Abstract
It remains important to investigate the changing and impact of routine blood values (RBVs) in order to predict mortality and follow an appropriate treatment in COVID-19 patients. In the study, the importance of RBVs in the mortality of patients with COVID-19 was investigated. The changes in the biochemical, hematological, and immunological parameters of patients who recovered (n = 4364) and died (n = 233) from COVID-19 over time and their relationship with the mortality of the disease were evaluated retrospectively. Odds ratios of the parameters affecting one-month mortality were calculated by running multiple-logistic-regression analysis. The cut off values and diagnostic efficiencies of the parameters that posed a risk for mortality were obtained via receiver operating curve analysis. It was determined that the C-reactive protein (CRP), D-dimer, procalcitonin, erythrocyte-sedimentation-rate (ESR), troponin values were at abnormal levels until death occurred in the patients who died. In addition, the procalcitonin levels were consistently high in patients who died. The patients who died generally had a sustained increase in their leukocyte and neutrophil levels and biochemical variables, and an ongoing decrease in lymphopenia and eosinopenia levels. Although significant changes were observed in liver function tests, cardiac troponin, hemogram values, kidney function tests and parameters related to inflammation in deceased patients, high ESR, international-normalized-ratio (INR), prothrombin-time (PT), CRP, D-dimer, ferritin and red-cell-distribution width (RDW) values, respectively, were the most effective predictive mortality risk biomarkers of COVID-19. In addition, neutrophilia, leukocytosis, thrombocytopenia, erythrocytopenia were other risk predictors of mortality. Indicators was found in this study can be successfully used to predict mortality from COVID-19.
Keywords: COVID-19, Mortality risk indicators, Routine laboratory characteristics, Hypercoagulation, Leukocytosis, Erythrocytopenia
1. Introduction
The new severe acute respiratory syndrome coronavirus (SARS-CoV-2), first identified in China at the end of 2019, quickly affected the world and caused a pandemic. Defined as coronavirus 2019 (COVID-19), the disease can cause severe pneumonia and fatal acute respiratory distress syndrome (ARDS) [1]. While the number of cases is increasing day by day, there is limited information about the hematological and laboratory findings of the disease [[2], [3], [39]]. It was reported that routine laboratory data showed significant changes in COVID-19, but could not be fully elucidated, especially in patients who died [[4], [5], [38]].
While the disease may be asymptomatic, severe ARDS is thought to be due to an inflammatory cytokine storm that may be encountered during this period [6]. As a matter of fact, it has been stated in other studies that this pathogen may cause a serious respiratory disorder that requires special management in intensive care units (ICUs) and causes death in some cases [6], [7], [8]. Most patients with SARS-CoV-2 infection have mild symptoms (such as fever, dry cough, dyspnea, myalgia, fatigue, and so on), but in severe cases, the disease can rapidly progress into ARDS, septic shock, bleeding, coagulation dysfunction, metabolic acidosis, and a high mortality rate [9].
Previous studies have shown that the change in routine blood parameters has certain clinical application value in predicting the progression of infectious diseases [[1], [10], [37]]. Indeed, many abnormalities have been reported in the peripheral blood of some infected patients [2], [6], [9], [11]. Mild patients have milder symptoms and a good prognosis, but severe and critically ill patients are difficult to treat and have a high mortality rate [12]. In some studies, high mortality rates were reported in severe COVID-19 patients despite intensive care treatment [12], [13].
However, information on the early predictive indicators for particularly severe and fatal cases is relatively limited and further research is needed [[9], [37]]. In addition, the rapid spread of the disease raises concerns about the need for intensive care that can overwhelm health care system resources [13].
Prognostic predictors of mortality in COVID-19 patients should be identified to help make optimal treatment decisions and assess the severity of the condition. Therefore, it has become the focus of research efforts to determine whether patients who are likely to benefit from early intervention with supportive care are at risk and how to identify them [11].
In addition, while examining the changes in routine blood values (RBVs) in COVID-19, the identification of severe and mild patients is a very important and clinically difficult process in terms of morbidity and mortality. In previous studies, the RBVs of COVID-19 patients were compared according to the treatment units, respiratory distress levels (respiratory rate), oxygen saturation levels at rest, and arterial blood oxygen partial pressures/oxygen concentration ratios [1], [9], [11].
In addition, in more effective studies conducted to determine the mortality of the disease, the patient sample was determined as those who died from COVID-19 and those who survived, and the changes in the RBVs were compared in these groups [2], [8], [12], [14]. However, the number of routine blood parameters evaluated in these studies and the number of patients who died and survived in these study samples were much less when compared to the study conducted herein. In addition, there have been no studies on the dynamic changes in the RBVs, especially in patients who died.
In this study, the biochemical, hematological and inflammatory tests performed in surviving and non-surviving COVID-19 patients when they first applied to the hospital were also compared with the tests performed in the following days during their hospitalization. In addition, ROC curves were obtained by determining the blood values affecting the mortality of the disease via multivariate logistic regression analysis. In doing so, the parameters affecting the mortality of the disease were determined and it was seen that the cut-off values of the parameters could be used effectively in the risk estimation of mortality. In addition, an open access data source was created for the data used in the study so that they can be used in studies on COVID-19 (http://covdats.duzce.edu.tr).
2. Material and methods
This retrospective single-center study was conducted in accordance with the 1989 Helsinki Declaration. Data matching our criteria were collected from Erzincan Binali Yıldırım University Mengücek Gazi Training and Research Hospital information system between January and June, 2021and were included in the study. The research only covered people over 18 years old. The tests performed when the patients first applied to the hospital and the tests performed on the following days during their hospitalization were recorded. In order for the data used in this study to be used in different studies too, our data set has been converted into an appropriate format and shared at “http://covdats.duzce.edu.tr” as an open access data source. Our data set will be shared with researchers upon their request.
2.1. Study design and participants criteria
The diagnosis of COVID-19 was defined only in cases detected as SARS-CoV-2 by real-time reverse-transcriptase polymerase chain reaction on nasopharyngeal or oropharyngeal swabs at the dates covered by this study in our hospital. This retrospective single-center study was conducted by analyzing the data collected between the specified dates. During this period, the information of 4597 patients who were treated at the hospital for COVID-19 could be accessed and the exit status of these patients was examined. According to the exit reports, n = 233 of the patients included in this study died, while n = 4364 recovered and were discharged. While data were being recorded, the laboratory findings and demographic characteristics of those who died from COVID-19 and those who recovered from COVID-19 were reported in two different groups. The demographic data and laboratory findings of the patients were obtained retrospectively from the electronic information system of the hospital.
The tests measured at the first admission (pre) to the hospital and the tests measured before discharge (post) or discharge (post) of patients who died and recovered from COVID-19 were compared within the patient group and between groups. This procedure was performed as follows: First of all, the routine blood pre-values of patients who died from COVID-19 were compared with the pre-values of patients who recovered. Similarly, the routine blood post-values of the two patient groups were compared. Then, the routine blood pre-values and post-values of patients who died from COVID-19 were compared. Similarly, the routine blood pre-values and post-values of patients recovering from COVID-19 were compared.
Also, the routine blood tests of the patients, which were measured at admission, were examined during the days following the hospitalization until exit and the dynamic changes of the tests were reported. Next, multivariate logistic regression was run to reveal mortality risk indicators. Moreover, receiver operating curve (ROC) analysis was run to calculate the cut-off values of the indicators affecting mortality. Finally, the diagnostic performances of indicators, a predictive predictor of COVID-19 mortality, were obtained.
2.2. Blood routine laboratory parameters used in the study
The Sysmex XN-1000 Hematology System (Sysmex Corporation, Kobe, Japan) was used to perform cell blood count of patients. Biochemical tests were analyzed from serum by spectrophotometric method using Beckman Coulter Olympus AU2700 Plus Chemistry Analyzer (Beckman Coulter, Tokyo, Japan). Ferritin was evaluated by a chemiluminescence immunoassay (Centaur XP, Siemens Healthcare, Germany). Prothrombin time (PT), activated partial prothrombin time (aPTT), and fibrinogen were determined with a fully digital coagulation device of Ceveron-Alpha (Diapharma Group Inc., West Chester, Canada). C-reactive protein (CRP) was measured by the nephelometric method on the BN ™ II System (Siemens, Munich, Germany). Procalcitonin (PCT), D-dimer and Troponin were analyzed from whole blood on the AQT90 flex Radiometer® (Bronshoj, Denmark).
2.3. Statistical analysis
Categorical variables were presented as frequency and percentage, while continuous variables were given as the mean, median and quartile. The Shapiro-Wilk test was used to verify the normality of the distributions of quantitative variables. For the analysis of the differences between the variables of survivor and non-survivor COVID-19 patients, the ındependent-samples t-test was used when the normal distribution hypothesis was provided, otherwise the Mann-Whitney U test was used. In addition, for the analysis of the pre and post values of the variables within the patient groups, the paired-samples t test was used when the normal distribution hypothesis was met, otherwise the Wilcoxon signed-rank test was used. Categorical variables were analyzed using the χ2 test. Odds ratios (ORs) were calculated by running multivariate logistic regression to determine the parameters associated with the mortality of the disease and to reveal the lethal risk indicators. Variables were selected in the regression model with the forward method. The dependent variable in the regression model comprised the living and dying status of the patient in the last data obtained. Finally, cut-off values were calculated to determine the fatal risk level of a predictive biomarker of mortality in this study. ROC analysis was performed to determine the lethal risk threshold values of the parameters affecting the mortality of the disease and to measure their diagnostic performance. IBM SPSS Statistics for Windows 20.0 (IBM Corp., Armonk, NY, USA) software was used for the statistical analysis, and P < 0.05 was considered as statistically significant.
3. Results
The demographic characteristics of the patients are summarized in Table 1 , where it can be seen that 233 (5.07%) of the 4597 patients died during the study period. The mean age of the non-surviving group was significantly higher than the average age of the surviving group (median: 76.00 versus 56.00). There was a significant difference in the male/female ratio in the non-surviving group (P < 0.05). Male gender was seen as a factor influencing fatality.
Table 1.
Comparison of demographical characteristics and biochemical variables between survivor and non-survivor groups.
| Survivor Covid-19 | Non-survivor Covid-19 | *p-value | ||
|---|---|---|---|---|
| Median (25 th quartile-75th quartile) | Median (25 th quartile-75th quartile) | |||
| Age | 56.00 (40.00–69.00) | 76.00 68.00 – 82.00) | 0.000 | |
| SexMale n(%)Female n(%) | 2228 (51.1)2136 (48.9) | 143 (61.4)90 (38.6) | 0.000 | |
| Parameters | Reference range | |||
| ALT (pre) U/L | 0–35 | 25.00 (16.00–42.00) | 21.00 (14.00–34.00) | 0.000 |
| ALT (post) U/L | 32.00 (19.00–64.00) | 24.00 (16.00–47.00) | 0.004 | |
| AST (pre) U/L | 0–50 | 26.00 (20.00–37.00) | 30.00 (20.00–47.00) | 0.002 |
| AST (post) U/L | 27.50 (21.00–41.00) | 32.00 (22.00–56.00) | 0.064 | |
| Albumin (pre) μmol/L | 527–782 | 582.30 (516.01–651.52) | 544.84 (476.83–605.63) | 0.000 |
| Albumin (post) μmol/L | 503.31 (454.41–586.82) | 455.91 (386.70–556.72) | 0.001 | |
| Alkaline Phosphatase (pre) μmol/s.L | 0.5–2.0 | 1.25 (1.03–1.57) | 1.42 1.10–3.13) | 0.000 |
| Alkaline Phosphatase (post) μmol/s.L | 1.09 (1.09–1.7) | 1.55 (1.20–2.00) | 0.020 | |
| Amylase (pre) μmol/s.L | 0.47–1.67 | 1.13 (0.85–1.50) | 1.12 (0.78–1.48) | 0.685 |
| Amylase (post) μmol/s.L | 1.46 (1.00–1.78) | 1.13 (1.13–1.53) | 0.058 | |
| CK - MB (pre) u/L | 0–24 | 15.70 (12.40–21.60) | 22.65 (15.60–31.40) | 0.000 |
| CK - MB (post) u/L | 16.90 (12.70–21.10) | 21.40 (16.10–32.00) | 0.000 | |
| Direct Bilirubin (pre) μmol/L | 0–3.4 | 1.88 (1.37–2.56) | 2.39 (1.54–4.10) | 0.000 |
| Direct Bilirubin (post) μmol/L | 1.88 (1.37 – 2.56) | 2.91 (1.88–4.61) | 0.000 | |
| GGT (pre) μmol/s.L | 0–0.63 | 0.51(0.30–0.94) | 0.55 (0.32–1.18) | 0.139 |
| GGT (post) μmol/s.L | 0.78 (0.42–1.32) | 0.77 (0.37–2.01) | 0.779 | |
| Glucose (pre) mmol/L | 4.11–5.55 | 5.99 (5.16–8.10) | 7.72 (6.22–10.77) | 0.000 |
| Glucose (post) mmol/L | 6.27 (5.16–10.05) | 8.21(6.27–13.82) | 0.000 | |
| HDL Cholesterol (pre) mmol/L | 1.03–1.55 | 0.35 (27.00–0.70) | 0.88 (0.80–1.24) | 0.234 |
| HDL Cholesterol (post) mmol/L | 0.93 (0.72 – 1.11) | 0.82 (0.72–1.03) | 0.427 | |
| Cholesterol (pre) mmol/L | 0–5.17 | 4.19 (3.49–5.04) | 4.26 (3.62–5.66) | 0.432 |
| Cholesterol (post) mmol/L | 4.60 (3.72–5.61) | 4.32 (3.49–5.35) | 0.370 | |
| Creatinine (pre) μmol/L | 74.26–110.50 | 80.44 (68.95–92.82) | 96.35 (75.14–133.48) | 0.000 |
| Creatinine (post) μmol/L | 83.09 (68.95–99.89) | 103.43 (82.21–152.93) | 0.000 | |
| Creatine Kinase (pre) u/L | 0–145 | 71.00 (44.00–106.00) | 101.00 (59.00–237.00) | 0.000 |
| Creatine Kinase (post) u/L | 44.00 (31.00–91.50) | 91.00 (50.00–171.00) | 0.000 | |
| LDH (pre) u/L | 0–248 | 233.00 (193.00–292.00) | 289.00 (223.00–367.00) | 0.000 |
| LDH (post) u/L | 264.00 (217.00–332.00) | 338.50 (255.00–484.50) | 0.000 | |
| Total Bilirubin (pre) μmol/L | 5.13–20.52 | 8.04 (5.98–10.94) | 9.74 (6.67–14.19) | 0.000 |
| Total Bilirubin (post) μmol/L | 8.38 (6.15–11.45) | 10.60 (7.18–15.73) | 0.003 | |
| Total Protein (pre) g/L | 66–83 | 68.10 (63.40–73.45) | 66.33 (59.71–70.24) | 0.000 |
| Total Protein (post) g/L | 64.60 (57.25–69.95) | 61.35 (54.70–68.50) | 0.071 | |
| Triglyceride (pre) mmol/L | 0–1.69 | 1.49 (1.08–2.10) | 1.26 (1.04–2.05) | 0.558 |
| Triglyceride (post) mmol/L | 1.71 (1.36–2.41) | 1.62 (0.84–2.89) | 0.510 | |
| eGFR (pre) | 85.10 (66.33–99.36) | 61.08 (40.86–83.83) | 0.000 | |
| eGFR (post) | 77.44 (56.04–92.94) | 55.48 (34.34–76.59) | 0.000 | |
| Urea (pre) mmol/L | 2.83–7.16 | 5.33 (3.99–7.61) | 8.88 (6.46–13.32) | 0.000 |
| Urea (post) mmol/L | 6.99 (4.99–10.32) | 11.57 (7.49–22.81) | 0.000 | |
| Uric acid (pre) mmol/L | 0.21–0.43 | 0.30 (0.24–0.39) | 0.40 (0.29–0.51) | 0.000 |
| Uric acid (post) mmol/L | 0.28 (0.20–0.38) | 0.33 (0.27–0.40) | 0.141 |
ALT: alanine aminotransaminase; AST: aspartate aminotransferase; CK-MB: creatine kinase myocardial band; GGT: Gamma Glutamyl Transferase enzyme; LDH: lactate dehydrogenase; eGFR: estimated glomerular filtration rate; IQR: interquartile range; The p values indicate the comparison of the survivor and non-survivor groups in pre-post measurements and they are bold when p < 0.05.
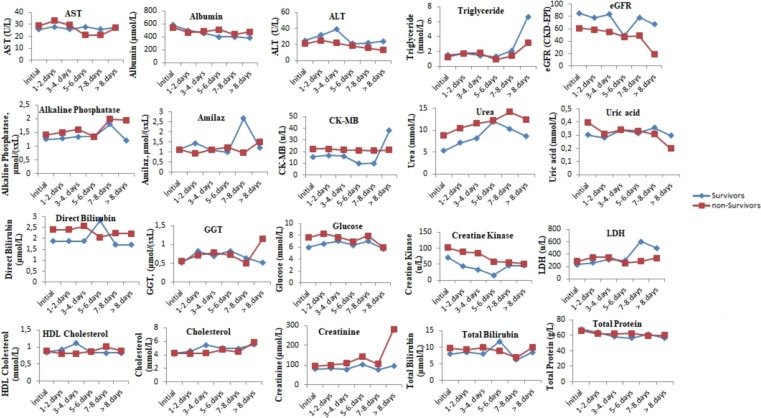
The comparison of the pre- and post-values of the biochemistry laboratory parameters of the surviving and non-surviving groups is presented in Table 1. These pre-values of the aspartate aminotransferase (AST), alkaline phosphatase, creatine kinase myocardial band (CK-MB), direct bilirubin, glucose, creatinine, creatinine kinase, lactate dehydrogenase (LDH), total bilirubin, urea and uric acid in the non-surviving group were higher, while the pre-values of albumin, total protein and were lower in the non-surviving group. In addition, the pre-post levels of alanine aminotransaminase (ALT), albumin and estimated glomerular filtration rate (eGFR) were lower in the non-surviving group, while the post-values of creatinine kinase, glucose, creatinine, CK-MB, urea, alkaline phosphatase, direct bilirubin, LDH and total bilirubin were higher in the non-surviving group.
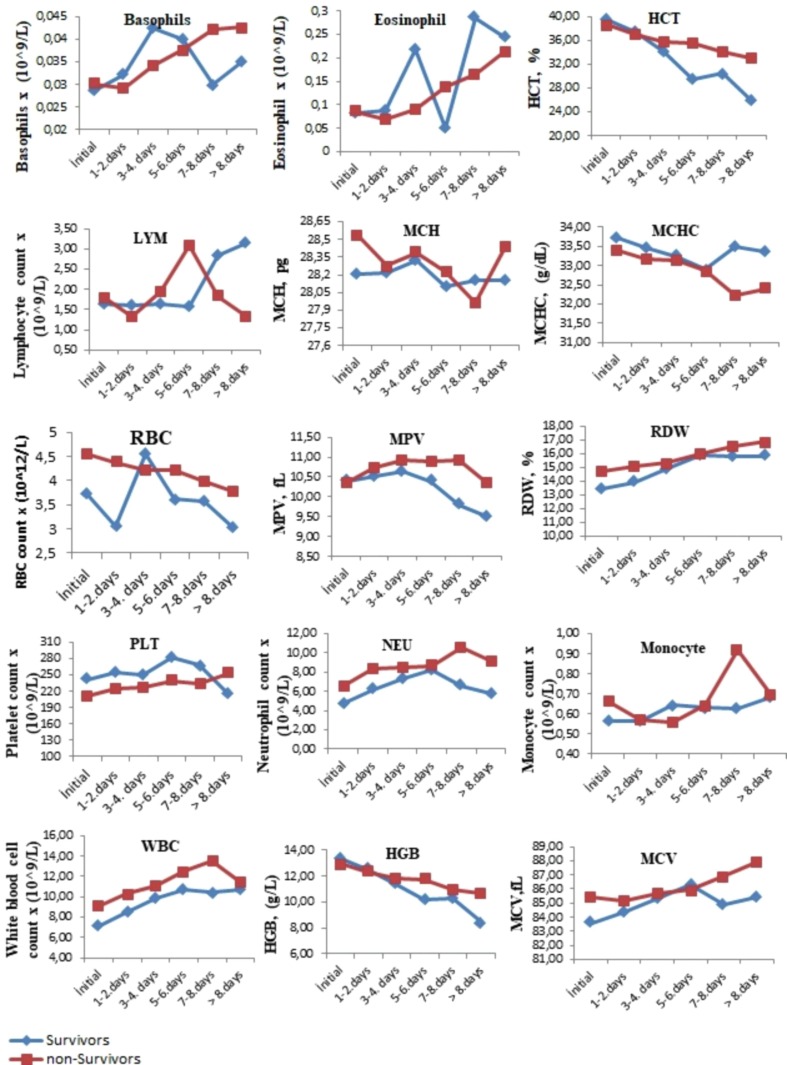
The comparison of the pre- and post-values of the hematological laboratory parameters of the surviving and non-survivor groups is presented in Table 2 . These pre-values of the hematocrit (HCT), hemoglobin (HGB), lymphocyte count (LYM), mean erythrocyte hemoglobin concentration (MCHC), platelet count (PLT) and red blood cells (RBC) in the non-surviving group were lower, while the pre-values of mean corpuscular hemoglobin (MCH), mean corpuscular volume (MCV), monocytes (MONO), neutrophil count (NEU), white blood cell count (WBC) and red cell distribution width (RDW) were higher in the non-survivor group. In addition, the post- levels of eosinophil count (ESO), HCT, HGB, LYM, MCHC, MONO, RBC, and PLT were lower in the non-survivor group, while the post-values of MCV, mean platelet volume (MPV), NEU, RDW, and WBC were higher in the non-survivor group.
Table 2.
Comparison of hematological variables between survivor and non-survivor groups.
| Survivor Covid-19 | Non-survivor Covid-19 | *p-value | ||
|---|---|---|---|---|
| Median (25 th quartile-75th quartile) | Median (25 th quartile-75th quartile) | |||
| Parameters | Reference range | |||
| BASO (pre) 109/L | 0.01–0.07 | 0.02 (0.01–0.04) | 0.02 (0.01–0.04) | 0.869 |
| BASO (post) 109/L | 0.03 (0.02–0.05) | 0.02 (0.01–0.05) | 0.262 | |
| ESO (pre) 109/L | 0.01–0.4 | 0.04 (0.01–0.12) | 0.03 (0.00–0.12) | 0.232 |
| ESO (post) 109/L | 0.05 (0.00–0.15) | 0.01 (0.00–0.08) | 0.000 | |
| HCT (pre) % | 36–48 | 39.55 (36.00–43.20) | 38.80 (34.90–42.30) | 0.041 |
| HCT (post) % | 37.10 (33.60–40.70) | 35.60 (31.20–39.90) | 0.022 | |
| HGB(pre) g/L | 13.5–17.5 | 13.30 (12.00–14.65) | 13.10 (11.50–14.50) | 0.016 |
| HGB (post) g/L | 12.50 (11.10–13.80) | 11.75 (9.90–13.30) | 0.002 | |
| LYM (pre) 109/L | 1–5 | 1.46 (0.99–2.03) | 1.32 (0.85–1.88) | 0.015 |
| LYM (post) 109/L | 1.24 (0.83–1.89) | 0.74 (0.42–1.23) | 0.000 | |
| MCH (pre) pg | 26–34 | 28.60 (27.30–29.60) | 28.80 (27.20–30.10) | 0.041 |
| MCH (post) pg | 28.60 (27.20–29.50) | 28.50 (26.90–29.70) | 0.989 | |
| MCHC (pre) g/dL | 33.80 (32.90–34.70) | 33.50 (32.40–34.60) | 0.004 | |
| MCHC (post) g/dL | 33.70 (32.60 – 34.40) | 33.20 (32.00–34.10) | 0.004 | |
| MCV (pre) fL | 80–96 | 83.90 (80.80–87.00) | 85.20 (81.80–88.90) | 0.000 |
| MCV (post) fL | 84.40 (81.20–87.30) | 85.55 (81.55–89.25) | 0.045 | |
| MONO (pre) 109/L | 0.1–1 | 0.51 (0.38–0.67) | 0.56 (0.44–0.72) | 0.000 |
| MONO (post) 109/L | 0.51 (0.37–0.68) | 0.42 (0.27–0.64) | 0.000 | |
| MPV (pre) fL | 9.1–11.9 | 10.30 (9.70–10.90) | 10.30 (9.55–11.00) | 0.613 |
| MPV (post) fL | 10.40 (9.90–11.00) | 11.00 (10.20–11.70) | 0.000 | |
| NEU (pre) 109/L | 1.8–6.98 | 4.05 (2.85–5.85) | 5.25 (3.98–7.65) | 0.000 |
| NEU (post) 109/L | 5.11 (3.53–7.99) | 8.59 (5.01–13.21) | 0.000 | |
| PLT (pre) 109/L | 150–450 | 229.00 (184.00–287.00) | 192.50 (138.00–204.00) | 0.000 |
| PLT (post) 109/L | 243.00 (182.00–311.00) | 197.50 (149.00–213.50) | 0.000 | |
| RBC (pre) 1012/L | 4.5–6 | 4.74 (4.36–5.14) | 3.21 (3.16–3.98) | 0.000 |
| RBC (post) 1012/L | 4.41 (3.99–4.84) | 3.19 (2.55–3.78) | 0.007 | |
| RDW (pre) % | 11–14 | 13.10 (12.50–13.90) | 14.00 (13.20–15.40) | 0.000 |
| RDW (post) % | 13.60 (12.80–14.70) | 14.95 (13.75–16.80) | 0.000 | |
| WBC(pre) 109/L | Male: 3.91–10.9Female: 4.49–12.68 | 6.5 (5.00–8.30) | 7.80 (6.20–10.10) | 0.000 |
| WBC (post) 109/L | 7.30 (5.60–10.10) | 10.15 (6.75–14.80) | 0.000 |
BASO: Basophil count; ESO: Eosinophil count; HCT: Hematocrit; HGB: Hemoglobin; LYM: Lymphocyte count; MCH: Mean corpuscular hemoglobin; MCHC: Mean erythrocyte hemoglobın concentratıon; MCV: Mean corpuscular volume; MONO: Monocyte count; MPV: Mean platelet volume; NEU: Neutrophil count; PLT: Platelet count; RBC: Red blood cells; RDW: Red cell distribution width; WBC: White blood cell count; IQR: interquartile range; The p values indicate the comparison of the survivor and non-survivor groups in pre-post measurements and they are bold when p < 0.05.
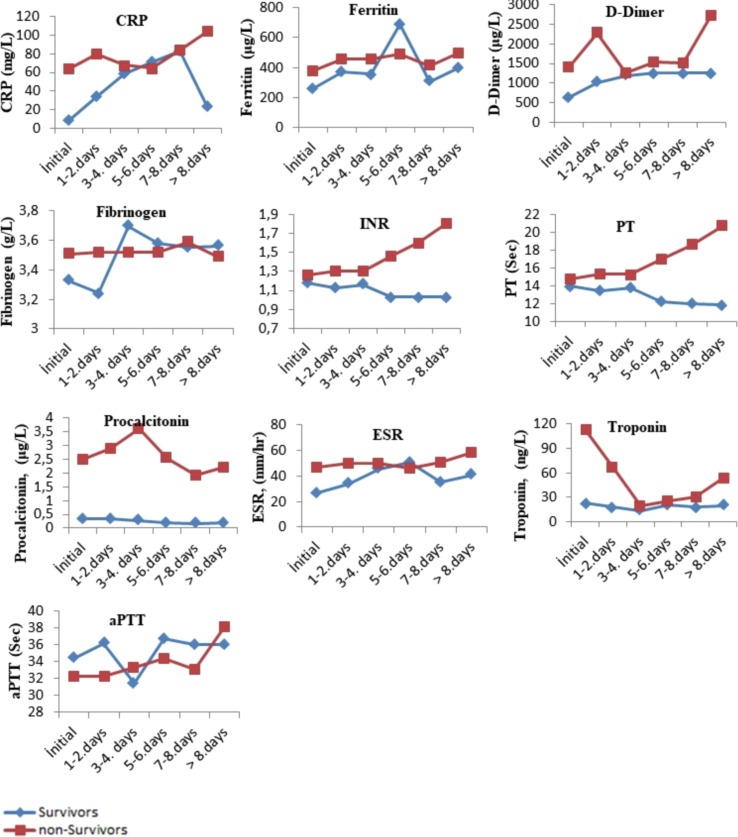
The comparison of the pre- and post-values of the immunological laboratory parameters of the surviving and non-survivor groups is presented in Table 3 . These pre-post values of C-reactive protein (CRP), D-dimer, ferritin, fibrinogen, international normalized ratio (INR), prothrombin time (PT), troponin, procalcitonin and erythrocyte sedimentation rate (ESR) were higher in the non-surviving group. In addition, the pre- and post-values of activated partial prothrombin time (aPTT) values were lower in the non-surviving group.
Table 3.
Comparison of inflammatory, cardiac and coagulation variables between survivor and non-survivor groups.
| survivor Covid-19 | non-survivor Covid-19 | *p-value | ||
|---|---|---|---|---|
| Median (25 th quartile-75th quartile) | Median (25 th quartile-75th quartile) | |||
| Parameters | Reference range | |||
| CRP (pre) mg/L | 0–5 | 6.76 (3.02–25.20) | 63.91 (17.10–72.90) | 0.000 |
| CRP (post) mg/L | 6.42 (3.02–41.00) | 97.61 (37.70–146.00) | 0.000 | |
| D-dimer (pre) μg/L | 80–583 | 441.00 (252.00–843.95) | 1390.00 (599.36 – 1992.00) | 0.000 |
| D-dimer (post) μg/L | 524.55 (232.00–1249.63) | 2214.19 (852.81–4460.00) | 0.000 | |
| Ferritin (pre) μg/L | Male: 22–322 Female: 10–291 | 125.95 (47.10–317.50) | 379.38 (282.50–395.00) | 0.000 |
| Ferritin (post) μg/L | 224.30 (75.90–436.40) | 527.18 (216.50–1650.00) | 0.000 | |
| Fibrinogen (pre) g/L | 18–145 | 3.211 (2.878–3.649) | 3.50 (3.50–3.50) | 0.000 |
| Fibrinogen (post) g/L | 3.24 (2.81–3.73) | 3.50 (3.50 – 3.50) | 0.002 | |
| INR (pre) | 0.6–1.2 | 1.10 (1.04–1.17) | 1.25 (1.20–1.44) | 0.000 |
| INR (post) | 1.10 (1.03–1.16) | 1.20 (1.20–1.20) | 0.000 | |
| PT (pre) Sec | 10.1–15.9 | 13.10 (12.40–13.90) | 14.83 (14.20–16.90) | 0.000 |
| PT (post) Sec | 13.10 (12.30–13.80) | 14.20 (14.20–14.20) | 0.000 | |
| Procalcitonin (pre) μg/L | < 0.15 | 0.12 (0.12–0.12) | 2.53 (2.53–3.00) | 0.000 |
| Procalcitonin (post) μg/L | 0.12 (0.12 – 0.19) | 3.46 (0.17–4.00) | 0.000 | |
| ESR (pre) mm/hr | 0–20 | 17.00 (70.00–38.00) | 47.05 (20.00–49.00) | 0.000 |
| ESR (post) mm/hr | 19.00 (8.00–55.50) | 52.36 (15.00–79.00) | 0.000 | |
| Troponin (pre) ng/L | 10–23 | 17.00 (10.00–22.51) | 57.00 (10.00–112.75) | 0.000 |
| Troponin (post) ng/L | 10.00 (10.00–10.00) | 112.00 (10.00–10.00) | 0.000 | |
| aPTT (pre) Sec | 22–45 | 35.34 (29.20–37.05) | 31.00 (30.00–32.00) | 0.000 |
| aPTT (post) Sec | 33.30 (28.10–38.80) | 32.00 (32.00–32.20) | 0.047 |
CRP: C-reactive protein; INR: international normalized ratio; PT: prothrombin time; ESR: erythrocyte sedimentation rate; aPTT: activated partial prothrombin time; IQR: interquartile range; The p values indicate the comparison of the Survivor and non-Survivor groups in pre-post measurements and they are bold when p < 0.05.
The pre- and post-values of the biochemical parameters of the surviving and non-surviving groups were compared within the groups (Table 4 ). The post-values of the ALT, GGT, LDH, and urea were higher than their pre-values in both groups, while the post-values of the albumin and total protein were lower than their pre-values in both groups. Moreover, the pre- and post-values of the AST, alkaline phosphatase, amylase, CK-MB, direct bilirubin, glucose, HDL cholesterol, cholesterol, creatine kinase, total bilirubin and triglyceride did not change in the non-surviving group. Furthermore, the pre- and post-values of creatinine and uric acid did not change in the surviving group, while the post-values of these parameters were lower than the pre-values in the non-surviving group.
Table 4.
Comparison of “pre” and “post” biochemical values in survivor and non-survivor groups.
| Survivor Covid-19 | *p value | Non-survivor Covid-19 | *p value | ||
|---|---|---|---|---|---|
| Median (25 th quartile-75th quartile) | Median (25 th quartile-75th quartile) | ||||
| Parameters | Reference range | ||||
| ALT (pre) U/L | 0–35 | 25.00 (16.00–42.00) | 0.000 | 21.00 (14.00–34.00) | 0.029 |
| ALT (post) U/L | 32.00 (19.00–64.00) | 24.00 (16.00–47.00) | |||
| AST (pre) U/L | 0–50 | 26.00 (20.00–37.00) | 0.072 | 30.00 (20.00–47.00) | 0.349 |
| AST (post) U/L | 27.50 (21.00–41.00) | 32.00 (22.00–56.00) | |||
| Albumin (pre) μmol/L | 527–782 | 582.30 (516.01–651.52) | 0.000 | 544.84 (476.83–605.63) | 0.000 |
| Albumin (post) μmol/L | 503.31 (454.41–586.82) | 455.91 (386.70–556.72) | |||
| Alkaline Phosphatase (pre) μmol/s.L | 0.5–2.0 | 1.25 (1.03–1.57) | 0.075 | 1.42 1.10–3.13) | 0.268 |
| Alkaline Phosphatase (post) μmol/s.L | 1.09 (1.09–1.7) | 1.55 (1.20–2.00) | |||
| Amylase (pre) μmol/s.L | 0.47–1.67 | 1.13 (0.85–1.50) | 0.025 | 1.12 (0.78–1.48) | 0.613 |
| Amylase (post) μmol/s.L | 1.46 (1.00–1.78) | 1.13 (1.13–1.53) | |||
| CK - MB (pre) u/L | 0–24 | 15.70 (12.40–21.60) | 0.672 | 22.65 (15.60–31.40) | 0.985 |
| CK - MB (post) u/L | 16.90 (12.70–21.10) | 21.40 (16.10–32.00) | |||
| Direct Bilirubin (pre) μmol/L | 0–3.4 | 1.88 (1.37–2.56) | 0.481 | 2.39 (1.54–4.10) | 0.053 |
| Direct Bilirubin (post) μmol/L | 1.88 (1.37 – 2.56) | 2.91 (1.88–4.61) | |||
| GGT (pre) μmol/s.L | 0–0.63 | 0.51(0.30–0.94) | 0.000 | 0.55 (0.32–1.18) | 0.031 |
| GGT (post) μmol/s.L | 0.78 (0.42–1.32) | 0.77 (0.37–2.01) | |||
| Glucose (pre) mmol/L | 4.11–5.55 | 5.99 (5.16–8.10) | 0.095 | 7.72 (6.22–10.77) | 0.178 |
| Glucose (post) mmol/L | 6.27 (5.16–10.05) | 8.21(6.27–13.82) | |||
| HDL Cholesterol (pre) mmol/L | 1.03–1.55 | 0.35 (27.00–0.70) | 0.090 | 0.88 (0.80–1.24) | 0.276 |
| HDL Cholesterol (post) mmol/L | 0.93 (0.72 – 1.11) | 0.82 (0.72–1.03) | |||
| Cholesterol (pre) mmol/L | 0–5.17 | 4.19 (3.49–5.04) | 0.010 | 4.26 (3.62–5.66) | 0.775 |
| Cholesterol (post) mmol/L | 4.60 (3.72–5.61) | 4.32 (3.49–5.35) | |||
| Creatinine (pre) μmol/L | 74.26–110.50 | 80.44 (68.95–92.82) | 0.090 | 96.35 (75.14–133.48) | 0.028 |
| Creatinine (post) μmol/L | 83.09 (68.95–99.89) | 103.43 (82.21–152.93) | |||
| Creatine Kinase (pre) u/L | 0–145 | 71.00 (44.00–106.00) | 0.000 | 101.00 (59.00–237.00) | 0.115 |
| Creatine Kinase (post) u/L | 44.00 (31.00–91.50) | 91.00 (50.00–171.00) | |||
| LDH (pre) u/L | 0–248 | 233.00 (193.00–292.00) | 0.000 | 289.00 (223.00–367.00) | 0.008 |
| LDH (post) u/L | 264.00 (217.00–332.00) | 338.50 (255.00–484.50) | |||
| Total Bilirubin (pre) μmol/L | 5.13–20.52 | 8.04 (5.98–10.94) | 0.354 | 9.74 (6.67–14.19) | 0.418 |
| Total Bilirubin (post) μmol/L | 8.38 (6.15–11.45) | 10.60 (7.18–15.73) | |||
| Total Protein (pre) g/L | 66–83 | 68.10 (63.40–73.45) | 0.000 | 66.33 (59.71–70.24) | 0.004 |
| Total Protein (post) g/L | 64.60 (57.25–69.95) | 61.35 (54.70–68.50) | |||
| Triglyceride (pre) mmol/L | 0–1.69 | 1.49 (1.08–2.10) | 0.007 | 1.26 (1.04–2.05) | 0.659 |
| Triglyceride (post) mmol/L | 1.71 (1.36–2.41) | 1.62 (0.84–2.89) | |||
| eGFR (pre) | 85.10 (66.33–99.36) | 0.000 | 61.08 (40.86–83.83) | 0.061 | |
| eGFR (post) | 77.44 (56.04–92.94) | 55.48 (34.34–76.59) | |||
| Urea (pre) mmol/L | 2.83–7.16 | 5.33 (3.99–7.61) | 0.000 | 8.88 (6.46–13.32) | 0.000 |
| Urea (post) mmol/L | 6.99 (4.99–10.32) | 11.57 (7.49–22.81) | |||
| Uric acid (pre) mmol/L | 0.21–0.43 | 0.30 (0.24–0.39) | 0.111 | 0.40 (0.29–0.51) | 0.031 |
| Uric acid (post) mmol/L | 0.28 (0.20–0.38) | 0.33 (0.27–0.40) |
ALT: alanine aminotransaminase; AST: aspartate aminotransferase; CK-MB: creatine kinase myocardial band; GGT: Gamma Glutamyl Transferase enzyme; LDH: lactate dehydrogenase; eGFR: estimated glomerular filtration rate; IQR: interquartile range; The p values indicate the comparison of the pre and post values of the parameters of each group and they are bold when p < 0.05.
The pre- and post-values of the hematological parameters of the surviving and non-surviving groups were compared within the groups (Table 5 ). The post-values of the HCT, HGB, LYM, MCHC, and RBC were lower than the pre-values in both groups, while the post-values of the MPV, NEU, PLT, RDW, WBC, and RDW in the non-surviving group were higher than in the pre-value. In addition, the post-values of ESO and MONO in the non-surviving group were lower than in the pre-value, while the pre- and post-values of these parameters did not change in the surviving group. The pre- and post-values of BASO, MCH, MCV, ESO, MPV, and MONO did not change in the surviving group.
Table 5.
Comparison of “pre” and “post” hemotological values in survivor and non-survivor groups.
| Survivor Covid-19 |
*p value |
Non-Survivor Covid-19 |
*p value |
||
|---|---|---|---|---|---|
| Median (25 th quartile-75th quartile) | Median (25 th quartile-75th quartile) | ||||
| Parameters | Reference range | ||||
| BASO (pre) 109/L | 0.01–0.07 | 0.02 (0.01–0.04) | 0.000 | 0.02 (0.01–0.04) | 0.191 |
| BASO (post) 109/L | 0.03 (0.02–0.05) | 0.02 (0.01–0.05) | |||
| ESO (pre) 109/L | 0.01–0.4 | 0.04 (0.01–0.12) | 0.282 | 0.03 (0.00–0.12) | 0.017 |
| ESO (post) 109/L | 0.05 (0.00–0.15) | 0.01 (0.00–0.08) | |||
| HCT (pre) % | 36–48 | 39.55 (36.00–43.20) | 0.000 | 38.80 (34.90–42.30) | 0.000 |
| HCT (post) % | 37.10 (33.60–40.70) | 35.60 (31.20–39.90) | |||
| HGB(pre) g/L | 13.5–17.5 | 13.30 (12.00–14.65) | 0.000 | 13.10 (11.50–14.50) | 0.000 |
| HGB (post) g/L | 12.50 (11.10–13.80) | 11.75 (9.90–13.30) | |||
| LYM (pre) 109/L | 1–5 | 1.46 (0.99–2.03) | 0.001 | 1.32 (0.85–1.88) | 0.000 |
| LYM (post) 109/L | 1.24 (0.83–1.89) | 0.74 (0.42–1.23) | |||
| MCH (pre) pg | 26–34 | 28.60 (27.30–29.60) | 0.736 | 28.80 (27.20–30.10) | 0.145 |
| MCH (post) pg | 28.60 (27.20–29.50) | 28.50 (26.90–29.70) | |||
| MCHC (pre) g/dL | 33.80 (32.90–34.70) | 0.022 | 33.50 (32.40–34.60) | 0.043 | |
| MCHC (post) g/dL | 33.70 (32.60 – 34.40) | 33.20 (32.00–34.10) | |||
| MCV (pre) fL | 80–96 | 83.90 (80.80–87.00) | 0.173 | 85.20 (81.80–88.90) | 0.961 |
| MCV (post) fL | 84.40 (81.20–87.30) | 85.55 (81.55–89.25) | |||
| MONO (pre) 109/L | 0.1–1 | 0.51 (0.38–0.67) | 0.628 | 0.56 (0.44–0.72) | 0.000 |
| MONO (post) 109/L | 0.51 (0.37–0.68) | 0.42 (0.27–0.64) | |||
| MPV (pre) fL | 9.1–11.9 | 10.30 (9.70–10.90) | 0.160 | 10.30 (9.55–11.00) | 0.000 |
| MPV (post) fL | 10.40 (9.90–11.00) | 11.00 (10.20–11.70) | |||
| NEU (pre) 109/L | 1.8–6.98 | 4.05 (2.85–5.85) | 0.000 | 5.25 (3.98–7.65) | 0.000 |
| NEU (post) 109/L | 5.11 (3.53–7.99) | 8.59 (5.01–13.21) | |||
| PLT (pre) 109/L | 150–450 | 229.00 (184.00–287.00) | 0.040 | 192.50 (138.00–204.00) | 0.000 |
| PLT (post) 109/L | 243.00 (182.00–311.00) | 197.50 (149.00–213.50) | |||
| RBC (pre) 1012/L | 4.5–6 | 4.74 (4.36–5.14) | 0.000 | 3.21 (3.16–3.98) | 0.000 |
| RBC (post) 1012/L | 4.41 (3.99–4.84) | 3.19 (2.55–3.78) | |||
| RDW (pre) % | 11–14 | 13.10 (12.50–13.90) | 0.000 | 14.00 (13.20–15.40) | 0.000 |
| RDW (post) % | 13.60 (12.80–14.70) | 14.95 (13.75–16.80) | |||
| WBC(pre) 109/L | Male: 3.91–10.9 Female: 4.49–12.68 |
6.5 (5.00–8.30) | 0.000 | 7.80 (6.20–10.10) | 0.000 |
| WBC (post) 109/L | 7.30 (5.60–10.10) | 10.15 (6.75–14.80) |
BASO: Basophil count; ESO: Eosinophil count; HCT: Hematocrit; HGB: Hemoglobin; LYM: Lymphocyte count; MCH: Mean corpuscular hemoglobin; MCHC: Mean erythrocyte hemoglobın concentratıon; MCV: Mean corpuscular volume; MONO: Monocyte count; MPV: Mean platelet volume; NEU: Neutrophil count; PLT: Platelet count; RBC: Red blood cells; RDW: Red cell distribution width; WBC: White blood cell count; IQR: interquartile range; The p values indicate the comparison of the pre and post values of the parameters of each group and they are bold when p < 0.05.
The pre- and post-values of the immunological parameters of the surviving and non-surviving groups were compared within the groups (Table 6 ). The post-value of ferritin was higher than the pre-value in both groups. In addition, the pre- and post-values of fibrinogen, INR, PT were not different between the groups. Moreover, the post-values of the CRP, D-dimer, procalcitonin, ESR, troponin, aPTT in the non-surviving group were higher than the pre-values, while the pre- and post-values of these parameters were not different in the surviving group.
Table 6.
Comparison of “pre” and “post” immunological values in survivor and non-survivor groups.
| survivor Covid-19 | *p value | non-survivor Covid-19 | *p value | ||
|---|---|---|---|---|---|
| Median (25 th quartile-75th quartile) | Median (25 th quartile-75th quartile) | ||||
| Parameters | Reference range | ||||
| CRP (pre) mg/L | 0–5 | 6.76 (3.02–25.20) | 0.424 | 63.91 (17.10–72.90) | 0.000 |
| CRP (post) mg/L | 6.42 (3.02–41.00) | 97.61 (37.70–146.00) | |||
| D-Dimer (pre) μg/L | 80–583 | 441.00 (252.00–843.95) | 0.498 | 1390.00 (599.36 – 1992.00) | 0.015 |
| D-Dimer (post) μg/L | 524.55 (232.00–1249.63) | 2214.19 (852.81–4460.00) | |||
| Ferritin (pre) μg/L | Male: 22–322 Female: 10–291 |
125.95 (47.10–317.50) | 0.002 | 379.38 (282.50–395.00) | 0.000 |
| Ferritin (post) μg/L | 224.30 (75.90–436.40) | 527.18 (216.50–1650.00) | |||
| Fibrinogen (pre) g/L | 18–145 | 3.211 (2.878–3.649) | 0.853 | 3.50 (3.50–3.50) | 0.610 |
| Fibrinogen (post) g/L | 3.24 (2.81–3.73) | 3.50 (3.50 – 3.50) | |||
| INR (pre) | 0.6–1.2 | 1.10 (1.04–1.17) | 0.704 | 1.25 (1.20–1.44) | 0.076 |
| INR (post) | 1.10 (1.03–1.16) | 1.20 (1.20–1.20) | |||
| PT (pre) Sec | 10.1–15.9 | 13.10 (12.40–13.90) | 0.753 | 14.83 (14.20–16.90) | 0.160 |
| PT (post) Sec | 13.10 (12.30–13.80) | 14.20 (14.20–14.20) | |||
| Procalcitonin (pre) μg/L | < 0.15 | 0.12 (0.12–0.12) | 0.235 | 2.53 (2.53–3.00) | 0.044 |
| Procalcitonin (post) μg/L | 0.12 (0.12 – 0.19) | 3.46 (0.17–4.00) | |||
| ESR (pre) mm/hr | 0–20 | 17.00 (70.00–38.00) | 0.184 | 47.05 (20.00–49.00) | 0.002 |
| ESR (post) mm/hr | 19.00 (8.00–55.50) | 52.36 (15.00–79.00) | |||
| Troponin (pre) ng/L | 10–23 | 17.00 (10.00–22.51) | 0.446 | 57.00 (10.00–112.75) | 0.005 |
| Troponin (post) ng/L | 10.00 (10.00–10.00) | 112.00 (10.00–10.00) | |||
| aPTT (pre) Sec | 22–45 | 35.34 (29.20–37.05) | 0.574 | 31.00 (30.00–32.00) | 0.003 |
| aPTT (post) Sec | 33.30 (28.10–38.80) | 32.00 (32.00–32.20) |
CRP: C-reactive protein; INR: international normalized ratio; PT: prothrombin time; ESR: erythrocyte sedimentation rate; aPTT: activated partial prothrombin time; IQR: interquartile range; The p values indicate the comparison of the pre and post values of the parameters of each group and they are bold when p < 0.05.
The comparison of the tests performed when the patients were first admitted to the hospital with the tests results performed on the following days during their hospitalization is presented in Fig. 1, Fig. 2, Fig. 3 . The levels of albumin (except for 1–4 days), alkaline phosphatase (except for 5–6 days), CK-MB (except for > 8 days), direct bilirubin (except for 5–6 days), glucose, creatinine, creatinine kinase, LDH (except for 5–8 days), total bilirubin (except for 5–6 days), total protein (except for > 7 days), urea (except for 5–6 days), uric acid (except for > 7 days), MCH (except for 7–8 days), RBC (except for 1–4 days), MPV (except for the initial values), RDW (except for 5–6 days), PLT (except for > 8 days), NEU, WBC, HGB (except for 1–4 days), MCV (except for 5–6 days), CRP (except for 5–6 days), ferritin (except for 5–6 days), D-dimer (except for 3–4 days), INR, PT, procalcitonin, ESR (except for 5–6 days) and troponin were higher in the non-surviving group. In addition, the levels of ALT, eGFR (except for 5–6 days), ESO (except for 5–6 days), LYM (except for 1–7 days), MCHC (except 5–6 days), PLT (except for > 8 days) and fibrinogen (1–4 for days) were lower in the non-surviving group.
Fig. 1.
The comparison of the biochemical tests performed when the surviving and non-surviving COVID-19 patients were first admitted to the hospital with the its results performed on the following days during their hospitalization.
Fig. 2.
The comparison of the hemotological tests performed when the surviving and non-surviving COVID-19 patients were first admitted to the hospital with the its results performed on the following days during their hospitalization.
Fig. 3.
The comparison of the immunological tests performed when the surviving and non– surviving COVID-19 patients were first admitted to the hospital with the its results performed on the following days during their hospitalization.
Multivariate logistic regression analysis was performed to identify the complete blood count parameters that constituted the risk indicators for mortality, and significant ORs were reported (Table 7 ). When the OR values were examined, the CRP (OR = 1.739, 95% CI = 1.221–1.958), D-dimer (OR = 1.610, 95% CI = 1.110–1.744), ESR (OR = 2.072, 95% CI = 1.435–2.110), INR (OR = 1.432, 95% CI = 1.163–1.507), PT (OR = 1.966, 95% CI = 0.961–3.916), RDW (OR = 1.534, 95% CI = 1.250–1.623), and PLT (OR = 0.795) , 95% CI = 0.580–0.811) were the most significant risk indicators for mortality. Furthermore, ferritin (OR = 1.186, 95% CI = 1.013–1.298), procalcitonin (OR = 1.068, 95% CI = 1.036–1.088), WBC (OR = 1.126, 95% CI = 1.030–1.490), NEU (OR = 1.149, 95% CI = 1.097–1.205), and RBC (OR = 0.911, 95% CI = 0.800–0.922) were other important risk indicators (Table 7).
Table 7.
Variables associated with COVID-19 mortality obtained as a result of multivariate logistic regression analysis.
| Independent variables | OR | 95 % CI | *p-value |
|---|---|---|---|
| CRP (mg/L) | 1.739 | 1.221–1.958 | 0.000 |
| D-Dimer (μg/L) | 1.610 | 1.110–1.744 | 0.006 |
| Ferritin (μg/L) | 1.186 | 1.013–1.298 | 0.022 |
| Procalcitonin (μg/L) | 1.068 | 1.036–1.088 | 0.000 |
| ESR (mm/hr) | 2.072 | 1.435–2.110 | 0.000 |
| INR | 1.432 | 1.163–1.507 | 0.000 |
| PT (Sec) | 1.966 | 0.961–3.916 | 0.007 |
| WBC (109/L) | 1.126 | 1.030–1.490 | 0.000 |
| RDW (%) | 1.534 | 1.250–1.623 | 0.000 |
| NEU (109/L) | 1.149 | 1.097–1.205 | 0.000 |
| RBC (1012/L) | 0.911 | 0.800–0.922 | 0.000 |
| PLT (109/L) | 0.795 | 0.580–0.811 | 0.000 |
CRP: C-reactive protein; ESR: erythrocyte sedimentation rate INR: international normalized ratio; PT: prothrombin time; WBC: White blood cell count; RDW: Red cell distribution width; NEU: Neutrophil count; RBC: Red blood cells; PLT: Platelet count; OR: Odss-raito; CI: Confidence interval; *p value < 0.05 was considered significant.
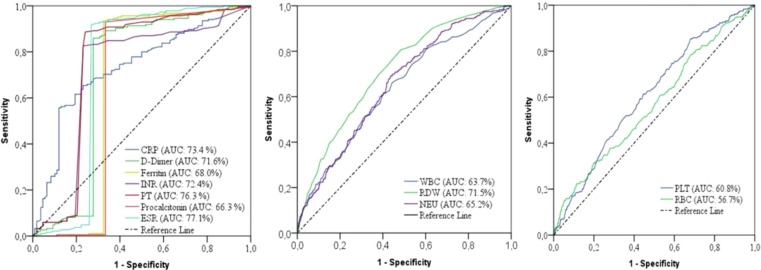
ROC analysis was performed to determine the cut-off values of the parameters that were risk indicators as a predictor of mortality in COVID-19 disease (Table 8 ). In addition, the survival status of the patients was determined accurately, to a large extent, by using the cut-off values that were calculated for the predictors (Fig. 4 ).
Table 8.
Diagnostic performances of risk parameters for Covid-19 mortality.
| Independent variables | Cut off | Specificity (%) (95 % CI) |
Sensitivity (%) (95 % CI) |
AUC (%) (95 % CI) |
*p |
|---|---|---|---|---|---|
| CRP (mg/L) | > 62.6 | 87.0 (82.4–95.4) |
76.2 (62.4–78.0) |
73.4 (71.0–76.4) |
0.000 |
| D-Dimer (μg/L) | > 1262.26 | 72.2 (70.2–82.5) |
85.9 (75.8–89.7) |
71.6 (68.3–72.6) |
0.000 |
| Ferritin (μg/L) | > 392.05 | 67.6 (56.9–70.1) |
93.5 (88.2–95.5) |
68.0 (67.2–71.9) |
0.000 |
| Procalcitonin (μg/L) | > 0.14 | 66.7 (56.2–69.3) |
92.0 (90.0–95.4) |
66.3 (64.1–68.1) |
0.000 |
| ESR (mm/hr) | > 48.50 | 73.1 (71.2–77.0) |
91.9 (90.1–94.2) |
77.1 (75.6–80.1) |
0.000 |
| INR | > 1.19 | 76.9 (75.4–79.8) |
82.7 (80.8–87.7) |
72.4 (69.5–74.1) |
0.000 |
| PT (Sec) | > 13.95 | 75.9 (71.5–78.6) |
88.6 (86.7–90.5) |
76.3 (75.4–80.0) |
0.000 |
| WBC (109/L) | > 12.25 | 87.9 (85.5–90.3) |
66.8 (61.4–68.1) |
63.6 (59.0–66.2) |
0.000 |
| RDW (%) | > 15.05 | 90.9 (88.7–93.2) |
68.8 (65.6–71.2) |
71.5 (70.8–72.4) |
0.000 |
| NEU (109/L) | > 10.38 | 84.6 (82.3–86.9) |
64.2 (58.0–67.1) |
65.2 (63.8–68.6) |
0.000 |
| PLT (109/L) | < 148.50 | 89.3 (87.0–94.1) |
66.7 (57.4–68.7) |
60.8 (58.6–64.5) |
0.000 |
| RBC 1012/L | < 3.10 | 92.3 (90.5–93.2) |
63.4 (60.8–68.3) |
56.7 (55.0–61.2) |
0.000 |
CRP: C-reactive protein; ESR: erythrocyte sedimentation rate; INR: international normalized ratio; PT: prothrombin time; WBC: White blood cell count; RDW: Red cell distribution width; NEU: Neutrophil count; PLT: Platelet count; RBC: Red blood cells; *p value < 0.05 was considered significant.
Fig. 4.
ROC curves of biomarkers for COVID-19 mortality.
According to the results of the ROC analysis, the cut-off values of the CRP, D-dimer, ferritin, procalcitonin, ESR, INR, PT, WBC, RDW, NEU, PLT, and RBC were > 62.6 mg/L, >1262.26 µg/L, >392.05 µg/L, >0.14 µg/L, >48.50 mm/h, >1.19, >13.95 s, >12.25 × 109/L, >15.05%, >10.38 × 109/L, >148.50 × 109/L, and < 3.10 × 1012/L for mortality, respectively.
Moreover, the diagnostic performances of the biomarkers that were predictive indicators of mortality were obtained (Table 8). The independent mortality biomarkers with the highest diagnostic performance values were as follows: ESR (AUC: 77.1, sensitivity: 91.9, specificity: 73.1), PT (AUC: 76.3, sensitivity: 88.6, specificity: 75.9), CRP (AUC: 73.4, sensitivity: 76.2, specificity: 87.0), INR (AUC: 72.4, sensitivity: 76.9, specificity: 82.7), D-dimer (AUC: 71.6, sensitivity: 85.9, specificity: 72.2), RDW (AUC: 71.5, sensitivity: 68.8, specificity: 90.9). The diagnostic performances of other effective biomarkers in the prediction of COVID-19 mortality were as follows: ferritin (AUC: 68.0, sensitivity: 93.5, specificity: 67.6), procalcitonin (AUC: 66.3, sensitivity: 92.0, specificity: 66.7), NEU (AUC: 65.2, sensitivity: 64.2, specificity: 84.6), WBC (AUC: 63.6, sensitivity: 66.8, specificity: 87.9), PLT (AUC: 60.8, sensitivity: 66.7, specificity: 89.3), and RBC (AUC: 56.7, sensitivity: 63.4, specificity: 92.3), respectively.
4. Discussion
COVID-19 spread rapidly around the world and has infected millions of people. Despite the abundance of publications, many are contradictory, and many pathological aspects of this disease remain unclear [15]. During the course of the disease, changes have been observed in many biochemical parameters, in addition to hematological abnormalities [15]. COVID-19 disease, caused by SARS-CoV-2, leads to high mortality with widespread inflammation and cytokine storm [16]. Due to the high death rates of this pandemic, a serious struggle against the disease continues all over the world [16].
It has been reported that the mortality rate in critically ill patients with COVID-19 can reach 61.5% [17]. Therefore, it is important to provide appropriate treatment and reduce the death rate from the serious and critical diseases that are caused by COVID-19 [16], [17]. In this study, the changes in the RBVs and clinical characteristics of the patients who died from COVID-19 infection and those who recovered from this infection were compared retrospectively, and significant differences were observed. To the best of our knowledge, this was the first study in the literature to monitor the dynamic changes of most of the routine biochemistry and hematological variables in non-surviving patients. In this study, the characteristics of the biochemical and hematological laboratory parameters in the living and deceased COVID-19 patients were determined and predictors of disease mortality were obtained. In addition, determining the groups as surviving and non-surviving patients made extremely important the findings of the study. The findings will provide important evidence to revise the diagnosis and treatment scheme of critically ill patients to improve clinical outcomes.
In this study, the mortality of the treated patients (n = 4597) was 5.07% (233/4597). This result was lower than that reported by Zhou et al. (28.3%) [18]. However, Guan et al. found the mortality of the disease as 1.4%, while this rate in another large-scale study was at 2.3% [6]. These different mortality results may have been due to the different sample sizes and case inclusion criteria used in the studies. In this study, the deceased patients were older than survivors, and male gender was found to be significant in the deceased patients. It was found to be consistent with the literature that the high age and male gender were indicators affecting mortality [11], [16]. Previous studies have indicated that reduced immunity and greater underlying comorbidities in older patients may have an impact on poor prognosis [2], [19], [20].
In many studies, it has been stated that the levels of serum AST, ALT and LDH, which are liver function tests, were higher in severe COVID-19 patients when compared to the mild COVID-19 patients [1], [5], [21]. Onur et al. [20] showed that the AST, ALT, and LDH activities were higher in deceased COVID-19 patients than in living patients. Guan et al [6] noted that the ALT and AST levels were higher in severe patients than in non-serious patients. In this manuscript, the pre- and post-levels of ALT were lower and the pre-levels of AST and pre- and post-levels of LDH were found to be higher in the non-surviving group when compared to the surviving group. These results indicated that severe liver dysfunction may have developed in those who died from COVID-19.
In a study, was found that the serum levels of CK-MB, bilirubin, urea, and creatinine, which are renal function tests, were higher and the eGFR level decreased in severe COVID-19 patients when compared to mildly infected patients [14]. Similarly, in another one study was found that the bilirubin level was higher in severe cases of COVID-19 than in non-serious cases [6]. In this study, the pre- and post-values, and dynamic changes in the direct bilirubin, alkaline phosphatase, CK-MB, bilirubin, urea, uric acid, creatinine, creatine kinase values were higher, while the values of eGFR and albumin in the non-surviving patients were lower than in the surviving patients. These results indicated that kidney dysfunction may have developed due to severe kidney involvement in those who died from COVID-19.
Viruses alter their host cell metabolism to provide optimal conditions for their rapid/efficient replication and spread. One of these changes is the deterioration in glucose metabolism [22]. One study suggested that high fasting blood glucose in COVID-19 was an independent predictor of mortality, even in nondiabetic subjects [1]. In this study, the pre- and post-values, and dynamic changes in the glucose levels were higher in the non-surviving patients than in the surviving patients.
CRP is an inflammatory marker that is widely used in clinical studies. High values of CRP indicate inflammation caused by various infections [16]. In many studies examining the changes in routine laboratory parameters in COVID-19, increased CRP levels were reported in severe patients when compared to mild patients [1], [5], [20]. In another study, it was stated that COVID patients who died had higher CRP levels than living infected patients [2]. In addition, Wang et al. [23] reported higher CRP levels in patients who died when compared to surviving patients during their 15-day follow-up after the diagnosis of COVID-19. In the current study, patients in the deceased group had higher pre and post CRP levels than the patients in the living group. Wu et al. [24] found that an increased high-sensitivity CRP value was significantly associated with higher risks of ARDS in COVID-19 patients. Zhang et al. [25] suggested that CRP is a marker of cytokine storm developing in COVID-19 patients and is associated with disease mortality. In the study, increased acute respiratory distress and cytokine storm due to high CRP levels can be shown as the cause of mortality in the deceased group.
Various studies have been conducted on the level of acute phase reactants in COVID-19. In one study, it was reported that ESR, D-dimer, and procalcitonin levels increased significantly in severe cases [1], [5]. In another study, ferritin and procalcitonin values were found to be high in non-surviving and severe COVID-19 patients. In another study, it was stated that laboratory changes, such as an increased PT and increased D-dimer were important predictors of ICU admission [26]. In the current study, the pre and post-values of the D-dimer, ESR, ferritin, fibrinogen, INR, PT, and procalcitonin were higher in the patients who died from COVID-19 than in those who survived (Table 3, Fig. 3). In addition, the procalcitonin levels of the patients who died were high when compared to the survivors until the last measurement, and it was the parameter that clearly showed the prognosis of the disease. It was reported in previous studies that the increase in these values was due to a secondary bacterial infection or an increased inflammatory response defined as the cytokine storm associated with COVID-19 infection [23], [27].
The major difference was D-dimer, a fibrin degradation product indicative of intravascular thrombosis. Because coagulation indicators and platelets are used to generate microthrombi, the significant increase in PT and decrease in PLT in this study supported the notion of possible coagulopathy [6], [23], [24], [25], [26], [27], [28], [29], [30]. In addition, the high values of PT and INR were interpreted in favor of hypercoagulation in a significant portion of the patients who died in this study. In another study, the prolonged PT, INR, and aPTT values were interpreted in favor of hypercoagulation in a significant number of the patients [1]. In the present study, coagulation indicators were significantly higher in the deceased group than in the living group. These results indicated that cardiovascular pathologies due to coagulation may have increased in the patients who died. In one study, the troponin values were found to be higher in patients with COVID-19 who died than in surviving patients [2]. In this study, the cardiac troponin values, a marker of heart muscle damage, were significantly higher in the deceased patients. The increase in the troponin values may suggest the risk of tissue death, organ failure, or heart attack in the patients who died.
In one study, it was stated that in the peripheral blood system of COVID-19 patients, lower levels of RBC, lymphocytes, platelets, HGB and higher neutrophils were observed [16]. In another study, it was stated that laboratory changes, such as leukocytosis, neutrophilia, and lymphopenia, were important determinants of admission to the ICU [25]. In another study, it was stated that these hematological changes in the deceased patients shortened the half-life of the erythrocytes or suppressed their production [1], [2]. It was also said that findings similar to changes in the MCV and RDW in the current study may indicate a reduction in erythrocytes size and anisocytosis [1]. Studies have also hypothesized that RBC damage occurs as a result of physical cell damage due to immune-mediated mechanisms and/or COVID-19 microangiopathy [16], [27], [31]. Another study reported that the loss of RBC biconcavity and complement activation observed in COVID-19 may facilitate RBC increase and spontaneous agglutination, possibly contributing to COVID-19-specific microvascular thrombosis [16].
In another study, Mertoglu et al. [14] stated that severe COVID-19 patients had the lowest levels of ESO, LYM, HGB, and RBC and highest levels of NEU, MONO, and WBC when compared to mildly infected patients. Onur et al. [20] reported that COVID-19 patients who died had the lowest values of LYM, HCT, and HGB, and highest values of WBC than surviving patients. In another study, Zhang et al. [25] stated that COVID-19 patients who died had higher the counts of leukocyte and neutrophil, and lower counts of platelet, lymphocyte, ESO, and MONO than surviving infected patients. As reported by previous studies, eosinopenia may be an indicator for SARS-CoV-2 infection, and the degree of eosinopenia has been associated with the severity of COVID-19 [25], [32]. In this study, the pre- and post-values of ESO of patients who died were lower than those who survived. Moreover, in this study, the pre- and post-values of the MCHC, HCT, HGB, LYM, and MONO were lower in the deceased patients than in living patients. Many studies have reported that leukocytosis and lymphopenia levels are independent predictors of in-hospital mortality [13], [15], [29], [27], [30]. In another study, it was reported that in severe cases, the PLT level decreased and was linked to the severity of the disease [15], [33]. Furthermore, only shortness of breath and leukocytosis were found to be independent risk indicators for death in critically ill COVID-19 patients [16]. In this study, while the EOS and other hematological values were not found to be a predictive risk factor for the mortality of COVID-19, low PLT and RBC, and high NEU, WBC, and RDW values were important risk indicators. A recent study showed an increased trend of eosinophils, lymphocytes, and platelets in surviving patients, but lower levels in non-surviving patients, which is consistent with the results determined herein [16], [34]. In this study, which included a comparison of the pre- and post-laboratory findings of deceased patients and their dynamic changes, the patients who died had a generally sustained increase in their leukocyte and neutrophil levels, and biochemical variables and an ongoing decrease in lymphopenia and eosinopenia levels.
In this study, the non-survivors had a sustained decline in HGB levels and a relatively more pronounced increase in PLTs than the survivors. This may have been due to the possible high doses of drugs given to the patients who died during the intensive care treatment process. However, the PLT was usually low until the last measurement in the deceased patients. In addition, the fact that the deceased group had the highest pre- and post-values of NEU and WBC when compared to the surviving group suggested the signs of leukocytosis and neutrophilia in the deceased group (Table 2). Although this was a reflection of a proinflammatory state, it may have been caused by the possible use of high-dose drugs (steroids, etc.) in the ICU [30]. In another study, it was stated that increased neutrophil and decreased LYM levels may reflect an imbalance in the inflammatory response and be considered a possible indicator of the severity of infectious diseases, such as sepsis and bacteremia [12], [35].
Similarly, one study noted that neutrophils play an important role in inflammation and are not typically elevated in viral infections [24]. Moreover, another study noted that the concentration of neutrophils (since neutrophils are the main producers of pro-inflammatory cytokines) is increased in COVID-19, and this increase contributes to the development of ARDS [24]. In yet another study, neutrophil was noted to be an independent predictor of severe disease and was associated with hypersensitivity pneumonia in SARS‐CoV-2 [24], [36].
Moreover, among the risk indicators for mortality in patients who died using the multivariate logistic regression method in this study, the most influential variables were high ESR, PT, CRP, INR, D-dimer, and RDW levels (Table 7). In addition, high ferritin, procalcitonin, WBC, and NEU levels, and low PLT and RBC levels were other risk indicators for COVID-19 mortality (Table 7). Although there were significant changes in many other hematological and biochemical parameters in the survivor and non-survivor groups, these parameters did not appear to be independent risk indicators for mortality in the COVID-19 patients.
ROC analysis was performed to determine the cut-off values and diagnostic performances of the predictive biomarkers of COVID-19 mortality. As a result of the ROC analysis (Fig. 4), the predictors with the highest AUC values were ESR (77.1%), PT (76.3%), CRP (73.4%), INR (72.4%), D-dimer (71.6), and RDW (71.5%), respectively, and the cut-off values of these variables were obtained (Table 8).
The surviving and non-surviving status of a large proportion of the COVID-19 patients was correctly determined according to the levels of ESR (>48.50 mm/h), PT (>13.95 s), CRP (>62.6 mg/L), INR (>1.19), D-dimer (>1262.26 µg/L), and RDW (>15.05%), respectively.
In this study, independent biomarkers ESR PT, CRP, INR, D-dimer, and RDW largely and accurately predicted the mortality of the COVID-19 patients. These results showed that ESR, PT, CRP, INR, D-dimer, and RDW are reliable and important biomarkers that predict mortality in COVID-19 according to the determined cut-off values. The cut-off values obtained can be evaluated as the survival risk limit of the patients in the COVID-19 epidemic, and can be used as a mortality predictive diagnostic tool for clinicians in the intervention.
5. Limitations
This study was a single center study conducted on a relatively small population. The data were obtained from an electronic record system that imposed limitations on the provision of incomplete or outdated information. In addition, the comorbidity data of the patients could not be accessed because the records were obtained retrospectively. Moreover, the study data may need to be confirmed by multicenter prospective studies, as retrospective studies naturally lack control of the variables.
6. Conclusion
In this research, the procalcitonin level was the parameter that most clearly showed the progression of the disease in the patients who died. The patients who died generally had a sustained increase in their leukocyte and neutrophil levels, and biochemical variables, and an ongoing decrease in their lymphopenia and eosinopenia levels. In addition, high ESR, INR, PT, CRP, D-dimer, ferritin and RDW values were the most effective independent mortality risk predictors of COVID-19. In addition, neutrophilia, leukocytosis, thrombocytopenia, and erythrocytopenia were other mortality risk predictors. The calculated cut-off values of these predictors can be used as predictive diagnostic biomarkers of mortality in COVID-19 patients at risk of death. In this way, earlier and more accurate medical intervention can be provided to patients and the health system can be assisted.
Credit authorship contribution statement
Mehmet Tahir Huyut: conceived and designed the study, scanned the literature, determined and implemented the method, organized, analyzed and interpreted data, wrote and revised the work. Zübeyir Huyut: reviewed the work, commented and revised in the discussion section. Fatih İlkbahar: organized the raw data and created the open access dataset. Cuma Mertoğlu: obtained the data, scanned the literature.
Informed consent
Informed consent was obtained from all individuals included in this study.
Ethical approval
The study protocol was approved by the Institutional Ethics Review Board of Erzincan Binali Yıldırım University after being approved by the Ministry of Health of the Republic of Turkey in accordance with the Declaration of the Helsinki World Medical Association (Ethics Committee Decision number: 2021/02–07).
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Amgalan A., Othman M. Hemostatic laboratory derangements in COVID-19 with a focus on platelet count. Platelets. 2020;31(6):740–746. doi: 10.1080/09537104.2020.1768523. [DOI] [PubMed] [Google Scholar]
- 2.Ardestani A., Azizi Z. Targeting glucose metabolism for treatment of COVID-19. Signal Transduction Targeted Therapy. 2021;6(1):112. doi: 10.1038/s41392-021-00532-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen R., Sang L., Jiang M., et al. Longitudinal hematologic and immunologic variations associated with the progression of COVID-19 patients in China. J. Allergy Clin. Immunol. 2020;146(1):89–100. doi: 10.1016/j.jaci.2020.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Doğanay F., Elkonca F., Seyhan A.U., et al. Shock index as a predictor of mortality among the Covid-19 patients. Am. J. Emerg. Med. 2021;40:106–109. doi: 10.1016/j.ajem.2020.12.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Erol A.T., Aşar S., Sabaz M.S., et al. Risk Factors for 28-day Mortality Among COVID-19 Patients in an Intensive Care Unit of a Tertiary Care Center in Istanbul. Med. J. Bakirkoy. 2021 doi: 10.5222/BMJ.2021.77200. [DOI] [Google Scholar]
- 6.Guan W., Ni Z., Hu Y., et al. Clinical characteristics of coronavirus disease 2019 in China. N. Engl. J. Med. 2020;382(18):1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.C. Huang, Y. Wang, X. Li, et al. Clinical characteristics of patients infected with the 2019 novel coronavirus in Wuhan, China. Lancet, 395 (2020), pp. 497–506. https://doi.org/10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed]
- 8.Huyut M.T., İlkbahar F. The Effectiveness of Blood Routine Parameters and Some Biomarkers as a Potential Diagnostic Tool in the Diagnosis and Prognosis of Covid-19 Disease. Int. Immunopharmacol. 2021;98 doi: 10.1016/j.intimp.2021.107838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jiang S.Q., Huang Q.F., Xie W.M., et al. The association between severe COVID-19 and low platelet count: evidence from 31 observational studies involving 7613 participants. Br. J. Haematol. 2020;190(1):e29–e33. doi: 10.1111/bjh.16817. [DOI] [PubMed] [Google Scholar]
- 10.Khinda J., Janjua N.Z., Cheng S., et al. Association between markers of immune response at hospital admission and COVID-19 disease severity and mortality: A meta-analysis and meta-regression. J. Med. Virol. 2021;93(2):1078–1098. doi: 10.1002/jmv.26411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li X., Wang L., Yan S., et al. Clinical characteristics of 25 death cases with COVID-19: A retrospective review of medical records in a single medical center, Wuhan, China. Int. J. Infect. Dis. 2020;94:128–132. doi: 10.1016/j.ijid.2020.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu Y., Yang Y., Zhang C., et al. Clinical and biochemical indexes from 2019-nCoV infected patients linked to viral loads and lung injury. Sci China Life. 2020;63(3):364–374. doi: 10.1007/s11427-020-1643-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.G. Lippi, M. Plebani, B. M. Henry. Thrombocytopenia is associated with severe coronavirus disease 2019 (COVID‐19) infections: a metaanalysis. Clin Chim Acta., 506 (2020), pp. 145‐148. https://doi.org/10.1016/j.cca.2020.03.022. [DOI] [PMC free article] [PubMed]
- 14.Mertoglu C., Huyut M.T., Arslan Y., et al. How do routine laboratory tests change in coronavirus disease 2019? Scand. J. Clin. Lab. Invest. 2021;81(1):24–33. doi: 10.1080/00365513.2020.1855470. [DOI] [PubMed] [Google Scholar]
- 15.Mikami T., Miyashita H., Yamada T., et al. Risk Factors for Mortality in Patients with COVID-19 in New York City. J. Gen. Intern. Med. 2020;36(1):17–26. doi: 10.1007/s11606-020-05983-z). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mo P., Xing Y., Xiao Y., et al. Clinical characteristics of refractory COVID-19 pneumonia in Wuhan, China. Clin. Infect. Dis. 2020;16:ciaa270. doi: 10.1093/cid/ciaa270. [DOI] [Google Scholar]
- 17.Moradi E.V., Teimouri A., Rezaee R., et al. Increased age. neutrophil-to-lymphocyte ratio (NLR) and white blood cells count are associated with higher COVID-19 mortality. Am. J. Emerg. Med. 2021;40:11–14. doi: 10.1016/j.ajem.2020.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou F., Yu T., Du R., et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395(10229):1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mousavi S.A., Rad S., Rostami T., et al. Hematologic predictors of mortality in hospitalized patients with COVID-19: a comparative study. Hematology. 2020;25(1):383–388. doi: 10.1080/16078454.2020.1833435. [DOI] [PubMed] [Google Scholar]
- 20.Onur S.T., Altın S., Sokucu S.N., et al. Could ferritin level be an indicator of COVID-19 disease mortality? J. Med. Virol. 2021;93(3):1672–1677. doi: 10.1002/jmv.26543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Salamanna F., Maglio M., Landini M.P., et al. Platelet functions and activities as potential hematologic parameters related to coronavirus disease 2019 (Covid-19) Platelets. 2020;31(5):627–632. doi: 10.1080/09537104.2020.1762852. [DOI] [PubMed] [Google Scholar]
- 22.Opal S.M., Girard T.D., Ely E.W. The immunopathogenesis of sepsis in elderly patients. Clin. Infect. Dis. 2005;41:504–512. doi: 10.1086/432007. [DOI] [PubMed] [Google Scholar]
- 23.Wang K., Zhang Z., Yu M., et al. 15-day mortality and associated risk factors for hospitalized patients with COVID-19 in Wuhan, China: an ambispective observational cohort study. Intensive Care Med. 2020;46(7):1472–1474. doi: 10.1007/s00134-020-06047-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu C., Chen X., Risk C.Y., et al. Factors associated with acute respiratory distress syndrome and death in patients with coronavirus disease 2019 pneumonia in Wuhan, China. JAMA Intern Med. 2020;180(7):934–943. doi: 10.1001/jamainternmed.2020.0994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.J. J. Zhang,,Y. y. Cao, G. Tan, et al. Clinical, radiological, and laboratory characteristics and risk factors for severity and mortality of 289 hospitalized COVID-19 patients. Allergy, 76(2) (2021), pp. 533–550. https://doi.org/10.1111/all.14496. [DOI] [PMC free article] [PubMed]
- 26.Tang N., Li D., Wang X., et al. Abnormal coagulation parameters are associated with poor prognosis in patients with novel coronavirus pneumonia. J. Thromb. Haemost. 2020;18(4):844–847. doi: 10.1111/jth.14768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun S., Cai X., Wang H., et al. Abnormalities of peripheral blood system in patients with COVID-19 in Wenzhou, China. Clin. Chim. Acta. 2020;507:174–180. doi: 10.1016/j.cca.2020.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.World Health Organization (WHO). Coronavirus disease (COVID-2019) situation reports. https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports. Accessed July 2, 2020.
- 29.Song H., Kim H.J., Park K.N., et al. Neutrophil to lymphocyte ratio is associated with in-hospital mortality in older adults admitted to the emergency department. Am. J. Emerg. Med. 2021;40:133–137. doi: 10.1016/j.ajem.2020.01.044. [DOI] [PubMed] [Google Scholar]
- 30.Xie G., Ding F., Han L., et al. The role of peripheral blood eosinophil counts in COVID-19 patients. Allergy. 2021;76(2):471–482. doi: 10.1111/all.14465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.A. P. Yang, J.p. Liu, W.q. Tao, et al. The diagnostic and predictive role of NLR, d-NLR and PLR in COVID-19 patients. Int Immunopharmacol., 84 (2020), pp. 106504. https://doi.org/10.1016/j.intimp.2020.106504. [DOI] [PMC free article] [PubMed]
- 32.Zhao L., Zhang Y.P., Yang X., Liu X. Eosinopenia is associated with greater severity in patients with coronavirus disease 2019. Allergy. 2021;76(2):562–564. doi: 10.1111/all.14455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.X. Yang, Y. Yu, J. Xu, et al. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan. China: a single-centered. retrospective. observational study. Lancet Respir Med., 8(5) (2020), pp. 475–481. https://doi.org/10.1016/S2213-2600(20)30079-5. [DOI] [PMC free article] [PubMed]
- 34.Zhao X., Li Y., Ge Y., et al. Evaluation of Nutrition Risk and Its Association With Mortality Risk in Severely and Critically Ill COVID-19 Patients. J. Parenteral Enteral Nutrit. 2021;45(1):32–42. doi: 10.1002/jpen.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhenga Y., Zhanga Y., Chia H., et al. The hemocyte counts as a potential biomarker for predicting disease progression in COVID-19: a retrospective study. Clin. Chem. Lab. Med. 2020;58(7):1106–1115. doi: 10.1515/cclm-2020-0377. [DOI] [PubMed] [Google Scholar]
- 36.N. Zhu, D. Zhang, W. Wang et al. A novel coronavirus from patients with pneumonia in China. 2019. New Engl J Med., 382(8) (2020), pp. 727–733. https://doi.org/10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed]
- 37.Huyut M.T., Huyut Z. Forecasting of Oxidant/Antioxidant levels of COVID-19 patients by using Expert models with biomarkers used in the Diagnosis/Prognosis of COVID-19. International Immunopharmacology. 2021;100(108127) doi: 10.1016/j.intimp.2021.108127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huyut M.T., Üstündağ H. Prediction of diagnosis and prognosis of COVID-19 disease by blood gas parameters using decision trees machine learning model: a retrospective observational study. Med Gas Res. 2022;12(2):60–66. doi: 10.4103/2045-9912.326002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mertoğlu C., Huyut M.T., Ölmez H., et al. COVID-19 is more dangerous for older people and its severity is increasing: a case-control study. Med Gas Res. 2022;12(2):51–54. doi: 10.4103/2045-9912.325992. [DOI] [PMC free article] [PubMed] [Google Scholar]