Abstract
Background
To provide information about pathogens’ coinfection prevalence with SARS‐CoV‐2 could be a real help to save patients’ lives. This study aims to evaluate the pathogens’ coinfection prevalence among COVID‐19 patients.
Method
In order to find all of the relevant articles, we used systematic search approach. Research‐based databases including PubMed, Web of Science, Embase, and Scopus, without language restrictions, were searched to identify the relevant bacterial, fungal, and viral coinfections among COVID‐19 cases from December 1, 2019, to August 23, 2021. In order to dig deeper, other scientific repositories such as Medrxiv were probed.
Results
A total of 13,023 studies were found through systematic search. After thorough analysis, only 64 studies with 61,547 patients were included in the study. The most common causative agents of coinfection among COVID‐19 patients were bacteria (pooled prevalence: 20.97%; 95% CI: 15.95–26.46; I 2: 99.9%) and less frequent were virus coinfections (pooled prevalence: 12.58%; 95% CI: 7.31–18.96; I 2: 98.7%). The pooled prevalence of fungal coinfections was also 12.60% (95% CI: 7.84–17.36; I 2: 98.3%). Meta‐regression analysis showed that the age sample size and WHO geographic region did not influenced heterogeneity.
Conclusion
We identified a high prevalence of pathogenic microorganism coinfection among COVID‐19 patients. Because of this rate of coinfection empirical use of antibacterial, antifungal, and antiviral treatment are advisable specifically at the early stage of COVID‐19 infection. We also suggest running simultaneously diagnostic tests to identify other microbiological agents’ coinfection with SARS‐CoV‐2.
Keywords: coinfection, coronavirus, COVID‐19, meta‐analysis, systematic review
Prevalence of microbial coinfection in COVID‐19 patients is essential for clinicians. Systematic search andmeta‐analysis showed a high prevalence of pathogenic microorganisms, especially bacteria, among COVID‐19 patients.
1. INTRODUCTION
COVID‐19 was declared as the new respiratory pandemic in March 2020. 1 Microbial pathogens coinfections always played an important role in increasing mortality and morbidity rate in pandemics. Their coinfection with SARS‐CoV‐2 is not an exception. While countries applied different measures to limit spread of the virus, new wave still striking and quickly mutate and gain new feature which has made it more dangerous than ever. 2
Viral, bacterial, and fungal coinfections alter the pathophysiology of disease, also the patient recovery outcome. 3 , 4 Respiratory viruses’ including hRV, hMPV, and RSV are associated with majority respiratory viral coinfection. 5 Also, immunosuppression and immunodeficiency condition such as HIV infection could effect on COVID‐19 disease. 6
Fungal infection plays a major threat to patient's life in intensive care units. 7 Fungal coinfections such as Aspergillus and Candida species could increase mortality rate, especially in critically ill patients. 8 One of the great challenges for clinicians is their detection. Fungal coinfection remained undetectable even after the death of the patients. 9 Similar clinical and radiological features between SARS‐CoV‐2 and fungal pathogens are the other difficulties that healthcare providers have to dealt with. 10
Among microbiological coinfections, bacterial pathogens are considered more important agents based on their previous record viral outbreaks and pandemics. 11 It also was reported people with bacterial coinfection showed high number of mortality. Critical ill patients showed greater percentage of coinfection compared to hospitalized patients. 12 One of the main importance of assessing bacterial coinfection prevalence is about applying empirical antibiotic treatment, in SARS‐CoV‐2 patients. Extensive use of antibiotics could lead to several such as antibacterial resistance. 13 , 14 Some of the respiratory bacterial pathogen such as pneumococcal, staphylococcal, and Klebsiella with SARS‐CoV‐2 have common clinical manifestation; therefore, antibiotics treatment would be more difficult than regular situation. 15 This study aims to evaluate the microbiological coinfection prevalence among COVID‐19 patients.
2. MATERIALS AND METHODS
We performed our research based on PRISMA guideline studies 16 we registered our article search protocol in the International Prospective Register of Systemic Reviews with CRD42021277142. We used related unique keywords to conduct our search strategy and retrieving all of the related articles.
2.1. Method of literature search
We explored the online scientific repositories without setting any language barrier. PubMed, Web of Science, Embase, and Scopus were probed to find the relevant articles about pathogens’ coinfection prevalence in COVID‐19‐infected persons between December 1, 2019, and August 23, 2021. Other knowledge‐based databases such as Medrxiv and SSRN were also used to gather the off‐the‐record articles. We chose the keywords in this article based on MeSH Terms. The PICOTS in our study are available in Appendix 1.
To find other off‐the‐record publication, we probed Google Scholar. A microbiologist was asked to identify and validate the related articles. Simultaneously, we hand‐searched our articles library to gather other relevant studies. We imported all of the gathered data to Endnote X6. The duplicated articles were removed. We scanned the remained studies in three distinguished steps. Firstly, we probed the articles based on their titles. Afterward, the abstract of the screened articles were reviewed, and the full text of the relevant ones were collected. We conducted the study selection procedure based on blinding and task separation. The mentioned procedure was done by two independent reviewers simultaneously. In case of any disagreement between reviewers (Inter‐rater discrepancies), another rater were asked to resolve the problem. The kappa coefficient for agreement between two raters was equal to 93%.
2.2. Inclusion and exclusion criteria
All the related studies including cross‐sectional, case series, and cohort studies evaluating the prevalence of viral, bacterial, and fungal coinfections among COVID‐19 cases were gathered. The case series and case report articles with <10 sample sizes did not reviewed in this study. We excluded the other types of articles including clinical trials, reviews, and case‐control articles.
2.3. Data extraction
We extracted the necessary data from all of the studies including authors’ name, study year, country, study design, sample size, gender, age, number, and type of coinfections.
2.4. Variable definition
Bacteria type were classified based on transmission way and clinical signs. Countries were categorized based on the latest WHO definition that includes the following six regions: Regional Office for Africa (AFRO), Regional Office for the Americas (AMRO), Regional Office for the Eastern Mediterranean (EMRO), Regional Office for Europe (EURO), Regional Office for South‐East Asia (SEARO), and the Regional Office for the Western Pacific (WPRO).
2.5. Quality assessment
Newcastle‐Ottawa Scale 17 evaluated the quality of the finalized studies. We assessed the studies based on three selection steps of this scale: 1‐Selection 2‐Confounder, and 3‐Exposure. Two independent reviewers examined the articles based on the Newcastle‐Ottawa criteria (RP and SS), and the total score for each study in the three steps was calculated. Afterward, the selected studies were categorized in the following groups: very good, good, satisfactory, and unsatisfactory studies. 18
2.6. Statistical analysis
All statistical tests in this study were performed with Stata 14.0. Just like previous researches, 18 , 19 , 20 , 21 the sample size, the coinfection prevalence in COVID‐19 cases, and the coinfection causative agent's types and species were extracted. We applied Cochran's Q test to determine the heterogeneity. We also quantified it with the I 2 index. I 2 values above 0.7 were determined as high heterogeneity based on the Higgins classification approach. 22 Metaprop package were used to calculate the pooled prevalence with 95% confidence interval. Random‐effects model was applied to estimate the pooled prevalence. This package applies double arcsine transformations to stabilize the variance in the meta‐analyses. The effect of sample size, age, and WHO geographic regions on the studies heterogeneity were analyzed by meta‐regression analysis. Publication bias evaluated by “metabias” command. In case of any publication bias, we adjusted the prevalence rate with “metatrim” command applying trim‐and‐fill approach. Statistical significance was considered 0.05.
3. RESULT
We collected 13,241 articles probing the mentioned databases. We also found 151 articles through other resources. By removing the duplicated articles, 8838 articles remained. The remained articles were screened in three distinguished steps. First, we exclude the 6542 studies by analyzing their titles. Then After reviewing the abstracts, 1924 studies were removed from the library. In the third step, the full text of the 372 remained articles was comprehensively studied, and we exclude the 308 studies. A total of 64 studies 3 , 5 , 18 , 23 , 24 , 25 , 26 , 27 , 28 , 29 , 30 , 31 , 32 , 33 , 34 , 35 , 36 , 37 , 38 , 39 , 40 , 41 , 42 , 43 , 44 , 45 , 46 , 47 , 48 , 49 , 50 , 51 , 52 , 53 , 54 , 55 , 56 , 57 , 58 , 59 , 60 , 61 , 62 , 63 , 64 , 65 , 66 , 67 , 68 , 69 , 70 , 71 , 72 , 73 , 74 , 75 , 76 , 77 , 78 , 79 with 61,547 total sample size were included in our study. Selection process flow chart is available in Figure 1, and Table 1 shows the studies’ characteristics. The highest studies number belonged to Western Pacific (25 studies) area, Southeast Asia (three studies), and Eastern Mediterranean Region (three studies) was the lowest one. All the included studies were published during 2020. The minimum and maximum age range of the subjects was for Wu et al. 28 article had the lowest age ranges (mean age = 6 years old) and Wang et al. 65 study (mean age = 73 years old), and D’Onofrio et al. 56 study (mean age = 73 years old) reported the highest age range. Twenty‐eight (43.75%) of studies were case series. There were also 29 (45.31%) cohort and 7 (10.94%) cross‐sectional.
FIGURE 1.
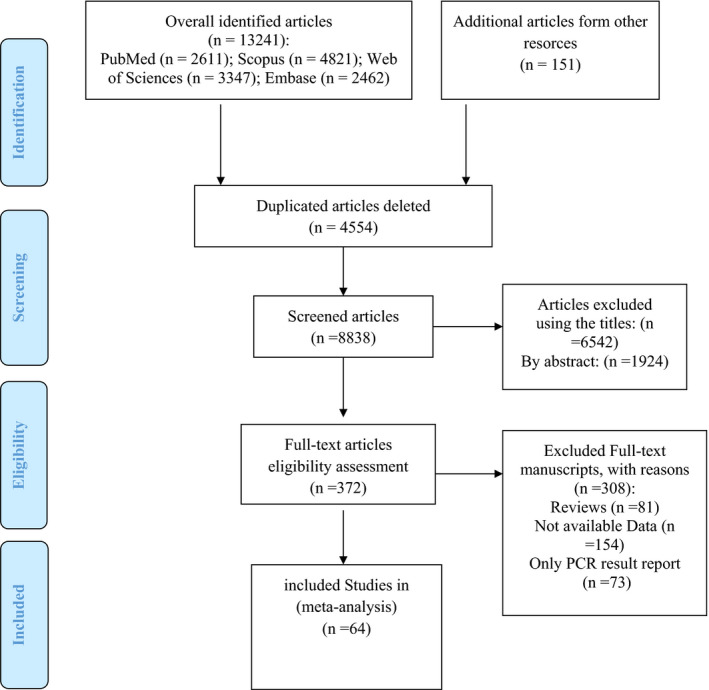
Study selection process based on PRISMA flow diagram
TABLE 1.
Characteristics of the studies included in this meta‐analysis
| Author | Country | Design | Publication year | Mean age | Sample size | Viral coinfections prevalence (95% CI) | Bacterial coinfections prevalence (95% CI) | Fungal coinfections prevalence (95% CI) |
|---|---|---|---|---|---|---|---|---|
| Zhu et al. 22 | China | Case series | 2020 | 51 | 257 | 31.52 (25.89–37.58) | 91.83 (87.78–94.87) | 23.35 (18.31–29) |
| Zheng et al. 75 | China | Case series | 2020 | 30.6 | 1001 | 0.40 (0.11–1.02) | NA | NA |
| Agrifoglio et al. 76 | Spain | Cohort | 2020 | 58.7 | 139 | NA | NA | 16.55 (10.79–23.79) |
| Blasco et al. 58 | Spain | Case series | 2020 | 64 | 183 | 0.55 (0.01–3.01) | 1.64 (0.34–4.72) | NA |
| Contou et al. 56 | France | Case series | 2020 | 61 | 92 | 14.13 (7.74–22.95) | 95.65 (89.24–98.8) | NA |
| Sarinoglu et al. 34 | Turkey | Cross‐sectional | 2020 | – | 30 | NA | 6.67 (0.82–22.07) | NA |
| Chauhdary et al. 57 | Brunei | Case series | 2020 | – | 141 | NA | 4.53 (3.01–6.52) | NA |
| Chen et al. 42 | China | Case series | 2020 | 52.5 | 326 | 6.13 (3.79–9.32) | NA | NA |
| Chen et al. 59 | China | Case series | 2020 | 51 | 123 | 12.2 (6.99–19.32) | NA | NA |
| Cheng et al. 42 | China | Cohort | 2020 | 36 | 62 | NA | 40.32 (28.05–53.55) | NA |
| Chowdhary et al. 77 | India | Cross‐sectional | 2020 | 596 | NA | NA | 4.53 (3.01–6.52) | |
| D’Onofrio et al. 55 | Belgium | Cohort | 2020 | 73 | 110 | NA | 2.73 (0.57–7.76) | NA |
| Luna et al. 60 | Brazil | Case series | 2020 | 115 | 11.3 (6.16–18.55) | NA | NA | |
| Ding et al. 61 | China | Case series | 2020 | 50.2 | 115 | 4.35 (1.43–9.85) | NA | NA |
| Ebrahim 62 | Saudi Arabia | Case series | 2020 | 44 | 99 | 0 (0–3.66) | NA | NA |
| Fu et al. 54 | China | Cohort | 2020 | 101 | NA | 4.95 (1.63–11.18) | NA | |
| Garcia‐Vidal et al. 53 | Spain | Cohort | 2020 | 62 | 989 | 0.61 (0.22–1.32) | 2.93 (1.97–4.18) | NA |
| Gayam et al. 52 | USA | Cohort | 2020 | 57 | 350 | NA | 1.71 (0.63–3.69) | NA |
| Gupta et al. 50 | India | Cohort | 2020 | 36 | 1073 | NA | 2.05 (1.29–3.09) | NA |
| Hashemi et al. 63 | Iran | Case series | 2020 | 105 | 21.9 (14.42–31.03) | NA | NA | |
| Hazra et al. 49 | USA | Cross‐sectional | 2020 | 459 | 3.7 (2.17–5.86) | 0.00 (0.00–0.80) | NA | |
| Hirotsu et al. 48 | Japan | Cross‐sectional | 2020 | 40 | NA | 0.00 (0.00–8.81) | NA | |
| Hughes et al. 47 | UK | Case series | 2020 | 69.5 | 836 | 0.00 (0.00–0.44) | 3.23 (2.14–4.66) | 0.36 (0.07–1.05) |
| Intra et al. 46 | Italy | Cohort | 2020 | 61 | NA | 68.85 (55.71–80.1) | 68.85 (55.71–80.1) | |
| Jiang et al. 64 | China | Case series | 2020 | 161 | 47.2 (39.3–55.22) | NA | NA | |
| Karami et al. 45 | Netherlands | Cohort | 2020 | 70 | 925 | NA | 0.86 (0.37–1.7) | NA |
| Kim et al. 5 | USA | Cross‐sectional | 2020 | 46.9 | 116 | 21.55 (14.46–30.15) | 0.00 (0.00–3.13) | NA |
| Kimmig et al. 44 | USA | Cohort | 2020 | 46.9 | 111 | NA | 37.84 (28.8–47.54) | NA |
| Leuzinger et al. 65 | Switzerland | Case series | 2020 | 49 | 825 | 12.97 (10.75–15.46) | NA | NA |
| Li et al. 27 | China | Cohort | 2020 | 66.2 | 1495 | NA | 20.6 (18.58–22.74) | NA |
| Li et al. 18 | China | Case series | 2020 | 57 | 32 | 15.63 (5.28–32.79) | 31.25 (16.12–50.01) | NA |
| Lin et al. 66 | China | Case series | 2020 | 92 | 6.52 (2.43–13.66) | NA | NA | |
| Lin et al. 67 | China | Case series | 2020 | 45 | 133 | 12.78 (7.63–19.67) | NA | NA |
| Liu et al. 43 | China | Case series | 2020 | 46.5 | 20 | NA | 20 (5.73–43.66) | NA |
| Lv et al. 42 | China | Cohort | 2020 | 62 | 354 | 0.28 (0.01–1.56) | 14.12 (10.67–18.19) | NA |
| Ma et al. 41 | China | Case series | 2020 | 45.5 | 250 | 8.8 (5.6–13.02) | 9.6 (6.25–13.95) | NA |
| Ma et al. 68 | China | Cross‐sectional | 2020 | 67 | 93 | 49.46 (38.93–60.03) | NA | NA |
| Massey et al. 40 | USA | Case series | 2020 | 62.3 | 790 | 34.18 (30.87–37.6) | 55.44 (51.9–58.95) | NA |
| Motta et al. 39 | Multi‐place a | Cohort | 2020 | 69 | 1.45 (0.04–7.81) | 7.25 (2.39–16.11) | NA | |
| Neto et al. 51 | USA | Cohort | 2020 | 66 | 242 | NA | 19.01 (14.27–24.53) | NA |
| Verroken et al. 28 | Netherlands | Cohort | 2020 | 32 | NA | 18.75 (7.21–36.44) | NA | |
| Nori et al. 38 | USA | Cohort | 2020 | 62 | 152 | NA | 44.08 (36.04–52.35) | NA |
| Nowak et al. 69 | USA | Case series | 2020 | 60.2 | 408 | 20.34 (16.54–24.58) | NA | NA |
| Pandey et al. 29 | India | Cross‐sectional | 2020 | 120 | NA | 13.33 (7.82–20.75) | NA | |
| Porretta et al. 37 | Italy | Cohort | 2020 | 67.4 | 331 | NA | 9.67 (6.71–13.37) | NA |
| Ripa et al. 36 | Italy | Cohort | 2020 | 64 | 731 | NA | 7.25 (5.48–9.38) | NA |
| Rothe et al. 35 | Germany | Cohort | 2020 | 63.5 | 140 | NA | 76.43 (68.52–83.19) | NA |
| Segrelles‐Calvo et al. 78 | Spain | Cohort | 2020 | 59.6 | 215 | NA | NA | 29.3 (23.31–35.88) |
| Sepulveda et al. 33 | USA | Cohort | 2020 | 28011 | NA | 3.8 (3.58–4.03) | NA | |
| Sharifipour et al. 3 | Iran | Case series | 2020 | 67.1 | 19 | NA | 100 (82.35–100) | NA |
| Sharov et al. 32 | Russia | Case series | 2020 | 147 | 59.86 (51.47–67.85) | 75.51 (67.74–82.22) | NA | |
| Sy et al. 31 | Philippine | Cohort | 2020 | 44.21 | 12513 | NA | 0.90 (0.74–1.08) | NA |
| Tadolini M et al. 30 | Global | Cohort | 2020 | 48 | 49 | NA | 85.71 (72.76–94.06) | NA |
| Teotonio et al. 70 | Brazil | Case series | 2020 | 44.55 | 112 | 38.39 (29.36–48.06) | NA | NA |
| Vaughn et al. 71 | USA | Cohort | 2020 | 64.7 | 1705 | 0.53 (0.24–1.00) | NA | NA |
| Wang J et al. 64 | China | Case series | 2020 | 73 | 104 | NA | NA | 7.69 (3.38–14.6) |
| Weissberg et al. 72 | Switzerland | Cohort | 2020 | 49 | 11 | 9.09 (0.23–41.28) | NA | NA |
| Wu et al. 27 | China | Case series | 2020 | 6 | 74 | 13.51 (6.68–23.45) | 47.3 (35.57–59.25) | NA |
| Youngs et al. 26 | UK | Cohort | 2020 | 59 | 36 | NA | 30.56 (16.35–48.11) | NA |
| Yu et al. 25 | Sweden | Cohort | 2020 | 2240 | NA | 10.09 (8.87–11.41) | NA | |
| Yu et al. 73 | China | Cohort | 2020 | 57 | 67 | 10.45 (4.3–20.35) | NA | NA |
| Yue et al. 74 | China | Case series | 2020 | 307 | 49.84 (44.11–55.57) | NA | NA | |
| Zha et al. 24 | China | Cohort | 2020 | 57 | 874 | NA | 2.52 (1.58–3.79) | NA |
| Zhang et al. 23 | China | Case series | 2020 | 64.76 | 38 | 15.79 (6.02–31.25) | 57.89 (40.82–73.69) | 10.53 (2.94–24.8) |
Abbreviation: CI, confidence interval.
Belgium, Brazil, France, Italy, Russia, Singapore, Spain, and Switzerland.
3.1. Pooled prevalence of coinfections in COVID‐19 patients
Table 1 exhibits all included studies coinfection prevalence. Figure 2 shows the coinfection prevalence forest plot. Minimum and maximum coinfection prevalence were in Hazra et al. study 50 (prevalence: 0.00%; 95%CI: 0.00–0.80) from the USA and Sharif pour et al. article 3 (prevalence: 100.00%:95% CI: 82.35–100.00) from Iran which were resulted from random‐effects model approach (available in Figure 2) respectively. Pooled estimate of coinfection prevalence was 16.98% (95% CI: 13.62–20.62). Therefore, from every 1000 COVID‐19‐infected person, 136 to 20.6 individuals infected with another types of pathogens have coinfections.
FIGURE 2.
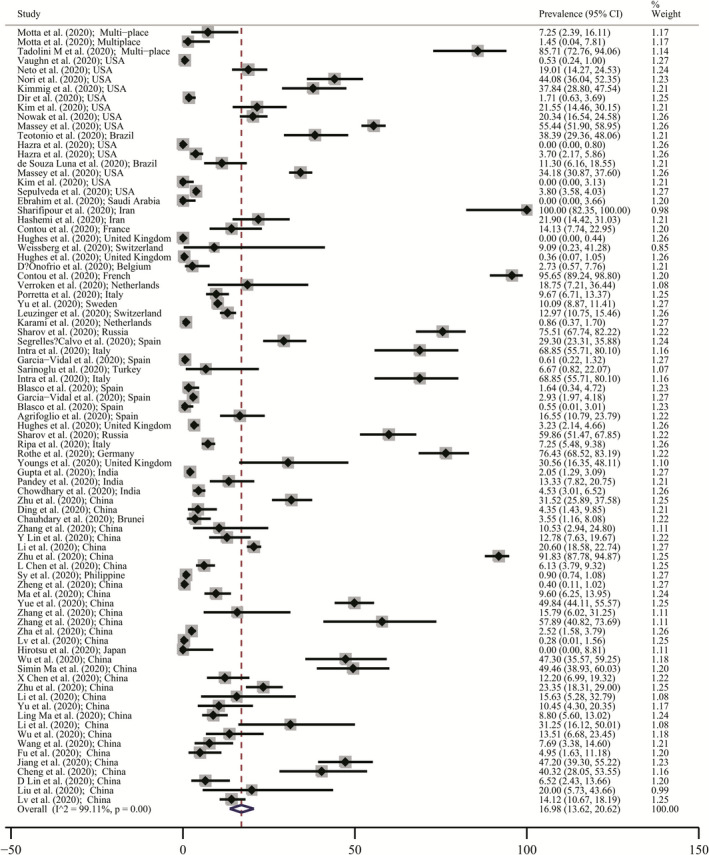
Prevalence of all‐type coinfections in patients with COVID‐19 Forest plot based on a random‐effects model. Each study identifies distinguished by their author (year) and countries. Each line segment's midpoint shows the prevalence estimate, length of line segment indicates 95% confidence interval (CI) in each study, and diamond mark illustrates the pooled estimate
3.2. Pooled prevalence of coinfections based on different subgroups
Pooled coinfection prevalence based on coinfections pathogens subtypes and regions are listed in Figure 3. Supplements 1–3 show the different pathogens species (bacterial, fungal, and viral coinfections) coinfection prevalence forest plot. The most prevalent subtype was bacteria (pooled prevalence: 20.97%; 95% CI: 15.95−26.46; I 2: 99.9%), and viral coinfections were the less frequent ones (pooled prevalence: 12.58%: 95% CI: 7.31–18.96; I 2: 98.7%). The pooled prevalence of fungal coinfections was 12.60% (95% CI: 7.84–17.36; I 2: 98.3%).
FIGURE 3.
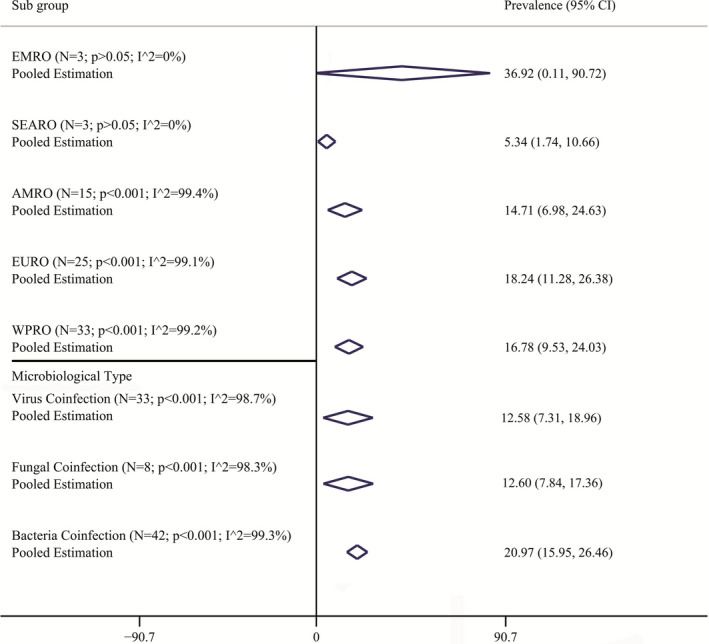
Pooled prevalence with 95% confidence interval [CI] and heterogeneity indexes of coinfections in COVID‐19 patients based on type of the coinfections and different regional places. The diamond mark illustrates the pooled prevalence and the length of the diamond indicates the 95% CI
Analysis showed that EMRO were the most coinfection regions 36.92% (95% CI: 0.00–90.72; I 2: 0%). least coinfections pooled prevalence were resulted from SEARO regions 5.34% (95% CI: 1.74–10.66; I 2: 0%): EURO, WPRO, and AMRO pooled prevalence are accessible in Figure 3.
3.3. Heterogeneity and meta‐regression
Heterogeneity results are available in Table 2. Cochran's Q test showed the included studies had high heterogeneity (p < 0.001). The I 2 index for total coinfections and pathogen subtypes were up to 90%. Meta‐regression analysis showed the age (Coefficient: −0. × 10−3; p: 0.777), sample size (Coefficient: −0.1 × 10−4; p: 0.192) and region (based WHO regional office) size (Coefficient: −0.034; p: 0.214) possess no significant effect on the studies heterogeneity (Figure 4A,B).
TABLE 2.
The univariate meta‐regression analysis on the hertogenisity of the determinants in included studies for coinfections in COVID‐19 patients
| Variables | Coefficient | 95% CI | p‐value |
|---|---|---|---|
| Age (year) | −0.7 × 10−3 | −5.6 × 10−3 to 4.2 × 10−3 | 0.777 |
| WHO region (score) | −0.034 | −0.087 to 0.019 | 0.214 |
| Sample size (Number) | −0.1 × 10−4 | −0.2 × 10−4 to 0.5 × 10−5 | 0.192 |
Coding of WHO region: 1 = EMRO; 2 = EURO; 3 = AMRO; 4 = WPRO; SEARO = 5.
Abbreviation: CI, confidence interval.
FIGURE 4.
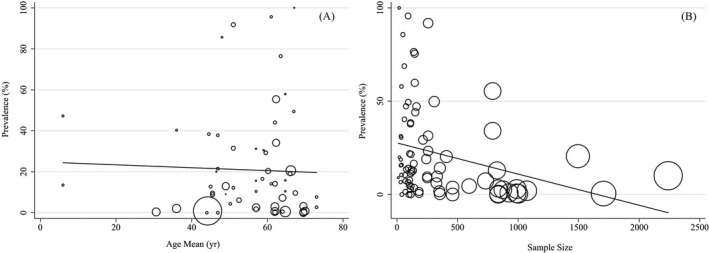
Association between prevalence of age (A) and sample size (B) with prevalence of coinfections by means of meta‐regression. The size of circles indicates the precision of each study. There is no significant association with respect to the prevalence of coinfections with age sample size
3.4. Publication bias
Egger's test results (coefficient: −0.41, p: 0.899) exhibited that there was not any significant publication bias in this meta‐analysis.
4. DISCUSSION
Our result elucidated that overall coinfection prevalence was 16.98. The lowest coinfection prevalence was reported in the USA and the highest level of coinfection was in Iran. As we expected between pathogenic microorganisms, bacterial agents were the most frequent and viral coinfection had the lowest coinfection rate in COVID‐19 patients. We also found out that EMRO region had the most prevalence of coinfection and compare to that SEARO region was the lowest coinfection area.
Respiratory viruses transmitted between different species and manifests clinical features similar to COVID‐19, which is a potential threat for COVID‐19‐infected cases. 80 , 81 A systematic review and meta‐analysis reported that influenza type A, rhinovirus, and non‐SARS‐CoV‐2 coronaviruses are the most frequent viruses among coinfected patients 82 Another systematic review showed that 11.6% of SARS‐CoV‐2 patients had viral coinfection. 83 Malekifar et al. 84 showed the prevalence of 12.58% viral coinfection among COVID‐19 patients.
Compare to other systematic review studies focused on coinfection question, we found a higher coinfection rate. Our result showed that 20.97%; of patients were infected with at least one bacterial pathogens which is much higher than other studies reported 7%–8% of coinfection prevalence among COVID‐19 patients. 81
The rate of bacterial coinfection prevalence among critically ill patients is one of the important issues during pandemic, which related to higher comorbidity. A meta‐analysis study showed 8.1% of coinfection among critically ill patients compared to 5.9% hospitalized ones. 85 Soltani et al. 86 showed the prevalence of 20.97 bacterial co‐infection in COVID‐19‐infected cases. Another important aspect of bacterial co‐infection prevalence is about empirical bacterial treatment (52). Several research articles concluded that the increasing antibiotic prescription among COVID‐19 cases would lead to antibiotic resistance in the next few years. 87 More than 70% of COVID‐19 cases received some kind of antibiotics agents including fluoroquinolones and third‐generation cephalosporins. 85
We identified 12.60% of fungal coinfection among COVID‐19‐infected individuals, which is also higher than other studies focused on similar question. A systematic review and meta‐analysis showed the prevalence of fungal coinfections and super infection, 4% and 8% respectively 82 another study reported of both the fungal coinfection and super infection 4%. Like other types of pathogens, which mentioned before fungal pathogens have similar laboratory manifestation with other respiratory viruses. This problem could be detrimental when it comes to patients’ clinical care and treatment. 15 For example, there were negative serology and cell culture test for Aspergillus coinfection in COVID‐19 patients. 88 Candida albicans is the most frequent candida species among COVID‐19 patients with critical conditions. 89 Aspergillus is the other frequent invasive fungal pathogens among the patients. 90
4.1. Strength, limitation, and suggestions for future studies
We faced some limitation in our study. One: we could not perform gender‐specific estimation because of primary studies little data;
Two: pooled prevalence in this study were analyzed based on WHO regional office; therefore, we wanted to conduct the spatial analysis in geographic regions, 91 , 92 , 93 , 94 but because of infrequent studies number, we would not sure about robust results. Performing a through‐full study probe search and estimating the different coinfections species pooled prevalence were our study's strengths. Because of increasing rate of pathogens coinfection prevalence in COVID‐19 patients, we suggest that a world registry will be developed in order to screen the pattern of coinfections. 95 , 96
5. CONCLUSION
In conclusion, we identified a higher level of pathogenic microorganism coinfection among COVID‐19 patients. Because of this rate of coinfection, we support the empirical use of antibacterial, antifungal, and antiviral treatment specifically at the onset of the COVID‐19 infection. We also encourage clinician to run diagnostic test for other pathogens simultaneously with SARS‐CoV‐2, which is important to properly patient's treatment.
CONFLICTS OF INTEREST
The authors report no conflicts of interest in this work.
AUTHOR CONTRIBUTIONS
Saber Soltani conceptualized and designed the review, Saber Soltani, Reza Pakzad, Pooneh Malekifar, Zainab Shateri, Milad Zandi and Abbas Farahani contributed to interpretation of data for the work, wrote the article, and final approval of the version to be published. Sara Akhavan Rezayat, Maral Soleymani, Mohammad Reza Karimi, and Seyed Esmaeil Ahmadi collected data and wrote the article. Ramin Shahbahrami, Iraj Pakzad, Mina Mobini Kesheh, Parastoo Hosseini, and Fatemeh Abdi supervised the collection of the data and wrote the article. All authors reviewed and approved the article.
6. ETHICAL APPROVAL
Not applicable.
Supporting information
Fig S1
Fig S2
Fig S3
ACKNOWLEDGMENTS
We are sincerely thankful to Hormozgan University of Medical Sciences and Tehran University of Medical Sciences, Iran (G. no. 4000407).
APPENDIX 1.
Population: COVID‐19 patients.
Intervention: None.
Comparison: None.
Outcome: Prevalence of coinfections.
Time: from December 1, 2019 until August 23, 2021.
Study design: Observational study.
The search strategy is described in Appendix 1 that is applied based on PICOTS for MEDLINE (MeSH) and then used in other databases.1
BOX 1. Search strategy based on PICO for MEDLINE (MeSH, Medical Subject Headings).
1. COVID‐19 [text word] OR COVID‐19 [Mesh term]
2. Coronavirus [text word] OR Coronavirus [Mesh term]
3. SARS‐CoV‐2 infection [text word] OR SARS‐CoV‐2 infection [Mesh term]
4. 1 OR 2 OR 3
5. Prevalence [text word] OR Prevalence [Mesh term]
6. Frequency [text word] OR Frequency [Mesh term]
7. Incidence [text word] OR Incidence [Mesh term]
8. 5 OR 6 OR 7
9. Coinfection [text word] OR Coinfection [Mesh term]
10. Mixed Infection [text word] OR Mixed Infection [Mesh term]
11. Polymicrobial Coinfection [text word] OR Polymicrobial Coinfection [Mesh term]
12. Bacterial Coinfection [text word] OR Bacterial Coinfection [Mesh term]
13. Viral Coinfection [text word] OR Viral Coinfection [Mesh term]
14. Fungal Coinfection [text word] OR Viral Coinfection [Mesh term]
15. 9 OR 10 OR 11 OR 12 OR 13 OR 14
16: 4 AND 8 AND 15
Pakzad R, Malekifar P, Shateri Z, et al. Worldwide prevalence of microbial agents’ coinfection among COVID‐19 patients: A comprehensive updated systematic review and meta‐analysis. J Clin Lab Anal.2022;36:e24151. doi: 10.1002/jcla.24151
Reza Pakzad and Pooneh Malekifar are co‐first (have contributed equally to this work).
Contributor Information
Abbas Farahani, Email: abbasfarahani25@yahoo.com.
Saber Soltani, Email: sabersoltani71@gmail.com, Email: s-soltani@razi.tums.ac.ir.
DATA AVAILABILITY STATEMENT
All data associated with this article are inclusive in this article.
REFERENCES
- 1. Blandenier E, Habibi Z, Kousi T, Sestito P, Flahault A, Rozanova L. Initial COVID‐19 outbreak: an epidemiological and socioeconomic case review of Iran. Int J Environ Res Public Health. 2020;17(24):9593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Güner R, Hasanoğlu İ, Aktaş F. COVID‐19: prevention and control measures in community. Turk J Med Sci. 2020;50(SI‐1):571‐577. doi: 10.3906/sag-2004-146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sharifipour E, Shams S, Esmkhani M, et al. Evaluation of bacterial co‐infections of the respiratory tract in COVID‐19 patients admitted to ICU. BMC Infect Dis. 2020;20(1):1‐7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Singh V, Upadhyay P, Reddy J, Granger J. SARS‐CoV‐2 respiratory co‐infections: incidence of viral and bacterial co‐pathogens. Int J Infect Dis. 2021;105:617‐620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kim D, Quinn J, Pinsky B, Shah NH, Brown I. Rates of co‐infection between SARS‐CoV‐2 and other respiratory pathogens. JAMA. 2020;323(20):2085‐2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gao Y, Chen Y, Liu M, Shi S, Tian J. Impacts of immunosuppression and immunodeficiency on COVID‐19: a systematic review and meta‐analysis. J Infect. 2020;81:e93‐e95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Vincent J‐L, Rello J, Marshall J, et al. International study of the prevalence and outcomes of infection in intensive care units. JAMA. 2009;302(21):2323‐2329. [DOI] [PubMed] [Google Scholar]
- 8. Song G, Liang G, Liu W. Fungal co‐infections associated with global COVID‐19 pandemic: a clinical and diagnostic perspective from China. Mycopathologia. 2020;185:599‐606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gangneux J‐P, Bougnoux M‐E, Dannaoui E, Cornet M, Zahar J. Invasive fungal diseases during COVID‐19: we should be prepared. J Mycol Med. 2020;30(2):100971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Feldman C, Anderson R. The role of co‐infections and secondary infections in patients with COVID‐19. Pneumonia. 2021;13(1):1‐15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rice TW, Rubinson L, Uyeki TM, et al. Critical illness from 2009 pandemic influenza A (H1N1) virus and bacterial co‐infection in the United States. Crit Care Med. 2012;40(5):1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Joseph C, Togawa Y, Shindo N. Bacterial and viral infections associated with influenza. Influenza Other Respir Viruses. 2013;7:105‐113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bengoechea JA, Bamford CG. SARS‐CoV‐2, bacterial co‐infections, and AMR: the deadly trio in COVID‐19? EMBO Mol Med. 2020;12(7):e12560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hsu J. How covid‐19 is accelerating the threat of antimicrobial resistance. BMJ. 2020;369:1–2. [DOI] [PubMed] [Google Scholar]
- 15. Chibabhai V, Duse A, Perovic O, Richards G. Collateral damage of the COVID‐19 pandemic: exacerbation of antimicrobial resistance and disruptions to antimicrobial stewardship programmes? S Afr Med J. 2020;110(7):1‐2. [DOI] [PubMed] [Google Scholar]
- 16. Moher D, Liberati A, Tetzlaff J, Altman DG, Group P . Preferred reporting items for systematic reviews and meta‐analyses: the PRISMA statement. PLoS Med. 2009;6(7):e1000097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wells GA, Shea B, O'Connell D, et al. The Newcastle‐Ottawa Scale (NOS) for Assessing the Quality of Nonrandomised Studies in Meta‐analyses. The Newcastle‐Ottawa Scale (NOS) for assessing the quality of non randomised studies in meta‐analyses. 2000. http://www.ohri.ca/programs/clinical_epidemiology/oxford [Google Scholar]
- 18. Hashemi H, Pakzad R, Yekta A, et al. Global and regional prevalence of age‐related cataract: a comprehensive systematic review and meta‐analysis. Eye. 2020;34(8):1357‐1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hallajzadeh J, Khoramdad M, Izadi N, et al. The association between metabolic syndrome and its components with systemic lupus erythematosus: a comprehensive systematic review and meta‐analysis of observational studies. Lupus. 2018;27(6):899‐912. [DOI] [PubMed] [Google Scholar]
- 20. Hashemi H, Pakzad R, Heydarian S, et al. Global and regional prevalence of strabismus: a comprehensive systematic review and meta‐analysis. Strabismus. 2019;27(2):54‐65. [DOI] [PubMed] [Google Scholar]
- 21. Soltani S, Tabibzadeh A, Zakeri A, et al. COVID‐19 associated central nervous system manifestations, mental and neurological symptoms: a systematic review and meta‐analysis. Rev Neurosci. 2021;32:351‐361. [DOI] [PubMed] [Google Scholar]
- 22. Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta‐analyses. BMJ. 2003;327(7414):557‐560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhu X, Ge Y, Wu T, et al. Co‐infection with respiratory pathogens among COVID‐2019 cases. Virus Res. 2020;285:198005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhang H, Zhang Y, Wu J, et al. Risks and features of secondary infections in severe and critical ill COVID‐19 patients. Emerg Microbes Infect. 2020;9(1):1958‐1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zha L, Shen J, Tefsen B, Wang Y, Lu W, Xu Q. Clinical features and outcomes of adult COVID‐19 patients co‐infected with Mycoplasma pneumoniae . J Infect. 2020;81:e12‐e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yu D, Ininbergs K, Hedman K, Giske CG, Strålin K, Özenci V. Low prevalence of bloodstream infection and high blood culture contamination rates in patients with COVID‐19. PLoS One. 2020;15(11):e0242533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Youngs J, Wyncoll D, Hopkins P, Arnold A, Ball J, Bicanic T. Improving antibiotic stewardship in COVID‐19: bacterial co‐infection is less common than with influenza. J Infect. 2020;81:e55‐e57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wu Q, Xing Y, Shi L, et al. Coinfection and other clinical characteristics of COVID‐19 in children. Pediatrics. 2020;146(1):e20200961. [DOI] [PubMed] [Google Scholar]
- 29. Verroken A, Scohy A, Gérard L, Wittebole X, Collienne C, Laterre P‐F. Co‐infections in COVID‐19 critically ill and antibiotic management: a prospective cohort analysis. Crit Care. 2020;24(1):1‐3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tiwari Pandey A, Pandey I, Zamboni P, et al. Traditional herbal remedies with a multifunctional therapeutic approach as an implication in COVID‐19 associated co‐infections. Coatings. 2020;10(8):761. [Google Scholar]
- 31. Tadolini M, Codecasa LR, García‐García J‐M, et al. Active tuberculosis, sequelae and COVID‐19 co‐infection: first cohort of 49 cases. Eur Respir J. 2020;56(1):2001398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sy KTL, Haw NJL, Uy J. Previous and active tuberculosis increases risk of death and prolongs recovery in patients with COVID‐19. Infect Dis. 2020;52(12):902‐907. [DOI] [PubMed] [Google Scholar]
- 33. Sharov KS. SARS‐CoV‐2‐related pneumonia cases in pneumonia picture in Russia in March‐May 2020: secondary bacterial pneumonia and viral co‐infections. J Glob Health. 2020;10(2):020504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sepulveda J, Westblade LF, Whittier S, et al. Bacteremia and blood culture utilization during COVID‐19 surge in New York City. J Clin Microbiol. 2020;58(8):e00875–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sarınoğlu RC, Sili U, Eryuksel E, Yildizeli SO, Cimsit C, Yagci AK. Tuberculosis and COVID‐19: an overlapping situation during pandemic. J Infect Dev Cntries. 2020;14(07):721‐725. [DOI] [PubMed] [Google Scholar]
- 36. Rothe K, Feihl S, Schneider J, et al. Rates of bacterial co‐infections and antimicrobial use in COVID‐19 patients: a retrospective cohort study in light of antibiotic stewardship. Eur J Clin Microbiol Infect Dis. 2020;40:859‐869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ripa M, Galli L, Poli A, et al. Secondary infections in patients hospitalized with COVID‐19: incidence and predictive factors. Clin Microbiol Infect. 2020;27:451‐457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Porretta AD, Baggiani A, Arzilli G, et al. Increased risk of acquisition of New Delhi metallo‐beta‐lactamase‐producing carbapenem‐resistant Enterobacterales (NDM‐CRE) among a cohort of COVID‐19 patients in a teaching hospital in Tuscany, Italy. Pathogens. 2020;9(8):635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Nori P, Cowman K, Chen V, et al. Bacterial and fungal coinfections in COVID‐19 patients hospitalized during the New York City pandemic surge. Infect Control Hosp Epidemiol. 2021;42(1):84‐88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Motta I, Centis R, D’Ambrosio L, et al. Tuberculosis, COVID‐19 and migrants: preliminary analysis of deaths occurring in 69 patients from two cohorts. Pulmonology. 2020;26(4):233‐240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Massey BW, Jayathilake K, Meltzer HY. Respiratory microbial co‐infection with SARS‐CoV‐2. Front Microbiol. 2020;11:2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ma L, Wang W, Le Grange JM, et al. Coinfection of SARS‐CoV‐2 and other respiratory pathogens. Infect Drug Resist. 2020;13:3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lv Z, Cheng S, Le J, et al. Clinical characteristics and co‐infections of 354 hospitalized patients with COVID‐19 in Wuhan, China: a retrospective cohort study. Microbes Infect. 2020;22(4):195‐199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Liu J, Zeng W, Cao Y, et al. Effect of a previous history of antiretroviral treatment on the clinical picture of patients with co‐infection of SARS‐CoV‐2 and HIV: a preliminary study. Int J Infect Dis. 2020;100:141‐148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kimmig LM, Wu D, Gold M, et al. Il‐6 inhibition in critically ill COVID‐19 patients is associated with increased secondary infections. Front Med. 2020;7:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Karami Z, Knoop BT, Dofferhoff AS, et al. Few bacterial co‐infections but frequent empiric antibiotic use in the early phase of hospitalized patients with COVID‐19: results from a multicentre retrospective cohort study in The Netherlands. Infect Dis. 2021;53(2):102‐110. [DOI] [PubMed] [Google Scholar]
- 47. Intra J, Sarto C, Beck E, Tiberti N, Leoni V, Brambilla P. Bacterial and fungal colonization of the respiratory tract in COVID‐19 patients should not be neglected. Am J Infect Control. 2020;48(9):1130‐1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hughes S, Troise O, Donaldson H, Mughal N, Moore LS. Bacterial and fungal coinfection among hospitalized patients with COVID‐19: a retrospective cohort study in a UK secondary‐care setting. Clin Microbiol Infect. 2020;26(10):1395‐1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hirotsu Y, Maejima M, Shibusawa M, et al. Analysis of Covid‐19 and non‐Covid‐19 viruses, including influenza viruses, to determine the influence of intensive preventive measures in Japan. J Clin Virol. 2020;129:104543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hazra A, Collison M, Pisano J, Kumar M, Oehler C, Ridgway JP. Coinfections with SARS‐CoV‐2 and other respiratory pathogens. Infect Control Hosp Epidemiol. 2020;41(10):1228‐1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Gupta N, Ish P, Gupta A, et al. A profile of a retrospective cohort of 22 patients with COVID‐19 and active/treated tuberculosis. Eur Respir J. 2020;56(5):2003408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Goncalves Mendes Neto A, Lo KB, Wattoo A, et al. Bacterial infections and patterns of antibiotic use in patients with COVID‐19. J Med Virol. 2021;93(3):1489‐1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Gayam V, Konala VM, Naramala S, et al. Presenting characteristics, comorbidities, and outcomes of patients coinfected with COVID‐19 and Mycoplasma pneumoniae in the USA. J Med Virol. 2020;92(10):2181‐2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Garcia‐Vidal C, Sanjuan G, Moreno‐García E, et al. Incidence of co‐infections and superinfections in hospitalized patients with COVID‐19: a retrospective cohort study. Clin Microbiol Infect. 2021;27(1):83‐88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Fu Y, Yang Q, Xu M, (eds) et al. Secondary bacterial infections in critical ill patients of COVID‐19. Open Forum Infect Dis. 2020;7:1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. D’Onofrio V, Van Steenkiste E, Meersman A, et al. Differentiating influenza from COVID‐19 in patients presenting with suspected sepsis. Eur J Clin Microbiol Infect Dis. 2020;40:987‐995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Contou D, Claudinon A, Pajot O, et al. Bacterial and viral co‐infections in patients with severe SARS‐CoV‐2 pneumonia admitted to a French ICU. Ann Intensive Care. 2020;10(1):119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Chauhdary WA, Chong PL, Mani BI, et al. Primary respiratory bacterial coinfections in patients with COVID‐19. Am J Trop Med Hyg. 2020;103(2):917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Blasco ML, Buesa J, Colomina J, et al. Co‐detection of respiratory pathogens in patients hospitalized with Coronavirus viral disease‐2019 pneumonia. J Med Virol. 2020;92(10):1799‐1801. doi: 10.1002/jmv.25922 [DOI] [PubMed] [Google Scholar]
- 60. Chen X, Jiang Q, Ma Z, et al. Clinical characteristics of hospitalized patients with SARS‐CoV‐2 and hepatitis B virus co‐infection. Virol Sin. 2020;35:842‐845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. de Souza Luna LK, Perosa AH, Conte DD, et al. Different patterns of Influenza A and B detected during early stages of COVID‐19 in a university hospital in São Paulo, Brazil. J Infect. 2020;81(2):e104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Ding Q, Lu P, Fan Y, Xia Y, Liu M. The clinical characteristics of pneumonia patients coinfected with 2019 novel coronavirus and influenza virus in Wuhan, China. J Med Virol. 2020;92(9):1549‐1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Ebrahim SH. Lack of MERS‐CoV Co‐infection among Hospitalized COVID‐19 Patients in Saudi Arabia. J Epidemiol Glob Health. 2020;10(3):191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Hashemi SA, Safamanesh S, Ghasemzadeh‐moghaddam H, Ghafouri M, Azimian A. High prevalence of SARS‐CoV‐2 and influenza A virus (H1N1) coinfection in dead patients in Northeastern Iran. J Med Virol. 2021;93(2):1008‐1012. [DOI] [PubMed] [Google Scholar]
- 65. Jiang S, Liu P, Xiong G, et al. Coinfection of SARS‐CoV‐2 and multiple respiratory pathogens in children. Clin Chem Lab Med. 2020;58(7):1160‐1161. [DOI] [PubMed] [Google Scholar]
- 66. Leuzinger K, Roloff T, Gosert R, et al. Epidemiology of severe acute respiratory syndrome coronavirus 2 emergence amidst community‐acquired respiratory viruses. J Infect Dis. 2020;222(8):1270‐1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Lin D, Liu L, Zhang M, et al. Co‐infections of SARS‐CoV‐2 with multiple common respiratory pathogens in infected patients. Sci China Life Sci. 2020;63:606‐609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Lin Y, Yuan J, Long Q, et al. Patients with SARS‐CoV‐2 and HBV co‐infection are at risk of greater liver injury. Genes Dis. 2020;8:484‐492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Ma S, Lai X, Chen Z, Tu S, Qin K. Clinical characteristics of critically ill patients co‐infected with SARS‐CoV‐2 and the influenza virus in Wuhan, China. Int J Infect Dis. 2020;96:683‐687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Nowak MD, Sordillo EM, Gitman MR, Paniz Mondolfi AE. Coinfection in SARS‐CoV‐2 infected patients: where are influenza virus and rhinovirus/enterovirus? J Med Virol. 2020;92(10):1699‐1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Teotônio IMSN, de Carvalho JL, Castro LC, et al. Clinical and biochemical parameters of COVID‐19 patients with prior or active dengue fever. Acta Trop. 2021;214:105782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Vaughn VM, Gandhi T, Petty LA, et al. Empiric antibacterial therapy and community‐onset bacterial co‐infection in patients hospitalized with COVID‐19: a multi‐hospital cohort study. Clin Infect Dis. 2020;72:e533‐e541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Weissberg D, Böni J, Rampini SK, et al. Does respiratory co‐infection facilitate dispersal of SARS‐CoV‐2? investigation of a super‐spreading event in an open‐space office. Antimicrob Resist Infect Control. 2020;9(1):1‐8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Yu R, Tan S, Dan Y, et al. Effect of SARS‐CoV‐2 coinfection was not apparent on the dynamics of chronic hepatitis B infection. Virology. 2021;553:131‐134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Yue H, Zhang M, Xing L, et al. The epidemiology and clinical characteristics of co‐infection of SARS‐CoV‐2 and influenza viruses in patients during COVID‐19 outbreak. J Med Virol. 2020;92(11):2870‐2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Zheng X, Wang H, Su Z, et al. Co‐infection of SARS‐CoV‐2 and Influenza virus in early stage of the COVID‐19 epidemic in Wuhan, China. J Infect. 2020;81:e128‐e129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Agrifoglio A, Cachafeiro L, Figueira J, Añón J, de Lorenzo AG. Critically ill patients with COVID‐19 and candidaemia: we must keep this in mind. J Mycol Med. 2020;30(4):101012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Chowdhary A, Tarai B, Singh A, Sharma A. Multidrug‐resistant Candida auris infections in critically Ill coronavirus disease patients, India, April–July 2020. Emerg Infect Dis. 2020;26(11):2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Segrelles‐Calvo G, Araújo GR, Llopis‐Pastor E, et al. Prevalence of opportunistic invasive aspergillosis in COVID‐19 patients with severe pneumonia. Mycoses. 2021;64(2):144‐151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Htar MTT, Yerramalla M, Moïsi J, Swerdlow D. The burden of respiratory syncytial virus in adults: a systematic review and meta‐analysis. Epidemiol Infect. 2020;148:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Lansbury L, Lim B, Baskaran V, Lim WS. Co‐infections in people with COVID‐19: a systematic review and meta‐analysis. J Infect. 2020;81(2):266‐275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Musuuza JS, Watson L, Parmasad V, Putman‐Buehler N, Christensen L, Safdar N. Prevalence and outcomes of co‐infection and superinfection with SARS‐CoV‐2 and other pathogens: a systematic review and meta‐analysis. PLoS One. 2021;16(5):e0251170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Davis B, Rothrock AN, Swetland S, Andris H, Davis P, Rothrock SG. Viral and atypical respiratory co‐infections in COVID‐19: a systematic review and meta‐analysis. J Am Coll Emerg Physicians Open. 2020;1(4):533‐548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Malekifar P, Pakzad R, Shahbahrami R, et al. Viral coinfection among COVID‐19 PATIENT groups: an update systematic review and meta‐analysis. Biomed Res Int. 2021;2021:1‐10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Langford BJ, So M, Raybardhan S, et al. Bacterial co‐infection and secondary infection in patients with COVID‐19: a living rapid review and meta‐analysis. Clin Microbiol Infect. 2020;26:1622‐1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Soltani S, Faramarzi S, Zandi M, et al. Bacterial co‐infection among COVID‐19 patient groups: an update systematic review and Meta‐analysis. New Microbes New Infect. 2021;43:100910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Goel N, Ahmad R, Fatima H, Khare SK. New threatening of SARS‐CoV‐2 co‐infection and strategies to fight the current pandemic. Med Drug Discov. 2021;10:100089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Blaize M, Mayaux J, Nabet C, et al. Fatal invasive aspergillosis and coronavirus disease in an immunocompetent patient. Emerg Infect Dis. 2020;26(7):1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Arastehfar A, Carvalho A, Nguyen MH, et al. COVID‐19‐associated candidiasis (CAC): an underestimated complication in the absence of immunological predispositions? J Fungi. 2020;6(4):211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. White L, Dhillon R, Cordey A, et al. A national strategy to diagnose COVID‐19 associated invasive fungal disease in the ICU. SSRN Electron J. 2020;e1634–e1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Mahdavifar N, Pakzad R, Ghoncheh M, Pakzad I, Moudi A, Salehiniya H. Spatial analysis of breast cancer incidence in Iran. Asian Pac J Cancer Prev. 2016;17:59‐64. [DOI] [PubMed] [Google Scholar]
- 92. Pakzad R, Moudi A, Pournamdar Z, et al. Spatial analysis of colorectal cancer in Iran. Asian Pac J Cancer Prev. 2016;17:53‐57. [DOI] [PubMed] [Google Scholar]
- 93. Pakzad R, Ghoncheh M, Pournamdar Z, et al. Spatial analysis of skin cancer incidence in Iran. Asian Pac J Cancer Prev. 2016;17:33‐37. [DOI] [PubMed] [Google Scholar]
- 94. Pakzad R, Khani Y, Pakzad I, et al. Spatial analysis of stomach cancer incidence in Iran. Asian Pac J Cancer Prev. 2016;17:27‐32. [DOI] [PubMed] [Google Scholar]
- 95. Kazemi‐Arpanahi H, Moulaei K, Shanbehzadeh M. Design and development of a web‐based registry for Coronavirus (COVID‐19) disease. Med J Islam Repub Iran. 2020;34:68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Shanbehzadeh M, Nopour R, kazemi‐arpanahi H. Determination of the most important diagnostic criteria for COVID‐19: a step forward to design an intelligent clinical decision support system. J Adv in Med Biomed Res. 2021;29(134):176‐182. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig S1
Fig S2
Fig S3
Data Availability Statement
All data associated with this article are inclusive in this article.


