Abstract
Background
Ralstonia solanacearum, one of the most devastating bacterial plant pathogens, is the causal agent of bacterial wilt. Recently, several studies on resistance to bacterial wilt have been conducted using the Arabidopsis-R. solanacearum system. However, the progress of R. solanacearum infection in Arabidopsis is still unclear.
Results
We generated a bioluminescent R. solanacearum by expressing plasmid-based luxCDABE. Expression of luxCDABE did not alter the bacterial growth and pathogenicity. The light intensity of bioluminescent R. solanacearum was linearly related to bacterial concentrations from 104 to 108 CFU·mL−1. After root inoculation with bioluminescent R. solanacearum strain, light signals in tomato and Arabidopsis were found to be transported from roots to stems via the vasculature. Quantification of light intensity from the bioluminescent strain accurately reported the difference in disease resistance between Arabidopsis wild type and resistant mutants.
Conclusions
Bioluminescent R. solanacearum strain spatially and quantitatively measured bacterial growth in tomato and Arabidopsis, and offered a tool for the high-throughput study of R. solanacearum-Arabidopsis interaction in the future.
Keywords: Ralstonia solanacearum, Arabidopsis, Bioluminescence, LuxCDABE, Bacterial wilt
Background
Bacterial wilt is a soil-borne bacterial disease caused by Ralstonia solanacearum, the second most devastating bacterium among plant pathogens. R. solanacearum can infect more than 250 plants, including tomato (Solanum lycopersicum), potato (S. tuberosum), banana (Musa nana), and other agriculturally important crops [1, 2]. After infection through wounds, root tips, or cracks in the lateral roots, R. solanacearum colonizes the root cortex, invades and multiplies in xylem vessels, and reaches the aerial parts of the plant, causing wilting symptoms and subsequent plant death [3, 4]. Control of bacterial wilt is a challenge because of the lack of available commercial resistant varieties and the long survival time of R. solanacearum in soil, water, and infected plant tissues.
Several studies on the molecular mechanism of disease resistance to R. solanacearum have been conducted using the Arabidopsis-R. solanacearum system owing to the availability of genetic resources and well-defined technology for this model plant species [5–10]. Study of a genetic mapping population from the cross between a susceptible (Col-5) and resistant ecotype (Nd-1) revealed that an atypical resistance gene (R gene) encoding RRS1 (resistance to Ralstonia solanacearum 1) confered resistance against R. solanacearum strain GMI1000 [5]. Interestingly, mutation in a neighboring gene encoding a well-known R protein (RPS4, resistance to Pseudomonas syringae 4) also impaired resistance to R. solanacearum [11]. Recently, using the Arabidopsis-R. solanacearum system, many regulatory genes, including suppressor of g2 allele of skp1 (SGT1), glutamate decarboxylases (GADs), LRR receptor-like kinases, such as ERECTA, CLAVATA1, and CLAVATA2, were found to function in resistance to R. solanacearum [7, 9, 10]. In addition, the involvement of phytohormone ethylene, salicylic acid (SA) and jasmonic acid signaling pathways in resistance to R. solanacearum was also studied using the Arabidopsis-R. solanacearum system [7, 12, 13]. Although these studies were conducted in Arabidopsis, the spatial distribution and infection progress of R. solanacearum in Arabidopsis is still unclear.
To guarantee the success of long-term breeding programs, it is crucial to develop a disease detection approach that allows easy tracking and quantification of bacterial colonization. A few methods are available for the detection of R. solanacearum, such as disease index, colony counting, and DNA-based amplification assay using polymerase chain reaction (PCR). Disease index is estimated by scoring the wilting severity; therefore, there is a possibility of human error in the evaluation of resistant varieties [14, 15]. The colony counting method can be relatively accurate in quantifying bacteria, but it is time-consuming and labor-intensive, and the sample heterogeneity in different location of tissues might cause errors as well [14, 15]. PCR assays are relatively sensitive to detect diseases, particularly diseases at an early stage; however, this method usually requires either expensive thermocycling equipment or expensive chemicals, and the laboratory skills to perform the technical procedures and data analysis [16]. In addition, none of these methods can systematically and comprehensively reflect the incidence of disease in the whole plant.
In recent years, bioluminescence imaging technology, mostly developed based on the expression of luxCDABE encoding autonomous light-emitting elements, has been successfully applied to engineer plant pathogenic bacteria [17, 18]. The genes luxA and luxB encode the heterodimeric enzyme luciferase, while luxC, luxD, and luxE are responsible for the synthesis of fatty aldehydes that serve as substrates for the luminescence reaction of luciferase, such that, the cells expressing luxCDABE operon will emit light autonomously [19]. The emitted light can be captured using a sensitive detector that converts light signals into electrical values, which are then digitalized and displayed on a computer monitor instantaneously. As the light signal produced by luxCDABE has a long life, high sensitivity, and strong specificity, the plant pathogens harboring luxCDABE can be directly monitored without destroying plant tissues after inoculation [20–22]. In addition, since the expression of luxCDABE is linearly correlated with the concentrations of living bacteria, the relative light intensity can be used to quickly quantify the levels of bacteria [23, 24]. Therefore, bioluminescent imaging technology provides a non-invasive method to study the spatial distribution and infection progress of pathogens, and quickly score the disease severity.
In this study, we generated a bioluminescent R. solanacearum strain (FJ91-LUX) by transforming a plasmid containing the luxCDABE operon into R. solanacearum FJ91, a strain isolated from Fujian, China. Transformation of the luxCDABE operon had no significant impact on R. solanacearum growth and pathogenicity in tomato and Arabidopsis. Light emission from the FJ91-LUX strain was detectable when concentration of the strain was above 104 CFU·mL−1 and was linearly related to bacterial concentrations below 108 CFU·mL−1. We monitored the infection progress in tomato and Arabidopsis after root inoculation with the FJ91-LUX strain and found that light signals were transmitted from roots to stems within 3 days and were mainly observed in the stems. We also compared the light intensity between Arabidopsis wild type and mutants defective in ethylene and SA signaling pathways after inoculation with FJ91-LUX, and it was found that the light intensity in mutants were significantly different from that in wild type. Thus, bioluminescent R. solanacearum can be used to study the defense mechanism and to facilitate isolation of disease-resistant varieties in the future.
Results
Generation of a bioluminescent R. solanacearum strain
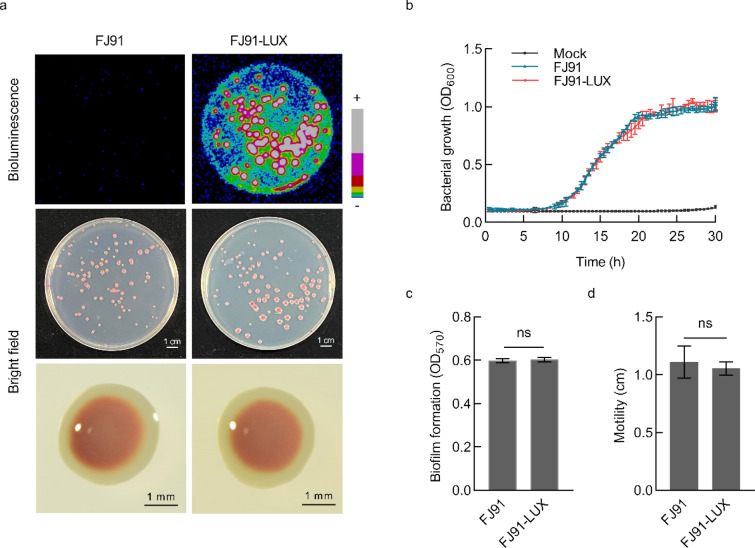
To generate a LUX-tagged R. solanacearum strain, we transformed a plasmid containing the luxCDABE operon into R. solanacearum FJ91, and the transformed strain is hereafter referred to as FJ91-LUX. R. solanacearum FJ91 was isolated from Fujian province of China. Clear bioluminescent signals were observed in the FJ91-LUX colonies under a photon camera (Photek HRPCS5), while no signals were detected in FJ91 under the same conditions (Fig. 1a). The shape of FJ91-LUX colonies was not significantly different from that of FJ91. The colonies showed a red-colored smooth circular shape on solid media containing triphenyl tetrazolium chloride (TTC), which was converted to red insoluble formazan products by bacterial dehydrogenases (Fig. 1a). To rule out the possibility that luxCDABE expression might alter the biological characteristics of R. solanacearum, we first examined the growth curve of FJ91-LUX. We found that, similar to the control (FJ91), FJ91-LUX started the logarithmic growth phase 7 h after culturing and reached a stable phase at approximately 22 h (Fig. 1b). Formation of biofilms through secretion of extracellular polysaccharides and other substances that adhere to the contact surface is an important mechanism for bacteria to adapt to the environment [25]. Therefore, we next compared the biofilms of FJ91-LUX and FJ91; however, no significant difference was observed between the two biofilms (Fig. 1c). Motility is also an indicator of bacterial growth and pathogenicity [26]. Therefore, we evaluated the motility of FJ91-LUX and found that it displayed the same motility as FJ91 (Fig. 1d). Collectively, these results suggest that the expression of the luxCDABE operon does not alter the biological characteristics of R. solanacearum.
Fig. 1.
Bacterial characteristics of Ralstonia solanacearum FJ91-LUX are not significantly different from those of FJ91. a Bacterial morphology and bioluminescence signals. Bioluminescent signals were detected using a photon-camera. + : Strong; −: Weak. b Bacterial growth curves. Bacterial growth was determined by measuring the optical density of media (OD600). Data are shown as the mean ± SD (n = 3). c Biofilm biomass. The amount of biofilm formed was quantified using crystal violet adherence assay. Data are shown as the mean ± SD (n = 4), and ns indicates no significant difference between FJ91 and FJ91-LUX (t-test). d Bacterial motility. Surface motility was evaluated by measuring the colony diameter of bacterial growth 48 h after inoculating the center of the plates. Data are shown as the mean ± SD (n = 4), and ns indicates no significant difference between FJ91 and FJ91-LUX (t-test)
Light intensity of FJ91-LUX is linearly related to the bacterial concentration
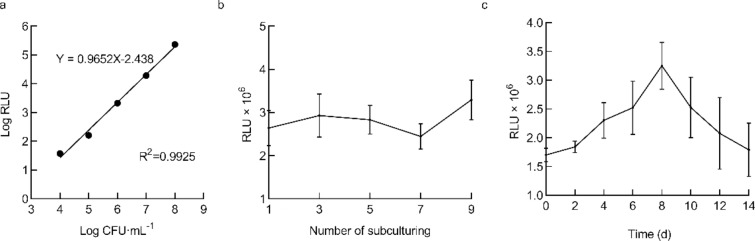
To investigate whether the bioluminescence of FJ91-LUX is related to its concentration, we measured the light signals at different bacterial concentrations (104, 105, 106, 107, and 108 CFU·mL−1) (Fig. 2a). We found that signal intensity had a linear logarithmic relationship with bacterial concentrations within the range of 104 to 108 CFU·mL−1 (Fig. 2a), and the correlation coefficient was R2 = 0.9925. Together, these results suggest that the light signals emitted from FJ91-LUX are a reflection of bacterial concentration.
Fig. 2.
The light intensity of Ralstonia solanacearum FJ91-LUX is linearly correlated to its concentration. a Regression analysis of bioluminescent signals versus bacterial concentrations. The bioluminescent signals of the bacteria at the indicated concentrations were measured. Data are shown as the mean ± SD (n = 3). b Bioluminescent signals of bacteria after successive subculturing. Data are shown as the mean ± SD (n = 3). c Light intensity of FJ91-LUX after continuous growth on solid media. Data are shown as the mean ± SD (n = 4)
Spontaneous loss of plasmid might be a potential problem during bacterial subculturing; therefore, we measured the light intensity for 9 generations of successive subculturing. We found that the light intensity of FJ91-LUX was not significantly reduced after subculturing (Fig. 2b). However, when FJ91-LUX was grown continuously on solid media for 14 d, its signals gradually increased until 8 d, and then decreased to basal levels at 14 d after culturing (Fig. 2c), probably because of the aging of bacteria. These results suggest that the aging of bacteria might reduce the light signals of FJ91-LUX; therefore, it is advisable to measure the signals in the early phase after culturing.
Real-time monitoring of the infection progress of FJ91-LUX in tomato
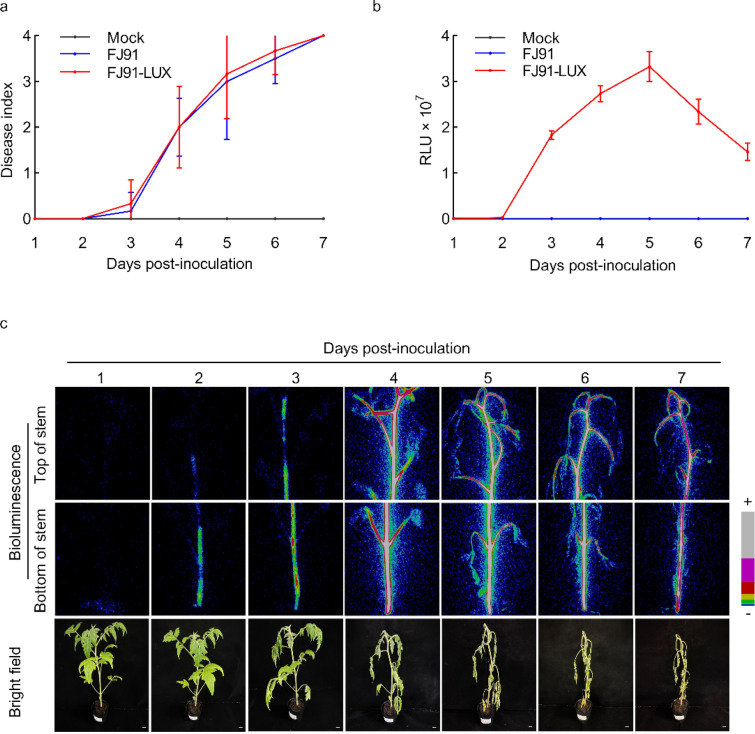
To evaluate whether the expression of luxCDABE affects the pathogenicity of R. solanacearum, we compared the disease index of tomato after inoculation with FJ91-LUX and FJ91. Tomato seedlings (28-d-old) were soil-soak inoculated with bacteria (OD600 = 1), and the disease index was scored every day after inoculation. The disease index was empirically categorized into five grades: 0 grade (no leaf wilting was observed), 1st grade (25% of the whole leaves were wilted), 2nd grade (50% of the whole leaves were wilted), 3rd grade (75% of the whole leaves were wilted), and 4th grade (100% of leaves were wilted or the entire plant died). Tomato seedlings started to wilt 3 d post inoculation (dpi), and the disease index reached 2nd grade at 4 dpi. More severe wilt symptoms were observed at 5 dpi, and all plants died by 7 dpi (Fig. 3a, b). The disease index after inoculation with FJ91-LUX was not significantly different from that of FJ91 (Fig. 3a). These results suggest that luxCDABE gene expression does not alter the pathogenicity of R. solanacearum.
Fig. 3.
Real-time monitoring of the infection progress of Ralstonia solanacearum FJ91-LUX in tomato. a Pathogenicity of R. solanacearum FJ91-LUX in tomato was not significantly different from that of FJ91. The disease index of tomato plants was scored after inoculation at the indicated time points. Disease index ranges from 0 to 4: 0 (no wilting), 1 (1–25% wilted), 2 (26–50% wilted), 3 (51–75% wilted), and 4 (76–100% wilted or dead). Data are shown as the mean ± SD (n = 6). b Light intensity of tomato plants after inoculation. Data are shown as the mean ± SD (n = 6). c Bioluminescent images of representative tomato plants. + : Strong; −: Weak. Bar = 1 cm
We next determined whether FJ91-LUX could be used to monitor the infection progress of R. solanacearum in tomato. We measured the light signals of tomato seedlings every day after inoculation with FJ91-LUX using a photon camera. We found that a weak signal was detected at the base of the tomato stem at 2 dpi, suggesting that it took one or two days for the bacteria to move from the root to the stem (Fig. 3c). Light signals were observed on the upper part of the stem at 3 dpi and continuously increased to reach maximal levels at 5 dpi. It has been reported that R. solanacearum mainly infects plant stems [27]. In agreement with these results, we observed light signals mainly in the stem region and rarely in the leaves (Fig. 3c). Even in the dying plants at 6–7 dpi, R. solanacearum did not seem to infect tomato leaf mesophyll cells. It is worth noting that fewer signals were detected at 6–7 dpi than at 5 dpi, indicating that the bacteria might lose their viability in the dying plants, consistent with the observation that aging bacteria reduced the light signals (Fig. 2c). Collectively, FJ91-LUX can be used for real-time monitoring of bacterial infection in tomato.
Real-time monitoring of the infection progress of FJ91-LUX in Arabidopsis
We next determined whether FJ91-LUX could be used to monitor the progression of R. solanacearum infection in Arabidopsis. Arabidopsis mature plants (24-d-old) were soil-soak inoculated with FJ91-LUX and FJ91 (OD600 = 1), and light signals were monitored every two days after inoculation. Consistent with the results in tomato, it was observed that FJ91-LUX exhibited the same level of pathogenicity in Arabidopsis, as did the strain FJ91 (Fig. 4a). Arabidopsis plants wilted and died at 15 dpi in cases of infection by either of the two strains of R. solanacearum (Fig. 4a). No light signal was detected at 1 dpi with FJ91-LUX, a weak signal was observed at 3 dpi, and then the light signals gradually increased from 3 to 9 dpi (Fig. 4b, c). The light signals were mainly found in the vein of rosette leaves before 9 dpi, and moved from rosette leaves to stems at 11 dpi, without diffusing in the mesophyll cells of rosette leaves (Fig. 4c). Compared with the light intensity at 9 dpi, the levels at 11 dpi were slightly decreased, probably because the rosette leaves withered at 11 dpi, leading to the reduced total signals. However, the signals in stems increased from 11 to 15 dpi (Fig. 4b, c). Collectively, our results suggest that FJ91-LUX can be used for real-time monitoring of bacterial infection progress in Arabidopsis.
Fig. 4.
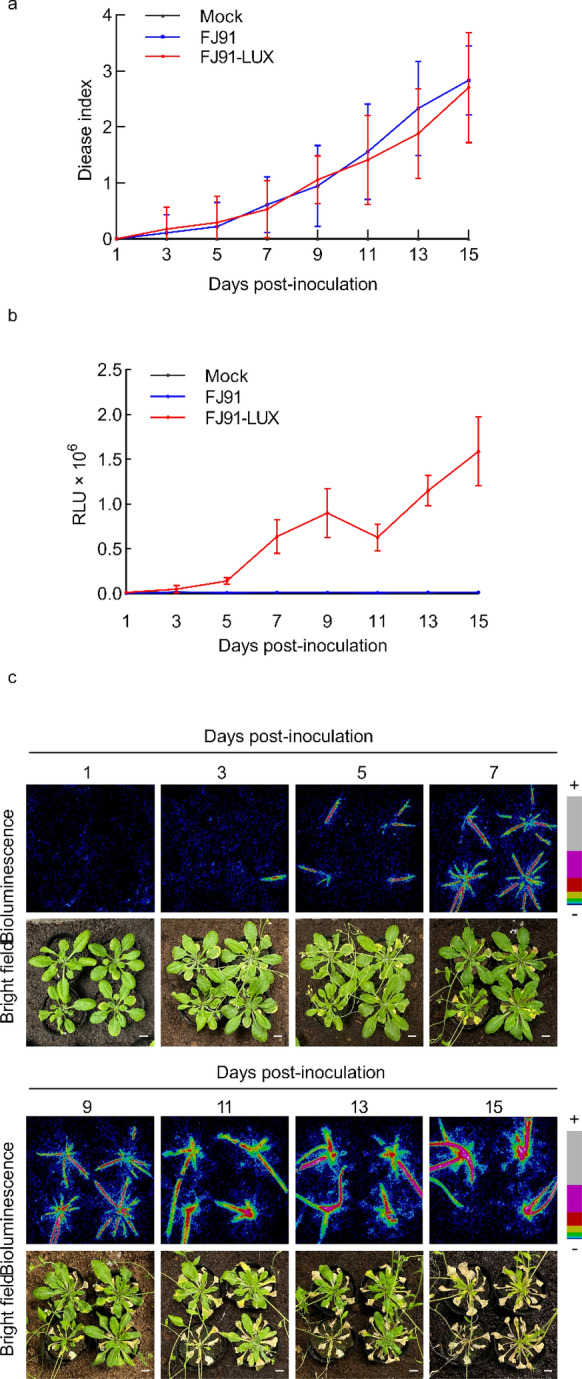
Real-time monitoring of the infection progress of Ralstonia solanacearum FJ91-LUX in Arabidopsis. a Pathogenicity of R. solanacearum FJ91-LUX in Arabidopsis is not significantly different from that of FJ91. R. solanacearum-inoculated Arabidopsis was scored every other day using a disease index ranging from 0 to 4: 0 (no wilting), 1 (1–25% wilted), 2 (26–50% wilted), 3 (51–75% wilted), and 4 (76–100% wilted or dead). Data are shown as the mean ± SD (n = 6). b Light intensity of Arabidopsis plants after inoculation. Data are shown as the mean ± SD (n = 6). c Bioluminescent images of representative Arabidopsis plants. + : Strong; −: Weak. Bar = 1 cm
Quantification of plant resistance to R. solanacearum using FJ91-LUX
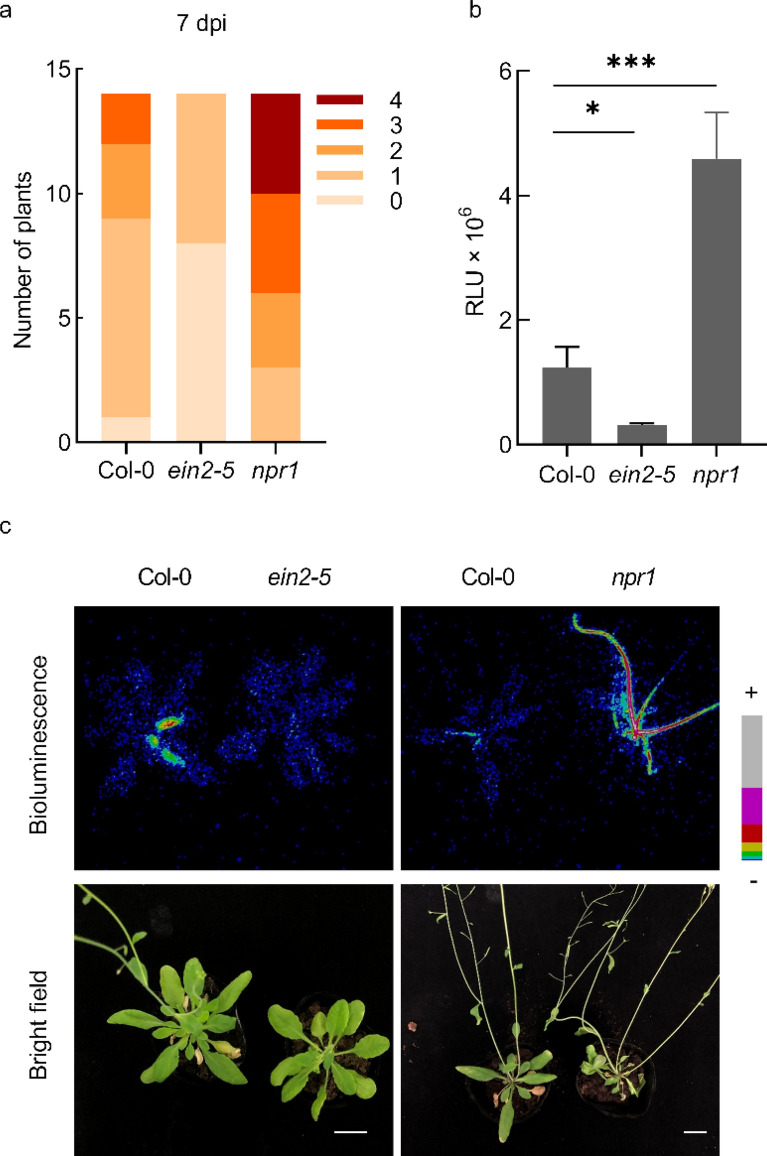
The light signals of FJ91-LUX were linearly correlated to their concentrations, when the infected plants were alive. We next examined whether FJ91-LUX could be used for the quantification of plant resistance to R. solanacearum. It has been reported that Arabidopsis ETHYLENE INSENSITIVE 2 (EIN2) gene plays a negative role in resistance to R. solanacearum, and ein2 mutants show delayed wilt symptoms [12]. Therefore, we inoculated Arabidopsis wild-type (Col-0) plants and ein2-5 mutants with FJ91-LUX and compared the light intensity at 7 dpi. Consistently, we found that ein2-5 mutants showed reduced disease index (Fig. 5a), and the light intensity in ein2-5 mutants was significantly lower than that in the wild type (Fig. 5b, c). In contrast, mutants with defects in NON EXPRESSER OF PATHOGENESIS RELATED 1 (NPR1, encoding a key transcriptional co-activator in SA signaling pathway) exhibited significantly stronger light intensity than wild type after inoculation with FJ91-LUX. Together, these results suggest that FJ91-LUX could be used for the quantification of plant resistance to R. solanacearum and possibly facilitate large-scale mutant screens in the future.
Fig. 5.
Light intensity of ein2 and npr1 mutants after inoculation with Ralstonia solanacearum FJ91-LUX. a Disease index of Col-0, ein2-5, and npr1 mutants inoculated with FJ91-LUX. R. solanacearum-inoculated Arabidopsis were scored 7 d post inoculation (dpi). Disease index: 0 (no wilting), 1 (1–25% wilted), 2 (26–50% wilted), 3 (51–75% wilted), and 4 (76–100% wilted or dead). b Bioluminescent signals of Col-0, ein2-5, and npr1 mutants inoculated with FJ91-LUX. Data are shown as the mean ± SE (n = 14, *P ≤ 0.05, ***P ≤ 0.001, t-test). c Bioluminescent images of representative plants inoculated with FJ91-LUX. + : Strong; −: Weak. Scale = 1 cm
Discussion
Bacterial wilt, caused by R. solanacearum, is one of the most devastating bacterial diseases worldwide. Owing to the limitations of the available methods, a fast real-time disease detection method for Arabidopsis is required. In this study, we generated a luxCDABE-labeled R. solanacearum, which allowed the use of bioluminescence imaging technology to detect bacterial wilt disease in tomato and Arabidopsis.
Numerous methods have been developed to determine plant disease resistance, such as disease index, bacterial growth by colony counting, detection of bacterial concentration by PCR or loop-mediated isothermal amplification (LAMP), serological assays (e.g. ELISA, enzyme-linked immunosorbent assay), hyperspectral imaging by remote sensors, or bioluminescence imaging technology [14, 15, 28]. Each method has its own advantages and potential pitfalls, and bioluminescence imaging technology provides the biggest advantage in terms of providing noninvasive, spatial, quantitative, and real-time monitoring of the infection progress [17]. This technology will provide a powerful tool for the study of disease-resistant mechanisms, as well as the breeding of disease-resistant varieties.
Bioluminescent bacterium can be generated by transforming a plasmid carrying the luxCDABE operon or inserting the luxCDABE operon directly into the genome of the bacterium [29, 30]. Expression of luxCDABE in plasmids is usually higher compared to its expression when inserted into the genome; therefore, bacteria carrying plasmid-expressed luxCDABE display stronger light signals and are easily detected. Although the loss of plasmids during successive subculturing is a potential pitfall, we found that the light signals of FJ91-LUX persisted even after 9 generations of subculturing. However, we cannot rule out the possibility of the loss of plasmid during infection in plant tissues. Therefore, insertion of the luxCDABE operon into the genome is an alternative method. To increase the expression of the luxCDABE core gene in the genome, other LUX components may be added; for example, luxF and luxG enhance the luminescence intensity of bacteria [31, 32]. Improving the luminous intensity of luxCDABE can further improve the sensitivity of the system.
The luxCDABE-tagged P. syringae strain has been used for large-scale screening of resistant ecotypes in Arabidopsis [20]. Here, we generated a luxCDABE-tagged R. solanacearum, which could also be used for large-scale screening of Arabidopsis mutants or ecotypes with altered resistance to R. solanacearum infection. Compared with wild type, ein2-5 and npr1 mutants showed altered light intensity and wilt symptom after inoculation with luxCDABE-tagged R. solanacearum. However, to date, the role of ethylene and SA signaling pathway in resistance to R. solanacearum remains poorly understood. Remarkably, we found that, in Arabidopsis, the light intensity in the stems at 15 dpi was 2 folds higher than that in rosette leaves at 9 dpi (Fig. 4c), suggesting that the developmental stage is critical for R. solanacearum proliferation. Therefore, the low light intensity in ein2-5 mutant might be due to its delayed bolting (Fig. 5c). In the case of SA signaling pathway, several lines of evidence indicate that NPR1 plays a positive role in resistance to R. solanacearum [33, 34], on the other hand, the Arabidopsis SA-deficient mutant sid2 (encoding isochorismate synthase) and NahG (encoding salicylate hydroxylase) transgenic lines do not display significant increase in bacterial proliferation [7, 13]. Therefore, function of ethylene and SA in resistance to R. solanacearum requires further analysis.
It is worth noting that the light intensity in tomato was reduced at the later stage because of bacterial death (Fig. 3c); therefore, it cannot reflect the disease severity at this stage for tomato. To avoid this, it is advisable to quantify the light signals at a relatively early stage after inoculation, or to monitor the kinetics first, to select the bacterial growth stage. On the other hand, this suggests that the luxCDABE operon provides a means of detecting bacterial viability and can be used to monitor chemical toxicity [29, 35]. Therefore, it is plausible that FJ91-LUX could be used in the future to facilitate the testing of bacterial antibiotics, DNA and membrane damage, or oxidative stress chemicals.
Conclusions
In this study, we generated a luxCDABE-labeled R. solanacearum strain that allows tracking of the bacterial movement and quick quantification of the disease intensity in a living host.
Methods
Bacterial strains and growth conditions
R. solanacearum FJ91 strain (CGMCC No. 1.12711) was isolated from Fujian Province, China [36]. R. solanacearum were cultivated in casamino acid-peptone-glucose (CPG) media containing TTC [37]. P. syringae and Escherichia coli DH5α strains were grown in Luria–Bertani medium. Bacterial concentrations were determined by measuring the absorbance at OD600.
Plasmid construction
A promoter-less luxCDABE operon was amplified from a P. syringae-LUX strain [20] by PCR using the primers luxF (5-CCGGAATTCATGACTAAAAAAATTTCATTCAT-3) and luxR (5-CGCGGATCCATCAACTATCAAACGCTTC-3), and ligated to the pBBR1MCS2 plasmid to generate pBBR1MCS2- luxCDABE plasmid. The inserted fragment was verified by sequencing.
Generation of FJ91-LUX strain
FJ91-LUX was generated by transforming the pBBR1MCS2-luxCDABE plasmid into R. solanacearum FJ91. Briefly, freshly grown R. solanacearum colonies were cultured in 5 mL of CPG medium at 28 °C for 16 h with constant shaking at 200 rpm. Aliquots (1 mL) were centrifuged at 3500×g at 4 °C for 5 min and resuspended in 300 mM sucrose solution. The cell pellet was washed twice and dissolved in 100 μL of 300 mM sucrose solution. The pBBR1MCS2-luxCDABE plasmid was then transformed into R. solanacearum competent cells by electroporation (V = 2.2 kV·cm−1, C = 25 μF, R = 400 Ω) to generate FJ91-LUX.
Bioluminescence assay
Bioluminescent signals were monitored using a photon camera, version HRPCS5 (Photek Ltd., UK). Images and data were processed using Image32 software, a program coupled with a photon camera. The major parameters of the camera setup were the binary slice and 10% ND filter. The photon counts were integrated over 5 min.
Growth curve determination
Bacteria in liquid media were collected after overnight cultures by centrifugation at 3000×g for 5 min, and the pellet was resuspended in sterile water. After washing three times with sterile water, the solution was adjusted to an OD600 of 0.005 using fresh liquid medium. The OD600 was measured every 0.5 h.
Biofilm assay
Bacterial biofilm formation was measured by the crystal violet staining method [38]. In brief, 100 μL of the bacterial culture (OD600 = 0.5) was cultivated in a 96-well cell culture plate for 72 h. The medium was removed, followed by gentle washing (three times) with 200 µL of sterile water. Then, 125 μL of 0.1% (w/v) crystal violet (Aladdin, China) solution was added for 30 min. The dye solution was discarded and the plates were dried at 24 °C, and the bound stain was dissolved in 1% sodium dodecyl sulfate. The absorbance was measured at 570 nm using a Multiskan FC microplate spectrophotometer (Thermo Fisher Scientific, USA).
Mobility measurement
The mobility assay was performed as described previously [39]. In brief, one drop of bacterial culture (5 μL, OD600 = 0.6) was placed at the center of a CPG plate with 0.3% agar and incubated at 28 °C after drying in a laminar airflow hood. The colony diameter was measured after 2 days.
Subculturing in liquid media
The bacteria were initially grown in liquid media to OD600 = 1, and then diluted 1000 folds with fresh media to continue growth. This passaging procedure was repeated 9 times, and bioluminescent signals were measured every time before dilution.
Plant materials and growth conditions
The seeds of Arabidopsis ein2-5 [40], npr1 [41] mutants and wild type Col-0 were sterilized with 10% bleach for 10 min, rinsed with sterile water three times, and grown on 1/2 Murashige-Skoog agar plates. Seedlings were transferred to a 6 × 6 cm pot containing peat-based compost (Sun Gro Horticulture, USA), and maintained in a growth chamber (Conviron, Canada) under conditions of 16 h photoperiod, 75% humidity, and 22 °C temperature. Tomato (Solanum lycopersicum) ecotype Zheza 809 (Zhejiang Academy of Agricultural Sciences, China) was used in this study. Seed sterilization was the same as that of Arabidopsis, except that it was treated with bleach for 20 min. Tomato plants were grown in a growth room under conditions of 16 h photoperiod, 75% humidity, and 25 °C temperature.
Inoculation of R. solanacearum
Plants were inoculated with R. solanacearum using a root-soaked approach [42, 43]. Bacterial overnight cultures were collected by centrifugation and adjusted to an OD600 of 0.1. For tomato inoculation, roots of four-week-old tomato seedlings were scratched with a blade at 1 cm from the hypocotyl, and then 50 mL of bacterial suspension was drenched around the root zone. For Arabidopsis inoculation, the pot with four-week-old Arabidopsis plants was cut off half from the bottom and the upper half was immediately submerged into a bacterial suspension. After incubation with bacteria for 20 min, the pots were carefully moved to a new tray with a thick layer of moist soil and covered with a dome to keep the moisture for 2 d.
Statistical analysis
Statistical analyses and data normality tests were performed using Excel, SPSS 20.0, and all graphs were generated using GraphPad Prism 8.
Acknowledgements
Not applicable.
Abbreviations
- CPG
Casamino peptone agar
- TTC
Triphenyl tetrazolium chloride
Authors’ contributions
YL conceived and designed the research. CX and LZ performed most of the experiments and analyzed the data. ZH and CL helped with the bacterial inoculation. JL helped with generation of constructs. XZ isolated R. solanacearum from Fujian Province. YL and CX wrote the article. All authors read and approved the final manuscript.
Funding
This work was supported by National Natural Science Foundation of China (31770263), Key Research and Development Program of Zhejiang Province (2021C02009 and 2021C02064-7), and the National Key Research and Development Program of China (2018YFD1000800).
Availability of data and materials
All data generated or analysed during this study are included in this published article.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Mansfield J, Genin S, Magori S, Citovsky V, Sriariyanum M, Ronald P, Dow M, Verdier V, Beer SV, Machado MA, et al. Top 10 plant pathogenic bacteria in molecular plant pathology. Mol Plant Pathol. 2012;13(6):614–629. doi: 10.1111/j.1364-3703.2012.00804.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Peeters N, Guidot A, Vailleau F, Valls M. Ralstonia solanacearum, a widespread bacterial plant pathogen in the post-genomic era. Mol Plant Pathol. 2013;14(7):651–662. doi: 10.1111/mpp.12038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McGarvey JA, Denny TP, Schell MA. Spatial-temporal and quantitative analysis of growth and EPS I production by Ralstonia solanacearum in resistant and susceptible tomato cultivars. Phytopathology. 1999;89(12):1233–1239. doi: 10.1094/PHYTO.1999.89.12.1233. [DOI] [PubMed] [Google Scholar]
- 4.Buddenhagen I, Kelman A. Biological and physiological aspects of bacterial wilt caused by pseudomonas solanacearum. Annu Rev Phytopathol. 1964;2(1):203–230. [Google Scholar]
- 5.Deslandes L, Olivier J, Theulières F, Hirsch J, Feng DX, Bittner-Eddy P, Beynon J, Marco Y. Resistance to Ralstonia solanacearum in Arabidopsis thaliana is conferred by the recessive RRS1-R gene, a member of a novel family of resistance genes. Proc Natl Acad Sci USA. 2002;99(4):2404–2409. doi: 10.1073/pnas.032485099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Digonnet C, Martinez Y, Denance N, Chasseray M, Dabos P, Ranocha P, Marco Y, Jauneau A, Goffner D. Deciphering the route of Ralstonia solanacearum colonization in Arabidopsis thaliana roots during a compatible interaction: focus at the plant cell wall. Planta. 2012;236(5):1419–1431. doi: 10.1007/s00425-012-1694-y. [DOI] [PubMed] [Google Scholar]
- 7.Hanemian M, Barlet X, Sorin C, Yadeta KA, Keller H, Favery B, Simon R, Thomma BP, Hartmann C, Crespi M, et al. Arabidopsis CLAVATA1 and CLAVATA2 receptors contribute to Ralstonia solanacearum pathogenicity through a miR169-dependent pathway. New Phytol. 2016;211(2):502–515. doi: 10.1111/nph.13913. [DOI] [PubMed] [Google Scholar]
- 8.Pfund C, Tans-Kersten J, Dunning FM, Alonso JM, Ecker JR, Allen C, Bent AF. Flagellin is not a major defense elicitor in Ralstonia solanacearum cells or extracts applied to Arabidopsis thaliana. Mol Plant–Microbe Interact. 2004;17(6):696–706. doi: 10.1094/MPMI.2004.17.6.696. [DOI] [PubMed] [Google Scholar]
- 9.Xian L, Yu G, Wei Y, Rufian JS, Li Y, Zhuang H, Xue H, Morcillo RJL, Macho AP. A bacterial effector protein hijacks plant metabolism to support pathogen nutrition. Cell Host Microbe. 2020;28(4):548–557. doi: 10.1016/j.chom.2020.07.003. [DOI] [PubMed] [Google Scholar]
- 10.Yu G, Xian L, Xue H, Yu W, Rufian JS, Sang Y, Morcillo RJL, Wang Y, Macho AP. A bacterial effector protein prevents MAPK-mediated phosphorylation of SGT1 to suppress plant immunity. PLoS Pathog. 2020;16(9):e1008933. doi: 10.1371/journal.ppat.1008933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Narusaka M, Shirasu K, Noutoshi Y, Kubo Y, Shiraishi T, Iwabuchi M, Narusaka Y. RRS1 and RPS4 provide a dual Resistance-gene system against fungal and bacterial pathogens. Plant J. 2009;60(2):218–226. doi: 10.1111/j.1365-313X.2009.03949.x. [DOI] [PubMed] [Google Scholar]
- 12.Hirsch J, Deslandes L, Feng DX, Balague C, Marco Y. Delayed symptom development in ein2-1, an Arabidopsis ethylene-insensitive mutant, in response to bacterial wilt caused by Ralstonia solanacearum. Phytopathology. 2002;92(10):1142–1148. doi: 10.1094/PHYTO.2002.92.10.1142. [DOI] [PubMed] [Google Scholar]
- 13.Hernandez-Blanco C, Feng DX, Hu J, Sanchez-Vallet A, Deslandes L, Llorente F, Berrocal-Lobo M, Keller H, Barlet X, Sanchez-Rodriguez C, et al. Impairment of cellulose synthases required for Arabidopsis secondary cell wall formation enhances disease resistance. Plant Cell. 2007;19(3):890–903. doi: 10.1105/tpc.106.048058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fang Y, Ramasamy RP. Current and prospective methods for plant disease detection. Biosensors (Basel) 2015;5(3):537–561. doi: 10.3390/bios5030537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alvarez AM. Integrated approaches for detection of plant pathogenic bacteria and diagnosis of bacterial diseases. Annu Rev Phytopathol. 2004;42:339–366. doi: 10.1146/annurev.phyto.42.040803.140329. [DOI] [PubMed] [Google Scholar]
- 16.Pastrik KH, Maiss E. Detection of Ralstonia solanacearum in potato tubers by polymerase chain reaction. J Phytopathol. 2000;148(11–12):619–626. [Google Scholar]
- 17.Waidmann MS, Bleichrodt FS, Laslo T, Riedel CU. Bacterial luciferase reporters: the Swiss army knife of molecular biology. Bioeng Bugs. 2011;2(1):8–16. doi: 10.4161/bbug.2.1.13566. [DOI] [PubMed] [Google Scholar]
- 18.Monteiro F, Genin S, van Dijk I, Valls M. A luminescent reporter evidences active expression of Ralstonia solanacearum type III secretion system genes throughout plant infection. Microbiology. 2012;158(8):2107–2116. doi: 10.1099/mic.0.058610-0. [DOI] [PubMed] [Google Scholar]
- 19.Meighen EA. Bacterial bioluminescence: organization, regulation, and application of the lux genes. FASEB J. 1993;7(11):1016–1022. doi: 10.1096/fasebj.7.11.8370470. [DOI] [PubMed] [Google Scholar]
- 20.Fan J, Crooks C, Lamb C. High-throughput quantitative luminescence assay of the growth in planta of Pseudomonas syringae chromosomally tagged with Photorhabdus luminescens luxCDABE. Plant J. 2008;53(2):393–399. doi: 10.1111/j.1365-313X.2007.03303.x. [DOI] [PubMed] [Google Scholar]
- 21.Xu X, Miller SA, Baysal-Gurel F, Gartemann KH, Eichenlaub R, Rajashekara G. Bioluminescence imaging of Clavibacter michiganensis subsp. michiganensis infection of tomato seeds and plants. Appl Environ Microbiol. 2010;76(12):3978–3988. doi: 10.1128/AEM.00493-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Du H, Chen B, Zhang X, Zhang F, Miller SA, Rajashekara G, Xu X, Geng S. Evaluation of Ralstonia solanacearum infection dynamics in resistant and susceptible pepper lines using bioluminescence imaging. Plant Dis. 2017;101(2):272–278. doi: 10.1094/PDIS-05-16-0714-RE. [DOI] [PubMed] [Google Scholar]
- 23.Shah N, Naseby DC. Bioluminescence-based measurement of viability of Pseudomonas aeruginosa ATCC 9027 harbouring plasmid-based lux genes under the control of constitutive promoters. J Appl Microbiol. 2014;117(5):1373–1387. doi: 10.1111/jam.12635. [DOI] [PubMed] [Google Scholar]
- 24.Tian X, Gao Y, Wang S, Adnan Hameed HM, Yu W, Fang C, Zhang J, Tan Y, Cao Z, Guan P, et al. Rapid visualized assessment of drug efficacy in live mice with a selectable marker-free autoluminescent Klebsiella pneumoniae. Biosens Bioelectron. 2021;177:112919. doi: 10.1016/j.bios.2020.112919. [DOI] [PubMed] [Google Scholar]
- 25.Branda SS, Vik S, Friedman L, Kolter R. Biofilms: the matrix revisited. Trends Microbiol. 2005;13(1):20–26. doi: 10.1016/j.tim.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 26.Corral J, Sebastià P, Coll NS, Barbé J, Aranda J, Valls M. Twitching and swimming motility play a role in Ralstonia solanacearum pathogenicity. mSphere. 2020;5(2):e00740. doi: 10.1128/mSphere.00740-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yasufumi H, Yuka M, Shiho I, Kazusa H, Kouhei O, Akinori K, Kenji K. Regulation involved in colonization of intercellular spaces of host plants in Ralstonia solanacearum. Front Plant Sci. 2017;8:967. doi: 10.3389/fpls.2017.00967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Caruso P, Gorris MT, Cambra M, Palomo JL, Collar J, Lopez MM. Enrichment double-antibody sandwich indirect enzyme-linked immunosorbent assay that uses a specific monoclonal antibody for sensitive detection of Ralstonia solanacearum in asymptomatic potato tubers. Appl Environ Microbiol. 2002;68(7):3634–3638. doi: 10.1128/AEM.68.7.3634-3638.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Billard P, DuBow MS. Bioluminescence-based assays for detection and characterization of bacteria and chemicals in clinical laboratories. Clin Biochem. 1998;31(1):1–14. doi: 10.1016/s0009-9120(97)00136-7. [DOI] [PubMed] [Google Scholar]
- 30.Stewart GS, Williams P. Lux genes and the applications of bacterial bioluminescence. J Gen Microbiol. 1992;138(7):1289–1300. doi: 10.1099/00221287-138-7-1289. [DOI] [PubMed] [Google Scholar]
- 31.Brodl E, Csamay A, Horn C, Niederhauser J, Weber H, Macheroux P. The impact of LuxF on light intensity in bacterial bioluminescence. J Photochem Photobiol B Biol. 2020;207:111881. doi: 10.1016/j.jphotobiol.2020.111881. [DOI] [PubMed] [Google Scholar]
- 32.Nijvipakul S, Wongratana J, Suadee C, Entsch B, Ballou DP, Chaiyen P. LuxG is a functioning flavin reductase for bacterial luminescence. J Bacteriol. 2008;190(5):1531. doi: 10.1128/JB.01660-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kawamura Y, Takenaka S, Hase S, Kubota M, Ichinose Y, Kanayama Y, Nakaho K, Klessig DF, Takahashi H. Enhanced defense responses in Arabidopsis induced by the cell wall protein fractions from Pythium oligandrum require SGT1, RAR1, NPR1 and JAR1. Plant Cell Physiol. 2009;50(5):924–934. doi: 10.1093/pcp/pcp044. [DOI] [PubMed] [Google Scholar]
- 34.Chen YY, Lin YM, Chao TC, Wang JF, Liu AC, Ho FI, Cheng CP. Virus-induced gene silencing reveals the involvement of ethylene-, salicylic acid- and mitogen-activated protein kinase-related defense pathways in the resistance of tomato to bacterial wilt. Physiol Plant. 2009;136(3):324–335. doi: 10.1111/j.1399-3054.2009.01226.x. [DOI] [PubMed] [Google Scholar]
- 35.Kurvet I, Ivask A, Bondarenko O, Sihtmae M, Kahru A. LuxCDABE–transformed constitutively bioluminescent Escherichia coli for toxicity screening: comparison with naturally luminous Vibrio fischeri. Sensors (Basel) 2011;11(8):7865–7878. doi: 10.3390/s110807865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen D, Liu B, Zhu Y, Zhang H, Chen Z, Zheng X, Xiao R, Chen Y. Complete genome sequence of Ralstonia solanacearum FJAT-91, a high-virulence pathogen of tomato wilt. Genome Announc. 2017;5(37):e00900–00917. doi: 10.1128/genomeA.00900-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kelman A. The relationship of pathogenicity of Pseudomonas solanacearum to colony appearance in a tetrazolium medium. Phytopathology. 1954;44(12):693–695. [Google Scholar]
- 38.Holmberg A, Lood R, Mörgelin M, Söderquist B, Holst E, Collin M, Christensson B, Rasmussen M. Biofilm formation by Propionibacterium acnes is a characteristic of invasive isolates. Clin Microbiol Infect. 2009;15(8):787–795. doi: 10.1111/j.1469-0691.2009.02747.x. [DOI] [PubMed] [Google Scholar]
- 39.Ha DG, Kuchma SL, O'Toole GA. Plate-based assay for swarming motility in Pseudomonas aeruginosa. Methods Mol Biol. 2014;1149:67–72. doi: 10.1007/978-1-4939-0473-0_8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Alonso JM, Hirayama T, Roman G, Nourizadeh S, Ecker JR. EIN2, a bifunctional transducer of ethylene and stress responses in Arabidopsis. Science. 1999;284(5423):2148–2152. doi: 10.1126/science.284.5423.2148. [DOI] [PubMed] [Google Scholar]
- 41.Shah J, Tsui F, Klessig DF. Characterization of a salicylic acid-insensitive mutant (sai1) of Arabidopsis thaliana, identified in a selective screen utilizing the SA-inducible expression of the tms2 gene. Mol Plant–Microbe Interact MPMI. 1997;10(1):69–78. doi: 10.1094/MPMI.1997.10.1.69. [DOI] [PubMed] [Google Scholar]
- 42.Morel A, Peeters N, Vailleau F, Barberis P, Jiang G, Berthomé R, Guidot A. Plant pathogenicity phenotyping of Ralstonia solanacearum strains. In: Medina C, López-Baena FJ, editors. Host–pathogen interactions: methods and protocols. New York: Springer; 2018. pp. 223–239. [DOI] [PubMed] [Google Scholar]
- 43.Kwak MJ, Kong HG, Choi K, Kwon SK, Song JY, Lee J, Lee PA, Choi SY, Seo M, Lee HJ, et al. Rhizosphere microbiome structure alters to enable wilt resistance in tomato. Nat Biotechnol. 2018;36:1100–1109. doi: 10.1038/nbt.4232. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analysed during this study are included in this published article.