Abstract
Background
Various nucleic acid amplification assays for the diagnosis of SARS‐CoV‐2 infection have been developed, and there is a need to assess their test performance relative to one another. The aim of this study was to compare the performance characteristics of the Biosewoom Real‐Q 2019‐nCoV assay targeting the E and RdRP genes to DaAn Gene 2019‐nCoV kit targeting the N gene and ORF1ab in the diagnosis of SARS‐CoV‐2.
Methods
We performed a diagnostic comparison study by testing nasopharyngeal samples for SARS‐CoV‐2 using the two reverse transcription polymerase chain reaction (RT‐PCR) assays. Assay agreement was assessed by overall percent agreement, negative percent agreement, positive percent agreement, and Cohen's kappa coefficient.
Results
A total of 48 nasopharyngeal samples were tested using the two assays. One sample was invalid, and three showed inconclusive results with Real‐Q; hence, 44 were included for the comparative analysis. Overall, percent agreement between the assays was 93.2% (95% CI 81.3%–98.6%), Positive percent agreement (PPA) was 86.4% (95% CI 65.1%–97.1%) and negative percent agreement (NPA) was 100% (95% CI 84.6%–100%). The kappa coefficient was 0.86 (95% CI 0.72–1.01). Three samples (6.8%) were positive with DaAn gene kit and negative with Real‐Q. The fluorescence intensity for Real‐Q reporter dyes was low.
Conclusion
The two kits showed high levels of concordance in their detection of SARS‐CoV‐2 despite having different gene targets. The Biosewoom kit can be improved through addressing the fluorescence intensity of the target dyes, and feedback was given to the manufacturer.
Keywords: coronavirus, COVID‐19, PCR, SARS‐CoV‐2, zimbabwe
Our study compared the analytical performance characteristics of the Real‐Q (BioSewoom) and Da An Gene assays for the detection of SARS‐CoV‐2 from nasopharyngeal swab samples. A total of 48 nasopharyngeal samples were tested using the two assays. The results suggests that the two kits have an overall percent agreement of 93.2%, positive percent agreement of 83.4% and negative percent agreement of 100%.

1. INTRODUCTION
The COVID‐19 pandemic, which started in Wuhan, China, is caused by the virus SARS‐CoV‐2, which is an enveloped single stranded RNA virus. 1 The COVID‐19 is a highly infectious disease, and after reporting the first case in China, it has rapidly covered the entire world and affected the life of millions of people. Due to the occurrence of many cases worldwide, on January 30th, 2020, the World Health Organization (WHO) declared a Public Health Emergency of International Concern. 2 Over 15 million cases have been reported globally with approximately 620,000 deaths as of July 22, 2020. 3
In Zimbabwe, the virus has spread to most parts of the country, and at present, there are 1820 cases and 26 deaths reported by the Ministry of Health and Child Care. 4 The virus is mainly associated with respiratory tract infections although infections in the gastrointestinal tract have been recorded. Most people infected with the COVID‐19 virus experience mild to moderate respiratory illness and recover without requiring special treatment. 5 Older people, and those with underlying medical problems like cardiovascular disease, diabetes, chronic respiratory disease, and cancer are more likely to develop serious illness. 5
Transmission of SARS‐CoV‐2 can occur through direct, indirect, or close contact with infected people through infected secretions such as saliva and respiratory secretions or their respiratory droplets. 6 Evidence suggests that SARS‐CoV‐2 RNA can be detected in people 1–3 days before their symptom onset, with the highest viral loads, as measured by reverse transcriptase polymerase chain reaction (RT‐PCR), observed around the day of symptom onset, followed by a gradual decline over time. 6 The duration of RT‐PCR positivity generally appears to be 1–2 weeks for asymptomatic persons, and up to 3 weeks or more for patients with mild‐to‐moderate disease. 7 In patients with severe COVID‐19 disease, it can be much longer. 7
In the clinical management and outbreak control of the COVID‐19 disease, rapid collection and laboratory testing of appropriate specimen from patients are very critical. The diagnosis of SARS‐CoV‐2 involves use of assays that detect viral nucleic acid or antigen from samples from the respiratory system (such as nasal swabs). Currently, RT‐PCR is the most suitable laboratory diagnostic test for COVID‐19 as other laboratory methods (viral culture, antigen testing, and serology) have various limitations. 8 , 9 , 10
To date, various commercial kits for the detection of SARS‐CoV‐2 using RT‐PCR have been developed. The PCR targets of the available kits include S, RdRP, N, and E genes as well as functionally important open reading frames 1a and 1b (ORF1ab). Most kits are multiplexed to detect at least two genes. 11 There is a need to understand the relative test performance of these assays to guide laboratories’ selection of assays and understand advantages and disadvantages of methods that may be used in parallel. In this study, the performance characteristics of the Biosewoom Real‐Q 2019‐nCoV RT‐PCR kit (E and RdRP genes) for the detection of SARS‐CoV‐2 was compared to those of DaAn Gene 2019‐nCoV kit (N gene and ORF1ab), which has been in use in Zimbabwe.
2. MATERIAL AND METHODS
2.1. Study subjects, ethical approvals, and sample collection
This was an operational laboratory‐based cross‐sectional study, where the Real‐Q 2019 nCoV (Biosewoom, Korea) kit was being evaluated before being adopted for testing. The DaAn gene kit, which has been use in Zimbabwe, was used as the non‐reference standard. Nasopharyngeal samples collected in viral transport medium (VTM) for SARS‐CoV‐2 testing were used.
2.2. DNA Extraction
Total nucleic acid from the nasopharyngeal swabs in viral transport medium (VTM) was extracted using a DaAn Gene nucleic acid extraction kit (DaAn Gene Co, Ltd., of Sun Yat‐sen University, China) as per manufacturer's instructions. The extracted DNA was stored at −20°C awaiting SARS‐CoV‐2 RT‐PCR.
2.3. SARS‐CoV‐2 RNA amplification by DaAn Gene 2019‐nCoV detection kit
The master mix was prepared by mixing 17 µl of NC (ORF1ab/N) PCR liquid A (reaction mix) and 3 µl of NC (ORF1ab/N) PCR reaction liquid B (enzyme). 5 µl of the extracted sample was added to make the PCRs final volume of 25 µl in a PCR plate on ice. The PCR tubes were immediately transferred to an ABI 7500 RT‐PCR machine (Applied Biosystems). The probe detection modes were set as: ORF1ab: VIC, Quencher: NONE, N‐Gene: FAM, Quencher: NONE, Internal Control: Cy5, Quencher: NONE, Passive reference: NONE. The PCR cycle was carried out on the following conditions: 1 cycle of 15 min at 50°C, 1 cycle of 15 min at 95°C, and 45 cycles of 94°C for 15 s and 55°C for 45 s (data collection).
2.4. SARS‐CoV‐2 RNA amplification by Real‐Q 2019‐nCoV detection kit
The master mix was prepared by mixing 12.5 µl of 2X PCR mixture, 0.07 µl of ROX Reference Dye, 3 µl of nCoV probe and primer mixture, 3 µl of RT‐PCR enzyme, and 3, 43 µl of water. 5 µl of the extracted sample was added to the master mixture to make the PCRs final volume of 25 µl in a PCR plate on ice. The PCR tubes were immediately transferred to an ABI 7500 RT‐PCR machine (Applied Biosystems). The probe detection modes were set as: Beta nCov E gene: VIC, Quencher: NONE, SARS‐CoV‐2 RdRP Gene: FAM, Quencher: NONE, Internal Control: Cy5, Quencher: NONE, Passive reference: NONE. The PCR cycle was carried out on the following conditions: 1 cycle of 30 min at 50°C, 1 cycle of 15 min at 95°C, and 40 cycles of 95°C for 15 s and 62°C for 45 s (data collection).
2.5. Data analysis
The results from PCR were recorded onto Excel Spreadsheet. Overall percent agreement, positive percent agreement (PPA), negative percent agreement (NPA), and associated 95% confidence intervals (CI) were performed with the DaAn gene kit serving as the non‐reference standard method. Cohen's kappa coefficient (κ) of qualitative results (detected/non‐detected) between the two assays with 95% CI was also calculated. The results were analyzed using the recommendations from FDA’s statistical guidance on reporting results from studies evaluating diagnostic tests handbook.
2.6. Permission
The permission to carry out the comparison using the remnant samples was granted by the National Microbiology Reference Laboratory as the study was used as the in‐house evaluation of the Real‐Q kit for adoption in national testing.
2.7. Ethical approval
All experiments were examined and approved by the appropriate Ethics Committee and have therefore been performed in accordance with the ethical standards laid down in the 1964 Declaration of Helsinki.
3. RESULTS
Forty‐eight nasopharyngeal swab samples were run using both methods for comparison and the positive and negative controls for each kit were valid. The DaAn gene kit interpreted 52.1% (25/48) specimens as detected for SARS‐CoV‐2 while the Real‐Q interpreted 39.6% (19/48) as detected. One sample (2%) was invalid with Real‐Q due to non‐amplification of internal control. Three samples (6%) were inconclusive using Real‐Q 2019‐nCoV‐2 according to the manufacturer's interpretation (two being RdRP positive, E gene negative, and one having invalid E gene and valid RdRP gene Ct values). Thus, four samples were excluded and forty‐four samples used for comparative analysis (Table 1).
TABLE 1.
Comparison of the Real‐Q (BioSewoom) and DaAn Gene assays for the detection of SARS‐CoV‐2
| Real‐Q 2019‐nCoV−2 (BioSewoom) (New Kit) | DaAn Gene SARS‐CoV2 (Non Reference Standard) | Total | |
|---|---|---|---|
| Positive | Negative | ||
| Positive | a (19) | b (0) | 19 |
| Negative | c (3) | d (22) | 25 |
| Total | a+c (22) | b+d (22) | 44 |
|
Positive Percent Agreement (PPA) = 100% × a/(a+c) = 100% × 19/(19+3) = 86.4% Negative Percent Agreement (NPA) = 100% × d/(b+d) = 100% × 22/(22+0) = 100% Overall Percent Agreement (OPA) = 100% × (a+d)/(a+b+c+d) = 19+22/(19+0+3+22) = 93.2% | |||
The detected samples included a wide range of quantification cycle (Cq)) values. DaAn gene had a Cq value median of 33 (Q1°32; Q3 = 34.1) for the N gene and 34 (Q1 = 33; Q3 = 36) for the ORF1ab. Real‐Q 2019‐nCoV‐2 had a Cq value median of 35.9 (Q1 = 33.3; Q3 = 36.5) for the E gene and 34.4 (Q1 = 32.2; Q3 = 35.5) for the RdRP gene. The overall percent agreement between the two assays was 93.2% (95% CI 81.3%–98.6%). The PPA was 86.4% (95% CI 65.1%–97.1%), and the NPA was 100% (95% CI 84.6%–100%). The kappa coefficient was 0.86 (95% CI 0.72–1.01), indicating an almost perfect agreement.
During RT‐PCR, fluorescent reporter molecule in each PCR produces a fluorescent signal with increasing intensity as the amount of PCR amplicon increases. Thus, in as much as the results from the two kits showed a high concordance rate, the results from the Real‐Q 2019‐nCoV‐2 (Biosewoom) kit showed that the dyes VIC (E gene) and Cy5 (Internal control) produced a low fluorescence intensity and fast reaching the plateau. When the threshold would be set to the values set by the manufacturer, most of the known positive samples would come out as negative. However, the samples would be positive when the threshold would be set to be automatically calculated by the machine, though the change in frequency (fluorescence intensity) remaining low. Figure 1 shows the amplification plots from the same sample processed with Real‐Q 2019‐nCoV‐2 (Biosewoom) and DaAn Gene kit, respectively.
FIGURE 1.
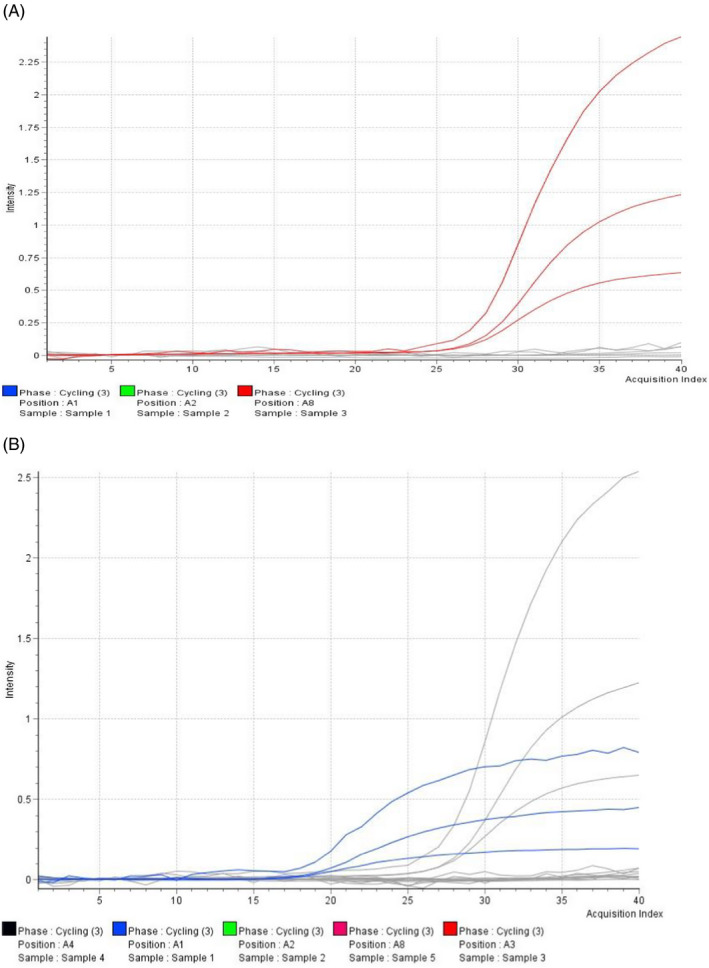
Appearance of the amplification plot on the same sample processed with (A) DaAn Gene 2019nCoV (red graph) and (B) Real‐Q 2019‐nCoV (blue graph) detection PCR kits
4. DISCUSSION
The global challenge posed by the SARS‐CoV‐2 virus has brought more challenges to the resource limited settings including Africa. Despite the efforts being made to set up molecular diagnostic laboratories, there is also the need to procure high‐quality diagnostic test kits. There are several kits that have managed to reach Zimbabwe, and hence, comparison of these kits is essential before they are adopted for use. With the increase in global demand for SARS‐CoV‐2 testing, there is a high likelihood of shortages in the preferred PCR testing kits and hence the need for switching to alternative PCR test kits. 12 In this operational research, the Real‐Q 2019‐nCoV‐2 (New test) was compared to the DaAn Gene PCR kit (Non‐Reference Standard) to determine their qualitative performance in the detection of SARS‐CoV‐2. DaAn gene kit was the best performing PCR kit, being the only kit with a 100% detection rate in level 4, lowest limit of detection of 500 copies/ml, and a positive detection rate of 100% when the kit was compared with other commonly used SARS‐CoV‐2 test kits in China. 13
All the used samples were properly extracted as evidence by the internal control Cq values being within range in both methods. The BioSewoom Real‐Q 2019‐nCoV Detection Kit gave similar results with the DaAn Gene detection kit on the samples used. There was 86.4% agreement in the analysis of SARS‐CoV‐2‐positive samples, which is to a greater extent reliable in testing and getting results for public health decisions under the current disaster situation. Other kits, which showed a similar agreement with DaAn gene, include BioPerfectus. 14 There was a 100% agreement in the negative samples between the two methods hence high likelihood that false positives will not occur. The overall percent agreement was 93.2% between the two methods; hence, a shift from one kit to the next has a 0.932 probability of having the same impact in detection and overall reporting of SARS‐CoV‐2 cases. However, the noted reduced fluorescence level with the BioSewoom kit may pose as a challenge in the interpretation of results from the ABI7500 analyzer. The graphs showed reduced level of fluorescence and hence not very clear sigmoid curves, which would need an experienced molecular scientist to identify. The most likely cause of this phenomenon from the literature is early primer depletion. 15 Feedback was given to the distributor for conveying to the manufacturer. The use of Cq value tables can however reduce the subjectivity associated with the prematurely declining amplification curves.
CONFLICT OF INTERESTS
The authors have declared that no competing interests exist.
AUTHOR CONTRIBUTIONS
TM and PC were responsible for the laboratory work and data acquisition and data analysis. TM, PC, DB, PM, TC, and CM participated in writing and review of the manuscript. All authors read and approved the final manuscript.
ACKNOWLEDGMENTS
We would like to acknowledge the African Institute of Biomedical Sciences (AiBST) for availing the equipment and infrastructure for laboratory work.
Marembo T, Chimbunde P, Chipendo T, Munemo C, Manangazira P, Bangure D. Comparison of Real‐Q 2019‐nCoV and DaAn Gene 2019‐nCoV polymerase chain reaction assays for the detection of SARS‐CoV‐2. J Clin Lab Anal.2022;36:e24161. doi: 10.1002/jcla.24161
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- 1. Sabino‐Silva R, Jardim ACG, Siqueira WL. Coronavirus COVID‐19 impacts to dentistry and potential salivary diagnosis. Clin Oral Invest. 2020;24(4):1619‐1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. COVID‐19 public health emergency of international concern (PHEIC) Global research and innovation forum [Internet]. [cited 2020 Jul 22]. Available from: https://www.who.int/publications/m/item/covid‐19‐public‐health‐emergency‐of‐international‐concern‐(pheic)‐global‐research‐and‐innovation‐forum
- 3. Coronavirus update (Live): 15,153,194 Cases and 621,153 deaths from COVID‐19 virus pandemic ‐ worldometer [Internet]. [cited 2020 Jul 22]. Available from: https://www.worldometers.info/coronavirus/?utm_campaign=homeAdvegas1?
- 4. Ministry of health and child care ‐ latest updates [Internet]. [cited 2020 Jul 22]. Available from: http://www.mohcc.gov.zw/
- 5. Rabaan AA, Al‐Ahmed SH, Haque S, et al. SARS‐CoV‐2, SARS‐CoV, and MERS‐COV: a comparative overview. Infez Med. 2020;28(2):174‐184. [PubMed] [Google Scholar]
- 6. Transmission of SARS‐CoV‐2: implications for infection prevention precautions [Internet]. [cited 2020 Jul 22]. Available from: https://www.who.int/news‐room/commentaries/detail/transmission‐of‐sars‐cov‐2‐implications‐for‐infection‐prevention‐precautions
- 7. Pan Y, Zhang D, Yang P, Poon LLM, Wang Q. Viral load of SARS‐CoV‐2 in clinical samples. Lancet Infect Dis. 2020;20(4):411‐412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chan JFW, Yip CCY, To KKW, et al. Improved molecular diagnosis of COVID‐19 by the novel, highly sensitive and specific COVID‐19‐RdRp/Hel real‐time reverse transcription‐PCR assay validated in vitro and with clinical specimens. J Clin Microbiol. 2020;58(5):e00310–e00320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Loeffelholz MJ, Tang YW. Laboratory diagnosis of emerging human coronavirus infections ‐ the state of the art. Emerg Microbes Infect. 2020;9(1):747‐756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pang J, Wang MX, Ang IYH, et al. Potential rapid diagnostics, vaccine and therapeutics for 2019 novel coronavirus (2019‐nCoV): a systematic review. J Clin Med. 2020;9(3):623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Iglói Z, leven M, Abdel‐Karem Abou‐Nouar Z, et al. Comparison of commercial realtime reverse transcription PCR assays for the detection of SARS‐CoV‐2. J Clin Virol. 2020;129:104510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fomsgaard AS, Rosenstierne MW. An alternative workflow for molecular detection of SARS‐CoV‐2 – escape from the NA extraction kit‐shortage, Copenhagen, Denmark, March 2020. Euro Surveill. 2020;25(14):2000398. doi: 10.2807/1560-7917.ES.2020.25.14.2000398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wang M, Chen D, Wu W, et al. Analytical performance evaluation of five RT‐PCR kits for severe acute respiratory syndrome coronavirus 2. J Clin Lab Anal. 2021;35(1):e23643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhuang QZ, Li ZZ, Chao Y, et al. Comparative performance of four nucleic acid amplification tests for SARS‐CoV‐2 virus. Clin Lab. 2021;67(7):201025. [DOI] [PubMed] [Google Scholar]
- 15. Jansson L, Hedman J. Challenging the proposed causes of the PCR plateau phase. Biomol Detect Quantif. 2019;17:100082. doi: 10.1016/j.bdq.2019.100082 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


