens included HP HP-Turkey/MN = clade 2.3.4.4 H5 HPAI A/Turkey/Minnesota/9845/2014 (H5N2) and HP-GYR/WA = A/Gyrfalcon/Washington/2014 (H5N8). North American H5 LP virus antigen included: LP-rgBWT/TX = reverse genetics-A/Blue-winged Teal/AI12-4150/Texas/2012 (ΔH5N2).
MN antigens included: LP-rgBWT/TX, HP-Chick/Neth = rg-A/Chicken/Netherland/EMC-3/2014 (ΔH5N8), and HP-SNOW/MO = rg-A/Snow Goose/Missouri/CC15-84a/2015 (ΔH5N2).
Wild bird samples with HI antibody titers twofold and onefold higher to clade 2.3.4.4 GsGD-HP-H5 IAV compared to North American lineage LP H5 IAV.
Wild bird samples that tested HI positive only for North American lineage LP H5 IAV.
Samples from Mallards experimentally infected with A/Mallard/Minnesota/355779/2000 (H5N2).
Control antisera (chicken) to A/Gyrfalcon/Washington/2014 (H5N8) (A/GYR/WA) provided by the National Veterinary Services Laboratories (NVSL), Veterinary Services, US Department of Agriculture.
DISCUSSION
Serologic testing of North American waterfowl sampled as part of this investigation provided limited evidence of previous infection with the clade 2.3.4.4 GsGD-HP-H5 IAV, which appears to contrast virologic results reported for wild birds inhabiting the Pacific Flyway during 2014–15. The low numbers of birds exhibiting serologic evidence of previous infections with clade 2.3.4.4 GsGD-HP-H5 IAV also is much lower than that reported for Europe, where antibody prevalence in birds tested from 2014 to 2017 ranged from 1% to 33% depending on species (Poen et al. 2018). Possible explanations for this include a spatial bias in sampling to eastern North American flyways and temporally after June 2015. Most of the reported isolates of clade 2.3.4.4 GsGD-HP-H5 IAV in North America were associated with waterfowl sampled in the Pacific and Central flyways (not including Texas) during the winter of 2014–15. Most of our samples from geese and ducks were collected from the Mississippi-Central flyway (including Texas; 51% of geese, 59% of ducks) and the Atlantic Flyway (38% of geese, 26% of ducks), where few clade 2.3.4.4 GsGD-HP-H5 viruses were reported from wild birds (Bevins et al. 2016). Unlike the European studies, our sampling also was primarily associated with a post-outbreak period, with 100% and 68% of sera from geese and ducks, respectively, collected after 1 June 2015. An additional consideration is the uncertainty associated with the actual prevalence of clade 2.3.4.4 GsGD-HP-H5 IAV in North American waterfowl. Although these viruses were commonly reported in waterfowl inhabiting the Pacific Flyway during 2014 and early 2015, prevalence estimates during the peak period of waterfowl detections were estimated at 0.8–1.3% (Bevins et al. 2016; Ramey et al. 2017). In contrast, few detections were reported from the Mississippi Flyway, where active surveillance often failed to detect evidence of infection in waterfowl despite numerous detections among commercial poultry flocks in this region (Jennelle et al. 2016).
Although sampling of wild waterfowl for this study was spatially and temporally biased, our results are consistent with previous inference (Bevins et al. 2016) regarding the spatiotemporal extent of the clade 2.3.4.4 GsGD-HP-H5 IAV outbreak in North America, particularly pertaining to wild birds. Most of our waterfowl samples originated from the Mississippi and Central flyways; all but one sample that met our criteria as antibody biased to a clade 2.3.4.4 GsGD-HP-H5 virus originated from these flyways. These putatively antibody-positive sera included samples from an AHY Canada Goose and a HY Mallard that were collected in counties in Minnesota where domestic turkeys had been previously infected, one HY Mallard that was sampled in northern Minnesota, and one HY Blue-winged Teal sampled in Texas. Clade 2.3.4.4 GsGD-HP-H5 IAV infections were detected in both Mallards and Canada Geese during the 2014–15 outbreak (Bevins et al. 2016). The lone sample outside of the Mississippi-Central flyway that met the two dilution bias criteria was from an AHY Mallard in western Pennsylvania (Atlantic Flyway). Although there were no detections of clade 2.3.4.4 GsGD-HP-H5 viruses from this flyway during the 2014–15 outbreak, this bird was sampled from a location on Lake Erie where the Mississippi and Atlantic flyways meet. Results from these few positive samples and the numerous negative samples are difficult to interpret but are consistent with a very low prevalence of exposure among wild birds within the central US during the outbreak and a dearth of evidence for the spread of GsGD-HP-H5 IAV to wild or domestic birds in the eastern US.
Based on the results from this study and previous work in Europe (Gilbert et al. 2014; Poen et al. 2016, 2018; Hill et al. 2019), serologic monitoring may have application to understanding if an introduction of an exotic IAV such as GsGD-HP-H5 IAV will be maintained in wild waterfowl populations. However, many unknowns related to data interpretation remain. Limitations include an unknown longevity of the detectable antibody response, potential nonspecific reactions, a variable antibody response to a specific subtype or strain, the effect of previous IAV infections on serologic response, and unknown effects related to antigenic diversity. All these limitations may be exacerbated by multiple exposures to diverse IAV subtypes, as often occurs in the normal life span of a waterfowl species.
Our results provide serologic evidence of previous infections with clade 2.3.4.4 GsGD-HP-H5 IAVs in North American waterfowl. Based on the limited detections in this study, the value of employing serologic surveillance for HP IAV exposure in wild birds on a broad or national scale is unclear. However, such an approach may have value if applied to wild bird species at specific sites, such as the testing of Eurasian Wigeon in Europe, where this species has been implicated in dispersal of GsGD-HP-H5 IAV (Poen et al. 2016), or the testing of intercontinental migrants in Beringia or the North Atlantic, where transcontinental movements of avian influenza viruses have been documented (Dusek et al. 2014; Ramey et al. 2015, 2018). This approach also may have application as a tool to support virologic testing to assess if an introduced virus such as clade 2.3.4.4 GsGD-HP-H5 persists in North American waterfowl populations. A third possible application relates to the potential use of serologic assays to determine existing levels of IAV immunity in wild bird populations (Hill et al. 2019). In the present study, some species had a high prevalence of antibodies as detected by bELISA, and antibodies to North American H5 IAV were detected using both HI and MNt. It has been previously demonstrated in field and experimental studies that prior infections with both homologous and heterologous IAV can prevent infection, greatly reduce the duration and extent of IAV shedding, and result in a higher infective dose required for subsequent infections (Latorre-Margalef et al. 2017; Segovia et al. 2018). Prevention and a reduction in viral shedding also have been reported with GsGD-HP-H5 IAV challenges of ducks and geese previously infected with LP IAV (Costa et al. 2011; Berhane et al. 2014). Unlike domestic poultry, wild waterfowl are routinely infected by multiple IAVs; it is possible that existing population immunity may limit the potential for a successful introduction and subsequent spread of an exotic HP IAV. Thus, serologic data, such as those used in this study, may be useful to determine the potential for an exotic HP IAV to cause mortality and become established in waterfowl populations (Krauss et al. 2016; Hill et al. 2019).
Supplementary Material
ACKNOWLEDGMENTS
The authors acknowledge the contributions of personnel from the state and federal wildlife agencies that aided in collecting serum samples and M. Kim, R. A. M. Fouchier, D. Kapczynski, and E. Spackman for supplying the control sera and antigens needed for serologic testing. The authors thank J. Bahl, J. Pearce, and two anonymous reviewers for suggestions on previous draft versions of this article. This project was funded by the National Institute of Allergy and Infectious Diseases, National Institutes of Health (NIH), Department of Health and Human Services, under contracts HHSN272201400006C and HHSN272201400008C, and by the US Geological Survey through the Wildlife Program of the Ecosystems Mission area. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIH. This report was reviewed and approved by USGS under the Fundamental Science Practices policy. Any use of trade, firm, or product names is for descriptive purposes only and does not imply endorsement by the US Government.
Footnotes
SUPPLEMENTARY MATERIAL
Supplementary material for this article is online at http://dx.doi.org/10.7589/2019-01-003.
LITERATURE CITED
- Berhane Y, Embury-Hyatt C, Leith M, Kehler H, Suderman M, Pasick J. 2014. Pre-exposing Canada Geese (Branta canadensis) to a low-pathogenic H1N1 avian influenza virus protects them against H5N1 HPAI virus challenge. J Wildl Dis 50:84–97. [DOI] [PubMed] [Google Scholar]
- Bevins SN, Dusek RJ, White CL, Gidlewski T, Bodenstein B, Mansfield KG, DeBruyn P, Kraege D, Rowan E, Gillin C, et al. 2016. Widespread detection of highly pathogenic H5 influenza viruses in wild birds from the Pacific Flyway of the United States. Sci Rep 6: 28980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JD, Luttrell MP, Berghaus RD, Kistler W, Keeler SP, Howey A, Wilcox B, Hall J, Niles L, Dey A, et al. 2010. Prevalence of antibodies to type A influenza virus in wild avian species using two serologic assays. J Wildl Dis 46:896–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa TP, Brown JD, Howerth EW, Stallknecht DE, Swayne DE. 2011. Homo- and heterosubtypic low pathogenic avian influenza exposure on H5N1 highly pathogenic avian influenza virus infection in wood ducks (Aix sponsa). PLoS One 6:e15987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dusek RJ, Hallgrimsson GT, Ip HS, Jónsson JE, Sreevatsan S, Nashold SW, TeSlaa JL, Enomoto S, Halpin RA, Lin X, et al. 2014. North Atlantic migratory bird flyways provide routes for intercontinental movement of avian influenza viruses. PLoS One 9:e92075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert M, Jambal L, Karesh WB, Fine A, Shiilegdamba E, Dulam P, Sodnomdarjaa R, Ganzorig K, Batch-uluun D, Tseveenmyadag N, et al. 2012. Highly pathogenic avian influenza virus among wild birds in Mongolia. PLoS One 7:e44097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert M, Koel BF, Bestebroer TM, Lewis NS, Smith DJ, Fouchier RAM. 2014. Serological evidence for non-lethal exposures of Mongolian wild birds to highly pathogenic avian influenza H5N1 virus. PLoS One 9:e113569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill SC, Hansen R, Watson S, Coward V, Russell C, Cooper J, Essen S, Everest H, Parag KV, Fiddaman S, et al. 2019. Comparative micro-epidemiology of pathogenic avian influenza virus outbreaks in a wild bird population. Philos Trans R Soc Lond B Biol Sci 374:20180259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ip HS, Torchetti MK, Crespo R, Kohrs P, DeBruyn P, Mansfield KG, Baszler T, Badcoe L, Bodenstein B, Shearn-Bochsler V, et al. 2015. Novel Eurasian highly pathogenic avian influenza A H5 viruses in wild birds, Washington, USA, 2014. Emerg Infect Dis 21:886–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennelle CS, Carstensen M, Hildebrand EC, Cornicelli L, Wolf P, Grear DA, Ip HS, Vandalen KK, Minicucci LA. 2016. Surveillance for highly pathogenic avian influenza virus in wild birds during outbreaks in domestic poultry, Minnesota, 2015. Emerg Infect Dis 22:1278–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krauss S, Stallknecht DE. Slemons RD, Bowman AS, Poulson RL, Nolting JM, Knowles JP, Webster RG. 2016. The enigma of the apparent disappearance of Eurasian highly pathogenic H5 clade 2.3.4.4 influenza A viruses in North American waterfowl. Proc Natl Acad Sci U S A 113:9033–9038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latorre-Margalef N, Brown JD, Fojtik A, Poulson RL, Carter D, Franca M, Stallknecht DE. 2017. Competition between influenza A virus subtypes through heterosubtypic immunity modulates re-infection and antibody dynamics in the mallard duck. PLoS Path 13:e1006419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DH, Bahl J, Torchetti MK, Killian ML, Ip HS, DeLiberto TJ, Swayne DE. 2016. Highly pathogenic avian influenza viruses and generation of novel reassortants, United States, 2014–2015. Emerg Infect Dis 22:1283–1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DH, Torchetti MK, Killian ML, DeLiberto TJ, Swayne DE. 2017. Reoccurrence of avian influenza A (H5N2) virus clade 2.3.4.4 in wild birds, Alaska, USA, 2016. Emerg Infect Dis 23:365–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DH, Torchetti MK, Winker K, Ip HS, Song CS, Swayne DE. 2015. Intercontinental spread of Asian-origin H5N8 to North America through Beringia by migratory birds. J Virol 89:6521–6524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasick J, Berhane Y, Joseph T, Bowes V, Hisanaga T, Handel K, Alexandersen S. 2015. Reassortant highly pathogenic influenza A H5N2 virus containing gene segments related to Eurasian H5N8 in British Columbia, Canada, 2014. Sci Rep 5:9484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez DR, Angel M, Gonzalez-Reiche AS, Santos J, Obadan A, Martinez-Sobrido L. 2017. Plasmid-based reverse genetics of influenza A virus. Methods Mol Biol 1602:251–273. [DOI] [PubMed] [Google Scholar]
- Poen MJ, Bestebroer TM, Vuong O, Scheuer RD, van der Jeugd HP, Kleyheeg E, Eggink D, Lexmond P, van den Brand JMA, Begeman L, et al. 2018. Local amplification of highly pathogenic avian influenza H5N8 viruses in wild birds in the Netherlands, 2016 to 2017. Euro Surveill 23:17–00449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poen MJ, Verhagen JH, Manvell RJ, Brown I, Bestebroer TM, van der Vliet S, Vuong O, Scheuer RD, van der Jeugd HP, Nolet BA, et al. 2016. Lack of virological and serological evidence for the continued circulation of highly pathogenic avian influenza H5N8 virus in wild birds in the Netherlands, 14 November 2014 to 31 January 2016. Euro Surveill 21:30349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramey AM, Hill NJ, Cline T, Plancarte M, De La Cruz S, Casazza ML, Ackerman JT, Fleskes JP, Vickers TW, Reeves AB, et al. 2017. Surveillance for highly pathogenic influenza A viruses in California during 2014–2015 provides insights into viral evolutionary pathways and spatiotemporal extent of viruses in the Pacific Americas Flyway. Emerg Microbes Infect 6: e80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramey AM, Reeves AB, Donnelly TF, Poulson RL, Stallknecht DE. 2018. Introduction of Eurasianorigin H8N4 influenza A virus into North America via migratory birds. Emerg Infect Dis 24:1950–1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramey AM, Reeves AB, Sonsthagen SA, TeSlaa JL, Nashold S, Donnelly T, Casler B, Hall JS. 2015. Dispersal of H9N2 influenza A viruses between East Asia and North America by wild birds. Virology 482: 79–83. [DOI] [PubMed] [Google Scholar]
- Ramey AM, Reeves AB, TeSlaa JL, Nashold S, Donnelly T, Bahl J, Hall JS. 2016. Evidence for common ancestry among viruses isolated from wild birds in Beringia and highly pathogenic intercontinental reassortant H5N1 and H5N2 influenza A viruses. Infect Genet Evol 40:176–185. [DOI] [PubMed] [Google Scholar]
- Saito T, Tanikawa T, Uchida Y, Takemae N, Kanehira K, Tsunekuni R. 2015. Intracontinental and intercontinental dissemination of Asian H5 highly pathogenic avian influenza virus (clade 2.3.4.4) in the winter of 2014–2015. Rev Med Virol 25:388–405. [DOI] [PubMed] [Google Scholar]
- Segovia KM, França MS, Leyson CL, Kapczynski DR, Chrzastek K, Bahnson CS, Stallknecht DE. 2018. Heterosubtypic immunity increases infectious dose required to infect mallard ducks with influenza A virus. PLoS One 13:e0196394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shriner SA, VanDalen KK, Root JJ, Sullivan HJ. 2016. Evaluation and optimization of a commercial blocking ELISA for detecting antibodies to influenza A virus for research and surveillance of mallards. J Virol Methods 228:130–134. [DOI] [PubMed] [Google Scholar]
- Tolf C, Latorre-Margalef N, Wille M, Bengtsson D, Gunnarsson G, Grosbois V, Hasselquist D, Olsen B, Elmberg J, Waldenström J. 2013. Individual variation in influenza A virus infection histories and long-term immune responses in mallards. PLoS One 8:e61201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- US Department of Agriculture. 2015. 2014–2015 HPAI outbreak https://www.aphis.usda.gov/aphis/ourfocus/animalhealth/animal-disease-information/avian/2014-2015-hpai-outbreak. Accessed July 2019.
- US Department of Agriculture. 2017. July 2016–June 2017 wild bird highly pathogenic avian influenza cases in the United States https://www.aphis.usda.gov/animal_health/downloads/animal_diseases/ai/uspositivecases17.pdf. Accessed July 2018.
- Wong JK, Wilcox BR, Fojtik A, Poulson RL, Stallknecht DE. 2016. Antibodies to influenza A viruses in wintering snow geese (Chen caerulescens) in Texas. Avian Dis 60:337–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.