Abstract
Melatonin is a biogenic amine that can be found in plants, animals and microorganism. The metabolic pathway of melatonin is different in various organisms, and biosynthetic endogenous melatonin acts as a molecular signal and antioxidant protection against external stress. Microbial synthesis pathways of melatonin are similar to those of animals but different from those of plants. At present, the method of using microorganism fermentation to produce melatonin is gradually prevailing, and exploring the biosynthetic pathway of melatonin to modify microorganism is becoming the mainstream, which has more advantages than traditional chemical synthesis. Here, we review recent advances in the synthesis, optimization of melatonin pathway. l-tryptophan is one of the two crucial precursors for the synthesis of melatonin, which can be produced through a four-step reaction. Enzymes involved in melatonin synthesis have low specificity and catalytic efficiency. Site-directed mutation, directed evolution or promotion of cofactor synthesis can enhance enzyme activity and increase the metabolic flow to promote microbial melatonin production. On the whole, the status and bottleneck of melatonin biosynthesis can be improved to a higher level, providing an effective reference for future microbial modification.
Keywords: Melatonin, Biosynthesis, Plants, Animals, Microorganisms, Metabolic modification, Enzyme catalysis mechanism
1. Introduction
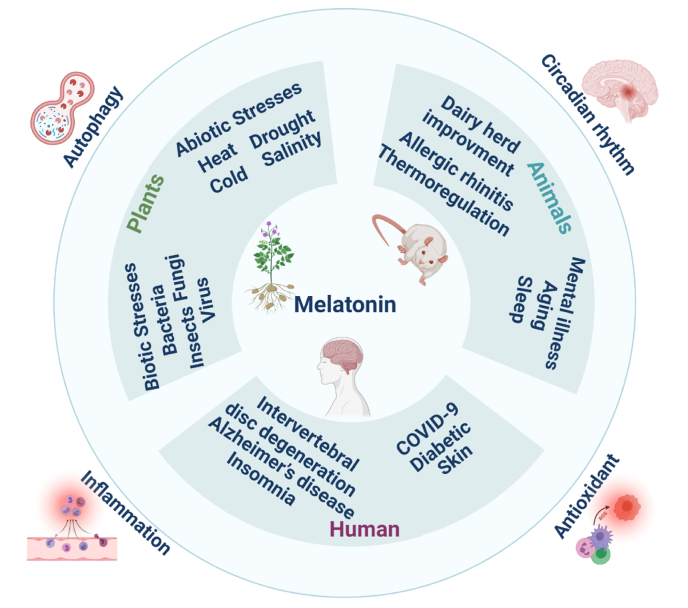
Melatonin (N-acetyl-5-methoxytryptamine) is an important indoleamine derivative. In 1967, Sagan speculated that melatonin was synthesized by mitochondria and chloroplasts, which at the time were considered to have evolved from precursors similar to Rhodospirillum rubrum and cyanobacteria [1,2]. Melatonin has anti-inflammatory, antioxidant and autophagic properties [3,4]. The level of melatonin in plants affects their response to biotic and abiotic stress [5]. In animals, melatonin can regulate sleep and body temperature [6] and improve dairy production [7]. In humans, it was demonstrated to protect lymphocytes against DNA damage induced by ionizing radiation [8], regulate the circadian rhythm [9], protect the skin [10], and ameliorate diabetes [11]. Very recently, melatonin was also investigated as a potential adjuvant treatment for slowing down the effects of COVID-19 [12] (Fig. 1). Nowadays, it is estimated that 50 to 70 million Americans chronically suffer from a sleep or circadian disorder, and the global market of melatonin was estimated at USD 1 billion in 2020, and it is expected to reach 3.4 billion USD by 2026 (https://marketresearchexpertz.com/report/global-melatonin-market-92220). Recently, it was reported that more than 300 million people in China suffer from sleep disorders. However, sales of melatonin products in the global market have soared, and demand exceeds production, so that it is necessary to further improve industrial approaches for melatonin synthesis. Commercial melatonin is mainly produced by chemical synthesis, which is highly polluting. The raw material supply from animal and plant extraction is small, and the extraction rate is low. In recent years, the rapid development of microbial fermentation methods has offered hope for more economic and environmentally friendly production. It is a promising method to design, optimize and introduce melatonin synthesis pathways into microorganisms.
Fig. 1.
The application value of melatonin.
Due to the complex metabolic network and numerous secondary metabolites of microorganisms, the synthesis pathway of melatonin has not been investigated in many species. Only a few strains have been found to produce melatonin by themselves, including yeast and pseudomonads, but the synthesis efficiency is limited [[13], [14], [15], [16]]. Pseudomonas fluorescens can synthesize melatonin by itself as a growth signal molecule or as a protective agent against ROS in the medium to promote early adaptation. However, the melatonin concentration reached a maximal value of only 1.32 μg/L [16]. Back et al. speculated that the melatonin synthesis pathway of yeast is similar to that of cyanobacteria or plants [17]. Melatonin is a secondary metabolite of yeast which can interact with glycolytic proteins and has a positive effect on fermentative metabolism by increasing resistance to oxidative stress and UV radiation [[18], [19], [20]]. Yeast follows the Ehrlich pathway during early alcohol fermentation and will use amino acids such as l-tryptophan as the source of nitrogen [21]. The melatonin production of Saccharomyces cerevisiae QA23 and Levucell SC20 (from animal nutrition) reached 1.35 ± 1.69 and 93.14 ± 85.86 ng/mL during alcoholic fermentation, respectively [14,22].
Heterologous expression systems can be widely employed to screen various putative melatonin synthesis genes from animals and plants to overproduce melatonin in various microorganisms [23]. There are few reports on the ectopic overproduction of melatonin in E. coli because of either the low or insoluble expression of melatonin biosynthesis genes [[23], [24], [25]]. Zhang et al. combined the physostigmine biosynthetic genes from Streptomyces albulus, a gene encoding phenylalanine 4-hydroxylase from Xanthomonas campestris and caffeic acid 3-O-methyltransferase (COMT) from Oryza sativa in E. coli, which resulted in a melatonin yield of 136.17 ± 1.33 mg/L in a shake flask and 0.65 ± 0.11 g/L in a 2L-bioreacorr [26]. When a melatonin-producing strain was assembled using the three plasmids ddc, aanat and asmt, encoding the cofactor pterin-4α-carbinolamine dehydratase and using glucose as the sole carbon source for tryptophan supply, the final melatonin yield reached a record level of over 2 g/L [27].
However, the synthesis of melatonin in microorganisms is complex and the yield is generally low. The metabolic engineering approaches are not systematic and there is a lack of research on related pathways in the prior art. Therefore, we need to explore metabolic engineering strategies to facilitate the large-scale production of melatonin by microorganisms.
2. Natural melatonin synthesis pathways
2.1. Melatonin synthesis pathway in plants
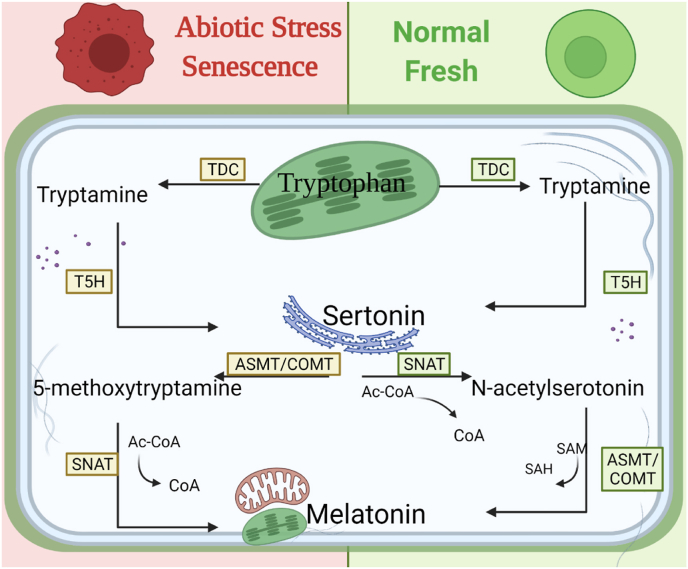
Melatonin was first reported in plants in 2000 and was found to be distributed in various plant tissues [28]. Mitochondria and chloroplasts are the main sites of melatonin synthesis in plants. From an evolutionary point of view, the precursor of mitochondria probably was similar to a purple nonsulfur bacterium, and chloroplasts are probably the descendants of cyanobacteria [29,30]. Studies have shown that l-tryptophan can be converted into melatonin via two pathways in plants, one in healthy plants and the other under senescence or biological stress [28,31].
Under normal plant growth, l-tryptophan is fist carboxylated by l-tryptophan decarboxylase (TDC) into tryptamine in cytoplasm. The second committed enzyme trptamine-5-hydroxylase (T5H), which catalyzes tryptamine into serotonin and requires acetyl-coenzyme A (Ac-CoA) on the endoplasmic reticulum. Usually in fresh plants, the third step is the acetylation of serotonin into N-acetylserotonin by serotonin N-acetyltransferase (SNAT) in the chloroplast. N-Acetylserotonin methyltransferase (ASMT) is believed to be the last enzyme, it catalyzes N-acetyl-serotonin into melatonin by methylation reaction in the cytoplasm. When plants under senescence and cadmium stress, the serotoin maybe accumulated so that the melatonin synthesis would favor use ASMT catalyzes serotonin into 5-methexytryptamine, and then synthesize melatonin by SNAT [32]. During the last step, the conversion of acetyl-5-hydroxytryptamine to melatonin is accompanied by the co-conversion of S-adenosyl-l-methionine (SAM) to S-adenosyl-l-homocysteine (SAH). Overall, when plants are subjected to biological stress and senescence, the same as the normal growth conditions of plants is that tryptophan is carboxylated and hydroxylated to produce serotonin. The difference is that serotonin is first methylated to produce 5-methyltryptamine, and then 5-methyltryptamine is acetylated to produce melatonin [17]. In plants, the existence of two melatonin metabolic pathways may be determined by the strict regulatory mechanisms of plants themselves (Fig. 2). However, the deeper regulation of enzymes that plants biosynthesize melatonin under different conditions needs to be explored.
Fig. 2.
Synthesis pathway of melatonin in plants
TDC, tryptophan decarboxylase; T5H, tryptamine-5-hydroxylase; ASMT, N
-acetylserotonin methyltransferase; COMT, acetylserotonin O
-methyltransferase; SNAT, serotonin N
-acetyltransferase.
2.2. Melatonin synthesis pathway in animals
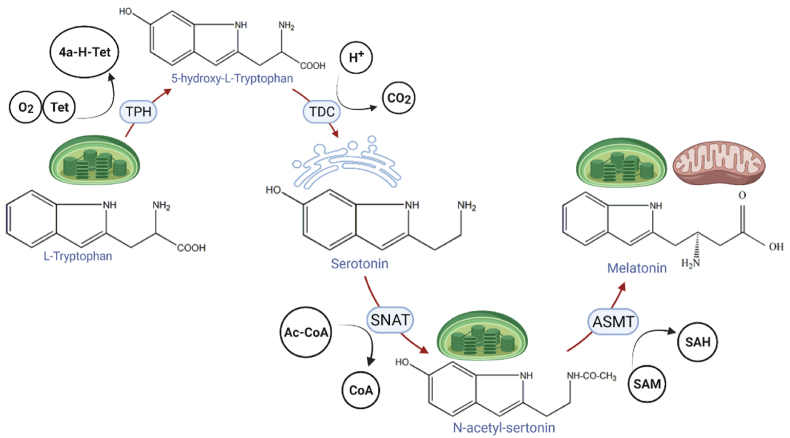
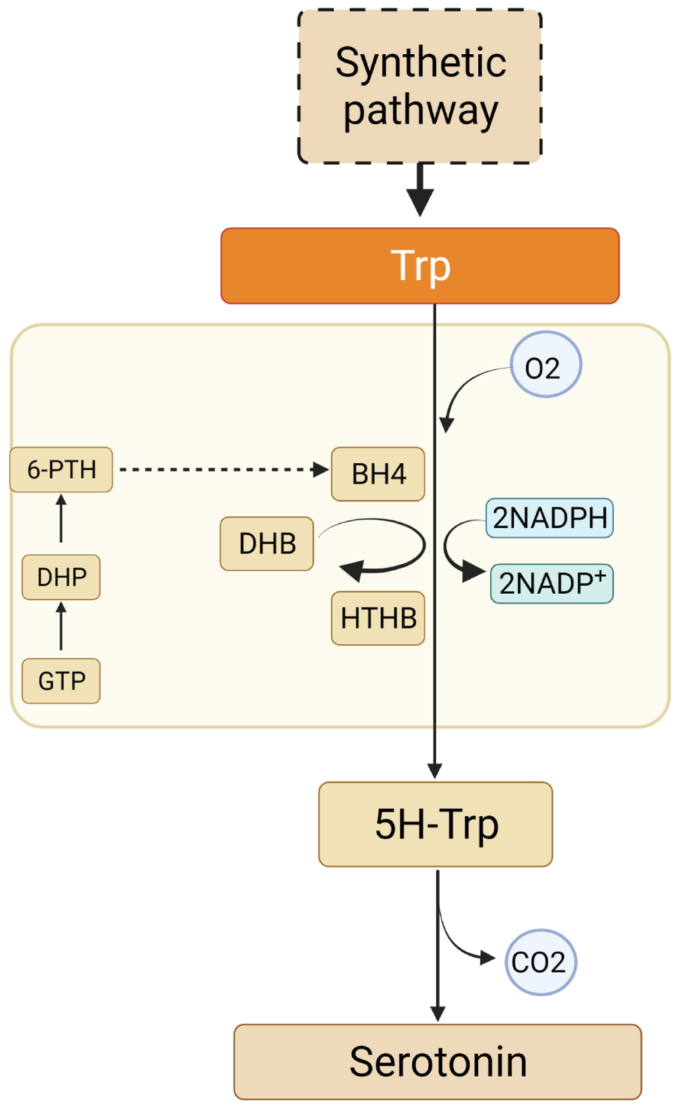
Animals have a single source of melatonin, mitochondria. Moreover, animals cannot synthesize l-tryptophan themselves, and it must be ingested externally. Therefore, animals have lower melatonin metabolism than plants. In animals, the synthesis of melatonin proceeds via a four-step pathway. First, tryptophan hydroxylase produces 5-hydroxytryptophan. This reaction requires the cofactor BH4 and oxygen. Next, tryptophan carboxylase converts 5-hydroxytryptophan into serotonin, which is the main precursor of melatonin synthesis. This reaction is accompanied by the release of carbon dioxide. Subsequently, aralkylamine N-acetyltransferase produces N-acetyl serotonin at the expense of acetyl-CoA is. Finally, N-acetyl-serotonin methyltransferase generates the final product melatonin accompanied by the conversion of the cofactor SAM into SAH [17,31] (Fig. 3).
Fig. 3.
The biosynthetic pathways of melatonin in all known living organisms.
3. Synthesis and modification pathways of melatonin in microorganisms
In addition to plants and animals, many microbes are also capable of melatonin synthesis [16,18,19,33]. The application of engineered microorganisms to synthesize melatonin is a potential industrial production method. The pathway through which microorganisms synthesize melatonin is similar to that of animals. It requires encompasses consecutive enzymatic reactions with the involvement of coenzymes or cofactors. However, there are few literature reports on the improvement of microbial melatonin production through metabolic engineering. According to the basic approaches of metabolic engineering, the production of melatonin can be improved by increasing the supply of precursors, optimizing the expression of pathway enzymes, or improving the utilization of cofactors and coenzymes.
3.1. l-tryptophan synthesis
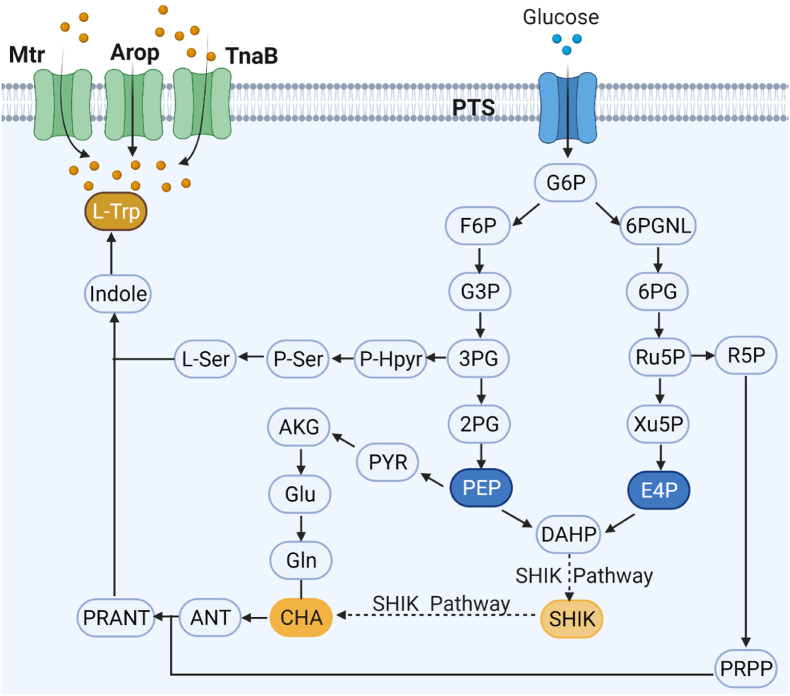
L-tryptophan is a precursor for the synthesis of melatonin [34]. Normally, glucose is transported through the phosphotransferase system to produce glucose hexaphosphate, which then enters the glycolytic pathway and the pentose phosphate pathway to produce phosphoenolpyruvate pyruvic acid (PEP) and erythrose 4-phosphate (E4P), respectively. Then, 3-deoxy-d-arabinoheptulose (DAHP) is produced by the condensation of PEP and E4P, and passes through the shikimic acid pathway to generate chorismate. The biosynthetic pathway of l-tryptophan is summarized in Fig. 4. In E. coli, l-tryptophan is synthesized by a long pathway of enzymes encoded by the trpEDCBA operon [35]. The main rate-limiting enzymes that have a major impact on l-tryptophan synthesis are anthranilate synthase encoded by trpE and DHAP synthase encoded by aroG. These enzymes are subject to feedback inhibition by tryptophan and phenylalanine, respectively. The transcriptional regulation of the synthesis and transport is mainly achieved by the repressor proteins encoded trpR. Weakening the TrpR repressor protein can relieve its inhibitory effect on the enzymes in the l-tryptophan synthesis pathway and increase the production of l-tryptophan [36,37]. Restraining the expression of tnaA (l-tryptophanase) was necessary to prevent the formation of serotonin and improve hydroxylation for increase the production of melatonin [38].
Fig. 4.
The biosynthetic pathway of l-tryptophan.
Abbreviations: G6P, glucose-6-phosphate; F6P, fructose-6-phosphate; 6PGNL
6-phosphoglucono-lactone, G3P, glyceraldehyde-3-phosphate; 3 PG
glycerate-3-phosphate; 3-Hpyr, 3-phosphonooxypruvate; PEP, phosphoenolpyruvate; E4P, erythrose-4-phosphate; Ru5P, ribulose-5-phosphate; R5P, ribose-5-phosphate; PRPP, phosphoribosyl pyr-ophosphate; PYR, pyruvate; AKG; α-ketoglutarate; Glu, glutamate; Gln, glutamine; P-Ser, 3-phosphoserine; L-Ser, serine;DAHP, 3-deoxy-d-arabi-noheptulosonate-7-phosphate; SHIK, shikimate; CHA, chorismate; ANT, anthranilate; PRANT, N-(5′-phosphoribosyl)-anthranilate; Trp, tryptophan.
Both E. coli and yeast can synthesize l-tryptophan, which can be used to synthesize melatonin. At present, most strains with high tryptophan yield were constructed from E. coli, indicating that it may be a better chassis for the production of melatonin. Zhu et al. expressed heterologous glutamine synthetase in the engineered E. coli strain KW001, followed by the overexpression of icd and gdhA, as well as the expression of mutated serA and thrA variants. Finally, sthA and pntAB, encoding transhydrogenase, were overexpressed to maintain cofactor balance. The engineered strain could produce 1.71 g/L l-tryptophan in shake-flask fermentation, which was 2.76-times higher than the titer of the parental strain [39]. Zeng and Chen rationally engineered wild-type E. coli W3110 by knocking out tryptophanase (tnaA), 5-methyltetrahydrofolate-homocysteine methyltransferase (MTR), and 3-deoxy-7-phosphoheptanonic acid synthase (aroFGH), as well as overexpressing D-3-phosphoglycerate dehydrogenase (serA), which resulted in a final l-tryptophan titer of 30–34 g/L in a 1.5 L bioreactor [40]. Xiong et al. first introduced phosphoketolase from Bifidobacterium adolescentis to strengthen E4P formation, after which the phosphotransferase system was substituted with PEP-independent glucose transport, meditated by a glucose facilitator from Zymomonas mobilis and native glucokinase. Finally, they rewired the PEP-pyruvate-oxaloacetate node. Fed-batch fermentation in a 5-L bioreactor produced 41.7 g/L l-tryptophan, which is the highest yield reported to date [41].
3.2. Serotonin synthesis
Serotonin is formed by a two-step conversion of l-tryptophan. First, l-tryptophan is hydroxylated to 5-hydroxyserotonin by TPH, which in turn is carboxylated by TDC to yield serotonin. Serotonin is mainly known for its role as a major neurotransmitter and mood regulator, leading to the epithet ‘hormone of happiness’ based on the relationship between low serotonin levels and depression [34]. Serotonin is synthesized in the central nervous system and gastrointestinal tract of mammals [42]. The presence of serotonin is strictly related to the synthesis of melatonin in plants and animals. It is a key intermediate in the melatonin synthesis pathway. The biosynthetic pathway of serotonin from l-tryptophan is summarized in Fig. 5.
Fig. 5.
Synthesis of serotonin from l-tryptophan.
In microbial production, serotonin is mainly synthesized from externally added l-tryptophan. Germann et al. used truncated H. sapiens HsTPH146-460, lacking both the N- and C-terminal regulatory regions in order to increase heterologous expression and enhance protein stability, along with H. sapiens DDC, Bos taurus AANAT and H. sapiens ASMT to produce 2.4 mg/L of serotonin [18]. Tryptophan hydroxylase has been expressed in E. coli with low enzymatic activity, as the solubility and stability of the enzyme seem to be affected in this host. A serotonin-producing strain of Saccharomyces cerevisiae was constructed by introducing l-tryptophan hydroxylase from Cupriavidus taiwanensis together with a cofactor reconstitution pathway, and the yield of serotonin reached 154.3 ± 14.3 mg/L [43]. Therefore, introducing heterologous enzymes is helpful for the improvement of 5HT production. The production of serotonin can also be improved by regulating the expression levels of relevant enzymes. Directed evolution of enzymes can increase their affinity for the substrate and reduce side-reactions. Improving the supply of cofactors and oxygen are also effective ways to improve the titer of serotonin.
3.2.1. l-tryptophan hydroxylase
Tryptophan hydroxylase (TPH) is a mononuclear non-heme iron enzyme, which catalyzes the reaction between tryptophan, O2, and tetrahydrobiopterin (BH4) to produce 5-hydroxytryptophan and 4a-hydroxytetrahydrobiopterin [44]. The role of TPH is to replace the H at the C5 position on the benzene ring of tryptophan with an OH radical, which as the first step in the synthesis of serotonin. The hydroxylation reaction is the first and rate-limiting step in the synthesis of serotonin [45,46]. Therefore, it is crucial to find a suitable enzyme to increase serotonin production.
Directed evolution can improve the catalytic activity of enzymes. Kino et al. found that L101F and W180F changes in the active center of Pseudomonas aeruginosa have an effect on substrate specificity and hydroxylase activity. The catalytic rate constant Kcat increased from 0.4 to 2.08 after mutation at the two sites [47,48]. Rosetta modeling is a useful tool for studying the effects of mutations on catalytic function in more detail. Generally, high-throughput screening based on biosensors can be used to identify effective mutations. Maxel et al. applied this selection platform to screen a NADPH-dependent-hydroxybenzoic acid hydroxylase that uses 3,4-dihydroxyphenyl acid as a substrate, and obtained variants with roughly 8-fold improved apparent catalytic efficiency (Kcat/Km) for 3,4-DHBA, compared to the wild type [49,50].
Rational design of enzymes is becoming the dominant approach. Therefore, it is necessary to elucidate the structure of the enzyme and modify it to enhance the specificity of substrate binding by library construction and rational in silico design [51]. Windahl et al. solved the first crystal structure of an aromatic amino acid hydroxylase, TPH. The substrate specificity of the TPH is controlled through interactions with the side chain of tryptophan. The side chain of the tryptophan is bound in a hydrophobic pocket lined by residues Tyr236, Thr266, Pro267, Glu268, Pro269, His273, Phe314, Phe319, and Ile367 [44]. Human TPH can be expressed in E. coli and human embryonic kidney cells (HEK293). With l-tryptophan as the substrate, the Km value of wild-type TPH is 41 μM, while the Km value of the TPH2 mutant R441H was 33 μM [52].(Table 1 and Table 2).
Table 1.
Melatonin synthesis enzymes from plants and their Km values.
| Enzymes | Organizm | Km[mM] | Enzymes | Organizm | Km[mM] |
|---|---|---|---|---|---|
| TDC | Catharanthus roseus | 0.075 [84] | SNAT | Homo sapiens | 0.06 [85] |
| Vinca minor | 1.3 [86] | 0.096 [85] | |||
| Withania coagulans | 1.7 [87] | 0.106 [85] | |||
| Gelatoporia subvermispora | 0.16 [88] | 1.23 [85] | |||
| 0.16 [88] | 1.35 [85] | ||||
| 0.29 [88] | Ovis aries | 0.125 [89] | |||
| 1.94 [88] | 0.64 [90] | ||||
| 1.94 [88] | Rattus norvegicus | 1.7 [90] | |||
| Bacillus sp. (in: Bacteria) | 0.3 [91] | 2 [92] | |||
| T5H | Homo sapiens | 0.0147 [93] | Sparus aurata | 0.05 [94] | |
| 0.015 [95] | 2.05 [94] | ||||
| 0.0217 [93] | Drosophila melanogaster | 0.16 [96] | |||
| 0.0249 [93] | Aedes aegypti | 0.23 [97] | |||
| 0.0265 [98] | 0.42 [97] | ||||
| 0.0289 [93] | Arabidopsis thaliana | 0.232 [69] | |||
| 0.0351 [98] | Oryza sativa | 0.371 [99] | |||
| 0.0423 [100] | Pyropia yezoensis | 0.467 [101] | |||
| 0.0518 [93] | Saccharopolyspora erythraea | 13 [102] | |||
| 0.0684 [93] | ASMT | Oncorhynchus tshawytscha | 0.005 [103] | ||
| 0.077 [104] | Ovibos moschatus | 0.015 [105] | |||
| . | 0.315 [98] | Bos taurus | 0.0291 [106] | ||
| Oryctolagus cuniculus | 0.0142 [107] | 0.054 [106] | |||
| 0.031 [108] | Rattus norvegicus | 0.04 [109] | |||
| 0.135 [107] | 0.1 [110] | ||||
| Mus musculus | 0.294 [111] | Oryza sativa | 0.243 [112] | ||
| Gallus gallus | 0.324 [113] | 0.864 [25] | |||
| Schistosoma mansoni | 0.0067 [114] | Arabidopsis thaliana | 0.456 [115] |
Table 2.
Melatonin synthesis enzymes from animals and their Km values.
| Enzymes | Organizm | Km[mM] | Enzymes | Organizm | Km[mM] |
|---|---|---|---|---|---|
| TPH | Homo sapiens | 0.0075 [116] | TPH | Mus musculus | 0.0075 [117] |
| 0.0078 [118] | 0.00873 [119] | ||||
| 0.0106 [120] | 0.0166 [117] | ||||
| 0.0131 [121] | 0.0192 [117] | ||||
| 0.0132 [122] | 0.031–0.033 [111] | ||||
| 0.015 [123] | Rattus norvegicus | 0.0865 [111] | |||
| 0.017 [124] | 0.119 [111] | ||||
| 0.0185 [116] | 0.119 [100] | ||||
| 0.0186 [52] | Cavia porcellus | 0.002 [125] | |||
| 0.02 [126] | 0.3 [125] | ||||
| 0.0228 [120] | Oryctolagus cuniculus | 0.0021 [127] | |||
| 0.0228 [123] | 0.0058 [107] | ||||
| 0.023 [126] | 0.032 | ||||
| 0.02426 [128] | 0.0479 [129] | ||||
| 0.0255 [52] | Gallus gallus | 0.0077 | |||
| 0.0282 | Bos taurus | 0.016 [130] | |||
| 0.033 [124] | Thunnus albacares | 0.023 [131] | |||
| 0.0334 [52] | Schistosoma mansoni | 0.022 [131] | |||
| 0.0403 [121] | TDC | Catharanthus roseus | 1.3 [132] | ||
| 0.0413 [52] | Gelatoporia subvermispora | 0.32 [88] | |||
| 0.0434 [120] | 0.32 [88] | ||||
| 0.0449 [100] | 0.35 [88] | ||||
| 0.0511 [52] | 1.72 [88] | ||||
| 0.0545 [100] | |||||
| 0.111 [120] | |||||
3.2.2. l-tryptophan decarboxylase
L-tryptophan decarboxylase (TDC) is a cytosolic type-II aromatic l-amino acid decarboxylase [[53], [54], [55], [56], [57]]. It catalyzes the final step in the microbial pathway of serotonin synthesis. TDC catalyzes two reactions, one of which produces tryptamine from l-tryptophan and the other serotonin from 5-hydroxytryptophan. Because TDC has an affinity for l-tryptophan, tryptamine is readily produced as a byproduct, which can affect the accumulation of serotonin. Therefore, to reduce the production of the by-product tryptamine, it is particularly important to select appropriate enzymes.
TDC has different affinity for l-tryptophan and 5-hydroxytryptamine. The Km value of TDC is generally higher in plants. Similar to the enzymes from rice and Ophiorrhiza pumila, Withania somnifera TDC is highly specific for tryptophan, and does not accept 5-hydroxytryptophan to produce serotonin [40]. The Km values of TDC enzymes from different sources are summarized in Table 1, Table 2 TDC is a homodimer with strict specificity to the substrate tryptophan. The affinity between the enzyme and substrate is not only related to the size of the active site pocket, but is also affected by the hydrophobicity of the surrounding residues [58]. In the synthesis of serotonin, TDC can convert both tryptophan and 5-hydroxytryptophan, but exhibits different catalytic efficiency [[58], [59], [60], [61]]. Zhou et al. used protein modeling to deduce that the bottom binding pocket and active cavity of TDC were mainly composed of hydrophobic residues of one subunit (W95, F103, F104 and L336) and adjacent subunits (V125, F127, L360 and V380) [58]. In contrast to 5-hydroxytryptophan, l-tryptophan lacks a hydroxyl group on the indole ring. Therefore, the potential reason why the TDC binding pocket can hold tryptophan is reduced steric hindrance and higher hydrophobicity. Thus, possible strategies to construct a TDC mutant that can use 5-hydroxytryptophan to initiate the synthesis of melatonin include increasing the size of the active site and promoting the formation of hydrogen bonds between the enzyme and the substrate. It was reported that H214 andY359 are crucial for substrate recognition and influence TDC activity [58,62,63]. The KOD-plus-neo PCR method can also be used for site-directed mutagenesis to change the size of the active pocket and achieve better binding of the substrate 5-hydroxytryptophan [58]. A optimization through promoter engineering and confirms the interaction model can be used which can also lift the rate-limiting bottleneck enzyme. Song Gao, using promoter regulation and directed evolution in the host Saccharomyces cerevisiae, established a high-throughput screening method to improve the catalytic efficiency of flavonoid 3′-hydroxylase, so that (2S)-eriodictyol reached the current highest titer of 3.3 g/L [64].
3.2.3. Cofactors
To increased product titers, it is necessary to not only overexpress pathway enzymes, but also boost the supply of cofactors such as BH4 and NADPH. The conversion of l-tryptophan into 5-hydroxytryptophan by TPH requires both oxygen and the cofactor BH4, which is produced from GTP [65]. Germann et al. constructed a strain of S. cerevisiae containing heterologous genes and increased the supply of cofactors to achieve a product titer of 14.5 mg/L [18,33]. However, E. coli cannot synthesize the cofactor BH4 by itself [47]. Directed metabolic pathway evolution was applied to achieve pterin-dependent hydroxylation of l-tryptophan in E. coli. Luo et at. used a minimum set of heterologous enzymes and a host folE (T198I) mutation for achieving pterin-dependent l-tryptophan hydroxylation by directed metabolic pathway evolution. More important, a heterologously introduced BH4 biosynthetic pathway was deleted during adaptive laboratory evolution, the host strain repurposed one of its native cofactors, but it remained unclear exactly which E. coli pterin was repurposed as an alternative cofactor [66]. Heterologous expression of enzymes and the enzyme mutation folE (T198I) in E. coli can be used to increase the production of GTP and BH4. A mutagenized TPH was expressed in E. coli, and 0.8 mM 5-HTP was synthesized after adding BH4 as a substrate. In addition, the BH4 regeneration pathway and glucose dehydrogenase of Bacillus subtilis were introduced to increase the utilization rate of BH4, and the production of 5-HTP was increased to 2.5 mM [38,47,66].
3.3. Melatonin synthesis
Melatonin produced by a two-step enzymatic conversion of serotonin. Serotonin is first converted by N-acetyltransferase, which replaces its hydrogen at the C3 position with N-acetyl-2-aminoethyl. Subsequently, it is methylated at hydroxy position [67]. In the pathway of melatonin synthesis from serotonin, ASMT is a rate-limiting enzyme. The biosynthetic pathway of melatonin from serotonin is summarized in Fig. 6.
Fig. 6.
Synthesis of melatonin from serotonin.
3.3.1. Arylalkylamine N-acetyltransferase
Arylalkylamine N-acetyltransferase (AANAT/SNAT) is an enzyme of the GNAT (GCN-5 related N-acetyltransferase) superfamily, which is generally considered to be a crucial enzyme for melatonin synthesis in vertebrates [68]. It also converts serotonin to N-acetyl-serotonin during microbial synthesis of melatonin [69].
Increasing the yield of N-acetyl serotonin requires increasing the catalytic activity of the enzyme. Therefore, we need to understand its catalytic mechanism. AANAT has a catalytic funnel combined with reactive groups (amine and AcCoA). The catalytic histidines (His120, His122) at the bottom of the active site form a proton channel, which deprotonates the amino group to initiate the acetyl transfer reaction. The residue corresponding to Ile160 in AANAT of sea bream (Sparus aurata) is naturally mutated to methionine, which enhances acetylation and reduces the Km value [70]. However, someone studies reported that the effect of the mutation at position 160 is difficult to characterize because the enzyme has the same residue at that position. At the same time, mutation at position 114 will also cause differences in catalytic performance. Therefore, it was speculated that the catalytic reaction requires by the synergistic effect of the two mutations [71].
In addition to complex enzyme modification, heterologous expression can also increase substrate conversion. During microbial melatonin synthesis, allogenic expression of enzymes from plants and animals (AANAT/SNAT) can increase the production of N-acetyl serotonin and melatonin. Ectopic expression of SNAT from Arabidopsis, alfalfa, tomato or apple leads to increased melatonin production in transgenic tomatoes and Arabidopsis [72,73]. When the heterologous AANAT from Bos taurus was introduced into Saccharomyces cerevisiae, the highest yield of N-acetyl-serotonin reached 9.1 mg/L in shake-flask cultures [18]. Overexpression of ovine AANAT in switchgrass increased the content of melatonin three times compared with the empty vector control [74]. The Km values of animal and plant enzymes are summarized in Table 1, Table 2
3.3.2. Acetylserotonin O-methyltransferase
N-acetyl-serotonin methyltransferase (ASMT/COMT) belongs to the O-methyltransferase family. The ASMT reaction seems to be the main bottleneck in the biosynthesis of melatonin [75]. The catalytic center of the enzyme exhibits different binding affinity for compounds with similar structures, resulting in variation of catalytic efficiency towards different substrates [76]. It can recognize both N-acetyl-serotonin and serotonin, and catalyzes the methylation reaction in the core pathway of O-melatonin biosynthesis. However, due to the presence of amide groups in the N-acetyl serotonin terminal chain, when the substrate enters the substrate binding pocket of Arabidopsis thaliana COMT, strong electrostatic repulsion will occur, preventing N-acetyl serotonin from binding to the catalytic pocket [77].
In order to improve the catalytic efficiency of ASMT with N-acetyl serotonin, we need to improve the substrate affinity of the enzyme. COMT has only three highly conserved amino acids (Met-230, Asp-233 and Met-386), among which the conserved hydrophobic residues Met-230 and Met-386 are isolated in the benzene ring, and the benzene ring faces the 5th position of the SAM aromatic ring provide the active hydroxyl. Based on notable differences in the terminal structure of caffeic acid and N-acetyl serotonin, mutants were designed to strengthen the interactions between the substrate binding pocket of the enzyme and the terminal structure of the unnatural substrate N-acetyl serotonin [77,78]. Rational design not only eliminated electrostatic repulsion (Gln310Leu), but also introduced an N single bond (Phe296),hydrogen-π interaction and hydrophobic interaction. These changes increased the catalytic efficiency in the conversion of N-acetyl serotonin to melatonin. The construction of COMT variant containing C303F and V321T mutations increased the production of melatonin 5-fold [26]. Molecular dynamics simulations and binding free energy analysis resulted in the construction of a triple mutant (C296F-Q310L-V314T) with a 9.5-fold increase of activity [77].
In addition to modifying enzymes, we can also express enzymes from different sources. ASMT/COMT orthologs from different plants and animals exhibit different activities. Compared with the enzymes from animals, ASMT orthologs from plants exhibit lower Km values (Table 1, Table 2) [77,78].
3.3.3. Coenzymes
In the synthesis process of serotonin to melatonin, the cofactors SAM and acetyl-CoA play a significant role. The conversion of serotonin to N-acetyl-serotonin requires the participation of acetyl-CoA [79]. In Saccharomyces cerevisiae, the supply of acetyl-CoA can be increased by overexpressing acetaldehyde dehydrogenase or by restricting oxygen [80]. However, anaerobic conditions will affect the upstream hydroxylation reactions. Kocharin et al. increased the acetyl-CoA supply by overexpressing acetaldehyde dehydrogenase, which enhanced the productivity of polyhydroxybutyrate approximately 16.5 times in bioreactor cultivations [81]. In addition, SAM is also an important cofactor for the last step of the pathway, in which ASMT methylates N-acetyl-serotonin to the final product melatonin. Thus, the conversion to melatonin is accompanied by the concomitant conversion of SAM to S-adenosyl-l-homocysteine (SAH). The SAM cycle is native and constitutively expressed in budding yeast [82]. Cofactor regulation also plays a vital role in constructing a highly efficient cell factory [39]. A comparison of the specific productivities of polyhydroxybutyrate producing strains revealed that employing the acetyl-CoA boost plasmid helps redirect carbon fluxes towards the polyhydroxybutyrate pathway [81,83].
4. Conclusions
In mammals, melatonin is released from the pineal gland into the third ventricle and from there into circulation. It is involved in regulating the body's sleep-wake cycle through its interactions with the suprachiasmatic nucleus of the hypothalamus and the retina, promoting sleep [133]. In today's stressful society, melatonin is a hot commodity. However, with the increase of annual demand, a green and efficient synthesis method is needed to meet the market demand, and the most promising approach is microbial fermentation. The synthesis pathways of melatonin differ between species. In animals and plants, the first two steps of melatonin synthesis are reversed and the Km of the involved enzymes are significantly different (Table 1, Table 2). Among microorganisms, the synthetic pathways of yeast and Pseudomonas are similar to those in animals. E. coli requires the expression of heterologous enzymes synthesize melatonin, such as TDC, SNAT, AANAT, ASMT and COMT. We summarized recent advances in the modification of microorganisms to produce melatonin. Therefore, recent and representative publications on the subject using different strategies were selected and discussed, providing a good reference for future microbial metabolic engineering projects for the biosynthesis of melatonin.
Microbial metabolic networks are complex, and intermediate metabolites participate in many different metabolic modules. If the supply of the precursor l-tryptophan or serotonin is insufficient, there will note be enough impetus to produce melatonin [39]. Therefore, to improve l-tryptophan production, we can enhance the supply of precursors, such as glutamine, l-serine, shikimic acid and phosphoribose pyrophosphate. Feedback inhibition of key enzymes such as DAHP synthase (aroGFH) and anthranilate synthase (trpED) can be removed [35,134]. For the biosynthesis of melatonin in E. coli, TDC is the rate-limiting enzyme. However, TDC generates the by-product tryptamine, and TPH has a low affinity for tryptamine, which leads to the accumulation of a large amount of tryptamine, which affects cell growth [27]. Hence, to increase the precursor supply of serotonin, we can improve the activity of TPH and TDC through site-directed mutation guided by rational design or through directed evolution based on high-throughput screening [51]. To improve TDC, it is necessary to enhance the specific binding between the enzyme and substrate [58]. To improve the ASMT/COMT and SNAT/AANAT enzymes that synthesize melatonin, it is necessary to understand their structure, analyze their catalytic mechanism, identify the key sites and carry out site-directed mutagenesis.
With the rapid development of computer technology, there are endless ways to modify enzymes based on in silico analysis. Some scholars have combined the relevant theories of proteomics and bioinformatics to use protein design software to improve enzyme activity. For example, DeepMind has been developing the Alpha-Fold system, which system improves the accuracy of protein structure prediction by integrating novel neural network architectures and training programs based on evolutionary, physical, and geometric constraints of protein structure, which could be used as a computer-aided tool for enzyme modification in the future [135]. The development of metabolic engineering has brought greater appreciation of its feasibility for the industrial production of melatonin, and significant progress is expected in the future.
Funding
This work was supported by the National Key R&D Program of China (2021YFC2100900), National Nature Science Foundation of China (32100062), Youth Innovation Promotion Association, CAS (2020182), Tianjin Synthetic Biotechnology Inno-vation Capacity Improvement Project (TSBICIP-CXRC-029).
Declaration of competing interest
We declare that we have no financial and personal relationships with other people or organizations that can inappropriately influence our work, there is no professional or other personal interest of any nature or kind in any product, service and/or company that could be construed as influencing the position presented in, or the review of, the manuscript entitled “Melatonin biosynthesis pathways in nature and its production in engineered microorganisms”.
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Contributor Information
Yumei Sun, Email: sunyumei62@163.com.
Dawei Zhang, Email: zhang_dw@tib.cas.cn.
References
- 1.Sagan L. On the origin of mitosing cells. J Theor Biol. 1967;14:255–274. doi: 10.1016/0022-5193(67)90079-3. [DOI] [PubMed] [Google Scholar]
- 2.Tan D.X., Reiter R.J. An evolutionary view of melatonin synthesis and metabolism related to its biological functions in plants. J Exp Bot. 2020;71:4677–4689. doi: 10.1093/jxb/eraa235. [DOI] [PubMed] [Google Scholar]
- 3.Cross K.M., Landis D.M., Sehgal L., Payne J.D. Melatonin in early treatment for COVID-19: a narrative review of current evidence and possible efficacy. Endocr Pract. 2021 doi: 10.1016/j.eprac.2021.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ma N., Zhang J., Reiter R.J., Ma X. Melatonin mediates mucosal immune cells, microbial metabolism, and rhythm crosstalk: a therapeutic target to reduce intestinal inflammation. Med Res Rev. 2020;40:606–632. doi: 10.1002/med.21628. [DOI] [PubMed] [Google Scholar]
- 5.Zhao D., Wang H., Chen S., Yu D., Reiter R.J. Phytomelatonin: an emerging regulator of plant biotic stress resistance. Trends Plant Sci. 2021;26:70–82. doi: 10.1016/j.tplants.2020.08.009. [DOI] [PubMed] [Google Scholar]
- 6.Mendes C., Gomes G., Belpiede L.T., Buonfiglio D.D., Motta-Teixeira L.C., Amaral F.G., Cipolla J. The effects of melatonin daily supplementation to aged rats on the ability to withstand cold, thermoregulation and body weight. Life Sci. 2021:265. doi: 10.1016/j.lfs.2020.118769. [DOI] [PubMed] [Google Scholar]
- 7.Wu H., Yao S., Wang T., Wang J., Ren K., Yang H., Ma W., Ji P., Lu Y., Ma H., et al. Effects of melatonin on dairy herd improvement (DHI) of holstein cow with high SCS. Molecules. 2021;26 doi: 10.3390/molecules26040834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Esmaely F., Mahmoudzadeh A., Cheki M., Shirazi A. The radioprotective effect of melatonin against radiation-induced DNA double-strand breaks in radiology. J Cancer Res Therapeut. 2020;16:S59–S63. doi: 10.4103/jcrt.JCRT_370_18. [DOI] [PubMed] [Google Scholar]
- 9.Pereira N., Naufel M.F., Ribeiro E.B., Tufik S., Hachul H. Influence of dietary sources of melatonin on sleep quality: a review. J Food Sci. 2020;85:5–13. doi: 10.1111/1750-3841.14952. [DOI] [PubMed] [Google Scholar]
- 10.Rusanova I., Martinez-Ruiz L., Florido J., Rodriguez-Santana C., Guerra-Librero A., Acuna-Castroviejo D., Escames G. Protective effects of melatonin on the skin: future perspectives. Int J Mol Sci. 2019;20 doi: 10.3390/ijms20194948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guo C., He J.Q., Deng X., Wang D., Yuan G.Y. Potential therapeutic value of melatonin in diabetic nephropathy: improvement beyond anti-oxidative stress. Arch Physiol Biochem. 2021 doi: 10.1080/13813455.2021.1933539. [DOI] [PubMed] [Google Scholar]
- 12.Yayici Koken O., Gultutan P., Gungoren M.S., Bayhan G.I., Yilmaz D., Gurkas E., Ozyurek H., Citak Kurt A.N. Impact of COVID-19 on serum melatonin levels and sleep parameters in children. Turk J Med Sci. 2021 doi: 10.3906/sag-2012-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hardeland R., Poeggeler B. Non-vertebrate melatonin. J Pineal Res. 2003;34:233–241. doi: 10.1034/j.1600-079x.2003.00040.x. [DOI] [PubMed] [Google Scholar]
- 14.Morcillo-Parra M.A., Beltran G., Mas A., Torija M.J. Effect of several nutrients and environmental conditions on intracellular melatonin synthesis inSaccharomyces cerevisiae. Microorganisms. 2020;8 doi: 10.3390/microorganisms8060853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Valera M.J., Morcillo-Parra M.A., Zagorska I., Mas A., Beltran G., Torija M.J. Effects of melatonin and tryptophol addition on fermentations carried out by Saccharomyces cerevisiae and non-Saccharomyces yeast species under different nitrogen conditions. Int J Food Microbiol. 2019;289:174–181. doi: 10.1016/j.ijfoodmicro.2018.09.013. [DOI] [PubMed] [Google Scholar]
- 16.Ma Y.E., Jiao J., Fan X.C., Sun H.S., Zhang Y., Jiang J.F., Liu C.H. Endophytic bacterium Pseudomonas fluorescens RG11 may transform tryptophan to melatonin and promote endogenous melatonin levels in the roots of four grape cultivars. Front Plant Sci. 2017;7 doi: 10.3389/fpls.2016.02068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Back K., Tan D.X., Reiter R.J. Melatonin biosynthesis in plants: multiple pathways catalyze tryptophan to melatonin in the cytoplasm or chloroplasts. J Pineal Res. 2016;61:426–437. doi: 10.1111/jpi.12364. [DOI] [PubMed] [Google Scholar]
- 18.Germann S.M., Baallal Jacobsen S.A., Schneider K., Harrison S.J., Jensen N.B., Chen X., Stahlhut S.G., Borodina I., Luo H., Zhu J., et al. Glucose-based microbial production of the hormone melatonin in yeast Saccharomyces cerevisiae. Biotechnol J. 2016;11:717–724. doi: 10.1002/biot.201500143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fernandez-Cruz E., Gonzalez B., Muniz-Calvo S., Morcillo-Parra M.A., Bisquert R., Troncoso A.M., Garcia-Parrilla M.C., Torija M.J., Guillamon J.M. Intracellular biosynthesis of melatonin and other indolic compounds in Saccharomyces and non-Saccharomyces wine yeasts. Eur Food Res Tech. 2019;245:1553–1560. [Google Scholar]
- 20.Morcillo-Parra M.A., Gonzalez B., Beltran G., Mas A., Torija M.J. Melatonin and glycolytic protein interactions are related to yeast fermentative capacity. Food Microbiol. 2020;87 doi: 10.1016/j.fm.2019.103398. [DOI] [PubMed] [Google Scholar]
- 21.Fernandez-Cruz E., Alvarez-Fernandez M.A., Valero E., Troncoso A.M., Garcia-Parrilla M.C. Melatonin and derived l-tryptophan metabolites produced during alcoholic fermentation by different wine yeast strains. Food Chem. 2017;217:431–437. doi: 10.1016/j.foodchem.2016.08.020. [DOI] [PubMed] [Google Scholar]
- 22.Sun T., Chen L., Zhang W. Microbial production of mammalian melatonin - a promising solution to melatonin industry. Biotechnol J. 2016;11:601–602. doi: 10.1002/biot.201500604. [DOI] [PubMed] [Google Scholar]
- 23.Byeon Y., Back K. Melatonin production in Escherichia coli by dual expression of serotonin N-acetyltransferase and caffeic acid O-methyltransferase. Appl Microbiol Biotechnol. 2016;100:6683–6691. doi: 10.1007/s00253-016-7458-z. [DOI] [PubMed] [Google Scholar]
- 24.Ben-Abdallah M., Bondet V., Fauchereau F., Beguin P., Goubran-Botros H., Pagan C., Bourgeron T., Bellalou J. Production of soluble, active acetyl serotonin methyl transferase in Leishmania tarentolae. Protein Expr Purif. 2011;75:114–118. doi: 10.1016/j.pep.2010.07.011. [DOI] [PubMed] [Google Scholar]
- 25.Park S., Byeon Y., Kim Y.S., Back K. Kinetic analysis of purified recombinant rice N-acetylserotonin methyltransferase and peak melatonin production in etiolated rice shoots. J Pineal Res. 2013;54:139–144. doi: 10.1111/j.1600-079X.2012.01019.x. [DOI] [PubMed] [Google Scholar]
- 26.Zhang Y.F., He Y.Z., Zhang N., Gan J.J., Zhang S., Dong Z.Y. Combining protein and metabolic engineering strategies for biosynthesis of melatonin in Escherichia coli. Microb Cell Factories. 2021:20. doi: 10.1186/s12934-021-01662-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luo H., Schneider K., Christensen U., Lei Y., Herrgard M.J., Palsson B.O. Microbial synthesis of human-hormone melatonin at gram scales. ACS Synth Biol. 2020;9:1240–1245. doi: 10.1021/acssynbio.0c00065. [DOI] [PubMed] [Google Scholar]
- 28.Sun C.L., Liu L.J., Wang L.X., Li B.H., Jin C.W., Lin X.Y. Melatonin: a master regulator of plant development and stress responses. J Integr Plant Biol. 2021;63:126–145. doi: 10.1111/jipb.12993. [DOI] [PubMed] [Google Scholar]
- 29.Zhao D., Yu Y., Shen Y., Liu Q., Zhao Z.W., Sharma R., Reiter R.J. Melatonin synthesis and function: evolutionary history in animals and plants. Front Endocrinol. 2019;10 doi: 10.3389/fendo.2019.00249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tan D.X., Manchester L.C., Liu X.Y., Rosales-Corral S.A., Acuna-Castroviejo D., Reiter R.J. Mitochondria and chloroplasts as the original sites of melatonin synthesis: a hypothesis related to melatonin's primary function and evolution in eukaryotes. J Pineal Res. 2013;54:127–138. doi: 10.1111/jpi.12026. [DOI] [PubMed] [Google Scholar]
- 31.Agathokleous E., Kitao M., Calabrese E.J. New insights into the role of melatonin in plants and animals. Chem Biol Interact. 2019;299:163–167. doi: 10.1016/j.cbi.2018.12.008. [DOI] [PubMed] [Google Scholar]
- 32.Ye T.T., Yin X.M., Yu L., Zheng S.J., Cai W.J., Wu Y., Feng Y.Q. Metabolic analysis of the melatonin biosynthesis pathway using chemical labeling coupled with liquid chromatography-mass spectrometry. J Pineal Res. 2019;66 doi: 10.1111/jpi.12531. [DOI] [PubMed] [Google Scholar]
- 33.Germann S.M., Jacobsen S.A.B., Schneider K., Harrison S.J., Jensen N.B., Chen X., Stahlhut S.G., Borodina I., Luo H., Zhu J.F., et al. Glucose-based microbial production of the hormone melatonin in yeast Saccharomyces cerevisiae. Biotechnol J. 2016;11:717–724. doi: 10.1002/biot.201500143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Negri S., Commisso M., Avesani L., Guzzo F. The case of tryptamine and serotonin in plants: a mysterious precursor for an illustrious metabolite. J Exp Bot. 2021;72:5336–5355. doi: 10.1093/jxb/erab220. [DOI] [PubMed] [Google Scholar]
- 35.Chen L., Zeng A.P. Rational design and metabolic analysis of Escherichia coli for effective production of L-tryptophan at high concentration. Appl Microbiol Biotechnol. 2017;101:559–568. doi: 10.1007/s00253-016-7772-5. [DOI] [PubMed] [Google Scholar]
- 36.Lawley B., Pittard A.J. Regulation of arol expression by tyrr protein and Trp repressor in escherichia-coli K-12. J Bacteriol. 1994;176:6921–6930. doi: 10.1128/jb.176.22.6921-6930.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Heatwole V.M., Somerville R.L. The tryptophan-specific permease gene, mtr, is differentially regulated by the tryptophan and tyrosine repressors in escherichia-coli K-12. J Bacteriol. 1991;173:3601–3604. doi: 10.1128/jb.173.11.3601-3604.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hara R., Kino K. Enhanced synthesis of 5-hydroxy-L-tryptophan through tetrahydropterin regeneration. Amb Express. 2013;3 doi: 10.1186/2191-0855-3-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li Z., Ding D., Wang H., Liu L., Fang H., Chen T., Zhang D. Engineering Escherichia coli to improve tryptophan production via genetic manipulation of precursor and cofactor pathways. Synth Syst Biotechnol. 2020;5:200–205. doi: 10.1016/j.synbio.2020.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen L., Zeng A.P. Rational design and metabolic analysis of Escherichia coli for effective production of L-tryptophan at high concentration. Appl Microbiol Biotechnol. 2017;101:559–568. doi: 10.1007/s00253-016-7772-5. [DOI] [PubMed] [Google Scholar]
- 41.Xiong B., Zhu Y., Tian D., Jiang S., Fan X., Ma Q., Wu H., Xie X. Flux redistribution of central carbon metabolism for efficient production of l-tryptophan in Escherichia coli. Biotechnol Bioeng. 2021;118:1393–1404. doi: 10.1002/bit.27665. [DOI] [PubMed] [Google Scholar]
- 42.Swami T., Weber H.C. Updates on the biology of serotonin and tryptophan hydroxylase. Curr Opin Endocrinol Diabetes Obes. 2018;25:12–21. doi: 10.1097/MED.0000000000000383. [DOI] [PubMed] [Google Scholar]
- 43.Mora-Villalobos J.A., Zeng A.P. Synthetic pathways and processes for effective production of 5-hydroxytryptophan and serotonin from glucose in Escherichia coli. J Biol Eng. 2018;12 doi: 10.1186/s13036-018-0094-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Windahl M.S., Petersen C.R., Christensen H.E.M., Harris P. Crystal structure of tryptophan hydroxylase with bound amino acid substrate. Biochemistry. 2008;47:12087–12094. doi: 10.1021/bi8015263. [DOI] [PubMed] [Google Scholar]
- 45.Xu J., Li Y., Lv Y., Bian C., You X., Endoh D., Teraoka H., Shi Q. Molecular evolution of tryptophan hydroxylases in vertebrates: a comparative genomic survey. Genes. 2019;10 doi: 10.3390/genes10030203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bader M. Inhibition of serotonin synthesis: a novel therapeutic paradigm. Pharmacol Ther. 2020;205:107423. doi: 10.1016/j.pharmthera.2019.107423. [DOI] [PubMed] [Google Scholar]
- 47.Liu X.X., Zhang B., Ai L.Z. Advances in the microbial synthesis of 5-hydroxytryptophan. Front Bioeng Biotechnol. 2021;9 doi: 10.3389/fbioe.2021.624503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kino K., Hara R., Nozawa A. Enhancement of L-tryptophan 5-hydroxylation activity by structure-based modification of L-phenylalanine 4-hydroxylase from Chromobacterium violaceum. J Biosci Bioeng. 2009;108:184–189. doi: 10.1016/j.jbiosc.2009.04.002. [DOI] [PubMed] [Google Scholar]
- 49.Maxel S., Aspacio D., King E., Zhang L., Acosta A.P., Li H. A growth-based, high-throughput selection platform enables remodeling of 4-hydroxybenzoate hydroxylase active site. ACS Catal. 2020;10:6969–6974. doi: 10.1021/acscatal.0c01892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Moriwaki Y., Yato M., Terada T., Saito S., Nukui N., Iwasaki T., Nishi T., Kawaguchi Y., Okamoto K., Arakawa T., et al. Understanding the molecular mechanism underlying the high catalytic activity of p-hydroxybenzoate hydroxylase mutants for producing gallic acid. Biochemistry. 2019;58:4543–4558. doi: 10.1021/acs.biochem.9b00443. [DOI] [PubMed] [Google Scholar]
- 51.Hamdan S.H., Maiangwa J., Ali M.S.M., Normi Y.M., Sabri S., Leow T.C. Thermostable lipases and their dynamics of improved enzymatic properties. Appl Microbiol Biotechnol. 2021 doi: 10.1007/s00253-021-11520-7. [DOI] [PubMed] [Google Scholar]
- 52.Winge I., McKinney J.A., Knappskog P.M., Haavik J. Characterization of wild-type and mutant forms of human tryptophan hydroxylase 2. J Neurochem. 2007;100:1648–1657. doi: 10.1111/j.1471-4159.2006.04290.x. [DOI] [PubMed] [Google Scholar]
- 53.You D.W., Feng Y., Wang C., Sun C.T., Wang Y., Zhao D.G., Kai G.Y. Cloning, characterization, and enzymatic identification of a new tryptophan decarboxylase from Ophiorrhiza pumila. Biotechnol Appl Biochem. 2021;68:381–389. doi: 10.1002/bab.1935. [DOI] [PubMed] [Google Scholar]
- 54.Pang X., Wei Y.P., Cheng Y., Pan L.Z., Ye Q.J., Wang R.Q., Ruan M.Y., Zhou G.Z., Yao Z.P., Li Z.M., et al. The tryptophan decarboxylase in Solanum lycopersicum. Molecules. 2018;23 doi: 10.3390/molecules23050998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Byeon Y., Park S., Lee H.Y., Kim Y.S., Back K. Elevated production of melatonin in transgenic rice seeds expressing rice tryptophan decarboxylase. J Pineal Res. 2014;56:275–282. doi: 10.1111/jpi.12120. [DOI] [PubMed] [Google Scholar]
- 56.Lei Q., Wang L., Tan D.X., Zhao Y., Zheng X.D., Chen H., Li Q.T., Zuo B.X., Kong J. Identification of genes for melatonin synthetic enzymes in 'Red Fuji' apple (Malus domestica Borkh. cv. Red) and their expression and melatonin production during fruit development. J Pineal Res. 2013;55:443–451. doi: 10.1111/jpi.12096. [DOI] [PubMed] [Google Scholar]
- 57.Zhao Y., Tan D.X., Lei Q., Chen H., Wang L., Li Q.T., Gao Y.A., Kong J. Melatonin and its potential biological functions in the fruits of sweet cherry. J Pineal Res. 2013;55:79–88. doi: 10.1111/jpi.12044. [DOI] [PubMed] [Google Scholar]
- 58.Zhou Y.Z., Liao L.J., Liu X.K., Liu B., Chen X.X., Guo Y., Huang C.L., Zhao Y.C., Zeng Z.X. Crystal structure of Oryza sativa TDC reveals the substrate specificity for TDC-mediated melatonin biosynthesis. J Adv Res. 2020;24:501–511. doi: 10.1016/j.jare.2020.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Park M., Kang K., Park S., Back K. Conversion of 5-hydroxytryptophan into serotonin by tryptophan decarboxylase in plants, Escherichia coli, and yeast. Biosc Biotech Biochem. 2008;72:2456–2458. doi: 10.1271/bbb.80220. [DOI] [PubMed] [Google Scholar]
- 60.Murch S.J., KrishnaRaj S., Saxena P.K. Tryptophan is a precursor for melatonin and serotonin biosynthesis in in vitro regenerated St. John's wort (Hypericum perforatum L. cv. Anthos) plants. Plant Cell Rep. 2000;19:698–704. doi: 10.1007/s002990000206. [DOI] [PubMed] [Google Scholar]
- 61.Kang S., Kang K., Lee K., Back K. Characterization of rice tryptophan decarboxylases and their direct involvement in serotonin biosynthesis in transgenic rice. Planta. 2007;227:263–272. doi: 10.1007/s00425-007-0614-z. [DOI] [PubMed] [Google Scholar]
- 62.Burkhard P., Dominici P., Borri-Voltattorni C., Jansonius J.N., Malashkevich V.N. Structural insight into Parkinson's disease treatment from drug-inhibited DOPA decarboxylase. Nat Struct Biol. 2001;8:963–967. doi: 10.1038/nsb1101-963. [DOI] [PubMed] [Google Scholar]
- 63.Williams B.B., Van Benschoten A.H., Cimermancic P., Donia M.S., Zimmermann M., Taketani M., Ishihara A., Kashyap P.C., Fraser J.S., Fischbach M.A. Discovery and characterization of gut microbiota decarboxylases that can produce the neurotransmitter tryptamine. Cell Host Microbe. 2014;16:495–503. doi: 10.1016/j.chom.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gao S., Xu X.Y., Zeng W.Z., Xu S., Lyv Y.B., Feng Y., Kai G.Y., Zhou J.W., Chen J. Efficient biosynthesis of (2S)-Eriodictyol from (2S)-Naringenin in Saccharomyces cerevisiae through a combination of promoter adjustment and directed evolution. ACS Synth Biol. 2020;9:3288–3297. doi: 10.1021/acssynbio.0c00346. [DOI] [PubMed] [Google Scholar]
- 65.Yamamoto K., Kataoka E., Miyamoto N., Furukawa K., Ohsuye K., Yabuta M. Genetic engineering of Escherichia coli for production of tetrahydrobiopterin. Metab Eng. 2003;5:246–254. doi: 10.1016/s1096-7176(03)00046-6. [DOI] [PubMed] [Google Scholar]
- 66.Luo H., Yang L., Kim S.H., Wulff T., Feist A.M., Herrgard M., Palsson B.O. Directed metabolic pathway evolution enables functional pterin-dependent aromatic-amino-acid hydroxylation in Escherichia coli. ACS Synth Biol. 2020;9:494–499. doi: 10.1021/acssynbio.9b00488. [DOI] [PubMed] [Google Scholar]
- 67.Wang S.Y., Shi X.C., Laborda P. Indole-based melatonin analogues: synthetic approaches and biological activity. Eur J Med Chem. 2020;185:111847. doi: 10.1016/j.ejmech.2019.111847. [DOI] [PubMed] [Google Scholar]
- 68.Scheibner K.A., De Angelis J., Burley S.K., Cole P.A. Investigation of the roles of catalytic residues in serotonin N-acetyltransferase. J Biol Chem. 2002;277:18118–18126. doi: 10.1074/jbc.M200595200. [DOI] [PubMed] [Google Scholar]
- 69.Lee H.Y., Lee K., Back K. Knockout of Arabidopsis serotonin N-Acetyltransferase-2 reduces melatonin levels and delays flowering. Biomolecules. 2019;9 doi: 10.3390/biom9110712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zilberman-Peled B., Bransburg-Zabary S., Klein D.C., Gothilf Y. Molecular evolution of multiple arylalkylamine N-acetyltransferase (AANAT) in fish. Mar Drugs. 2011;9:906–921. doi: 10.3390/md9050906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cazamea-Catalan D., Magnanou E., Helland R., Besseau L., Boeuf G., Falcon J., Jorgensen E.H. Unique arylalkylamine N-acetyltransferase-2 polymorphism in salmonids and profound variations in thermal stability and catalytic efficiency conferred by two residues. J Exp Biol. 2013;216:1938–1948. doi: 10.1242/jeb.080960. [DOI] [PubMed] [Google Scholar]
- 72.Wang X., Zhang H., Xie Q., Liu Y., Lv H., Bai R., Ma R., Li X., Zhang X., Guo Y.D., Zhang N. SlSNAT interacts with HSP40, a molecular chaperone, to regulate melatonin biosynthesis and promote thermotolerance in tomato. Plant Cell Physiol. 2020;61:909–921. doi: 10.1093/pcp/pcaa018. [DOI] [PubMed] [Google Scholar]
- 73.Hwang O.J., Back K. Simultaneous suppression of two distinct serotonin N-acetyltransferase isogenes by RNA interference leads to severe decreases in melatonin and accelerated seed deterioration in rice. Biomolecules. 2020;10 doi: 10.3390/biom10010141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yuan S., Huang Y.H., Liu S.J., Guan C., Cui X., Tian D.Y., Zhang Y.W., Yang F.Y. RNA-seq analysis of overexpressing ovine AANAT gene of melatonin biosynthesis in switchgrass. Front Plant Sci. 2016;7 doi: 10.3389/fpls.2016.01289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Johnston J.D., Bashforth R., Diack A., Andersson H., Lincoln G.A., Hazlerigg D.G. Rhythmic melatonin secretion does not correlate with the expression of arylalkylamine N-acetyltransferase, inducible cyclic amp early repressor, period1 or cryptochrome1 mRNA in the sheep pineal. Neuroscience. 2004;124:789–795. doi: 10.1016/j.neuroscience.2004.01.011. [DOI] [PubMed] [Google Scholar]
- 76.Moinuddin S.G.A., Jourdes M., Laskar D.D., Ki C., Cardenas C.L., Kim K.W., Zhang D., Davin L.B., Lewis N.G. Insights into lignin primary structure and deconstruction from Arabidopsis thaliana COMT (caffeic acid O-methyl transferase) mutant Atomt1. Org Biomol Chem. 2010;8:3928–3946. doi: 10.1039/c004817h. [DOI] [PubMed] [Google Scholar]
- 77.Wang W.Y., Su S.S., Wang S.Z., Ye L.D., Yu H.W. Significantly improved catalytic efficiency of caffeic acid O-methyltransferase towards N-acetylserotonin by strengthening its interactions with the unnatural substrate's terminal structure. Enzym Microb Technol. 2019;125:1–5. doi: 10.1016/j.enzmictec.2019.02.005. [DOI] [PubMed] [Google Scholar]
- 78.Wei Y., Liu G., Chang Y., Lin D., Reiter R.J., He C., Shi H. Melatonin biosynthesis enzymes recruit WRKY transcription factors to regulate melatonin accumulation and transcriptional activity on W-box in cassava. J Pineal Res. 2018;65 doi: 10.1111/jpi.12487. [DOI] [PubMed] [Google Scholar]
- 79.Voisin P., Namboodiri M.A., Klein D.C. Arylamine N-acetyltransferase and arylalkylamine N-acetyltransferase in the mammalian pineal gland. J Biol Chem. 1984;259:10913–10918. [PubMed] [Google Scholar]
- 80.Wolfe A.J. The acetate switch. Microbiol Mol Biol Rev. 2005;69:12–50. doi: 10.1128/MMBR.69.1.12-50.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kocharin K., Chen Y., Siewers V., Nielsen J. Engineering of acetyl-CoA metabolism for the improved production of polyhydroxybutyrate in Saccharomyces cerevisiae. Amb Express. 2012;2 doi: 10.1186/2191-0855-2-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Xu J., Wang L.L., Dammer E.B., Li C.B., Xu G., Chen S.D., Wang G. Melatonin for sleep disorders and cognition in dementia: a meta-analysis of randomized controlled trials. Am J Alzheimers Dis Other Demen. 2015;30:439–447. doi: 10.1177/1533317514568005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wei Y.X., Liu G.Y., Bai Y.J., Xia F.Y., He C.Z., Shi H.T. Two transcriptional activators of N-acetylserotonin O-methyltransferase 2 and melatonin biosynthesis in cassava. J Exp Bot. 2017;68:4997–5006. doi: 10.1093/jxb/erx305. [DOI] [PubMed] [Google Scholar]
- 84.Noe W., Mollenschott C., Berlin J. Tryptophan decarboxylase from Catharanthus roseus cell suspension cultures: purification, molecular and kinetic data of the homogenous protein. Plant Mol Biol. 1984;3:281–288. doi: 10.1007/BF00017782. [DOI] [PubMed] [Google Scholar]
- 85.Szewczuk L.M., Tarrant M.K., Sample V., Drury W.J., Zhang J., Cole P.A. Analysis of serotonin N-acetyltransferase regulation in vitro and in live cells using protein semisynthesis. Biochemistry. 2008;47:10407–10419. doi: 10.1021/bi801189d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Molchan O., Romashko S., Yurin V. L-tryptophan decarboxylase activity and tryptamine accumulation in callus cultures of Vinca minor L. Plant Cell Tissue Organ Cult. 2012;108:535–539. [Google Scholar]
- 87.Jadaun J.S., Sangwan N.S., Narnoliya L.K., Tripathi S., Sangwan R.S. Withania coagulans tryptophan decarboxylase gene cloning, heterologous expression, and catalytic characteristics of the recombinant enzyme. Protoplasma. 2017;254:181–192. doi: 10.1007/s00709-015-0929-8. [DOI] [PubMed] [Google Scholar]
- 88.Kalb D., Gressler J., Hoffmeister D. Active-site engineering expands the substrate profile of the basidiomycete L-tryptophan decarboxylase CsTDC. Chembiochem. 2016;17:132–136. doi: 10.1002/cbic.201500438. [DOI] [PubMed] [Google Scholar]
- 89.Ferry G., Loynel A., Kucharczyk N., Bertin S., Rodriguez M., Delagrange P., Galizzi J.P., Jacoby E., Volland J.P., Lesieur D., et al. Substrate specificity and inhibition studies of human serotonin N-acetyltransferase. J Biol Chem. 2000;275:8794–8805. doi: 10.1074/jbc.275.12.8794. [DOI] [PubMed] [Google Scholar]
- 90.Ferry G., Ubeaud C., Dauly C., Mozo J., Guillard S., Berger S., Jimenez S., Scoul C., Leclerc G., Yous S., et al. Purification of the recombinant human serotonin N-acetyltransferase (EC 2.3.1.87): further characterization of and comparison with AANAT from other species. Protein Expr Purif. 2004;38:84–98. doi: 10.1016/j.pep.2004.07.004. [DOI] [PubMed] [Google Scholar]
- 91.Buki K.G., Vinh D.Q., Horvath I. Partial-Purification and some properties of tryptophan decarboxylase from a Bacillus strain. Acta Microbiol Hung. 1985;32:65–73. [PubMed] [Google Scholar]
- 92.Deguchi T. Characteristics of serotonin-acetyl coenzyme a N-acetyltransferase in pineal-gland of rat. J Neurochem. 1975;24:1083–1085. doi: 10.1111/j.1471-4159.1975.tb03682.x. [DOI] [PubMed] [Google Scholar]
- 93.Winge I., McKinney J.A., Knappskog P.M., Haavik J. Characterization of wild-type and mutant forms of human tryptophan hydroxylase 2. J Neurochem. 2007;100:1648–1657. doi: 10.1111/j.1471-4159.2006.04290.x. [DOI] [PubMed] [Google Scholar]
- 94.Zilberman-Peled B., Benhar I., Coon S.L., Ron B., Gothilf Y. Duality of serotonin-N-acetyltransferase in the gilthead seabream (Sparus aurata): molecular cloning and characterization of recombinant enzymes. Gen Comp Endocrinol. 2004;138:139–147. doi: 10.1016/j.ygcen.2004.05.007. [DOI] [PubMed] [Google Scholar]
- 95.Tenner K., Walther D., Bader M. Influence of human tryptophan hydroxylase 2 N- and C-terminus on enzymatic activity and oligomerization. J Neurochem. 2007;102:1887–1894. doi: 10.1111/j.1471-4159.2007.04664.x. [DOI] [PubMed] [Google Scholar]
- 96.Dempsey D.R., Jeffries K.A., Bond J.D., Carpenter A.M., Rodriguez-Ospina S., Breydo L., Caswell K.K., Merkler D.J. Mechanistic and structural analysis of Drosophila melanogaster arylalkylamine N-acetyltransferases. Biochemistry. 2014;53:7777–7793. doi: 10.1021/bi5006078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Mehere P., Han Q., Christensen B.M., Li J. Identification and characterization of two arylalkylamine N-acetyltransferases in the yellow fever mosquito, Aedes aegypti. Insect Biochem Mol Biol. 2011;41:707–714. doi: 10.1016/j.ibmb.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Windahl M.S., Boesen J., Karlsen P.E., Christensen H.E. Expression, purification and enzymatic characterization of the catalytic domains of human tryptophan hydroxylase isoforms. Protein J. 2009;28:400–406. doi: 10.1007/s10930-009-9207-y. [DOI] [PubMed] [Google Scholar]
- 99.Byeon Y., Lee H.Y., Back K. Cloning and characterization of the serotonin N-acetyltransferase-2 gene (SNAT2) in rice (Oryza sativa) J Pineal Res. 2016;61:198–207. doi: 10.1111/jpi.12339. [DOI] [PubMed] [Google Scholar]
- 100.Winge I., McKinney J.A., Ying M., D'Santos C.S., Kleppe R., Knappskog P.M., Haavik J. Activation and stabilization of human tryptophan hydroxylase 2 by phosphorylation and 14-3-3 binding. Biochem J. 2008;410:195–204. doi: 10.1042/BJ20071033. [DOI] [PubMed] [Google Scholar]
- 101.Byeon Y., Yool Lee H., Choi D.W., Back K. Chloroplast-encoded serotonin N-acetyltransferase in the red alga Pyropia yezoensis: gene transition to the nucleus from chloroplasts. J Exp Bot. 2015;66:709–717. doi: 10.1093/jxb/eru357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Pan Q., Zhao F.L., Ye B.C. Eis, a novel family of arylalkylamine N-acetyltransferase (EC 2.3.1.87) Sci Rep. 2018;8:2435. doi: 10.1038/s41598-018-20802-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Birks E.K., Ewing R.D. Characterization of hydroxyindole-O-methyltransferase (HIOMT) from the pineal gland of chinook salmon (Oncorhynchus tshawytscha) Gen Comp Endocrinol. 1981;43:269–276. doi: 10.1016/0016-6480(81)90284-7. [DOI] [PubMed] [Google Scholar]
- 104.Tenner K., Walther D., Bader M. Influence of human tryptophan hydroxylase 2 N- and C-terminus on enzymatic activity and oligomerization. J Neurochem. 2007;102:1887–1894. doi: 10.1111/j.1471-4159.2007.04664.x. [DOI] [PubMed] [Google Scholar]
- 105.Tedesco S.C., Morton D.J., Reiter R.J. Hydroxyindole-O-methyltransferase activity in the pineal gland of the muskox (Ovibos moschatus) J Pineal Res. 1994;16:121–126. doi: 10.1111/j.1600-079x.1994.tb00091.x. [DOI] [PubMed] [Google Scholar]
- 106.Kuwano R., Yoshida Y., Takahashi Y. Purification of bovine pineal hydroxyindole O-methyltransferase by immunoadsorption chromatography. J Neurochem. 1978;31:815–824. doi: 10.1111/j.1471-4159.1978.tb00116.x. [DOI] [PubMed] [Google Scholar]
- 107.Moran G.R., Daubner S.C., Fitzpatrick P.F. Expression and characterization of the catalytic core of tryptophan hydroxylase. J Biol Chem. 1998;273:12259–12266. doi: 10.1074/jbc.273.20.12259. [DOI] [PubMed] [Google Scholar]
- 108.Friedman P.A., Kappelman A.H., Kaufman S. Partial purification and characterization of tryptophan hydroxylase from rabbit hindbrain. J Biol Chem. 1972;247:4165–4173. [PubMed] [Google Scholar]
- 109.Itoh M.T., Ishizuka B., Kudo Y., Fusama S., Amemiya A., Sumi Y. Detection of melatonin and serotonin N-acetyltransferase and hydroxyindole-O-methyltransferase activities in rat ovary. Mol Cell Endocrinol. 1997;136:7–13. doi: 10.1016/s0303-7207(97)00206-2. [DOI] [PubMed] [Google Scholar]
- 110.Nir I., Hirschmann N., Sulman F.G. Inhibition of pineal hydroxyindole-O-methyl transferase by pyridoxal-5'-phosphate. Biochem Pharmacol. 1976;25:581–583. doi: 10.1016/0006-2952(76)90391-9. [DOI] [PubMed] [Google Scholar]
- 111.Hasegawa H., Ichiyama A. Tryptophan 5-monooxygenase from mouse mastocytoma: high-performance liquid chromatography assay. Methods Enzymol. 1987;142:88–92. doi: 10.1016/s0076-6879(87)42013-2. [DOI] [PubMed] [Google Scholar]
- 112.Byeon Y., Choi G.H., Lee H.Y., Back K. Melatonin biosynthesis requires N-acetylserotonin methyltransferase activity of caffeic acid O-methyltransferase in rice. J Exp Bot. 2015;66:6917–6925. doi: 10.1093/jxb/erv396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Nielsen M.S., Petersen C.R., Munch A., Vendelboe T.V., Boesen J., Harris P., Christensen H.E. A simple two step procedure for purification of the catalytic domain of chicken tryptophan hydroxylase 1 in a form suitable for crystallization. Protein Expr Purif. 2008;57:116–126. doi: 10.1016/j.pep.2007.10.016. [DOI] [PubMed] [Google Scholar]
- 114.Hamdan F.F., Ribeiro P. Characterization of a stable form of tryptophan hydroxylase from the human parasite Schistosoma mansoni. J Biol Chem. 1999;274:21746–21754. doi: 10.1074/jbc.274.31.21746. [DOI] [PubMed] [Google Scholar]
- 115.Byeon Y., Lee H.J., Lee H.Y., Back K. Cloning and functional characterization of the Arabidopsis N-acetylserotonin O-methyltransferase responsible for melatonin synthesis. J Pineal Res. 2016;60:65–73. doi: 10.1111/jpi.12289. [DOI] [PubMed] [Google Scholar]
- 116.McKinney J., Knappskog P.M., Pereira J., Ekern T., Toska K., Kuitert B.B., Levine D., Gronenborn A.M., Martinez A., Haavik J. Expression and purification of human tryptophan hydroxylase from Escherichia coli and Pichia pastoris. Protein Expr Purif. 2004;33:185–194. doi: 10.1016/j.pep.2003.09.014. [DOI] [PubMed] [Google Scholar]
- 117.Nakamura K., Hasegawa H. Developmental role of tryptophan hydroxylase in the nervous system. Mol Neurobiol. 2007;35:45–54. doi: 10.1007/BF02700623. [DOI] [PubMed] [Google Scholar]
- 118.Wang L., Erlandsen H., Haavik J., Knappskog P.M., Stevens R.C. Three-dimensional structure of human tryptophan hydroxylase and its implications for the biosynthesis of the neurotransmitters serotonin and melatonin. Biochemistry. 2002;41:12569–12574. doi: 10.1021/bi026561f. [DOI] [PubMed] [Google Scholar]
- 119.Naoi M., Maruyama W., Takahashi T., Ota M., Parvez H. Inhibition of tryptophan hydroxylase by dopamine and the precursor amino acids. Biochem Pharmacol. 1994;48:207–211. doi: 10.1016/0006-2952(94)90243-7. [DOI] [PubMed] [Google Scholar]
- 120.McKinney J., Knappskog P.M., Haavik J. Different properties of the central and peripheral forms of human tryptophan hydroxylase. J Neurochem. 2005;92:311–320. doi: 10.1111/j.1471-4159.2004.02850.x. [DOI] [PubMed] [Google Scholar]
- 121.McKinney J., Knappskog P.M., Haavik J. Different properties of the central and peripheral forms of human tryptophan hydroxylase. J Neurochem. 2005;92:311–320. doi: 10.1111/j.1471-4159.2004.02850.x. [DOI] [PubMed] [Google Scholar]
- 122.Ogawa S., Ichinose H. Effect of metals and phenylalanine on the activity of human tryptophan hydroxylase-2: comparison with that on tyrosine hydroxylase activity. Neurosci Lett. 2006;401:261–265. doi: 10.1016/j.neulet.2006.03.031. [DOI] [PubMed] [Google Scholar]
- 123.Windahl M.S., Boesen J., Karlsen P.E., Christensen H.E.M. Expression, purification and enzymatic characterization of the catalytic domains of human tryptophan hydroxylase isoforms. Protein J. 2009;28:400–406. doi: 10.1007/s10930-009-9207-y. [DOI] [PubMed] [Google Scholar]
- 124.McKinney J., Teigen K., Froystein N.A., Salaun C., Knappskog P.M., Haavik J., Martinez A. Conformation of the substrate and pterin cofactor bound to human tryptophan hydroxylase. Important role of Phe313 in substrate specificity. Biochemistry. 2001;40:15591–15601. doi: 10.1021/bi015722x. [DOI] [PubMed] [Google Scholar]
- 125.Ichiyama A., Nakamura S., Nishizuka Y., Hayaishi O. Enzymic studies on the biosynthesis of serotonin in mammalian brain. J Biol Chem. 1970;245:1699–1709. [PubMed] [Google Scholar]
- 126.Kowlessur D., Kaufman S. Cloning and expression of recombinant human pineal tryptophan hydroxylase in Escherichia coli: purification and characterization of the cloned enzyme. Biochim Biophys Acta. 1999;1434:317–330. doi: 10.1016/s0167-4838(99)00184-3. [DOI] [PubMed] [Google Scholar]
- 127.Moran G.R., Phillips R.S., Fitzpatrick P.F. Influence of steric bulk and electrostatics on the hydroxylation regiospecificity of tryptophan hydroxylase: characterization of methyltryptophans and azatryptophans as substrates. Biochemistry. 1999;38:16283–16289. doi: 10.1021/bi991983j. [DOI] [PubMed] [Google Scholar]
- 128.Hasegawa H., Ichiyama A. Distinctive iron requirement of tryptophan 5-monooxygenase: TPH1 requires dissociable ferrous iron. Biochem Biophys Res Commun. 2005;338:277–284. doi: 10.1016/j.bbrc.2005.09.045. [DOI] [PubMed] [Google Scholar]
- 129.Moran G.R., Daubner S.C., Fitzpatrick P.F. Expression and characterization of the catalytic core of tryptophan hydroxylase. J Biol Chem. 1998;273:12259–12266. doi: 10.1074/jbc.273.20.12259. [DOI] [PubMed] [Google Scholar]
- 130.Nukiwa T., Tohyama C., Okita C., Kataoka T., Ichiyama A. Purification and some properties of bovine pineal tryptophan 5-monooxygenase. Biochem Biophys Res Commun. 1974;60:1029–1035. doi: 10.1016/0006-291x(74)90416-1. [DOI] [PubMed] [Google Scholar]
- 131.Nagai T., Hamada M., Kai N., Tanoue Y., Nagayama F. Characterization of yellowfin tuna (Thunnus albacares, Scombroidei) tryptophan hydroxylase. Compar Biochem Physiol B-Biochem Mol Biol. 1997;116:161–165. doi: 10.1016/s0305-0491(96)00184-8. [DOI] [PubMed] [Google Scholar]
- 132.Noe W., Mollenschott C., Berlin J. Tryptophan decarboxylase from catharanthus-roseus cell-suspension cultures - purification, molecular and kinetic data of the homogenous protein. Plant Mol Biol. 1984;3:281–288. doi: 10.1007/BF00017782. [DOI] [PubMed] [Google Scholar]
- 133.Savage R.A., Zafar N., Yohannan S., Miller J.M.M. StatPearls. 2021. Melatonin. Treasure Island (FL) [Google Scholar]
- 134.Minliang C., Chengwei M., Lin C., Zeng A.P. Integrated laboratory evolution and rational engineering of GalP/Glk-dependent Escherichia coli for higher yield and productivity of L-tryptophan biosynthesis. Metab Eng Commun. 2021;12 doi: 10.1016/j.mec.2021.e00167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., Tunyasuvunakool K., Bates R., Zidek A., Potapenko A., et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]