Summary
Background:
Incidence and mortality of gastric cancer are high in Mongolia despite Helicobacter pylori in the Mongolian population being less virulent.
Aim:
We aimed to evaluate gastric bacterial microbiota profiles in patients with gastric cancer and its precursor histological conditions.
Methods:
We conducted a case-control study among 48 gastric cancer and 120 non-cancer patients (20 normal gastric mucosa [control], 20 gastritis, 40 with atrophy, and 40 intestinal metaplasia [IM]). We performed 16S rRNA gene amplicon sequencing and compared taxonomic and functional prediction profiles based on the diagnosis group and H. pylori infection status.
Results:
The highest overall bacterial alpha diversity metrics were observed in the control group, followed by the IM and cancer groups. The gastritis and atrophy groups had the least diversity. Lactobacilli and Enterococci were the dominant genus in several cancer patients especially in the absence of H. pylori. In addition, Carnobacterium, Glutamicibacter, Paeniglutamicibacter, Fusobacterium, and Parvimonas were associated with gastric cancer regardless of H. pylori infection. Firmicutes were decreased in the gastritis and atrophy groups and increased in the IM and cancer groups. The functional metabolic activity of the Embden-Meyerhof-Parnas pathway and the utilization of sugar, were significantly increased in cancer group compared with the non-cancer group.
Conclusion:
Microbial factors other than H pylori may play a role in Mongolian gastric cancer. We identified novel associations between gastric cancer and the genera Enterococcus, Lactobacillus, Carnobacterium, Glutamicibacter, Paeniglutamicibacter, Fusobacterium, and Parvimonas.
1. INTRODUCTION
Gastric cancer (GC) is the fifth most common cancer and the third leading cause of cancer death globally (GLOBOCAN 2018; http://gco.iarc.fr/). Mongolia has the highest GC mortality in the world, with an age-standardized mortality rate of 25 per 100,000 in 2018. Epidemiological studies showed that 74–89% of GC patients were infected with H. pylori globally.1 2 By contrast, our previous study showed that only 57.4% of GC patients in Mongolia were infected with H. pylori, which was lower than the non-cancer control group (74.1%).3 While it is well-known that H. pylori disappear in cases of severe gastric atrophy and IM, our previous study showed that atrophy and intestinal metaplasia (IM) were significantly milder among Mongolian patients compared with Japanese patients.4 In addition, we reported that virulent strains with East-Asian-type CagA were rare in Mongolia.4 These data indicate that non-H. pylori factors could be involved in the development of GC in Mongolia.
These data warrant evaluating gastric microbiota beyond determining H. pylori status in high GC risk regions such as Mongolia. Using a 16S rDNA amplicon sequencing approach, Yang et al. examined the gastric mucosal microbiota among dyspeptic individuals in two Colombian cities where GC incidence was 25-fold different despite similar background H. pylori prevalence. They found regionally specific microbiota profiles,5 which raises the possibility that different gastric microbiota can affect the development of GC. A recent study among 63 healthy volunteers using fecal samples revealed that the Mongolian gut microbiome was unique and more diverse compared with that from Han Chinese participants, potentially reflecting the different environmental conditions of the subjects.6 However, to our knowledge, there are no published studies describing the gastric microbiota of a Mongolian population except for one study from inner Mongolian population in China, but the population is mixed with Chinese population.7 Therefore, we investigated the bacterial microbiota profile of gastric mucosa in Mongolian patients with GC, GC precursor diseases (atrophy and IM), and gastritis and normal controls.
2. MATERIALS AND METHODS
2.1. Study population
We conducted a cross-sectional study of patients prospectively recruited between November 2014 and August 2016 from different areas in Mongolia, Ulaanbaatar (capital city), Uvs Province (Western Mongolia), Khuvsgul Province (Northern), Umnugovi Province (Southern), and Khentii Province (Eastern). Study subjects were volunteers age ≥18 year with dyspepsia who were recruited through community-based advertisement. In addition, patients with GC were enrolled from the National Cancer Center hospital in Ulaanbaatar between August 2015 to August 2016. We excluded patients with recent antibiotic or proton pump inhibitor use, history of gastrectomy, reflux esophagitis (GERD grades A–D), or gastroduodenal ulcers. Experienced endoscopists performed all endoscopic examinations and obtained biopsy specimens. Written informed consent was obtained from all participants, and this study was approved by the Mongolian Ministry of Health, Mongolian National University of Medical Sciences (Mongolia), and Oita University Faculty of Medicine (Japan).
2.2. Histological evaluation
Biopsies for histology from non-GC participants were taken from the angulus corpus-antrum junction, specifically the greater curvatures the corpus (8–10 cm from the esophagogastric junction) and antrum (approximately 2 cm from pyloric ring).8 All biopsy specimens were fixed in 10% buffered formalin and embedded in paraffin. Serial sections were stained with hematoxylin and eosin and May-Giemsa stains and then examined by an experienced pathologist (TU). The polymorphonuclear cell (PMN) infiltration, mononuclear cell (MN) infiltration, atrophy, and IM scores were classified into four grades: 0 “normal”, 1 “mild”, 2 “moderate”, and 3 “marked” based on the updated Sydney system.9 Scores ≥ 1 were considered to be positive. Based on histological diagnosis, we assembled two comparison groups (GC vs non-cancer) to find cancer-specific microbial candidates. The non-cancer group was further subdivided into four subgroups (normal, gastritis, atrophy, and IM) to understand how stomach microbiota change along the normal-to-cancer cascade. We defined the normal group as all parameters (PMN, MN, atrophy, and IM) being grade 0; the gastritis group as having only a PMN or MN ≥ grade 1; the atrophy group as having atrophy scores ≥1 without IM, regardless of PMN or MN scores; and the IM group where IM scores were ≥ 1 regardless of the remaining parameter scores. The GC group was defined as having GC confirmed by histological evaluation regardless of other histological scores. Among GC patients, biopsies were collected from cancer tissue (for histology to confirm cancer diagnosis) and non-cancer tissue (from the antrum for microbiota study, rapid urease test, H. pylori culture, and histology diagnosis; from the angulus and corpus for histology). We also evaluated the atrophy grade by Operative Links of Gastric Atrophy (OLGA) 10 and IM grade by Operative Links of Gastric Intestinal Metaplasia (OLGIM). 11
2.3. Determination of H. pylori subtype using immunohistochemistry (IHC)
The histological determination of H. pylori infection was performed for H. pylori, CagA, and East-Asian-type CagA (α-EAS) using IHC as previously described.12 Briefly, following antigen retrieval and inactivation of endogenous peroxidase activity, samples were incubated with anti-H. pylori antibody (DAKO, Denmark), anti-CagA antibody (b-300 Santa Cruz, USA), or α-EAS antibody at 4°C for overnight, and followed by biotinylated goat anti-rabbit or anti-rat IgG (Nichirei Co., Japan).
2.4. DNA extraction and 16S rRNA gene amplicon sequencing
The biopsy materials for DNA extraction were kept in Allprotect Tissue Reagent buffer (KIAGEN Cat No./ID: 76405). Biopsy specimen for DNA were immediately stabilized in the buffer and kept –20°C until transporting to Japan, and then kept at –80°C until we extracted the DNA.
Gastric mucosal specimens were homogenized with 1 mL of phosphate-buffered saline. DNA was extracted using the DNeasy Blood & Tissue Kit (QIAGEN, Hilden, Germany) and concentrated using a DNA Clean & Concentrator (Zymo Research, CA, USA). We followed Illumina’s 16S rRNA gene DNA library preparation protocol (https://support.illumina.com/content/dam/illuminasupport/documents/documentation/chemistry_documentation/16s/16s-metagenomic-library-prep-guide-15044223-b.pdf). The 16S rRNA gene was amplified using primers 341F 5’-CCTACGGGNGGCWGCAG-3’ and 805R 5’-GACTACNVGGGTATCTAATCC-3’ targeting the V3–V4 hypervariable regions13 and sequenced using the Illumina MiSeq platform according to manufacturer’s instructions. PCR amplification was performed using KAPA HiFi HotStart ReadyMix (KAPA Biosystems Inc., Wilmington, MA, USA). Samples underwent denaturation at 95°C for 3 minutes; 35 cycles at 95°C for 30 seconds, 55°C for 30 seconds, and 72°C for 30 seconds; and a final elongation at 72°C for 5 minutes. Nextera XT Index kit (Illumina Inc., San Diego, CA) was added, followed by an additional eight cycles of PCR. Amplicons were purified using Agencourt AMPure XP magnetic beads (Beckman Coulter, Brea, CA, USA). We used a bioanalyzer MCE-202 MultiNA system (Shimadzu, Kyoto, Japan) and QuantFluor dsDNA system (Promega Corporation, Madison WI, USA) to validate the DNA library. Finally, the pooled 5 pM DNA library was denatured with 0.2 N NaOH and mixed with PhiX Control v3 (Illumina Inc., San Diego, CA) at 15% of the final concentration as described in the Illumina procedure. Paired-end sequencing were conducted using a next-generation sequencer MiSeq platform (Illumina Inc.) with MiSeq Reagent Kit version 3 (2×300 bp Paired-End Reads, Illumina Inc.).
2.5. Sequence curation and analysis
Trimming and quality filtering of the 16S rRNA gene sequence data were performed using the CLC genomic workbench 8.5.1. Operational Taxonomic Units (OTUs) with relative abundance (RA) ≤0.05% were removed. Sequences were matched to the SILVA v128 reference database. The amplicon sequencing taxonomic and statistical analysis were performed using web-interfaced Calypso software version 8.72 (http://cgenome.net/calypso/14) and R software.
Alpha diversity was determined based on the Simpson’s index, which is assessed using an ANOVA test. Microbial diversity was visualized using discriminant analysis of principal components (DAPC) based on diagnosis group. Taxonomic discovery analysis was performed using linear discriminant analysis effect size (LEfSe),15 and the Wilcoxon test was used for biomarker discovery. Identification of co-occurring and mutual exclusive bacteria were checked using network analysis. All analyses considered a p value <0.05 to be statistically significant. P values were adjusted using false discovery rate (FDR), Bonferroni, or area under curve correction. All figures contained statistical values based on 2×2 comparisons, except the multivariate analysis and the overview of RA. For network analysis, only FDRs < 0.05 are presented as colored lines (yellow, positive associations; blue negative associations). It showed the identification of mutual exclusive bacteria and clusters of co-occurring bacteria. Taxa are represented as nodes, taxa abundance as node size, and edges depict positive and negative associations. Nodes are coloured by the genus of the represented bacterial taxon or based on their association to environmental variables. Networks are generated by first computing associations between taxa using Pearson’s correlation. The resulting pairwise correlations are used to ordinate nodes in a two dimensional plot by PCoA. Correlating nodes are placed in close proximity and anti-correlating nodes are placed at distant locations. Nodes of correlating taxa are connected by edges.
2.6. Functional metagenome predictions
Reconstruction of the metagenome was performed using the amplicon sequencing approach in Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt; http://www.microbiomeanalyst.ca).16 Annotated KEGG Orthology (KO) ID tables were further annotated based on Functional Ontology Assignments for Metagenomes (FOAM) using the FuncTree 2 automatic annotation server (https://bioviz.tokyo/functree2).17 The statistical differences between diagnosis group were examined in the different FOAM Brite levels.
3. RESULTS
3.1. Patient demographics
We initially obtained samples from 259 patients who met the inclusion criteria; 64 were excluded due to inadequate biopsy specimens for histological diagnosis or failed DNA extraction, leaving 196 patients (48 in the GC group, 148 in the non-cancer group). The non-cancer patients included 22 with normal mucosa, 20 with gastritis, 66 with atrophy, and 40 with IM. We analyzed all patients with GC, gastritis, and IM, and randomly chose 40 patients with atrophy and 20 patients with normal mucosa (Supplementary figure 1). The overall mean age was 46.4 years old (standard deviation [SD] 16.4) and 70% were male. There were no significant age or gender differences among groups. The tumor was located in the lower, middle and upper part of stomach in 23.9%, 27.1% and 49% of cases, respectively. The histological cancer classification was diffuse (47.9%), intestinal (47.9%) or mixed (4.2%). The background gastric mucosa was atrophic in 43.8% and had IM in 50% of cases.
3.2. Detection of OTUs
We identified a total of 4466 unique OTUs among all samples. Among these, 137 OTUs were filtered based on OTU abundance. The minimum read count was 1850 and maximum read was 284,332 across the dataset. Helicobacter was the dominant genus in most samples, and its RA reached up to 99.9% at the genus level in some cases. Helicobacter RA was highest in the gastritis subgroup (mean ± SD, 69.7±36.6%), followed by the atrophy subgroup (74.3±32.8%), IM subgroup (32.2±38.8%), GC group (26.7±37.5%), and normal subgroup (6.9±19.6%) (Figure 1). The two other dominant bacterial genera in GC patients were Enterococcus (RA 68.0–99.9%) and Lactobacillus (RA 35.2–97.0%). Among all GC patients, the number of cases containing these dominant bacterial genera were 5/48 (10.4%) for both Enterococcus and Lactobacillus. (Figure 1).
Figure 1. Microbial relative abundance and case numbers by comparison group.
Microbial relative abundance (RA) percentages of the gastric microbiome are shown at the genus level by comparison group (normal, gastritis, atrophy, intestinal metaplasia and gastric cancer). The number of cases based on Helicobacter infection status (Helicobacter RA ≥2% is considered positive and RA <2% is considered negative) and dominant bacterial genera (RA of Enterococcus [E] >68.0%, Lactobacillus [L] >35.2%).
The mean ± SD of Enterococcus and Lactobacillus RA was lowest in the normal subgroup (0.002±0.004% and 0.005±0.01%, respectively), gastritis (1.3±5.9% and 0.08±0.3%, respectively) and atrophy (0% and 0.002±0.007%, respectively) group and increased along the spectrum of disease severity, from gastritis (1.3±5.9% and 0.08±0.3%, respectively) to atrophy (0% and 0.002±0.007%, respectively), IM (0.9±4.1% and 1.1±6.7%, respectively), and GC (9.6±28.4% and 7.1±21.4%, respectively). For Enterococcus infection we could stain the morphological evidence in gastric mucosa among several GC patients (Supplementary figure 2). The both OTU number of abundance for Lactobacillus (p<0.0001) and Enterococcus (p<0.0001) were significantly highest in the GC group followed by IM group by Kruskal-Wallis test. The mean and standard deviation of OTU number of abundance for Lactobacillus and Enterococcus is shown in Supplementary figure 3.
3.3. Multivariate taxonomic analysis of gender and environmental factors
We examined gastric microbiota by gender among the non-cancer and GC groups. There were no sex-related differences in alpha diversity in both groups. At the genus level only Campylobacter was higher in female than male group among GC patients (Supplementary figure 4). We examined several parameters indicative of alpha diversity according to environmental factors (salt, tobacco smoking and alcohol). There were no significant differences for alcohol drinking (No vs Yes), excessive amount of salt ingestion (No vs Yes) or tobacco smoking (No vs Yes) in our data set (Supplementary figure 5). Finally we examined the alpha diversities by geographic region. The highest OTU was found for samples of individuals coming from the provinces followed by cancer group irrespective of geographic location and the lowest OTU was in city group. DAPC graph represents the individuals as dots and the clusters as three different groups. In the LEfSe analysis the specifically enriched genera are shown in each group (Supplementary figure 6).
3.4. Multivariate taxonomic analysis of GC and its precursor diseases
Previous researchers defined a positive H. pylori infection as having an RA ≥2%.18 Therefore, we categorized H. pylori infection status as follows: Helicobacter RA ≥2% was considered positive and RA <2% was considered negative. We then analyzed by diagnosis status when including all cases and when excluding cases with dominant bacterial genera (i.e., excluding Enterococcus: RA >68% or Lactobacillus: RA>35.2%) or Helicobacter-positive status. DAPC graphical analysis showed each diagnosis group was separated into different clusters. Statistical significance was observed between these groups when including all cases (Figure 2A) or excluding cases with dominant bacterial genera or Helicobacter-positive status (Figure 2B).
Figure 2. Discriminant analysis of principal components (DAPC).
DAPC plot at the OTU level shows distinct clustering of the overall groups (normal, gastritis, atrophy, intestinal metaplasia [IM], and gastric cancer).
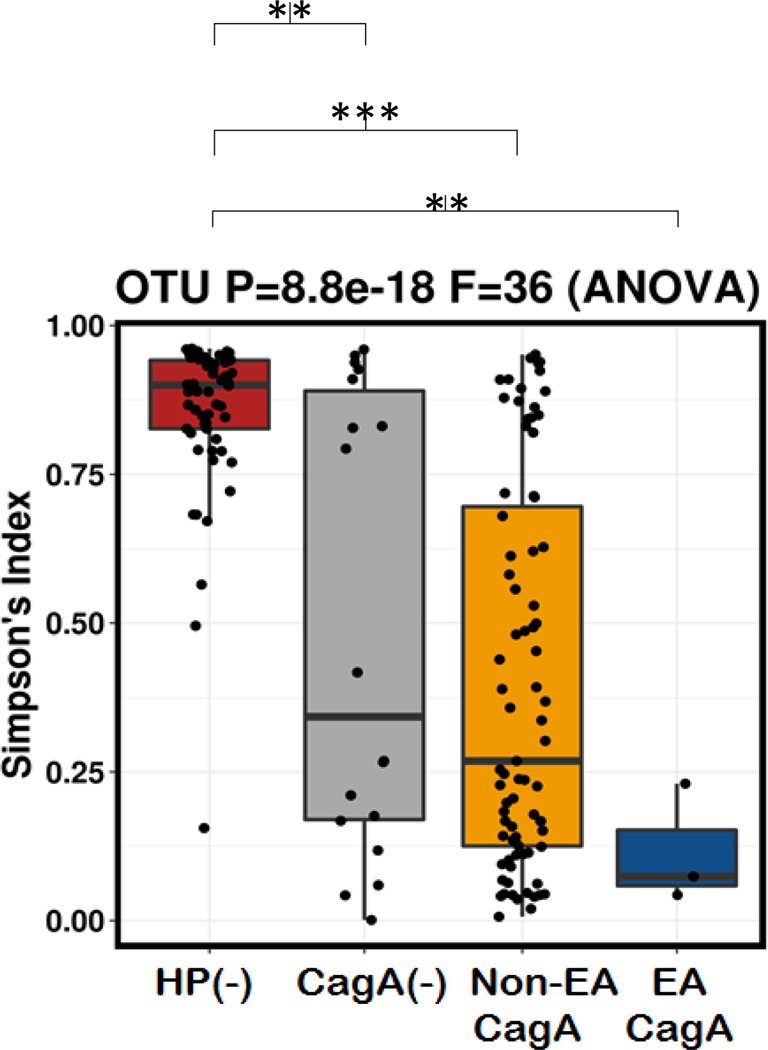
The overall bacterial alpha diversity, including Simpson index, between diagnosis statuses was significantly different at the species level. The bacterial diversity was highest in the normal subgroup followed by the IM subgroup and GC group. Lower bacterial diversity was found in the gastritis and atrophy subgroups (Figure 3A). Following exclusion of all cases that were H. pylori positive or had a dominant genera (Enterococcus and Lactobacillus), the statistically significant differences persisted only in the IM and cancer groups (Figure 3B). In addition to Simpson’s index; the evenness, richness and Shanon indices are shown in Supplementary figure 7.
Figure 3. Alpha diversity (Simpson’s index) by comparison group.
Simpson’s index is shown in all cases (A) and when excluding dominant bacterial genera and Helicobacter-positive cases (B) based on comparison groups. Each statistically significant 2×2 comparisons is shown on the top of the figure. *: p< 0.05, **: p< 0.01, ***: p< 0.001, ****: p< 0.0001.
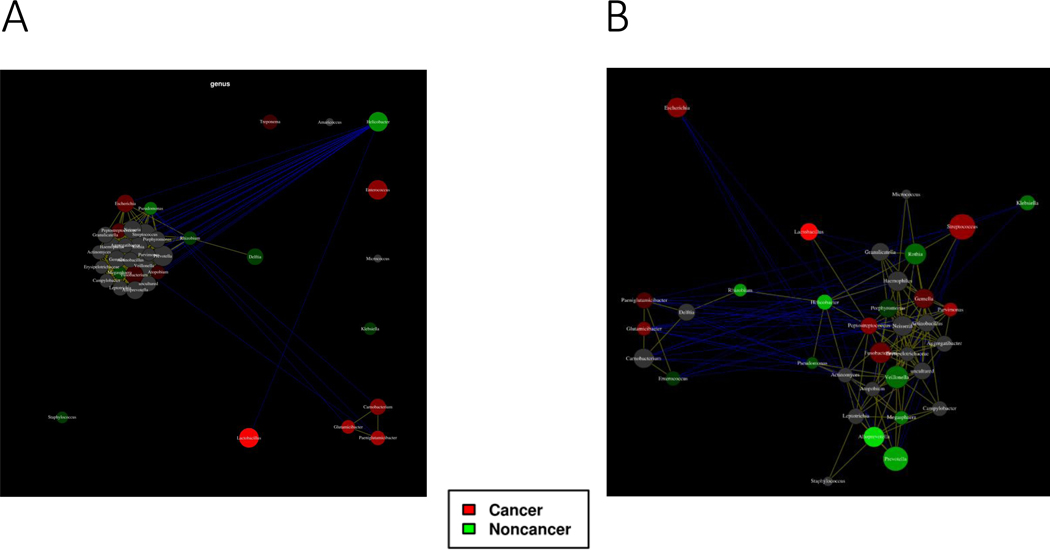
We performed network analysis for bacterial interactions comparing the GC vs non-cancer group. Correlating nodes are placed in close proximity and anti-correlating nodes are placed at distant locations in the network using the Javascript D3.js library. Nodes can be connected with edges based on Pearson’s correlation or Spearman’s rho correlation with p<0.05 are presented by an edge. Helicobacter was enriched in non-cancer group. The presence of Helicobacter was dominant and negatively correlated with the most other genera. In the GC group, Enterococcus, Lactobacillus, Glutamicibacter, Paeniglutamicibacter, Carnobacterium, and Granulicatella were specifically enriched. Lactobacillus was negatively correlated with Helicobacter. Glutamicibacter, Carnobacterium, and Paeniglutamicibacter were positively correlated with each other (Figure 4A). After removing dominant bacterial genera and Helicobacter-positive cases, Lactobacillus, Glutamicibacter, and Paeniglutamicibacter were still present in the microbiota of patients in the GC group. Parvimonas, Streptococcus, Peptostreptococcus, Fusobacterium, Glutamicibacter, and Escherichia were also enriched in GC group (Figure 4B).
Figure 4. Network analysis comparing gastric cancer and non-cancer groups.
The co-occurrence and disease-specific bacterial interactions at the genus level are shown in all cases (A) and when excluding cases with a dominant bacterial genera or Helicobacter-positive status (B) by comparison group. Taxa are represented as nodes, taxa abundance is represented as node size, and edges represent positive associations (positive inter-node correlations are yellow, negative correlations are blue). Nodes can be colored based on their association with selected environmental variables based on Pearson’s correlation.
We determined OLGA and OLGIM grades (Supplementary figure 8). For OLGA, the highest OTU was in stage 0 (non-atrophic) group and the lowest OTU was in stage III group. For OLGIM, the highest OTU was in stage I group followed by stage III group and the lowest OTU was in stage 0 group (non-metaplastic). There were no cases of OLGA/OLGIM stage IV.
3.5. Biomarker discovery analysis
LEfSe biomarker analysis showed that Enterococcus, Lactobacillus, Glutamicibacter, Paeniglutamicibacter, Escherichia, and Carnobacterium corresponded to GC group (Figure 5A). After removing the cases containing dominant bacterial genera (Enterococcus and Lactobacillus) and Helicobacter-positive cases, Parvimonas corresponded to the GC group (Figure 5B).
Figure 5. Linear discriminant analysis (LEfSe) comparing the gastric cancer vs non-cancer group.
LEfSe test at the phylum level for all cases (A) and when excluding cases with a dominant bacterial genera or Helicobacter-positive status (B) are shown by comparison group.
All significant bacterial genera for the GC and non-cancer groups were summarized at the genus level based on Wilcoxon tests that include all cases (Table 1A) and those excluding cases with dominant bacterial genera or Helicobacter-positive status (Table 1B). The results were consistent with those of the LEfSe for both comparisons. Species level results that include all cases and those that exclude cases with dominant bacterial genera or Helicobacter-positive status are shown in Supplementary Tables 1A and 1B.
Table 1A.
Wilcoxon test of biomarker discovery analysis for the gastric cancer group compared with the non-cancer group at the genus level
| Taxa | p value | Odds Ratio | LowerCI | Upper CI | FoldChange | Mean Cancer | Mean Non-cancer |
|---|---|---|---|---|---|---|---|
| Lactobacillus | 0.00000000058 | 13.84 | 2.15 | 269.14 | 17.88 | 7.07 | 0.40 |
| Paeniglutamicibacter | 0.000061 | 4.95 | 1.42 | 19.72 | 2.73 | 0.65 | 0.24 |
| Glutamicibacter | 0.00009 | 3.93 | 1.19 | 13.92 | 2.11 | 0.37 | 0.18 |
| Enterococcus | 0.00046 | 4.53 | 1.07 | 22.87 | 17.89 | 9.59 | 0.54 |
| Carnobacterium | 0.0015 | 2.64 | 0.60 | 11.59 | 2.44 | 1.39 | 0.57 |
| Helicobacter | 0.00045 | 0.39 | 0.18 | 0.78 | −1.8 | 26.69 | 48.09 |
Table 1B.
Wilcoxon test of biomarker discovery analysis for the gastric cancer group compared with the non-cancer group at the genus level after excluding cases with dominant bacterial genera or Helicobacter-positive status
| Taxa | p value | Odds Ratio | Lower CI | Upper CI | Fold Change | Mean Cancer | Mean Non-cancer |
|---|---|---|---|---|---|---|---|
| Parvimonas | 0.045 | 15 | 2.1 | 306.34 | 8.37 | 0.8 | 0.1 |
| Lactobacillus | 0.000067 | 2.23 | 0.25 | 20.31 | −3.7 | 0.38 | 1.42 |
| Alloprevotella | 0.0042 | 0.09 | 0 | 0.52 | −4.94 | 0.92 | 4.54 |
| Helicobacter | 0.02 | 0.27 | 0.05 | 1.04 | −2.77 | 0.24 | 0.67 |
| Prevotella | 0.03 | 0.42 | 0.11 | 1.46 | −1.61 | 12.62 | 20.31 |
3.6. Univariate analysis after excluding dominant genera
Based on biomarker discovery analysis, we examined ANOVA and rank tests based on diagnosis groups to observe any trends in each bacterial genera. Enterococcus, Glutamicibacter, Paeniglutamicibacter, Carnobacterium, and Lactobacillus RA were highest in the GC group followed by IM subgroup compared with the normal, gastritis, and atrophy subgroups). After excluding dominant bacterial genera and Helicobacter-positive cases, Parvimonas, Streptococcus, and Fusobacterium were highest in the GC group compared with the other subgroups (normal, gastritis, atrophy, and IM). Importantly, Helicobacter was significantly lower in the GC group than in the other subgroups, except for the normal subgroup (Figure 6).
Figure 6. Univariate analysis for selected genera by comparison group.
Relative abundance at the phylum level for all cases (A-F) and when excluding cases with dominant bacterial genera or Helicobacter-positive status (G-I) are shown based on comparison groups. Each statistically significant 2×2 comparison is shown on the top of the figure (A-I). *: p< 0.05, **: p< 0.01, ***: p< 0.001, ****: p< 0.0001.
3.7. Gastric microbiota status according to H. pylori genotyping
The frequency and percentages of gastric diseases (normal, gastritis, atrophy, IM and GC) according to H. pylori status and CagA subtyping (CagA negative, East Asian type CagA and non-East Asian type CagA) are shown in Supplementary figure 9. The RA of H. pylori was highest in patients infected with East-Asian-type CagA strains (93.9%), followed by those infected with non-East-Asian-type CagA strains (64.6%), those infected with CagA-negative strains (57.1%), and H. pylori-negative patients (4.1%) (Kruskal-Wallis test: p< 0.0001). The bacterial diversity was highest in H. pylori-negative patients, followed by patients infected with CagA-negative strains, those infected with East-Asian-type CagA strains, and those infected with non-East-Asian-type CagA strains (Figure 7).
Figure 7. Alpha diversity (Simpson’s index) by H. pylori infection status.
Simpson’s index is shown based on H. pylori-negative and -positive status. H. pylori-positive status is divided into those cases with CagA-negative and -positive status. CagA-positive status can be further subdivided into East-Asian-type and Non-East-Asian-type CagA based on immunohistochemistry. Each statistically significant 2×2 comparison is shown on the top of the figure. *: p< 0.05, **: p< 0.01, ***: p< 0.001, ****: p< 0.0001.
3.8. Functional analysis
Using bacterial taxonomy, we examined functional differences between the GC and non-cancer groups. At the level of FOAM layer 1 (Figure 8) and 2 (Supplementary Table 2), most parameters were significantly increased in the GC group compared with the non-cancer group, with the exception of hydrogen metabolism, methylotrophy, methanogenesis, nitrogen cycle, sulfur compound metabolism, and transporters. For nitrosating enzyme richness, the nfrA (Nitrate reductase cytochrome c-552) was higher in GC group than non-cancer group (p<0.05) (Supplementary figure 10).
Figure 8. Functional analysis comparing the gastric cancer and non-cancer group.
The functional analysis comparing the gastric cancer and non-cancer groups is performed at the FOAM layer 1 level. The statistical significance value is shown on the top each bar. *: p< 0.05, **: p< 0.01, ***: p< 0.001, ****: p< 0.0001.
4. DISCUSSION
We examined gastric mucosal microbiota in GC and several of its precursor histological conditions in a Mongolian population where the burden of GC mortality is the highest in the world. There were significantly different microbiota profiles between the GC group and various subgroups compared with the normal subgroup. As expected, Epsilonbacteraeota (H. pylori) was increased in the gastritis and atrophy subgroups. But we also found that Firmicutes (Lactobacillus and Enterococcus) were increased in the IM subgroup and dramatically so in the GC group. Proteobacteria, Bacteroidetes, and Actinobacteria were increased in the normal subgroup compared with the other subgroups.
It is possible that non- Helicobacter bacterial genera could play a role in GC development. GC-associated, non-H. pylori bacterial species described in previous studies have been summarized in Supplementary Table 3. We highlighted several novel GC-associated candidates, including Enterococcus, Carnobacterium, Glutamicibacter and Paeniglutamicibacter genera which are did not described previously. Among Mongolian individuals with dyspepsia, the most dominant gastric microbial genus was Helicobacter. The RA of Helicobacter infection was highest in the gastritis and atrophy subgroups and relatively decreased in the IM subgroup and GC group. Conversely, the overall bacterial diversity was higher in the IM subgroup and GC group than in the gastritis and atrophy subgroups. Bacterial diversity was lowest in patients infected with the most virulent East-Asian-type CagA strains, followed by those infected with non-East-Asian-type CagA strains and those infected with CagA-negative strains, which is shown in Figure 7. This observation can be explained by dramatic alterations and reductions in gastric microbiota diversity once the stomach is colonized by Helicobacter, especially strains containing the CagA virulence gene.19 20
H. pylori colonization gradually decreases due to an atrophied niche, which reduces gastric acidity, while other non-H. pylori species, especially those with nitrosating bacteria,20–22 survive in the stomach and possibly increase GC risk. Interestingly, similar to Helicobacter infection, Enterococcus and Lactobacillus genera strongly dominated other microbiota in several GC cases. Specifically, their RAs were 35.2–97% for Lactobacillus and 68.0–99% for Enterococcus. Previous studies reported that Lactobacillus and Enterococcus are commensal bacteria in healthy stomachs with RAs up to 30% for Lactobacillus and 51% for Enterococcus. Our study suggest that exceeding these limits could be associated with an increased risk for GC.23 24
Because it is well known that H. pylori is the major etiology of GC and that it reduces gastric microbial diversity,19 20 We focused on the 58 cases with H. pylori-negative cases (normal, gastritis, atrophy, IM and GC). In addition to H. pylori, Lactobacillus and Enterococcus genera strongly dominated other microbiota, so we excluded 10 cases with dominant Lactobacillus (n=5; RA >35.2%) and Enterococcus (n=5; RA >68%). Among the remaining cases, Fusobacterium, Parvimonas, and Streptococcus genera were enriched in GC group. In previous studies, Parvimonas and Streptococcus genera were enriched among GC patients, including inner Mongolia where the population is mixture of Mongolian and Chinese population.7 25 In our current study, Lactobacillus track closely with both cancer and IM with no cancer, but less so with atrophy. Similar to our results, several studies found increases in the RA of lactic acid-producing bacteria (Lactococcus and Lactobacillus) in GC patients.26–30 Certain species of Lactobacillus (e.g., reuteri, acidophilus, and fermentum) are used as probiotics;31 32 however, several other studies using a 16S rRNA sequencing approach reported that Lactobacillus was associated with GC in agreement with our study. For example, a clinical study from Taiwan found the presence of Lactobacillus reuteri and gasseri species in GC patients.25 Since Lactobacilli are not acid resistant, the abundance of Lactobacilli can be expected in the stomachs of subjects with IM and GC.
Our network analysis showed the presence of Carnobacterium, Glutamicibacter, and Paeniglutamicibacter in GC patients. The genus Carnobacterium encompasses 11 species that have been isolated in cold and temperate environments, from the gastrointestinal tract of animals, and from foods of animal origin, such as seafood, meat, and dairy products.33 34 Carnobacterium has been isolated from vacuum-packaged bologna sausage and was responsible for causing green discoloration of the product after package opening.35 Glutamicibacter and Paeniglutamicibacter were isolated from bad hygiene environments, and their clinical importance is unknown.36
In our study, the functional prediction analysis identified several functional parameters, including the Embden Meyerhof-Parnas pathway and the utilization of sugar, that were significantly increased in GC group compared with the non-cancer group. Previous researchers have found that lactate can serve as an energy source for tumor cells by inducing the expression of glycolytic enzymes to increase the ATP supply. Lactate can also promote inflammation and stimulate tumor angiogenesis.37–40
For the GC precursor diseases, we previously reported that the extension of IM in the corpus was associated with an increased GC risk in Mongolian population.3 41 In our current study, H. pylori was the predominant species in the gastritis and atrophy groups but was lower in IM group. Further, non-H. pylori bacteria, especially those associated with GC species, were higher in the IM group. Therefore, it seems likely that the development of IM is a precursor condition for the shift in gastric microbiota.
In conclusion, our results identifying non-H. pylori, GC-associated bacteria Lactobacillus, Parvimonas, Escherichia, and Streptococcus were consistent with those of other studies. We additionally found that Enterococcus, Carnobacterium, Glutamicibacter, and Paeniglutamicibacter were GC-associated genera among a Mongolian population in which the GC incidence is very high.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank the study participants and wish to thank Matsuhisa T, Davaadorj D, Tserentogtokh T, Battulga A, Bira N, Azzaya D, Byambajav TS, Enkh-Amar A, Bolor O, and Bilguudei B for collecting samples.
Funding information: This work was supported by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology (MEXT) of Japan (15H02657, 16H06279, 16H05191, and 18KK0266; Yamaoka Y; 18K16182; Matsumoto T), the Special Coordination Funds for Promoting Science and Technology from MEXT (Yamaoka Y), and a National Institutes of Health grant (DK62813; Yamaoka Y). This material is based upon work supported by a Cancer Prevention & Research Institute of Texas grant (RP150587) (El-Serag HB) and by the Center for Gastrointestinal Development, Infection, and Injury (NIDDK P30 DK56338) (El-Serag HB). Gantuya B is a doctoral student supported by the MEXT Scholarship Program for 2014.
Footnotes
Disclosures: The authors declare no conflicts of interest.
REFERENCES
- 1.Plummer M, Franceschi S, Vignat J, et al. Global burden of gastric cancer attributable to Helicobacter pylori. International journal of cancer 2015;136:487–90. [DOI] [PubMed] [Google Scholar]
- 2.Huang JQ, Zheng GF, Sumanac K, et al. Meta-analysis of the relationship between CagA seropositivity and gastric cancer. Gastroenterology 2003;125:1636–44. [DOI] [PubMed] [Google Scholar]
- 3.Gantuya B, Bolor D, Oyuntsetseg K, et al. New observations regarding Helicobacter pylori and gastric cancer in Mongolia. Helicobacter 2018:e12491. [DOI] [PMC free article] [PubMed]
- 4.Matsuhisa T, Yamaoka Y, Uchida T, et al. Gastric mucosa in Mongolian and Japanese patients with gastric cancer and Helicobacter pylori infection. World Journal of Gastroenterology: WJG 2015;21:8408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang I, Woltemate S, Piazuelo MB, et al. Different gastric microbiota compositions in two human populations with high and low gastric cancer risk in Colombia. Scientific reports 2016;6:18594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu W, Zhang J, Wu C, et al. Unique features of ethnic Mongolian gut microbiome revealed by metagenomic analysis. Scientific reports 2016;6:34826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coker OO, Dai Z, Nie Y, et al. Mucosal microbiome dysbiosis in gastric carcinogenesis. Gut 2018;67:1024–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sharaf RN, Shergill AK, Odze RD, et al. Endoscopic mucosal tissue sampling. Gastrointestinal endoscopy 2013;78:216–24. [DOI] [PubMed] [Google Scholar]
- 9.Dixon MF, Genta RM, Yardley JH, et al. Classification and grading of gastritis: the updated Sydney system. The American journal of surgical pathology 1996;20:1161–81. [DOI] [PubMed] [Google Scholar]
- 10.Rugge M, Meggio A, Pennelli G, et al. Gastritis staging in clinical practice: the OLGA staging system. Gut 2007;56:631–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Capelle LG, de Vries AC, Haringsma J, et al. The staging of gastritis with the OLGA system by using intestinal metaplasia as an accurate alternative for atrophic gastritis. Gastrointestinal endoscopy 2010;71:1150–58. [DOI] [PubMed] [Google Scholar]
- 12.Uchida T, Kanada R, Tsukamoto Y, et al. Immunohistochemical diagnosis of the cagA‐gene genotype of Helicobacter pylori with anti‐East Asian CagA‐specific antibody. Cancer science 2007;98:521–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klindworth A, Pruesse E, Schweer T, et al. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic acids research 2013;41:e1–e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zakrzewski M, Proietti C, Ellis JJ, et al. Calypso: a user-friendly web-server for mining and visualizing microbiome–environment interactions. Bioinformatics 2016;33:782–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Segata N, Izard J, Waldron L, et al. Metagenomic biomarker discovery and explanation. Genome biology 2011;12:R60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dhariwal A, Chong J, Habib S, et al. MicrobiomeAnalyst: a web-based tool for comprehensive statistical, visual and meta-analysis of microbiome data. Nucleic acids research 2017;45:W180–W88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Uchiyama T, Irie M, Mori H, et al. FuncTree: functional analysis and visualization for large-scale omics data. PloS one 2015;10:e0126967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li J. PGIP. Is There a Role for the Non-Helicobacter pylori bacteria in the risk of developing gastric cancer? Int J Mol Sci 2018;19:1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klymiuk I, Bilgilier C, Stadlmann A, et al. The human gastric microbiome is predicated upon infection with Helicobacter pylori. Frontiers in microbiology 2017;8:2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brawner KM, Morrow CD, Smith PD. Gastric microbiome and gastric cancer. Cancer journal (Sudbury, Mass) 2014;20:211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Noto JM, Peek RM Jr. The gastric microbiome, its interaction with Helicobacter pylori, and its potential role in the progression to stomach cancer. PLoS pathogens 2017;13:e1006573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dias-Jácome E, Libânio D, Borges-Canha M, et al. Gastric microbiota and carcinogenesis: the role of non-Helicobacter pylori bacteria: a systematic review. Revista Española de Enfermedades Digestivas 2016;108:530–40. [DOI] [PubMed] [Google Scholar]
- 23.Zhang C, Powell SE, Betel D, et al. The gastric microbiome and its influence on gastric carcinogenesis: current knowledge and ongoing research. Hematology/Oncology Clinics 2017;31:389–408. [DOI] [PubMed] [Google Scholar]
- 24.Delgado S, Cabrera-Rubio R, Mira A, et al. Microbiological survey of the human gastric ecosystem using culturing and pyrosequencing methods. Microbial ecology 2013;65:763–72. [DOI] [PubMed] [Google Scholar]
- 25.Hsieh Y-Y, Tung S-Y, Pan H-Y, et al. Increased abundance of Clostridium and Fusobacterium in gastric microbiota of patients with gastric cancer in Taiwan. Scientific reports 2018;8:158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aviles-Jimenez F, Vazquez-Jimenez F, Medrano-Guzman R, et al. Stomach microbiota composition varies between patients with non-atrophic gastritis and patients with intestinal type of gastric cancer. Scientific reports 2014;4:4202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Castaño-Rodríguez N, Goh K-L, Fock KM, et al. Dysbiosis of the microbiome in gastric carcinogenesis. Scientific reports 2017;7:15957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eun CS, Kim BK, Han DS, et al. Differences in gastric mucosal microbiota profiling in patients with chronic gastritis, intestinal metaplasia, and gastric cancer using pyrosequencing methods. Helicobacter 2014;19:407–16. [DOI] [PubMed] [Google Scholar]
- 29.Li TH, Qin Y, Sham PC, et al. Alterations in gastric microbiota after H. pylori eradication and in different histological stages of gastric carcinogenesis. Scientific reports 2017;7:44935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang L, Zhou J, Xin Y, et al. Bacterial overgrowth and diversification of microbiota in gastric cancer. European journal of gastroenterology & hepatology 2016;28:261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ryan KA, Jayaraman T, Daly P, et al. Isolation of lactobacilli with probiotic properties from the human stomach. Letters in applied microbiology 2008;47:269–74. [DOI] [PubMed] [Google Scholar]
- 32.Oh NS, Joung JY, Lee JY, et al. Probiotic and anti-inflammatory potential of Lactobacillus rhamnosus 4B15 and Lactobacillus gasseri 4M13 isolated from infant feces. PloS one 2018;13:e0192021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chaillou S, Chaulot-Talmon A, Caekebeke H, et al. Origin and ecological selection of core and food-specific bacterial communities associated with meat and seafood spoilage. The ISME journal 2015;9:1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Iskandar CF, Borges F, Taminiau B, et al. Comparative genomic analysis reveals ecological differentiation in the genus Carnobacterium. Frontiers in microbiology 2017;8:357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Holley RA, Guan TY, Peirson M, et al. Carnobacterium viridans sp. nov., an alkaliphilic, facultative anaerobe isolated from refrigerated, vacuum-packed bologna sausage. International journal of systematic and evolutionary microbiology 2002;52:1881–85. [DOI] [PubMed] [Google Scholar]
- 36.Busse H-J. Review of the taxonomy of the genus Arthrobacter, emendation of the genus Arthrobacter sensu lato, proposal to reclassify selected species of the genus Arthrobacter in the novel genera Glutamicibacter gen. nov., Paeniglutamicibacter gen. nov., Pseudoglutamicibacter gen. nov., Paenarthrobacter gen. nov. and Pseudarthrobacter gen. nov., and emended description of Arthrobacter roseus. International journal of systematic and evolutionary microbiology 2016;66:9–37. [DOI] [PubMed] [Google Scholar]
- 37.Doherty JR, Cleveland JL. Targeting lactate metabolism for cancer therapeutics. The Journal of clinical investigation 2013;123:3685–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kennedy KM, Scarbrough PM, Ribeiro A, et al. Catabolism of exogenous lactate reveals it as a legitimate metabolic substrate in breast cancer. PloS one 2013;8:e75154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sonveaux P, Copetti T, De Saedeleer CJ, et al. Targeting the lactate transporter MCT1 in endothelial cells inhibits lactate-induced HIF-1 activation and tumor angiogenesis. PloS one 2012;7:e33418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sonveaux P, Végran F, Schroeder T, et al. Targeting lactate-fueled respiration selectively kills hypoxic tumor cells in mice. The Journal of clinical investigation 2008;118:3930–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gantuya B, Oyuntsetseg K, Bolor D, et al. Evaluation of serum markers for gastric cancer and its precursor diseases among high incidence and mortality rate of gastric cancer area. Gastric Cancer 2019;22:104–12. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.