Abstract
CRISPR/Cas9 gene-editing technology allows researchers to study protein function by specifically introducing double-stranded breaks in the gene of interest then analyze its subsequent loss in sensitive biological assays. To help characterize one of a series of highly potent, conditionally active, T cell engaging bispecific molecules called COBRA™, the human EpCAM gene was disrupted in HT29 cells using CRISPR/Cas9 and guide RNA targeting its Exon 2. Although a commercially available antibody indicated loss of cell-surface expression, the EpCAM targeting bispecific COBRA was still able to lyse these cells in a T cell dependent cellular cytotoxicity assay. RT-PCR sequence analysis of these cells showed a major alternative transcript generated after CRISPR/Cas9, with Exon 1 and 3 spliced together in-frame, skipping Exon 2 completely, to express a truncated cell-surface receptor recognized by the EpCAM-COBRA. Researchers who use CRISPR/Cas9 must be cognizant of this potential to express alternative versions of their proteins and use sensitive orthogonal detection methods to ensure complete gene disruption.
Keywords: CRISPR, Bispecific, COBRA, EpCAM, Exon-skipping
Highlights
-
•
CRISPR/Cas9 allows researchers to target gene disruption and analyze its loss of expression.
-
•
After CRISPR/Cas9 targeting the EpCAM gene, no protein was detected after staining with a commercial antibody.
-
•
An in-house EpCAM T-cell engager was able to lyse these supposedly EpCAM KO cells.
-
•
cDNA analysis showed an alternative mRNA transcript arose after CRISPR/Cas9, creating a truncated version on the cell's surface.
-
•
Scientists using CRISPR must be aware of the potential to make alternate forms and try other means to ensure no expression.
1. Introduction
Clustered regularly interspaced short palindromic repeat (CRISPR) associated protein (Cas9) technology has become an indispensable tool, allowing researchers the ability to target gene disruption and analyze its loss of expression [1,2]. By co-transfecting cells with Cas9 enzyme and guide RNA (gRNA) with complimentary sequence to the gene of interest, double-stranded breaks are introduced within that region of homology [[3], [4], [5], [6]]. Natural attempts to repair this break result in deletions or insertions, creating premature termination codons (PTC), non-frameshift, or missense mutations [7]. Highly sensitive and specific detection methods are then used to confirm and isolate cells that exhibit complete loss of protein expression.
The ability to silence or knock-out (KO) tumor only expressing genes is particularly useful in cancer drug research. By abrogating its expression on transformed cells, scientists can both study its loss-of-function phenotype and assess the activity of their antibody-based drugs specific towards that protein [[8], [9], [10], [11], [12]]. One such class of therapeutics are T cell engaging bispecific antibodies which engage two distinct binding sites, one specifically targets an antigen on the surface of the tumor cell and the other binds an activating receptor on the surface of a T cell. The first therapeutic molecules of this type have proven to be very potent in directing cytotoxic T cell responses to specific target cells both in vitro and in vivo [[13], [14], [15], [16], [17]]. Because of their high potency and potential for off-target toxicity, every effort is made to ensure their specificity towards its expected target.
Maverick Therapeutics has developed a platform of conditionally active bispecific T cell engager molecule, called COBRA™, which may target a variety of cell-surface tumor antigens [17]. To characterize a novel COBRA molecule targeting human Epithelial Cell Adhesion Molecule (EpCAM; CD326) [18] for in vivo tumor efficacy studies, the HT29 human cancer cell line's EpCAM gene was subjected to CRISPR/Cas9 targeted disruption. Cas9 enzyme and gRNA against human EpCAM's Exon 2 were transiently transfected to prevent stable insertion and expression of foreign transcripts and a pool of EpCAM negative cells were sorted by flow cytometry to retain similar diversity to the original parental line. However, even after several rounds of negative sorting, the final population was still sensitive to T cell-dependent cellular cytotoxicity (TDCC) by the EpCAM-COBRA bispecific. Subsequent RT-PCR analysis shows an alternative splice variant of EpCAM, with Exon 2 deleted, was expressed on these cells after gene-disruption which was missed by the commercially available antibody but was still targeted by the EpCAM-COBRA bispecific molecule.
2. Results
2.1. CRISPR/Cas9 targeted disruption of HT29 cell's EpCAM gene
To generate cells lacking EpCAM via CRISPR/Cas9, HT29 cells were transfected with a guide RNA (gRNA) targeting Exon 2 (Ex2) (Fig. 1). Ex2 encodes the N terminus of the mature EpCAM protein and creating deletions and/or frameshift mutations in this region would be expected to prevent the generation of a functional cell-surface protein. This strategy was successfully employed in knocking out other cell-surface receptors, including B7H3 (Supplemental Fig. 1). As expected, the EpCAM knock-out (KO) efficiency using the Ex2 gRNA was not 100%, as determined by cell-surface staining with an EpCAM antibody (Fig. 2A). Thus, the cells not stained with the EpCAM antibody were FACS sorted out of the transfected population to yield the Ex2 KO cells (Fig. 2B).
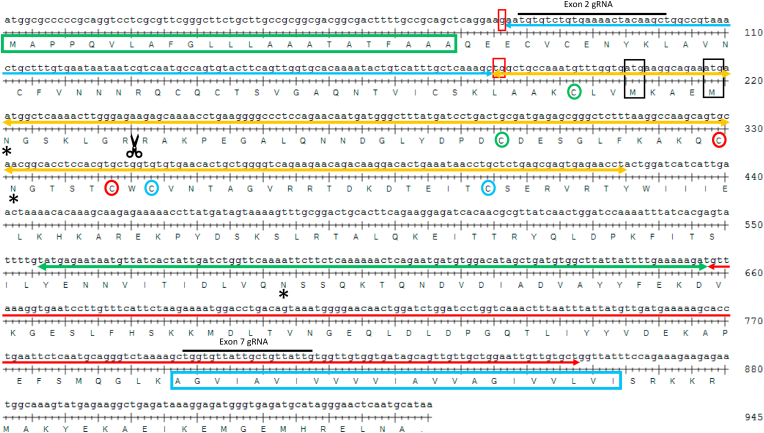
Fig. 1.
Coding sequence of human EpCAM (BC014785)
Green box: Signal Peptide; Blue box: Transmembrane Domain; Red Boxes: GTG/Val residue spliced between Exon 1 and 3; Black Boxes: Potential in-frame downstream ATG/Met start codons 69 and 73; Matching colored circles: Disulfide pairs; Asterisks: N-glycosylation sites; Scissors: protease cleavage site; Blue line arrow: Exon 2; Orange line arrow: Exon 3; Green line arrow: Exon 6; Red line arrow: Exon 7; Black overscores: gRNAs used. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
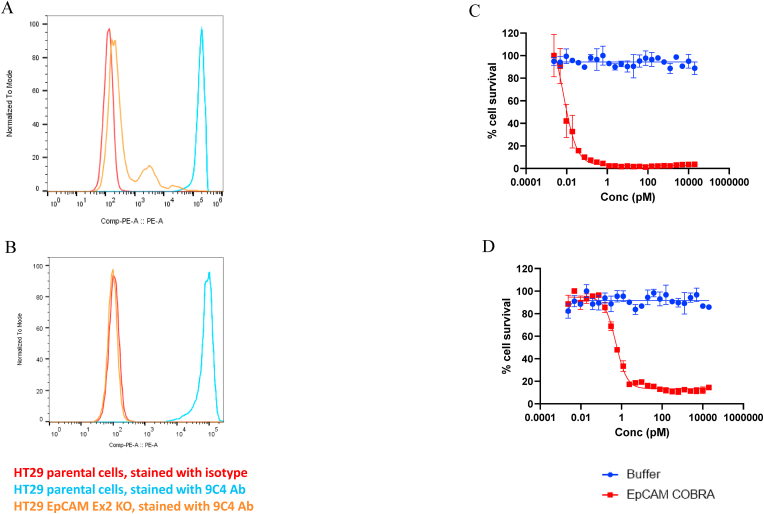
Fig. 2.
FACS and TDCC analysis of parental and EpCAM Exon 2 targeted CRISPR KO HT29 cells before and after sorting with commercial antibody. (A) After CRISPR/Cas9 treatment with gRNA targeting Ex2, HT29 cells were stained with an EpCAM antibody by flow cytometry to evaluate KO efficiency. (B) After sorting for cells not stained with the 9C4 antibody, the EpCAM KO HT29 cell pool was stained and compared with parental cells. HT29 parental cells stained with isotype control (red), HT29 parental cells stained with commercial antibody 9C4 (blue), and HT29 EpCAM KO cells with gRNA targeting EpCAM Ex2 stained with 9C4 (orange). TDCC was evaluated on parental HT29 cells (C), HT29 CRISPR EpCAM Ex2 KO cells, negatively sorted for 9C4 antibody staining (D) The EC50 of EpCAM COBRA on HT29 cells and Ex2 KO cells after sorting on 9C4 antibody were 0.007 pM and 0.53 pM, respectively. . (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
The Ex2 KO cells were subsequently evaluated for their resistance to T cell-dependent cellular cytotoxicity (TDCC) with an EpCAM-targeting bispecific T cell engager: EpCAM-COBRA (further described in Methods and Discussion). Unexpectedly, the Ex2 KO cells remained sensitive to killing, although the potency of the bispecific on these cells was decreased by 75-fold, compared to its potency on parental HT29 cells (Fig. 2C and D). Flow cytometry analysis of EpCAM-COBRA binding to the Ex2 KO cells revealed that the bispecific bound to a subset of these cells that the commercial antibody did not bind (Fig. 3A). A second round of FACS sorting, this time with the EpCAM-COBRA, on the cells negative for staining yielded cells that remained sensitive to killing via TDCC, although the potency of the COBRA on these cells was reduced by more than 400-fold, compared to its potency on cells negatively sorted with the EpCAM antibody (Fig. 3B). Since most Ex2 KO cells were sensitive to EpCAM-COBRA killing, these data suggested that most Ex2 KO cells were expressing a very low level of cell-surface EpCAM, to which the bispecific could apparently bind, but was not detected by flow cytometry using either the commercial EpCAM antibody or the EpCAM-COBRA. As an alternate strategy to eliminate EpCAM from the surface of HT29 cells, CRISPR/Cas9 was performed using a different gRNA, targeting Exon 7 (Ex7) (Fig. 1). Exon 7 encodes the transmembrane domain; thus, disrupting these sequences was predicted to result in a secreted form of EpCAM. After negative sorting on the cells that did not bind the EpCAM-COBRA, cells transfected with the Ex7 gRNA expressed undetectable levels of cell-surface EpCAM by flow cytometry when stained with the EpCAM-COBRA (Fig. 3A). Unexpectedly, these cells were sensitive in TDCC with EpCAM-COBRA; however, the potency of this bispecific on the Ex7 KO cells was more than 2000-fold lower than on parental HT29 cells (Fig. 3C). The Ex7 gRNA was also transfected into the Ex2 KO cells, and these cells showed no cell surface staining of EpCAM and were resistant to TDCC with the bispecific (Fig. 3D). Thus, only cells targeted with Ex2 and Ex7 gRNAs were resistant to T cell killing with an EpCAM COBRA.
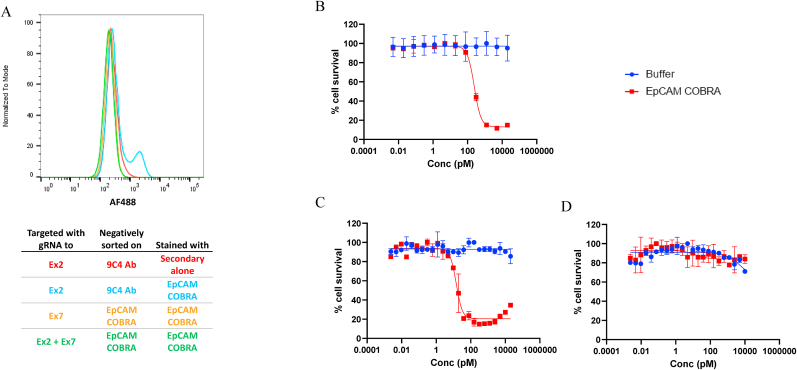
Fig. 3.
FACS and TDCC analysis of HT29 EpCAM KO cells targeted with different gRNAs. (A) EpCAM expression was evaluated by FACS on HT29 CRISPR EpCAM KO cells with gRNA targeting EpCAM Ex2 only and negatively sorted on 9C4 antibody staining (blue), Ex7 only and negatively sorted on EpCAM-COBRA staining (orange), and Ex2 & Ex7 and negatively sorted on EpCAM-COBRA staining (green), all stained with EpCAM-COBRA. HT29 EpCAM Ex2 KO cells stained with secondary alone (red) are included as control. TDCC was evaluated on Ex2 KO cells, negatively sorted for EpCAM-COBRA staining (B), Ex7 KO cells, negatively sorted for EpCAM-COBRA staining (C), and Ex2/Ex7 KO cells, negatively sorted for EpCAM-COBRA staining (D). The EC50 of Ex2 KO cells after EpCAM-COBRA sorting, and Ex7 KO cells were 243.8 pM, and 14.5 pM, respectively. HT29 CRISPR EpCAM KO cells with gRNAs targeting both EpCAM Ex2 and Ex7 were resistant to killing by activated EpCAM-COBRA. T = 48h. . (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
2.2. Analysis of HT29 EpCAM CRISPR/Cas9 KO mRNA transcripts
To understand what could still be expressed after CRISPR/Cas9 KO and 9C4 or EpCAM-COBRA negative cell-sorting, HT29 parental cells and HT29 cells transfected with EpCAM Ex2, EpCAM Ex7, EpCAM Ex2 and Ex7, and B7H3 gRNAs were subjected to total RNA extraction and 5′RACE. A 1.4 kb product was observed in all cell lines after amplifying for EpCAM cDNA, albeit the relative intensity of the bands appeared lower from the EpCAM KO cells (data not shown). Subsequent subcloning and sequencing of 8 random clones each confirmed that the 1.4 kb product encoded EpCAM cDNA and that both the parental and B7H3 gRNA transfected HT29 cells were still transcribing the complete wild-type mRNA (BC014785, Fig. 1).
On the other hand, 8 random clones each from the three EpCAM gRNA CRISPR/Cas9 transfected lines had deletions ranging from 2 to over 300 bp, all resulting in either out-of-frame truncations or significant deletions disrupting normal EpCAM protein translation. Specifically, for Ex2 KO cells negatively sorted on the 9C4 antibody, all 8 clones had various deletions after the signal sequence that would result in PTCs and truncated proteins. However, there are several downstream in-frame ATG/Met that may initiate translation and produce a membrane-associated protein. The 8 clones sequenced from Ex7 KO cells that were negatively sorted on the EpCAM-COBRA had deletions at the N-terminal half of the transmembrane domain, but 4 had relatively short deletions which could still translate a soluble variant of EpCAM since the rest of the 5’ coding region was still intact. However, cells transfected with both Ex2 and Ex7 gRNA encoded cDNA with deletions around both the signal sequence and transmembrane domains, resulting in the complete disruption of EpCAM protein, including any possible membrane-associated and soluble variants.
To further elucidate what the EpCAM-COBRA could be targeting on cells transfected with the CRISPR/Cas9 Ex2 gRNA, Ex2 KO cells, initially negatively sorted for EpCAM Ab binding, were subsequently positively sorted for EpCAM-COBRA binding in hopes of enriching the population for that specific moiety, and the total RNA extraction, 5′RACE and EpCAM cDNA amplification were repeated. Of the eight clones randomly subcloned and sequenced, four had deletions after the signal peptide region resulting in PTCs, like the previously described Ex2 KO, 9C4 negative-sorted cells. One clone had a 15 bp deletion/substitution resulting in a molecule with 5 amino-acids deleted in-frame near the 3′ end of Exon 2. Most interesting, the other three clones encoded identical deletion mutants in which the entire Exon 2 was replaced by an in-frame GTG/Val residue. It appears that some Ex2 KO cells created EpCAM transcripts that splice Exon 1 and Exon 3 in-frame together, skipping Exon 2 completely. This predominant, 108 nt/36 aa shorter, EpCAM variant, henceforth referred to as ΔEx2, still has the wild-type 5′ UTR, in-frame translation start, signal sequence, extracellular domain (ECD) from Exons 3–7, transmembrane domain, intracellular domain, stop codon, and the rest of the 3′ UTR. Furthermore, this deletion retains the protease cleavage site, all the N-glycosylation sites, and does not disrupt any of the other remaining disulfide bridges normally found in the rest of the ECD (Fig. 1) [18].
2.3. Western blot analysis of EpCAM CRISPR/Cas9 KO HT29 cells
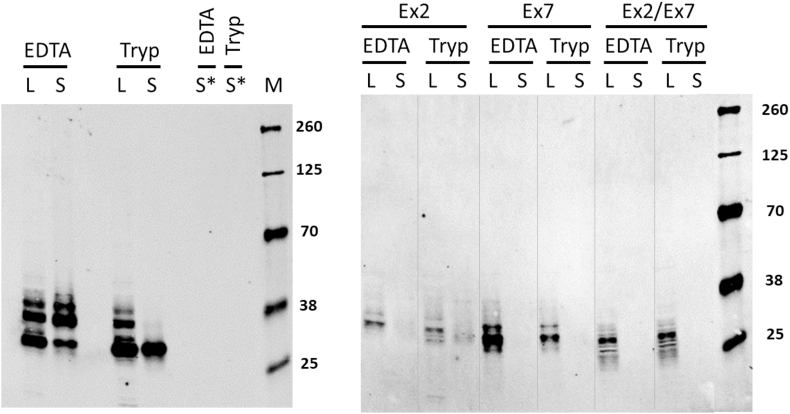
Having observed that HT29 cells transfected with the Ex2 gRNA remained sensitive to TDCC with EpCAM-COBRA, Western analysis was performed to evaluate the EpCAM protein products generated in cells after CRISPR/Cas9. Both total lysates and partially purified lysates after cell surface biotinylation were evaluated. In addition, because the ECD of EpCAM has previously been shown to contain a protease cleavage site (Fig. 1), lysates were prepared from cells incubated with or without trypsin prior to lysis to induce proteolytic cleavage of cell-surface EpCAM. Total lysates of HT29 parental cells contained three EpCAM products, consistent with glycosylated full-length mature EpCAM (∼40 kD and ∼36 kD) and glycosylated transmembrane EpCAM cleavage product (∼30 kD) as observed by others [18] (Fig. 4). As expected, in cells treated with trypsin, only cleaved EpCAM was detected on the cell surface. HT29 cells transfected with the Ex2 gRNA produced substantially less EpCAM protein than parental cells. In these cells, bands of ∼34 kD, ∼31 kD, and ∼29 kD were detected; a small amount of protein was detected on the cell surface, and at least some of that cell-surface protein was found to be sensitive to cleavage by trypsin (Supplemental Fig. 3). The findings that the EpCAM protein in Ex2 KO cells remained sensitive to protease cleavage and that the R80/R81 protease cleavage site is in Exon 3 are consistent with the results described above that several the cDNAs purified from Ex2 KO cells are ΔEx2 splice variants that simply remove Exon 2. Cells transfected with Exon 7 gRNA or with both Exon 2 and Exon 7 gRNAs expressed detectable levels of EpCAM protein by Western, with Ex7 KO cells expressing much more protein than Ex2/Ex7 KO cells, as similar levels of EpCAM protein were detected in 10 μg of Ex2/Ex7 KO lysate and 1 μg of Ex7 KO lysate (Fig. 4). EpCAM protein was not detected on the cell surface of either of these cells, and the protein produced in these cells was not sensitive to trypsin. However, as expected from targeting the transmembrane domain, cells transfected with Exon 7 gRNA alone produced relatively high levels of soluble EpCAM (Supplemental Fig. 3).
Fig. 4.
Western and cell-surface analysis of HT29 CRISPR cells.
EpCAM expression was analyzed by Western in parental HT29 cells (left panel) and after CRISPR with gRNAs targeting EpCAM Ex2 and/or Ex7 (right panel). For each cell line, cells were removed with either EDTA or trypsin, as indicated, and both total cell lysate (L) and the biotinylated fraction after cell surface biotinylation (S) were analyzed. For parental HT29 cells, mock biotinylated cells were also analyzed (S*). For parental HT29 cells, 3 μg lysate or equivalent was loaded per lane; for Ex2 and Ex2/Ex7 targeted cells, 10 μg lysate or equivalent was loaded per lane; for Ex7 targeted cells, 1 μg lysate or equivalent was loaded per lane.
2.4. Expression and characterization of EpCAM KO alternative transcripts
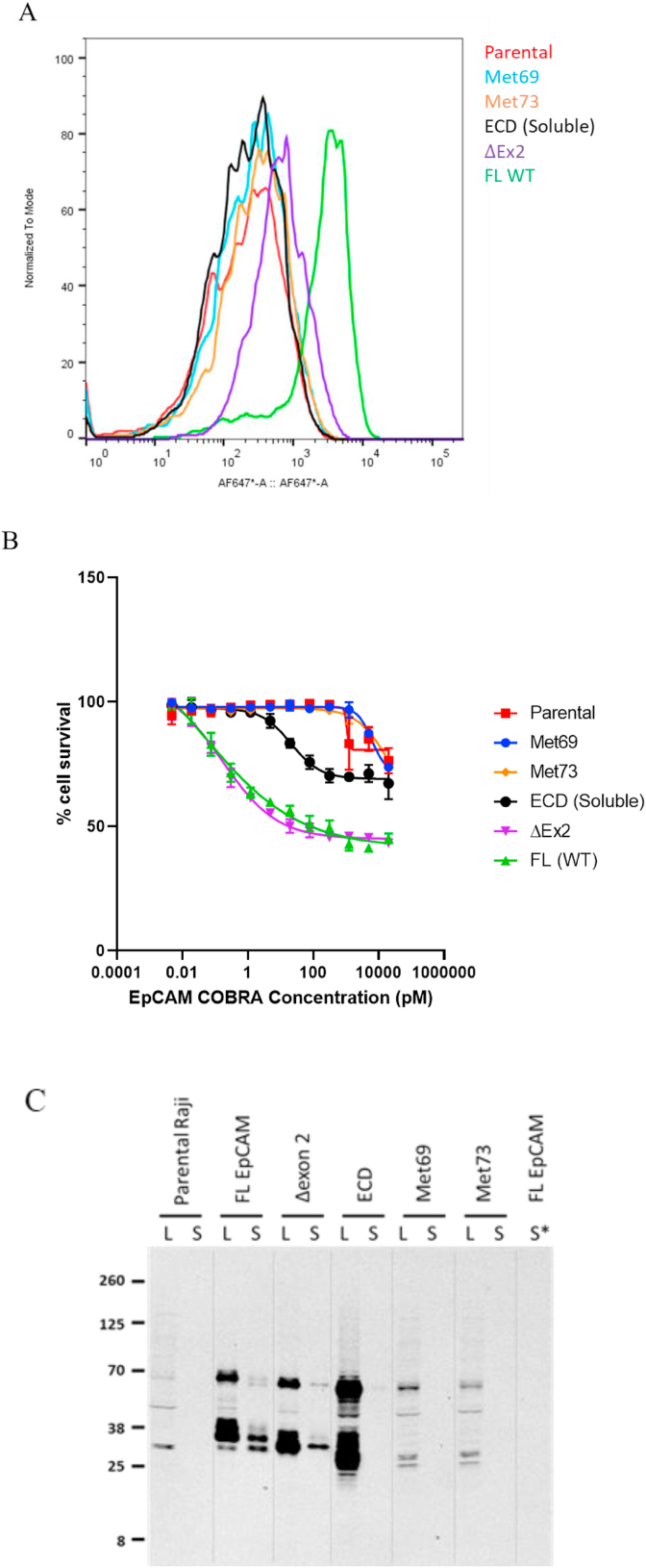
To test which potential deletion/truncation variants the EpCAM-COBRA was targeting to enable TDCC, pLenti expression vectors were constructed to express full-length human EpCAM (BC014785) (WT, positive control), ECD soluble variant (1–265 aa), Exon 2 deletion mutant (ΔEx2), and potential downstream Met69 and Met73 start versions (Supplemental Fig. 4), all with an in-frame C-terminal P2A-GFP marker. These transcripts will initiate translation with the N-terminal EpCAM protein variants first, when upon arriving the P2A signal, induce ribosomal skipping, creating a peptide break, then resume translating through the C-terminal GFP. As a result, transduced cells will express and fluoresce with GFP only if the EpCAM proteins were initially translated. Raji cells transduced with all five packaged lentiviruses expressed GFP, indicating that all five EpCAM variants were being translated (Supplemental Fig. 5). After sorting for GFP expression, the lines were subsequently characterized.
The GFP pool sorted cells were first stained with EpCAM-COBRA and analyzed by FACS (Fig. 5A). The full-length WT expressed the most cell surface EpCAM, followed by the Exon 2 deleted mutant, ΔEx2. The ECD soluble, Met69, and Met73-transduced cells showed similar staining as parental Raji cells, suggesting that these variants expressed little-to-no EpCAM on the cell surface. TDCC sensitivity reflected the FACS staining results, where cells expressing either the full-length WT or the ΔEx2 variant were highly sensitive to killing with EpCAM-COBRA. Interestingly, cells expressing the soluble ECD exhibited some sensitivity to killing with the bispecific, although at a much lower potency than on WT- or ΔEx2-expressing cells (Fig. 5B).
Fig. 5.
FACS, TDCC, and Western analysis of EpCAM-P2A-GFP molecules in Raji cells. (A) EpCAM expression was evaluated by FACS by staining various EpCAM-P2A-GFP molecules in Raji cells with EpCAM COBRA. (B) TDCC was evaluated on these EpCAM P2A GFP molecules in Raji cells. The EC50 of the EpCAM COBRA on Raji cells expressing full-length WT, the ΔEx2 variant, and EpCAM ECD were 0.05 pM, 0.17 pM, and 21.01 pM, respectively. T = 96h. (C) EpCAM expression in Raji cells transfected with the indicated EpCAM variants was analyzed by Western analysis. For each cell line, both total cell lysate (L) and the biotinylated fraction after cell surface biotinylation (S) were analyzed. For Raji cells transfected with full-length EpCAM, mock biotinylated cells were also analyzed (S*). For Raji cells transfected with full-length EpCAM and ΔEx2 EpCAM, 1 μg lysate or equivalent was loaded per lane. For parental Raji cells or Raji cells transfected with the extracellular domain, Met69, or Met73 variants, 10 μg lysate or equivalent was loaded per lane.
Raji cells ectopically expressing EpCAM protein variants were also analyzed by Western (Fig. 5C). In cells transduced with full-length EpCAM, major bands of ∼40 kD, ∼36 kD, and ∼30 kD bands, similar in size to those produced in HT29 parental cells, were detected. A substantial fraction of the protein in these cells was detected on the cell surface, as detected by cell surface biotinylation. In addition, a protein of ∼70 kD also was detected in these cells and in all cells transduced with EpCAM variants, the sizes of which are consistent with incomplete cleavage at the P2A site. In cells transduced with EpCAM ΔEx2, protein products of ∼31 and ∼34 kD were observed in total lysates and on the cell surface. These proteins are similar in size to those expressed in the Exon 2 gRNA transfected cells. In cells expressing the ECD, Met69, or Met73 variants, multiple protein products were detected in total cell lysates, but no protein was detected on the cell surface.
3. Discussion
CRISPR/Cas9 technology is a revolutionary tool that has greatly facilitated biologists’ ability to study gene and protein function. However, as more researchers employ this methodology, one unexpected by-product has frequently emerged: exon skipping [19]. Although most examples of exon skipping result in PTCs, in this instance, an in-frame exon deletion variant is a significant product, resulting in expression of a truncated human EpCAM protein on the cell surface that was recognized by an in-house EpCAM-COBRA bispecific, not by a commercial antibody.
This commercially available antibody, 9C4, was used to negative cell-sort for supposed loss of EpCAM surface expression from CRISPR/Cas9 treated HT29 cells. However, even after several rounds of negative-sorting with this antibody, the EpCAM-COBRA was still able to induce cytotoxicity of this population by T cells. The single domain antibody (sdAb) in this bispecific is a different entity from the anti-EpCAM VH/VL within the 9C4 antibody and these two binders likely recognize different epitopes. Indeed, the EpCAM-COBRA was found to bind the ΔEx2 version of EpCAM and was able to induce TDCC of cells expressing this variant (Fig. 5B). In contrast, the 9C4 antibody was found not to bind to this variant (Supplemental Fig. 6). In addition, the EpCAM-COBRA has sub-pM TDCC potency, a much more sensitive assay than FACS staining (Fig. 5). Taken together, these results most likely explain why the EpCAM-COBRA was still able to target the population of cells expressing the ΔEx2 version of EpCAM even though negatively sorting with 9C4 failed to remove them.
We have used Synthego's recommended guide RNA's targeting Exon 2 for B7H3 and another Type I receptor and in both cases saw complete loss of cell surface expression and subsequent lack of TDCC by our COBRA bispecifics (Supplemental Fig. 1 and data not shown). In every case where we sequenced CRISPR/Cas9 treated and negative-sorted cell's mRNA, we observed deletions around the gRNA location. Including the fact that EpCAM expression was ultimately knocked out after transfecting Exon 7 gRNA indicates that there was nothing inherently wrong with either their gRNA sequence algorithm or our application of CRISPR/Cas9 deletion technology.
There was the possibility that this alternative ΔEx2 transcript had already existed in HT29 cells before CRISPR/Cas9 treatment. However, this product was not observed in parental HT29 cDNA when a primer specific for this message, straddling Exon 1 and 3 spliced joining site, was used in 5′RACE RT-PCR (data not shown). Furthermore, this specific transcript has so far not shown up in GenBank's Expressed Sequence Tags database. Nevertheless, it is still possible this novel splice variant exists in other transformed cells or cell lines with extensive genomic instability.
When working with novel antibodies, such as the EpCAM sdAb in the EpCAM-COBRA molecule, specificity of binding is a critical question. This sdAb was found not to bind its closest family member, Trop2, either as recombinant protein or when expressed on the surface of Raji cells, analogous to the cells generated and described in Fig. 5. In addition, specificity of the EpCAM-COBRA for its target antigen was confirmed in the Retrogenix human cell microarray system and only bound to cells expressing human EpCAM (data not shown). EpCAM-COBRA's specificity was further confirmed in this work when the target was ultimately knocked out of HT29 cells by gRNAs against Exon 2 and Exon 7 of the human EpCAM gene and became resistant to binding and killing by this bispecific T cell engager (Fig. 3).
The purpose of establishing the HT29 EpCAM KO pool is to evaluate the dependence of EpCAM-COBRA's in vivo anti-tumor efficacy on EpCAM expression on tumor cells. Because clones of cells can have very different growth properties in vivo, the gRNAs and Cas9 enzymes were transiently transfected to prevent stable insertion and expression of exogenous transcripts and a negative pool was sorted to generate a diverse and heterogeneous population similar to that of the original target-expressing, wild-type cell line. On the other hand, Tsaktanis, et al. [20] disrupted the human EpCAM gene in their human cancer cell line by stably transfecting plasmids containing gRNA, Cas9, and GFP then isolated green fluorescing single cell clones. They transfected gRNAs against Exon 2 and Exon 3 in order to establish their KO cell clones, mirroring our need to target both Exon 2 and a downstream exon (in this case, Exon 7) to create our KO pool. Tsaktanis et al. do not disclose their gRNA sequences nor explain the purpose of using two different gRNA to completely disrupt the EpCAM gene in their human tumor cell line. They also do not observe any impact in cell-matrix and cell-cell adhesion in their EpCAM KO clones when compared to the parental cells.
Similarly, Yang, et al. [21] also stably transfected gRNA, Cas9 enzyme, and GFP expressing plasmids and sorted for green fluorescing cells when creating their EpCAM KO human cancer cell pool, but only targeted Exon 6. When only Exon 7 was targeted here, half of the KO clone's sequenced cDNAs had mutations that had the potential to produce a soluble portion of EpCAM, which appeared to be detected in cell supernatants (Supplemental Fig. 3). This product still allowed these cells to be lysed by the highly potent EpCAM-COBRA (Fig. 3C), perhaps because of cell-surface attachment of this adhesion molecule's secreted variant. It is not known whether Yang et al. observed a similar soluble, perhaps smaller, moiety. Unlike Tsaktanis et al., they observed significant reduction in matrix adhesion and colony formation from their EpCAM KO cell pools.
There are several unique aspects of EpCAM's genomic (NG_012352) and protein structure that allow this novel truncated receptor to be expressed after targeted disruption of Exon 2. First is the distance between Exon 1 and Exon 2 is approximately 4 Kb, as a result, a CRISPR/Cas9 induced deletion must be at least 4.2 kb to completely disrupt Exon 1 and Exon 3 from potentially splicing together, a loss that is significantly larger than the 9–600 bp average seen [22]. Second is Exon 1's splice donor can only join with Exon 3's splice acceptor to produce an in-frame protein fusion, all other Exons from 4 to 9 will result in PTCs. Third is this truncated ΔEx2 message can translate a receptor with most of EpCAM's key features still intact: all the N-glycosylation sites, the protease cleavage site, remaining normal disulfide bridges, and intracellular signaling domain (Fig. 1). These residual intact elements may not only contain the epitope for the EpCAM-COBRA, but they may also retain some inherent EpCAM structure and function. It is perhaps these combined features that allow the EpCAM ΔEx2 mRNA to be transcribed and its translated receptor arrive on the cell-surface, and neither be destroyed by the natural degradative mechanisms that target incomplete messages or malformed proteins.
Others have also observed alternative proteins arise after CRISPR/Cas9 treatment; however, the same antibody was used before and after gene-disruption to detect these mutants [23,24]. This post-CRISPR/Cas9 EpCAM ΔEx2 alternative protein transcript was detected by a unique in-house sdAb associated with a highly potent bispecific molecule. It is recommended that other researchers, likewise, use alternative and sensitive means to detect and confirm complete CRISPR/Cas9 knock-out of their target gene. Relying on only one methodology may provide a false sense of success and lead to incorrect analysis of the gene's loss-of-function phenotype.
4. Materials and methods
4.1. Cells
Cell lines were purchased from ATCC: HT29 colorectal cell line and Raji lymphoma cell line. Cells were propagated in the media recommended by the vendor, supplemented with 10% FBS. Human pan-T cells were isolated from fresh Leukopaks (Stemcell Technologies, #70500) using an immunomagnetic negative selection kit (Stemcell Technologies, 17951). Cells were analyzed on FACS for purity and viability and frozen into single use vials.
4.2. Knock-out cell line transfection
Cells were seeded 2 days before transfection. Ribonucleoprotein (RNP) complexes were formed with guide RNAs (EpCAM Exon 2 gRNA: UGUCUGUGAAAACUACAAGC, EpCAM Exon 7 gRNA: UGGUGUUAUUGCUGUUAUUG, and B7H3 Exon 2 gRNA: UGCCCACCAGUGCCACCACU) and Cas9 enzyme (Synthego). RNP complexes were then mixed with cells in Nucleofector™ Solution (Lonza) This mixture was transferred to a Nucelocuvette™ and placed in the Nucleofector using the SF Cell Line Protocol (Lonza).
4.3. EpCAM-COBRA generation
The general COBRA structure is described in Panchal et al. [17]. In the COBRA platform, two single domain antibodies (sdAbs), or nanobodies, that bind target cells are located on either side of a constrained CD3 Vh/Vl motif. sdAbs against EpCAM were generated by immunizing llamas with soluble human EpCAM protein at the VIB Nanobody Core Facility. These sdAbs were selected by ELISA and were further characterized for binding affinity towards the EpCAM protein and potency for inducing T cell-dependent cytotoxicity against human EpCAM transduced Raji cells. In addition, because EpCAM and Trop2 share more than 60% identity in their extracellular domains, the sdAbs were screened for lack of binding and inert activity toward cells expressing Trop2.
4.4. FACS
Cells were removed from the flask with TrypLE Express, washed once with RPMI+10% FBS, and once with Cell Staining Buffer (BioLegend). Cells were then stained with either PE anti-human EpCAM clone 9C4 (BioLegend) or EpCAM-COBRA, followed by AlexaFluor488-conjugated or AlexaFluor647-conjugated 13H4 or 7A8, antibodies that bind to the CD3 VH region in COBRA molecules [17]. Cells were incubated on ice, after which unbound antibodies were removed by several washes in Cell Staining Buffer. After washing, the cells were pool sorted on a BD FACSMelody™ Cell Sorter (BD Biosciences). Gates were set by using isotype control antibodies and set at the first 10% of the negative population. Cells were then sorted, and data were analyzed using Flow Jo (BD Biosciences) and GraphPad Prism version 8.1.2 for Windows (GraphPad Software).
4.5. TDCC (T cell-dependent cellular cytotoxicity)
Cells were engineered to constitutively express firefly luciferase (Biosettia) via lentivirus transduction and puromycin selection. Cells were removed from the flask with TrypLE Express, centrifuged and resuspended in culture medium. Purified human pan T cells were also thawed, centrifuged, and resuspended in culture medium. Target cells and T cells were counted and mixed at a ratio of 1:10, respectively. The co-culture of cells was then added to an assay plate. Serial dilutions of COBRAs were prepared separately in culture medium and transferred to the assay plate. Cells were incubated at 37°C for 48 h (HT29 cells) or 96 h (Raji cells), after which luciferase levels were measured using an Envision luminometer (PerkinElmer) by adding an equal volume of SteadyGlo (Promega). Data were analyzed using GraphPad Prism version 8.1.2 for Windows (GraphPad Software).
4.6. Western
Adherent cells were removed from their flasks with trypsin or 20 mM EDTA in PBS, then washed several times in PBS before lysing in 1x Cell Lysis Buffer (Cell Signaling Technology; Danvers, MA) containing 1x protease/phosphatase inhibitors (Cell Signaling Technology) Suspension cells were similarly washed in PBS before lysis. Lysates were reduced with 10% 2-mercaptoethanol and run on 4–20% Tris-Glycine SDS-PAGE, transferred to PVDF membranes, then probed with a goat polyclonal anti-EpCAM antibody (R&D, Cat# AF960), followed by a donkey anti-goat 800CW antibody (Licor), after which the membranes were scanned on an Odyssey (Licor).
For cell surface biotinylation experiments, biotinylation was performed on cells after removal from the flask, using the Pierce Cell Surface Biotinylation and Isolation kit (ThermoFisher). Cells not incubated with biotin were processed in parallel as controls. After biotinylation, cells were lysed according to the manufacturer's instructions, and protein concentrations in lysates were quantified prior to purification of biotinylated proteins with NeutrAvidin agarose. The proportion of purified protein equivalent to the original mass of total lysate was run on SDS-PAGE, then probed with antibodies as described above.
4.7. RNA Extraction and 5′RACE RT-PCR
Fresh cells were gently pelleted, and their total RNA was extracted using Qiagen's RNAeasy Plus Mini. After quantifying the final yield, 1 μg of total RNA was used for first strand and 5′ RACE cDNA synthesis using Takara's SMARTer RACE 5′ Kit. EpCAM transcript was amplified from the cDNA with New England Biolab's Q5® Hot Start High-Fidelity 2X Master Mix using Takara's Universal Primer A Mix (5′) and a human EpCAM gene-specific-primer (TGATTTGTGATTGAAAGCTGCC) within the 3′ untranslated region (UTR) (NG_012352). Amplified products were subcloned into pRACE via Takara's In-Fusion and sequenced with M13 forward and reverse primers.
4.8. Plasmid Construction and lentivirus packaging
All human EpCAM (BC014785) P2A-Green Fluorescent Protein (GFP) fusion protein inserts were synthesized by Integrated DNA Technologies and subcloned into pLenti vector by Takara's In-Fusion. Lentivirus was packaged using 293 TA cells and GeneCopoeia's Lenti-Pac™ HIV Expression Packaging Kit. Raji cells were spinoculated with concentrated lentivirus.
Author contributions
A.B.: Research and Investigation. P.A.C.: Research and Investigation, Supervision, Conceptualization. R.B.D.: Conceptualization. T.T.C.: Research and Investigation, Writing Original Draft, and Supervision.
Funding
The work was funded by Maverick Therapeutics Inc., a wholly owned subsidiary of Takeda Pharmaceutical Company Limited.
Declaration of interests
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests:
All authors are currently full-time employees of Takeda Pharmaceuticals.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bbrep.2022.101205.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A., Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mali P., Yang L., Esvelt K.M., Aach J., Guell M., DiCarlo J.E., Norville J.E., Church G.M. RNA-guided human genome engineering via Cas9. Science. 2013;339:823. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lin S., Staahl B.T., Alla R.K., Doudna J.A. Enhanced homology-directed human genome engineering by controlled timing of CRISPR/Cas9 delivery. Elife. 2014;3 doi: 10.7554/eLife.04766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim S., Kim D., Cho S.W., Kim J., Kim J.S. Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res. 2014;24(6):1012–1019. doi: 10.1101/gr.171322.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zuris J.A., Thompson D.B., Shu Y., Guilinger J.P., Bessen J.L., Hu J.H., Maeder M.L., Joung J.K., Chen Z.Y., Liu D.R. Cationic lipid-mediated delivery of proteins enables efficient protein-based genome editing in vitro and in vivo. Nat. Biotechnol. 2015;33(1):73–80. doi: 10.1038/nbt.3081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schumann K., Lin S., Boyer E., Simeonov D.R., Subramaniam M., Gate R.E., Haliburton G.E., Ye C.J., Bluestone J.A., Doudna J.A., Marson A. Generation of knock-in primary human T cells using Cas9 ribonucleoproteins. Proc. Natl. Acad. Sci. U.S.A. 2015;112:10437–10442. doi: 10.1073/pnas.1512503112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ran F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013;8(11):2282–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Capone E., Giansanti F., Ponziani S., Lamolinara A., Iezzi M., Cimini A., Angelucci F., Sorda R., Laurenzi V., Natali P.G. EV20-Sap, a novel anti-HER-3 antibody-drug conjugate, displays promising antitumor activity in melanoma. Oncotarget. 2017;8:95412–95424. doi: 10.18632/oncotarget.20728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nielsen C.F., van Putten S.M., Lund I.K., Melander M.C., Norregaard K.S., Jurgensen H.J., et al. The collagen receptor uPARAP/Endo180 as a novel target for antibody-drug conjugate mediated treatment of mesenchymal and leukemic cancers. Oncotarget. 2017;8:44605–44624. doi: 10.18632/oncotarget.17883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Von Neubeck B., Gondi G., Riganti C., Pan C., Damas A.P., Scherb H., Ertürk A., Zeidler R. An inhibitory antibody targeting carbonic anhydrase XII abrogates chemoresistance and significantly reduces lung metastases in an orthotopic breast cancer model in vivo. Int. J. Cancer. 2018;143:2065–2075. doi: 10.1002/ijc.31607. [DOI] [PubMed] [Google Scholar]
- 11.Huhe M., Lou J., Zhu Y., Zhao Y., Shi Y., Wang B., Sun X., Zhang X., Zhang Y., Chen Z.-N. A novel antibody-drug conjugate, HcHAb18-DM1, has potent anti-tumor activity against human non-small cell lung cancer. Biochem. Biophys. Res. Commun. 2019;513:1083–1091. doi: 10.1016/j.bbrc.2019.04.046. [DOI] [PubMed] [Google Scholar]
- 12.Alijaj N., Moutel S., Gouveia Z.L., Gray M., Roveri M., Dzhumashev D., Weber F., Meier G., Luciani P., Rössler J.K., Schafer B.W., Perez F., Bernasconi M. Novel FGFR4-targeting single-domain antibodies for multiple targeted therapies against Rhabdomyosarcoma. Cancers. 2020;12:3313. doi: 10.3390/cancers12113313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dreier T., Baeuerle P.A., Fichtner I., Grun M., Schlereth B., Lorenczewski G., Kufer P., Lutterbuse R., Reithmuller G., Gjorstrup P., Bargou R.C. T cell costimulus-independent and very efficacious inhibition of tumor growth in mice bearing subcutaneous or leukemic human B cell lymphoma xenografts by a CD19-/CD3- bispecific single-chain antibody construct. J. Immunol. 2003;170:4397–4402. doi: 10.4049/jimmunol.170.8.4397. [DOI] [PubMed] [Google Scholar]
- 14.Huehls A.M., Coupet T.A., Sentman C.L. Bispecific T-cell engagers for cancer immunotherapy. Immunol. Cell Biol. 2015;93:290–296. doi: 10.1038/icb.2014.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jitschin R., Saul D., Braun M., Tohumeken S., Volkl S., Kischel R., Lutteropp M., Dos Santos C., Mackensen A., Mougiakakos D. CD33/CD3-bispecific T-cell engaging (BiTE) antibody construct targets monocytic AML myeloid-derived suppressor cells. J. Immunother. Cancer. 2018;6:116. doi: 10.1186/s40425-018-0432-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fu M., He Q., Guo Z., Zhou X., Li H., Zhao L., Tang H., Zhou X., Zhu H., Shen G., He Y., Lei P. Therapeutic bispecific T-cell engager antibody targeting the transferrin receptor. Front. Immunol. 2019;10:1396. doi: 10.3389/fimmu.2019.01396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Panchal A., Seto P., Wall R., Hillier B.J., Zhu Y., Krakow J., Datt A., Pongo E., Bagheri A., Chen T.T., Degenhardt J.D., Culp P.A., Dettling D.E., Vinogradova M.V., May C., DuBridge R.B. COBRA™: a highly potent conditionally active T cell engager engineered for the treatment of solid tumors. mAbs. 2020;12(1) doi: 10.1080/19420862.2020.1792130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schnell U., Cirulli V., Giepmans B.N.G. EpCAM: structure and function in health and disease. Biochemica et Biophysica Acta. 2013;1828:1989–2001. doi: 10.1016/j.bbamem.2013.04.018. [DOI] [PubMed] [Google Scholar]
- 19.Sharp J.J., Cooper T.A. Unexpected consequences: exon skipping cause by CRISPR-generated mutations. Genome Biol. 2017;18:109. doi: 10.1186/s13059-017-1240-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tsaktanis T., Kremling H., Pavsic M., von Stackelberg R., Mack B., Fukumori A., Steiner H., Vielmuth F., Spinder V., Huang Z., Jakubowski J., Stoecklein N.H., Luxenburger E., Lauber K., Lenarcic B., Gires O. Cleavage and cell adhesion properties of human Epithelial cell adhesion molecule (HEPCAM) J. Biol. Chem. 2015;290(40):24574–24591. doi: 10.1074/jbc.M115.662700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang J., Isaji T., Zhang G., Qi F., Duan C., Fududa T., Gu J. EpCAM associates with integrin and regulates cell adhesion in cancer cells. Biochem. Biophys. Res. Commun. 2020;522(4):903–909. doi: 10.1016/j.bbrc.2019.11.152. Feb 19. [DOI] [PubMed] [Google Scholar]
- 22.Shin H.Y., Wang C., Lee H.K., Yoo K.H., Zeng X., Kuhns T., Yang C.M., Mohr T., Liu C., Hennighousen L. CRISPR/Cas9 targeting events cause complex deletions and insertions at 17 sites in the mouse genome. Nat. Commun. 2017;8:15464. doi: 10.1038/ncomms15464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mou H., Smith J.L., Peng L., Yin H., Moore J., Zhang X.-O., Song C.-Q., Sheel A., Wu Q., Ozata D.M., Li Y., Anderson D.G., Emerson C.P., Sontheimer E.J., Moore M.J., Weng Z., Xue W. CRISPR/Cas9-mediated genome editing induces exon skipping by alternative splicing or exon deletion. Genome Biol. 2017;18:108. doi: 10.1186/s13059-017-1237-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tuladhar R., Yeu Y., Piazza J.T., Tan Z., Clemenceau J.R., Wu X., Barrett Q., Herbert J., Mathews D.H., Kim J., Hwang T.H., Lum L. CRISPR-Cas9-based mutagenesis frequently provokes on-target mRNA misregulation. Nat. Commun. 2019;10:4056. doi: 10.1038/s41467-019-12028-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.