Abstract
The interaction of bacteria with plants can result in either a positive, negative, or neutral association. The rhizobium-legume interaction is a well-studied model system of a process that is considered a positive interaction. This process has evolved to require a complex signal exchange between the host and the symbiont. During this process, rhizobia are subject to several stresses, including low pH, oxidative stress, osmotic stress, as well as growth inhibiting plant peptides. A great deal of work has been carried out to characterize the bacterial response to these stresses. Many of the responses to stress are also observed to have key roles in symbiotic signaling. We propose that stress tolerance responses have been co-opted by the plant and bacterial partners to play a role in the complex signal exchange that occurs between rhizobia and legumes to establish functional symbiosis. This review will cover how rhizobia tolerate stresses, and how aspects of these tolerance mechanisms play a role in signal exchange between rhizobia and legumes.
Keywords: rhizobium, legume, symbiosis, stress, pH, osmolarity, oxygen, ROS
Introduction
Rhizobia-legume symbiosis is a well-studied interaction which results in the formation of a plant derived organelle for the purposes of symbiotic nitrogen fixation. Establishment of this interaction occurs through a complex signal exchange which is initiated by the secretion of plant derived flavonoids that are then recognized by compatible rhizobia species (Oldroyd and Downie, 2008; Oldroyd et al., 2011). Recognition of flavonoids results in the production of a lipo-chito-oligosaccharide termed Nod factor (NF) which can be perceived by the host legume (Barnett and Fisher, 2006). This triggers calcium spiking in the inner plant cortical cells, resulting in the division of cells which will form the nodule primordia (Ehrhardt et al., 1996; Shaw and Long, 2003). Nod factor recognition also leads to root hair curling which can trap attached rhizobia and form a curled colonized root hair (Fournier et al., 2008). Infection thread formation can be observed after signals, such as exopolysaccharides or lipopolysaccharides, are recognized. This structure penetrates down toward nodule primordial cells where rhizobia become endocytosed into the cells and enclosed in a symbiotic membrane (Jones et al., 2007). Rhizobia then become bacteroids, which may or may not be terminally differentiated, that functionally serve as a plant organelle to reduce atmospheric nitrogen into ammonia which is subsequently utilized by the host legume.
While the signaling events that lead to an effective symbiosis are complex, other factors also play a major role in the establishment of an effective symbiosis. During the infection and differentiation process, rhizobia are challenged by numerous stresses, both in the rhizosphere and in planta (Figure 1). To tolerate the stresses that are encountered, bacteria produce compounds or change their lifestyle in order to permit survival. In numerous cases, these changes are correlated with the ability to establish a functional symbiosis. Molecules involved in plant pathogen recognition may also be necessary for symbiotic establishment, and in fact may serve as a signal to the bacteria to produce symbiotic signals. The focus of this work is to review aspects of rhizobia and plant responses to stress, and how elements of these responses may have become co-opted as signals involved in establishing a functional symbiosis.
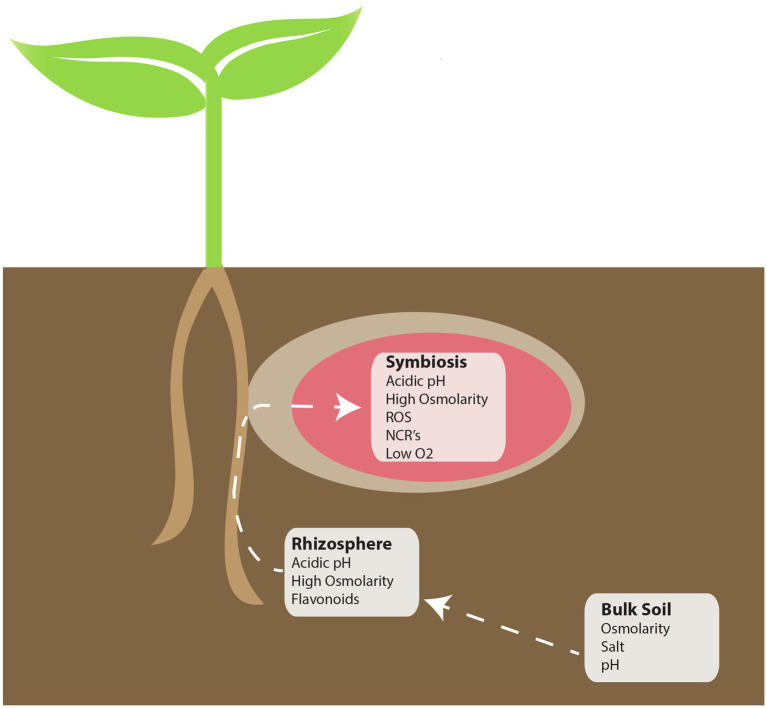
Figure 1.
Locations of perceived stress during the rhizobia-legume symbiosis. During symbiosis, there are three distinct environments that symbiotic bacteria must contend with: Bulk soil, the rhizosphere, and in planta. Each white box indicates potential “perceived” stressors that may be encountered in each of the indicated environments.
Flavonoids
The symbiotic interaction between legumes and rhizobia initiates when flavonoids are recognized by bacteria. The biosynthesis of flavonoids in plants is well understood (Ferrer et al., 2008), and to date, thousands of different flavonoids have been isolated. The biochemical diversity of flavonoids is achieved through modification of a limited number of base structures. These molecules play diverse roles in plant biology ranging from affecting flower color, auxin transport, and anti-microbial defenses (Winkel-Shirley, 2001).
Flavonoids are a known anti-microbial (Hassan and Mathesius, 2012) and represent one of the first directed challenges from plant toward bacteria. The production of these molecules is known to be induced in response to pathogen invasion and has been shown to be directly involved in the plant defense response (Cramer et al., 1985). One subgroup of flavonoids called the iso-flavonoids are found exclusively in legumes (Hirsch et al., 2001). Iso-flavonoids were originally thought to be involved in the defense response against fungi and were shown to also have toxic effects on some isolated bacteria. However, it has also been shown that iso-flavonoids play a role in rhizobium-legume symbiosis. Rhizobia which were exposed to purified iso-flavonoids from plant root exudates were shown to induce the transcription of nodABC, which encode proteins necessary for NF synthesis (Hartwig et al., 1990).
Secretion of the flavone luteolin which is produced by Medicago sativa has been shown to occur in distinct areas of the developing root where symbiotic interactions may occur (Ehrhardt et al., 1992). Flavones and isoflavones were also found to induce transcription of the nod genes in other rhizobia, and recognition of specific flavonoids was shown to play a role in plant-host specificity during symbiosis. Collectively, it seems that the initial role for flavonoids and iso-flavonoids secreted by plants was to be an anti-microbial (Cowan, 1999). However, rhizobia have been able to utilize very specific portions of flavonoids, the iso-flavonoids, as a signal indicating the presence of a compatible host and respond through the production of Nod factor to initiate symbiotic signaling.
Nod Factor
Signaling between host plants and symbiotic rhizobia or mycorrhizae share a common subset of genes and follow a similar pathway (Duc et al., 1989; Oldroyd, 2013). Each organism produces a lipo-chito-oligosaccharide (myc/nod factor) that is recognized by LysM type receptors on the plant (Chabaud et al., 2002; Geddes and Oresnik, 2016). Mycorrhizal symbiosis is thought to be an ancient process and able to occur with most land-based plants, with the oldest symbiotic interaction known to be with the phylum Glomeromycota (Oldroyd, 2013). The secretion of myc factor, which is structurally similar to nod factor, is essential to its symbiotic interaction with its host plants. Comparatively, rhizobial symbiosis is relatively new and only occurs with legumes and Parasponia plants through recognition of NF by LysM type receptors (Pueppke and Broughton, 1999; Madsen et al., 2003). The similarities of the signaling pathway, and insights into Parasponia symbiotic signaling, has led to the hypothesis that the use of Nod factor for symbiosis evolved from myc factor signaling in mycorrhiza (Streng et al., 2011).
The structure of NF is comprised of 3–5 β(1–4)-linked N-acetylglucosamine residues, with a fatty acid tail on the first residue, and can have various modifications to the N-acetylglucosamine residues (Mylona et al., 1995). Nod factor is structurally similar to fungal cell wall chitin, which is a known activator of the plant immune response (Pusztahelyi, 2018). In addition, both Nod factor and chitin are recognized by LysM type receptors which are thought to have evolved from an ancestral LysM receptor (Zipfel and Oldroyd, 2017). The key difference being that Nod factor contains shorter N-acetylglucosamine chain lengths. In the S. meliloti – M. truncatula model NF is recognized by the LysM receptors MtNFP and MtLYK3 (Oldroyd, 2013). This recognition induces numerous responses from M. truncatula which are necessary for successful symbiotic establishment. Transcriptomic studies have also revealed that Nod factor recognition regulates genes involved in the plant immune response (El-Yahyaoui et al., 2004). Studies have also shown that isolated Nod factor from rhizobia can modulate the immune response of both legumes and non-legumes. When Arabidopsis thaliana is exposed to the known pathogen associated molecular pattern flg22, the innate immune response of the plant becomes induced. Interestingly, when purified NF isolated from B. japonicum was applied, in addition to flg22, a more attenuated immune response was observed (Liang et al., 2013). This suggested that Nod factor-mediated suppression of the plants innate immune system may be necessary for successful colonization of plants.
Nod factor is also known to activate the innate defense responses from plants. Transcriptomic studies indicate that the plant immune response is initially activated due to S. meliloti inoculum (Lohar et al., 2006). Purified Nod factor has also been shown to induce the production of ROS in the nodulation zone of M. truncatula roots (Ramu et al., 2002). Further studies revealed that increased production of H2O2 can be observed to occur around root hair tips during Nod factor exposure (Cardenas et al., 2008). Interestingly, S. meliloti mutant strains over-expressing catalase also exhibited slower nodulation and malformed infection threads (Jamet et al., 2007). This suggests that while the suppression of the plant immune system is necessary for growth of rhizobia during symbiotic establishment, the initial immune response may bring about changes to both plant and bacteria that promote symbiosis. It has been suggested that the presence of H2O2 might be necessary for stabilizing the formation of infection threads or promoting a physiological change in rhizobia which would promote symbiosis (Jamet et al., 2007). These observations from early signaling involving the interplay of flavonoids and Nod factor production are a clear example of how a potential stress, flavonoids, induce a bacterial signal, Nod factor, that have become a key component for symbiotic interaction.
Environmental Conditions
The aforementioned topics on symbiotic signaling and regulation of the plant immune response provide a clear example of how tolerance of a stress has become intertwined in signaling. However, it is also important to consider how physiological conditions during symbiotic establishment may also promote symbiosis. Bacteria encounter a wide variety of conditions in both bulk soil and in planta. Bulk soil conditions can vary significantly around the world. In addition, the environment in planta can challenge bacteria with changes in osmolarity, oxygen content and oxidative stress, decreasing pH, and further plant peptide challenges. The following points investigate how these conditions can promote physiological changes necessary for stress tolerance that end up influencing symbiotic establishment or nitrogen fixation (Table 1).
Table 1.
Bacterial and plant changes due to perceived stress and their role in symbiosis.
| Stress | Response | Symbiotic relevance |
|---|---|---|
| Bacteria/Pathogen | Flavonoid | Nod factor induction |
| NCRs | Bacteroid differentiation | |
| Innate immune response | Oxidative burst (see ROS) | |
| Flavonoid | Nod factor | Calcium spiking |
| Salt/Ion stress | Nod factor | Calcium spiking |
| EPS-I | IT development | |
| Osmotic | cyclic β(1–2) glucans | Attachment/ IT formation |
| Intracellular Potassium concentration | Nitrogenase induction | |
| Acidic pH | actR/S | fixK/nifA |
| exoR/S/I | EPS-I | |
| Nod factor profile | Legume host range | |
| Reactive oxygen species | EPS-I | IT development |
| Membrane crosslinking | IT development | |
| Low oxygen | Intracellular potassium | Nitrogenase induction |
| Fix genes | Nitrogenase induction | |
| LPS modification | Legume host range |
Osmotic Stress
Genes involved in adaptation to varying osmotic conditions have been shown to be critical for the establishment of a functional symbiotic relationship. Osmotic conditions before symbiosis are fully dependent on salts and exudates present in the soil. Bulk soil is generally assumed to be an area of low osmolarity (Miller and Wood, 1996). However, the area of the rhizosphere is predicted to have a higher osmolarity due to plant root exudate and water uptake from both plants and bacteria (Jungk, 2002; Miller-Williams et al., 2006). While the osmotic conditions throughout symbiosis in the rhizobia-legume interaction are unknown, current research is consistent with the hypothesis that both high and low osmolarity conditions exist throughout symbiosis (Botsford and Lewis, 1990; Dylan et al., 1990a). The area of the rhizosphere having high osmolarity is particularly interesting as these conditions have been linked with inducing genes necessary for symbiosis. The presence of high osmotic conditions has been shown to induce transcription of genes involved in nodulation and nitrogen fixation (nod, nif, and fix genes) through NodD2 in Rhizobium tropici CIAT 899 (Del Cerro et al., 2019). This same pattern of regulation of these genes through NodD2 is also observed when R. tropici is exposed to increased salt stress present in the rhizosphere (Pérez-Montaño et al., 2016). In addition, increased salt concentrations are known to regulate exopolysaccharide production in S. meiloti (Miller-Williams et al., 2006). These observations provide the most direct link between osmotic stress recognition and promoting symbiosis.
The major link between osmotic stress tolerance and physiological changes with importance to symbiosis is the accumulation of periplasmic glucans in the presence of hypotonic stress. The majority of the organisms in the family Rhizobiaceae produces a cyclic β(1–2)-linked glucan (York et al., 1980; Lelpi et al., 1990; Breedveld and Miller, 1994). Accumulation of these periplasmic glucans can be observed when grown under hypotonic conditions (Miller et al., 1986; Dylan et al., 1990a; Breedveld and Miller, 1995). Further study of cyclic β(1–2) glucans in S. meliloti determined that the inability to produce this polysaccharide, by mutating the gene ndvB, resulted in sensitivity to hypotonic conditions, and abolished nodule formation on M. truncatula (Dylan et al., 1990b). It was hypothesized that extracellular cyclic β(1–2) may be involved in root attachment, but addition of purified cyclic β(1–2) was unable to restore symbiosis with M. truncatula to a ndvB mutant strain (Dylan et al., 1990b). Pseudorevertants of the ndvB mutant strain that still did not produce the cyclic glucan have been isolated and were found to be able to establish a functional symbiosis with M. truncatula. However, these suppressors were still heavily impacted in infection thread formation and were sensitive to hypoosmotic stress (Dylan et al., 1990b). This suggested that while cyclic β(1–2) glucan production is important for osmotic stress tolerance and can be linked to infection thread formation, their role in symbiosis extends past stress tolerance. Suppression of the symbiotic phenotype of ndvB mutants was later linked to the production the symbiotically important polysaccharide succinoglycan (Nagpal et al., 1992).This lead to the suggestion that production of succinoglycan might provide just enough osmoprotectant in the form of low molecular weight succinoglycan to allow for survival in the absence of the cyclic glucans. In addition, succinoglycan may provide or mask a signal necessary for symbiosis in the absence of cyclic glucans (Abe et al., 1982; Nagpal et al., 1992). Overall, osmolarity is involved in regulating cyclic β(1–2) glucans which have a role in symbiosis that extends past stress tolerance.
Another mechanism rhizobia and other bacteria utilize to tolerate high osmolarity is the accumulation of ions, such as potassium (Yancey et al., 1982; Csonka, 1989; Botsford and Lewis, 1990; Smith et al., 1994; Miller and Wood, 1996). Interestingly, increased potassium levels lead to an increase in nitrogenase activity in Bradyrhizobium sp. 32H1 when grown under low oxygen conditions (Gober and Kashket, 1987). As the bacteroid is predicted to be an area of elevated osmotic stress (Miller and Wood, 1996), this provides a link showing that osmotic stress tolerance may be a signal for the regulation of nitrogenase in the bacteroid through the regulation of potassium concentration.
Low Oxygen Content
During symbiotic establishment, rhizobia encounter areas of low oxygen concentration in the nodule. Control of oxygen concentration is important for symbiosis since oxygen inhibits the activity of nitrogenase (Hunt and Layzell, 1993). Oxygen levels are controlled through a diffusion barrier to create optimal oxygen concentrations for nitrogen fixation (Hunt et al., 1987). Tight regulation of oxygen concentration in bacteroids also leads to a number of signaling and physiological changes in bacteria, which promote symbiosis and nitrogen fixation. It has been well documented that a low oxygen concentration activates the two-component system FixJL, which in turn increases the transcription of the majority of genes involved in nitrogen fixation (David et al., 1988; Virts et al., 1988). Recent work has shown that there are 3 proteins that act oxygen sensors in Rhizobium leguminosarum; hFixL, FnrN, and NifA (Dixon and Kahn, 2004; Zamorano-Sánchez and Girard, 2015; Reyes-González et al., 2016). These proteins are tightly temporally controlled, with hFixL inducing expression of FnrN in zones I and II (meristem zone and invasion zone, respectively) of indeterminate nodules. FnrN then induces expression of fixNOQP in zone III (nitrogen fixing zone) when oxygen concentration is near anaerobic (Rutten et al., 2021). The induction the genes necessary for production of nitrogenase in near anaerobic conditions is necessary for function of the protein and is also a clear example of how microaerobic stress acts as a signal for symbiosis.
Oxygen concentration has also been shown to regulate lipopolysaccharide (LPS) synthesis and decoration (Kannenberg and Brewin, 1989; Tang and Hollingsworth, 1998). This is thought to have a role in adaptation to the low oxygen environment. Production and modification of LPS are strain specific and are involved in determining host range for symbiosis in some rhizobia (Via et al., 2016). The ability to produce, or properly modify, LPS has been linked to defects in symbiotic establishment (Keating et al., 2002). As LPS content and decoration are dynamic based upon its environment, it is expected that LPS modification would change during symbiosis. Recent work has also shown that flavonoids can induce changes in decoration of LPS and that these changes are necessary for symbiosis (Broughton et al., 2006). It is possible that low oxygen concentration might contribute to bringing about a change in LPS production and decoration which is necessary for both symbiosis and survival in these conditions.
Reactive Oxygen Species
In addition to low oxygen concentration, rhizobia encounter reactive oxygen species as part of the innate immune response of the plant, and it can be found throughout symbiotic compartments ranging from the IT to mature nodules (Santos et al., 2001). Formation of ROS from the plant immune response has been shown to be beneficial for symbiotic establishment (Puppo et al., 2013). ROS are generated upon Nod factor recognition and are thought to predominantly occur from the activity of NADPH oxidase (Lohar et al., 2007). Rhizobia utilize a number of mechanisms to deal with potential damage from ROS (Boscari et al., 2013). The importance of ROS scavenging during symbiosis is highlighted by the finding that strains which carry mutations in the genes katB/C, which encode for catalases, are impaired in forming bacteroids (Jamet et al., 2003). However, a positive role for ROS in symbiosis has also been observed. When catalase is over-expressed in S. meliloti, aberrant IT formation and delayed nodule development are observed (Jamet et al., 2007). While it is unknown exactly how ROS may contribute to symbiosis, two main suggestions have been made; either ROS plays a role in IT development, or ROS induces physiological changes in rhizobium that are necessary for symbiosis (Pauly et al., 2006). Recent work has investigated this further and has shown that ROS produced by PvRbohB in Phaseolus vulgaris is important for symbiosis. Cultivars of P. vulgaris silenced in expression of PvRbohB displayed abortive infection threads when inoculated with R. tropici (Fonseca-García et al., 2021). RNAseq data also revealed changes in carbon metabolism and cell cycle control; both of which can be linked with symbiosis (Geddes and Oresnik, 2016; Fonseca-García et al., 2021).
Consistent with the hypothesis that ROS may act as a signal to bacteria for symbiotic establishment, it has been shown B. japoncium exposed to oxidative stress produces an increased amount of exopolysaccharides (Donati et al., 2011). The production of exopolysaccharides (EPS) has long been suggested to be involved in the tolerance of various stresses encountered by bacteria. In S. meliloti and Pseudomonas syringae, mutants unable to produce EPS have been observed to be sensitive to ROS (Király et al., 1997; Lehman and Long, 2013). Furthermore, it was shown that low molecular weight succinoglycan (EPS-I) is the responsible fraction which scavenges H2O2 from media in S. meliloti (Lehman and Long, 2013). Taken together, oxidative stress is seen to promote the production of exopolysaccharides which are necessary for the tolerance of ROS and critical for symbiotic establishment. Since plants are observed to produce H2O2 in response to symbiotic establishment, this provides a potential example of how in planta conditions promote production of a symbiotic signal.
Ph Stress
The ability to tolerate acidic pH conditions has largely been studied from the perspective of tolerating acidic soils in the environment. The area of the rhizosphere is predicted to be an area of increased acidic stress, as throughout their life cycle, plants can excrete acidic compounds into the surrounding soil, decreasing the pH of the soil by as much as 2 pH units (Faget et al., 2013). This occurs from the secretion of protons to maintain the net charge across the root membrane and from the secretion of organic compounds (Jones et al., 2003). During the symbiotic interaction between rhizobia and legumes, it has been hypothesized that many plant derived compartments have an acidic pH. The bacteroid and peri-bacteroid space have been predicted to be an acidic compartment, reaching a pH of 4.5 (Fedorova et al., 1999; Pierre et al., 2013). Studies have also determined that the curled colonized root hair is an area of localized acidic pH stress (Geddes et al., 2014). These findings are particularly important as S. meliloti is known to have poor survival when medium pH decreases below six (Hellweg et al., 2009; Hawkins et al., 2017).
Transcriptomic studies addressing the response of rhizobium to acidic pH and have revealed large networks regulating multiple genes in response to acidic pH (Hellweg et al., 2009; Guerrero-Castro et al., 2018). The response of rhizobia to acidic pH is primarily regulated though two-component systems, actR/S and chvI/exoS/exoR (Dilworth et al., 2000; Fenner et al., 2004). These systems ultimately control the regulation of cytoplasmic pH, or the production of and modification of extracellular elements for pH tolerance components (Cunningham and Munns, 1984; Chen et al., 1993). Regulation of potassium efflux proteins is important for pH tolerance. The potassium efflux system in R. tropici has been shown to be regulated by glutathione, since mutants in glutathione synthesis were unable to accumulate intracellular potassium (Riccillo et al., 2000). Potassium concentrations have been shown to regulate nitrogenase activity so this accumulation of K+ in acidic conditions may act as a symbiotic signal (Gober and Kashket, 1987). In addition, glutathione is involved in tolerating many environmental stressors, including pH and ROS stress, and has been shown to be produced in increased amounts under acidic conditions (Riccillo et al., 2000; Muglia et al., 2007). Mutations in the synthesis pathway for glutathione are known to result in either a fix− or delayed nodulation phenotype (Harrison et al., 2005).
One physiological response of S. meliloti to low pH is the production of the symbiotically important exopolysaccharide EPS-I (Hawkins et al., 2017). Acidic pH is known to be present throughout the symbiotic process, being present in the rhizosphere all the way to bacteroids. Mutants which are unable to produce succinoglycan are unable to establish functional symbiosis with alfalfa. Further investigation has revealed that the succinylation of EPS-I is the critical component of the symbiotic interaction (Mendis et al., 2016). Production of EPS-I is also important for tolerance of low pH and contributes to survival in nodules (Hawkins et al., 2017; Maillet et al., 2020). However, symbiotic defects observed in exo mutant strains are likely due to a combination of a loss of pH stress tolerance and loss of proper symbiotic signaling. S. meliloti strains that lack exoK produce a succinylated high molecular weight EPS-I still exhibit high sensitivity to acidic pH, but only display minor symbiotic defects (Maillet et al., 2020). Collectively, these data suggest that EPS-I plays a role in both stress tolerance as well as symbiotic signaling.
It has also been observed that acid tolerant strains of rhizobia produce more exopolysaccharides than acid sensitive strains under non-stress conditions (Cunningham and Munns, 1984). Interestingly, mutations that resulted in an increased production of exopolysaccharide in R. leguminosarum and S. meliloti did not result in an increased tolerance to acidic media (Howieson et al., 1988; Reeve et al., 1997). These observations suggest that in terms of stress tolerance, the production of exopolysaccharides may serve an on/off function rather than a gradient of tolerance, and that the increased production of exopolysaccharides due to pH stress may have another role.
The response to low pH is largely mediated through the ExoR/ExoS/ChvI (RSI) system, which has been shown to be upregulated due to acidic pH in S. meliloti (Hellweg et al., 2009; Draghi et al., 2016). The RSI system is well studied for its ability to regulate the production of EPS-I and flagella (Cheng and Walker, 1998b; Heavner et al., 2015). It is long known that the production of EPS-I is important for symbiotic interaction (González et al., 1996; York and Walker, 1997; Cheng and Walker, 1998a). The protein ExoS acts as a sensor kinase which directly phosphorylates the response regulator ChvI in response to a signal (Cheng and Walker, 1998b; Yao et al., 2004). This system is regulated through direct binding of the repressor ExoR to ExoS in the periplasm (Chen et al., 2008). Homologs of this system in Agrobacterium tumefaciens have been shown to be involved in gene regulation due to acidic pH, and it has been suggested that acidity is a key signal in establishing virulence with plants (Li et al., 2002). Further study of the RSI regulon in A. tumefaciens has revealed that at acidic pH the repressor ExoR is degraded, resulting in increased EPS-I synthesis (Heckel et al., 2014). A mechanism for degradation of ExoR in S. meliloti has also been shown (Lu et al., 2012). Degradation of ExoR could account for the increase in transcription of exoR at lower pH. Taken together, this suggests that the acidic conditions found in the curled colonized root hair leads to the production of EPS-I which is necessary for symbiotic signaling and stress tolerance, making pH a key environmental regulator for symbiosis. Overall, these works suggest that low pH induces the production of glutathione and succinoglycan which are both involved in stress tolerance and symbiosis.
Nodule-Specific Cysteine Rich Peptides
Recently, there has been interest in a subsect of plant produced anti-microbial peptides (AMPs) called nodule-specific cysteine rich (NCR) peptides for their role in symbiotic establishment (Alunni and Gourion, 2016). AMPs are well studied for their anti-microbial activity (Maroti et al., 2011). The mechanism of action of AMPs involves the disruption of bacterial membranes through interaction with the cell surface and ribosome inactivation. In addition to their anti-microbial activity, it has been suggested that certain AMPs play a role in signaling (Schopfer, 1999).
NCRs are structurally and functionally similar to AMPs; they are predicted to be around 100 amino acids long, contain the conserved cysteine residues for disulfide bridge formation, and are predicted to be largely cationic (Mergaert et al., 2003). These peptides have also been shown to have anti-microbial activity against several organisms, including rhizobia (Haag et al., 2011). However, the presence of the protein BacA in S. meliloti, a transporter for AMPs, is observed to be involved in tolerating the challenge with NCRs, whereas mutants in bacA were observed to be hypersensitive to the anti-microbial activity in planta (Haag et al., 2011).
In M. truncatula, there are predicted to be upwards of 300 different NCRs produced by around 600 different genes (Mergaert et al., 2003; Zhou et al., 2013). Only legumes of the inverted-repeat lacking clade (IRLC) are observed to produce NCRs (Mergaert et al., 2006). In these legumes, symbiotic bacteria become terminally differentiated into bacteroids in plant nodules and cannot revert to normal functioning bacteria. Non-IRLC legumes, such as L. japonicus, do not produce NCRs, and symbiotic bacteria do not become terminally differentiated (Mergaert et al., 2003). This has led to the suggestion that NCRs are directly involved in the terminal differentiation of symbiotic bacteria. However, it is worth noting that examples of bacteroid differentiation outside of the IRLC legumes are starting to be found. Nodules formed in the Aeschynomene – Bradyrhizobium symbiotic relationship are found to house differentiated bacteroids with a polyploid genome (Czernic et al., 2015). While Aeschynomene sp. do not produce NCRs they have been shown to produce NCR-like peptides that likely play a role in differentiation of bacteroids. Silencing the homolog of dnf1 in Aeschynomene evenia, which is necessary for cleavage of NCRs for transport to the symbiosome and is essential for symbiosis, results in deformed bacteroids (Czernic et al., 2015). In addition, the protein BclA was identified in Bradyrhizobium sp. as having weak homology to BacA. BclA was shown to be necessary for formation of bacteroids and was observed to be able to transport the peptide NCR247 from M. truncatula (Guefrachi et al., 2015). Taken together, there is good indirect evidence that these NCR-like peptides are used for bacteroid differentiation.
The localization of NCRs suggests much about their role in symbiosis. When NCRs are expressed in the nodule, they are targeted to the symbiotic membrane by the plant secretory system and can also be found within the cytoplasm of bacteroids (Van de Velde et al., 2010). In the same study, it was also shown that a mutation in M. truncatula dnf-1 prevents targeting of NCRs to the bacteroid and prevented bacteroids from terminally differentiating. Also, when NCR035 from M. truncatula was expressed in L. japonicum, which is deficient in NCR production, it localized to the symbiosome of bacteroids resulting in the production of a single elongated bacteroid indicative of terminal differentiation (Alunni et al., 2007; Van de Velde et al., 2010). This highlighted the importance of NCRs for symbiotic establishment in the IRLC legumes. More recent studies on NCRs have shown that a mutation in the gene dnf7, which encodes for a protein involved in the production of NCR169, is unable to perform BNF in M. truncatula (Horváth et al., 2015). Nodules in this mutant were impaired in elongation and triggered early senescence. This was fully complemented by overexpression of NCR169. These studies show the necessity of NCRs in regulating bacteroid differentiation and symbiotic nitrogen fixation.
Microarray analysis has also revealed that NCR recognition may play a role in the bacterial stress response, as well as preventing cell division during symbiosis (Penterman et al., 2014). After exposure of S. meliloti to NCR247, the expression of genes involved in bacterial stress response and cellular division was found to be altered in transcription. This includes increased transcription of rpoH1, which is involved in regulating genes for acid and heat tolerance, and the two-component systems exoS-chvI and feuP-feuQ, which are responsible for regulating EPS and cyclic β(1–2) glucan production (Reuber et al., 1990; Griffitts et al., 2008). In line with this, NCR247 has been shown to induce transcription of the exo genes for EPS-I production, and high molecular weight EPS-I has been shown to aid survival when exposed to NCR247 (Arnold et al., 2017, 2018). Decreased transcription of cell cycle regulators ctrA and gcrA was also observed (Penterman et al., 2014). These observations led to the conclusion that NCR recognition may be a bacterial signal that allows for adaptation to in planta conditions and increase the production of polysaccharides necessary for symbiosis in addition to its role in bacteroid differentiation. This shows that NCRs may have evolved in plants from simply being an AMP produced as a response to bacterial invasion, to also being involved in symbiotic establishment as a signal which induces physiological and morphological changes in the bacteria necessary for nitrogen fixation.
Discussion
The establishment of the rhizobium – legume symbiotic interaction is often described as a direct complex signal exchange between both the bacteria and the plant, with emphasis placed on how a molecule from one induces changes in the other or invokes a signaling response. However, little emphasis has been placed on how environmental conditions and stress tolerance play into the interaction. Here, we provide evidence that the tolerance of environmental conditions and challenges by the plant immune system result in alterations of bacterial physiology which promotes establishment of symbiosis between plant and bacteria. This broadens our assumptions of the signaling cross-talk between legume and rhizobia which is largely considered from the perspective of secreted signal and direct response. In addition, we should consider overall physiological changes in bacteria due to conditions in the soil from root exudate or in planta as part of the signal exchange symbiosis in addition to the role in surviving the stress conditions. If molecules produced as part of the stress response by bacteria and plant are examined it can be seen that stress plays an important role from the start of symbiosis, all the way to nitrogen fixation (Figure 2).
Figure 2.
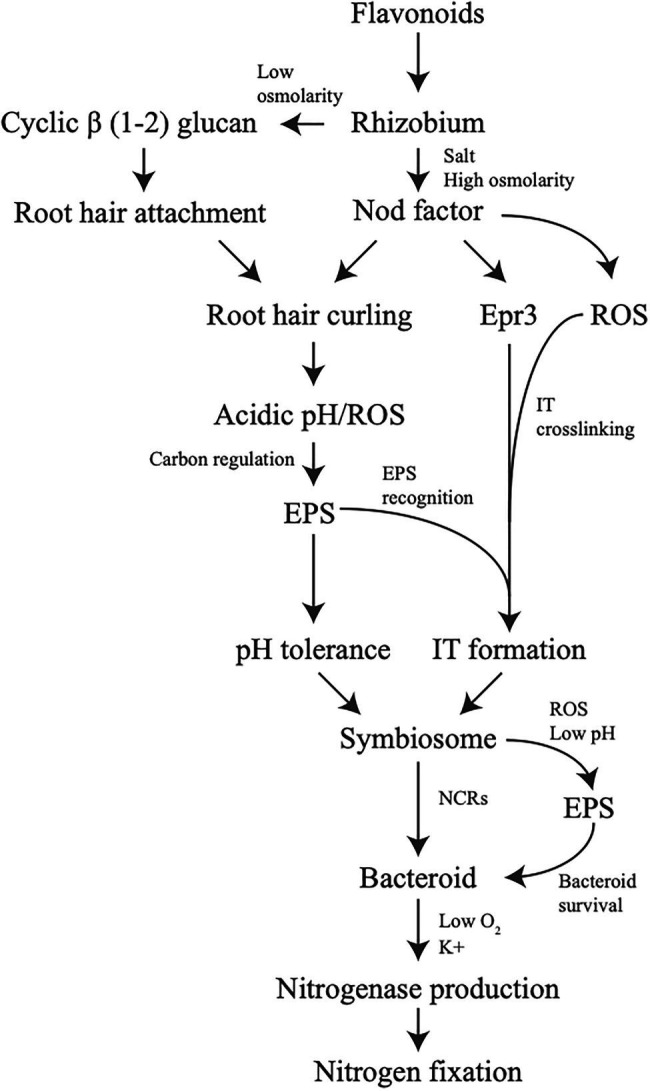
Stress tolerance involvement in symbiotic signaling. Changes in production of molecules or overall physiology due to stress can be observed to affect symbiosis all throughout the process. Each line indicates how a potential stress, or a response from a stress, influences the next step in the symbiotic process.
Some of the examples used are already well studied for their role specifically in symbiosis. This includes iso-flavonoids, Nod factor, and NCRs. While these molecules now have roles directly in symbiotic signaling, their overall origin in this process comes from the immune response of the plant. What likely originated as a stress challenge of anti-microbials for bacteria and fungi with flavonoids and NCRs has turned into critical signals to initiate the symbiotic process or for forming terminally differentiated bacteroids. Cell wall chitin from fungi which is recognized as a PAMP has become inherited by rhizobia in the form of Nod factor, which is now the critical signaling molecule secreted by bacteria to establish symbiosis. The similarities of the responses between either plant immunity or symbiosis are significant and stretch much further past what is discussed here (Berrabah et al., 2015; Tóth and Stacey, 2015; Zipfel and Oldroyd, 2017). It is quite likely that as more detailed mechanisms of each of these responses are uncovered, it will be seen that there is significant cross-talk or similarities linking stress responses and symbiosis. In a number of cases, the difference between either killing the bacteria or establishing a functional symbiosis seems to be based upon the strength of the plant immune response to the organism. A strong response to repel an invader, or an attenuated one to induce physiological changes in a potential symbiont.
Other examples of how potential conditions bacteria may be exposed to in the soil or in planta are less directly tied to symbiosis, but the link is still quite clear. A large part of the stress response of symbiotic rhizobia revolves around production or modification of various polysaccharides, such as cyclic β(1–2) glucans, lipopolysaccharides, and succinoglycan. These polysaccharides are also intrinsically linked to symbiotic establishment across a number of different rhizobia-legume interactions. While the role of cyclic β(1–2) glucans in symbiosis is yet unclear, production and proper decoration of LPS and succinoglycan are suggested to be critical signaling molecules to avoid the full activation of the plant immune response (Ojeda et al., 2013; Kawaharada et al., 2015; Maillet et al., 2020). Additionally, there are around 17 different hypothetical operons for polysaccharide production in S. meliloti so it is plausible to think other polysaccharides that are yet unclassified may play an important role in the stress tolerance/symbiosis picture as well.
Aside from polysaccharide production, these adverse conditions encountered also changes cell physiology in terms of ion uptake, glutathione production, and shifts in carbon metabolism which can all be linked in some regards to the symbiotic process. It is not hard to imagine that symbiotic bacteria may have evolved its responses over time to stress conditions present in root exudate or in planta to start adjusting its physiology for a symbiotic lifecycle. In addition, it is understandable why a plant would evolve to promote certain conditions using root exudate and use an altered immune response if the eventual gain becomes a symbiotic nitrogen fixing bacteria.
One of the major overall goals of nitrogen fixation research is to eventually bring the symbiotic relationship between legumes and rhizobia to non-legume plants, such as the cereal crops. The potential impact this could have in reducing use of nitrogen fertilizers, and for overall growth of plants where fertilizers are not available, is quite significant. While research in this area is new and ongoing, it largely focuses on adjusting and tuning directly observed signaling between rhizobia and these plants. It is important to remember that aside from signaling and adjusting the plant’s immune response to go from immunogenic to symbiotic, the overall environment in the rhizosphere and in planta may also play a key role for symbiosis and have to be accounted for. At the end of the day, it is always said that stress, unfortunately, is a great motivator in life. This also seems to be true with respect to the rhizobium-legume symbiosis.
Author Contributions
JPH and IJO conceived and wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by a Natural Sciences and Engineering Research Council of Canada Discovery grant (RGPIN-2018-04966) to IJO.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
- Abe M., Amemura A., Higashi S. (1982). Studies on cyclic β-1,2-glucan obtained from periplasmic space of rhizobium trifolii cells. Plant Soil 64, 315–324. doi: 10.1007/BF02372514 [DOI] [Google Scholar]
- Alunni B., Gourion B. (2016). Terminal bacteroid differentiation in the legume - rhizobium symbiosis: nodule-specific cysteine-rich peptides and beyond. New Phytol. 211, 411–417. doi: 10.1111/nph.14025, PMID: [DOI] [PubMed] [Google Scholar]
- Alunni B., Kevei Z., Redondo-Nieto M., Kondorosi A., Mergaert P., Kondorosi E. (2007). Genomic organization and evolutionary insights on GRP and NCR genes, two large nodule-specific gene families in Medicago truncatula. Mol. Plant Micro.Interact. 20, 1138–1148. doi: 10.1094/MPMI-20-9-1138, PMID: [DOI] [PubMed] [Google Scholar]
- Arnold M. F. F., Penterman J., Shabab M., Chen E. J., Walker C. (2018). Important late-stage symbiotic role of the Sinorhizobium meliloti exopolysaccharide Succinoglycan. J. Bacteriol. 200:e00665-17. doi: 10.1128/JB.00665-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold M. F. F., Shabab M., Penterman J., Boehme K. L., Griffitts J. S., Walker C. (2017). Genome-wide sensitivity analysis of the Microsymbiont Sinorhizobium meliloti to symbiotically important, Defensin-Like host peptides. MBio 8:e01060-17. doi: 10.1128/mBio.01060-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnett M. J., Fisher R. F. (2006). Global gene expression in the rhizobial-legume symbiosis. Symbiosis 42, 1–24. doi: 10.1111/j.1574-6968.1999.tb13650.x [DOI] [Google Scholar]
- Berrabah F., Ratet P., Gourion B. (2015). Multiple steps control immunity during the intracellular accommodation of rhizobia. J. Exp. Bot. 66, 1977–1985. doi: 10.1093/jxb/eru545, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boscari A., Meilhoc E., Castella C., Bruand C., Puppo A., Brouquisse R. (2013). Which role for nitric oxide in symbiotic N2-fixing nodules: toxic by-product or useful signaling/metabolic intermediate? Front. Plant Sci. 4:384. doi: 10.3389/fpls.2013.00384, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botsford J. L., Lewis T. A. (1990). Osmoregulation in rhizobium meliloti: production of glutamic acid in response to osmotic stress. Appl. Environ. Microbiol. 56, 488–494. doi: 10.1128/aem.56.2.488-494.1990, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breedveld M. W., Miller K. J. (1994). Cyclic β-glucans of members of the family Rhizobiaceae. Microbiol. Rev. 58, 145–161. doi: 10.1128/mr.58.2.145-161.1994, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breedveld M. W., Miller K. J. (1995). Synthesis of glycerophosphorylated cyclic (1,2)-β-glucans in rhizobium meliloti strain 1021 after osmotic shock. Microbiology 141, 583–588. doi: 10.1099/13500872-141-3-583, PMID: [DOI] [PubMed] [Google Scholar]
- Broughton W. J., Hanin M., Relic B., Kopcinska J., Golinowski W., Simsek S., et al. (2006). Flavonoid-inducible modifications to rhamnan O antigens are necessary for rhizobium sp. strain NGR234-legume symbioses. J. Bacteriol. 188, 3654–3663. doi: 10.1128/JB.188.10.3654-3663.2006, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardenas L., Martinez A., Sanchez F., Quinto C. (2008). Fast, transient and specific intracellular ROS changes in living root hair cells responding to nod factors (NFs). Plant J. 56, 802–813. doi: 10.1111/j.1365-313X.2008.03644.x, PMID: [DOI] [PubMed] [Google Scholar]
- Chabaud M., Venard C., Defaux-Petras A., Bécard G., Barker D. G. (2002). Targeted inoculation of Medicago truncatula in vitro root cultures reveals MtENOD11 expression during early stages of infection by arbuscular mycorrhizal fungi. New Phytol. 156, 265–273. doi: 10.1046/j.1469-8137.2002.00508.x [DOI] [PubMed] [Google Scholar]
- Chen H., Richardson A. E., Rolfe B. G. (1993). Studies of the physiological and genetic basis of acid tolerance in rhizobium leguminosarum biovar trifolii. Appl. Environ. Microbiol. 59, 1798–1804. doi: 10.1128/aem.59.6.1798-1804.1993, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen E. J., Sabio E. A., Long S. R. (2008). The periplasmic regulator ExoR inhibits ExoS/ChvI two- component signaling in Sinorhizobium meliloti. Mol. Microbiol. 69, 1290–1303. doi: 10.1038/jid.2014.371, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H. P., Walker G. C. (1998a). Succinoglycan is required for initiation and elongation of infection threads during nodulation of alfalfa by rhizobium meliloti. J. Bacteriol. 180, 5183–5191. doi: 10.1128/JB.180.19.5183-5191.1998, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H. P., Walker G. C. (1998b). Succinoglycan production by rhizobium meliloti is regulated through the ExoS-ChvI two-component regulatory system. J. Bacteriol. 180, 20–26. doi: 10.1128/JB.180.1.20-26.1998, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowan M. M. (1999). Plant products as antimicrobial agents. Clin. Microbiol. Rev. 12, 564–582. doi: 10.1128/CMR.12.4.564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramer C. L., Ryder T. B., Bell J. N., Lamb C. J. (1985). Rapid switching of plant gene expression induced by fungal elicitor. Science 227, 1240–1243. doi: 10.1126/science.227.4691.1240, PMID: [DOI] [PubMed] [Google Scholar]
- Csonka L. N. (1989). Physiological and genetic responses of bacteria to osmotic stress. Microbiol. Rev. 53, 121–147. doi: 10.1128/mr.53.1.121-147.1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham S. D., Munns D. N. (1984). The correlation between extracellular polysaccharide production and acid tolerance in rhizobium. Soil Sci. Soc. Am. J. 48, 1273–1276. doi: 10.2136/sssaj1984.03615995004800060014x [DOI] [Google Scholar]
- Czernic P., Gully D., Cartieaux F., Moulin L., Guefrachi I., Patrel D., et al. (2015). Convergent evolution of endosymbiont differentiation in Dalbergioid and inverted repeat-lacking clade legumes mediated by nodule-specific cysteine-rich peptides. Plant Physiol. 169, 1254–1265. doi: 10.1104/pp.15.00584, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- David M., Daveran M. L., Batut J., Dedieu A., Domergue O., Ghai J., et al. (1988). Cascade regulation of Nif gene-expression in rhizobium meliloti. Cell 54, 671–683. doi: 10.1016/S0092-8674(88)80012-6, PMID: [DOI] [PubMed] [Google Scholar]
- Del Cerro P., Megías M., López-Baena F. J., Gil-Serrano A., Pérez-Montaño F., Ollero F. J. (2019). Osmotic stress activates nif and fix genes and induces the rhizobium tropici CIAT 899 nod factor production via NodD2 by up-regulation of the nodA2 operon and the nodA3 gene. PLoS One 14:e0213298. doi: 10.1371/journal.pone.0213298, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dilworth M. J., Tiwari R. P., Reeve W. G., Glenn A. R. (2000). Legume root nodule bacteria and acid pH. Sci. Prog. 83, 357–389. PMID: [PubMed] [Google Scholar]
- Dixon R., Kahn D. (2004). Genetic regulation of biological nitrogen fixation. Nat. Rev. Microbiol. 2, 621–631. doi: 10.1038/nrmicro954 [DOI] [PubMed] [Google Scholar]
- Donati A. J., Jeon J. M., Sangurdekar D., So J. S., Chang W. S. (2011). Genome-wide transcriptional and physiological responses of Bradyrhizobium japonicum to paraquat-mediated oxidative stress. Appl. Environ. Microbiol. 77, 3633–3643. doi: 10.1128/AEM.00047-11, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draghi W. O., Del Papa M. F., Hellweg C., Watt S. A., Watt T. F., Barsch A., et al. (2016). A consolidated analysis of the physiologic and molecular responses induced under acid stress in the legume-symbiont model-soil bacterium Sinorhizobium meliloti. Sci. Rep. 6:e29278. doi: 10.1038/srep29278, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duc G., Trouvelot A., Gianinazzi-Pearson V., Gianinazzi S. (1989). First report of non-mycorrhizal plant mutants (Myc-) obtained in pea (Pisum sativum L.) and fababean (Vicia faba L.). Plant Sci. 60, 215–222. doi: 10.1016/0168-9452(89)90169-6 [DOI] [Google Scholar]
- Dylan T., Helinski D. R., Ditta G. S. (1990a). Hypoosmotic adaptation in rhizobium meliloti requires beta-(1-2)-glucan. J. Bacteriol. 172, 1400–1408. doi: 10.1128/jb.172.3.1400-1408.1990, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dylan T., Nagpal P., Helinski D. R., Ditta G. S. (1990b). Symbiotic pseudorevertants of rhizobium meliloti ndv mutants. J. Bacteriol. 172, 1409–1417. doi: 10.1128/jb.172.3.1409-1417.1990, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrhardt D. W., Atkinson E. M., Long S. R. (1992). Depolarization of alfalfa root hair membrane potential by rhizobium meliloti nod factors. Science 256, 998–1000. doi: 10.1126/science.10744524, PMID: [DOI] [PubMed] [Google Scholar]
- Ehrhardt D. W., Wais R., Long S. R. (1996). Calcium spiking in plant root hairs responding to rhizobium nodulation signals. Cell 85, 673–681. doi: 10.1016/S0092-8674(00)81234-9, PMID: [DOI] [PubMed] [Google Scholar]
- El-Yahyaoui F., Kuster H., Amor B. B., Hohnjec N., Pu A., Becker A., et al. (2004). Expression profiling in Medicago truncatula pdentifies more than 750 genes differentially expressed during nodulation, including many potential regulators of the symbiotic program. Plant Physiol. 136, 3159–3176. doi: 10.1104/pp.104.043612.the, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faget M., Blossfeld S., von Gillhaussen P., Schurr U., Temperton V. M. (2013). Disentangling who is who during rhizosphere acidification in root interactions: combining fluorescence with optode techniques. Front. Plant Sci. 4:392. doi: 10.3389/fpls.2013.00392, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedorova E., Thomson R., Whitehead L. F., Maudoux O., Udvardi M. K., Day D. A. (1999). Localization of H+-ATPase in soybean root nodules. Planta 209, 25–32. doi: 10.1007/s004250050603, PMID: [DOI] [PubMed] [Google Scholar]
- Fenner B. J., Tiwari R. P., Reeve W. G., Dilworth M. J., Glenn A. R. (2004). Sinorhizobium medicae genes whose regulation involves the ActS and/or ActR signal transduction proteins. FEMS Microbiol. Lett. 236, 21–31. doi: 10.1016/j.femsle.2004.05.016 [DOI] [PubMed] [Google Scholar]
- Ferrer J. L., Austin M. B., Stewart C., Noel J. P. (2008). Structure and function of enzymes involved in the biosynthesis of phenylpropanoids. Plant Physiol. Biochem. 46, 356–370. doi: 10.1016/j.plaphy.2007.12.009, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonseca-García C., Nava N., Lara M., Quinto C. (2021). An NADPH oxidase regulates carbon metabolism and the cell cycle during root nodule symbiosis in common bean (Phaseolus vulgaris). BMC Plant Biol. 21, 274–290. doi: 10.1186/s12870-021-03060-z, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fournier J., Timmers A. C. J., Sieberer B. J., Jauneau A., Chabaud M., Barker D. G. (2008). Mechanism of infection thread elongation in root hairs of Medicago truncatula and dynamic interplay with associated rhizobial colonization. Plant Physiol. 148, 1985–1995. doi: 10.1104/pp.108.125674, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geddes B. A., González J. E., Oresnik I. J. (2014). Exopolysaccharide production in response to medium acidification is correlated with an increase in competition for nodule occupancy. Mol. Plant-Microbe Interact. 27, 1307–1317. doi: 10.1094/MPMI-06-14-0168-R, PMID: [DOI] [PubMed] [Google Scholar]
- Geddes B. A., Oresnik I. J. (2016). “The mechanism of symbiotic nitrogen fixation,” in The Mechanistic Benefits of Microbial Symbionts. Advances in Environmental Microbiology. Vol. 2. Hurst (Ed.) C. J. (Switzerland: Springer International Publishing; ), 69–97. [Google Scholar]
- Gober J. W., Kashket E. R. (1987). K+ regulates bacteroid-associated functions of Bradyrhizobium. Proc. Natl. Acad. Sci. U. S. A. 84, 4650–4654. doi: 10.1073/pnas.84.13.4650, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- González J. E., Reuhs B. L., Walker G. C. (1996). Low molecular weight EPS II of rhizobium meliloti allows nodule invasion in Medicago sativa. Proc. Natl. Acad. Sci. U. S. A. 93, 8636–8641. doi: 10.1073/pnas.93.16.8636, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffitts J. S., Carlyon R. E., Erickson J. H., Moulton J. L., Barnett M. J., Toman C. J., et al. (2008). A Sinorhizobium meliloti osmosensory two-component system required for cyclic glucan export and symbiosis. Mol. Microbiol. 69, 479–490. doi: 10.1111/j.1365-2958.2008.06304.x, PMID: [DOI] [PubMed] [Google Scholar]
- Guefrachi I., Pierre O., Timchenko T., Alunni B., Barrière Q., Czernic P., et al. (2015). Bradyrhizobium BclA is a peptide transporter required for bacterial differentiation in Symbiosis with Aeschynomene legumes. Mol. Plant-Microbe Interact. 28, 1155–1166. doi: 10.1094/MPMI-04-15-0094-R, PMID: [DOI] [PubMed] [Google Scholar]
- Guerrero-Castro J., Lozano L., Sohlenkamp C. (2018). Dissecting the acid stress response of rhizobium tropici CIAT 899. Front. Microbiol. 9:846. doi: 10.3389/fmicb.2018.00846, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haag A. F., Baloban M., Sani M., Kerscher B., Pierre O., Farkas A., et al. (2011). Protection of Sinorhizobium against host cysteine-rich antimicrobial peptides is critical for symbiosis. PLoS Biol. 9:e1001169. doi: 10.1371/journal.pbio.1001169, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison J., Jamet A., Muglia C. I., Sype G., De Aguilar O. M., Puppo A., et al. (2005). Glutathione plays a fundamental role in growth and symbiotic capacity of Sinorhizobium meliloti. J. Bacteriol. 187, 168–174. doi: 10.1128/JB.187.1.168, PMID: . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwig U. A., Maxwell C. A., Joseph C. M., Phillips D. A. (1990). Effects of alfalfa nod gene-inducing flavonoids on nodABC transcription in rhizobium meliloti strains containing different nodD genes. J. Bacteriol. 172, 2769–2773. doi: 10.1128/jb.172.5.2769-2773.1990, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan S., Mathesius U. (2012). The role of flavonoids in root-rhizosphere signalling: opportunities and challenges for improving plant-microbe interactions. J. Exp. Bot. 63, 3429–3444. doi: 10.1093/jxb/err430, PMID: [DOI] [PubMed] [Google Scholar]
- Hawkins J. P., Geddes B. A., Oresnik I. J. (2017). Succinoglycan directly contributes to pH tolerance in Sinorhizboium meliloti Rm1021. Mol. Plant-Microbe Interact. 30, 1009–1019. doi: 10.1094/MPMI-07-17-0176-R, PMID: [DOI] [PubMed] [Google Scholar]
- Heavner M. E., Qiu W. G., Cheng H. (2015). Phylogenetic co-occurrence of ExoR, ExoS, and ChvI, components of the RSI bacterial invasion switch, suggests a key adaptive mechanism regulating the transition between free-living and host-invading phases in Rhizobiales. PLoS One 10:e0135655. doi: 10.1371/journal.pone.0135655, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckel B. C., Tomlinson A. D., Morton E. R., Choi J. H., Fuqua C. (2014). Agrobacterium tumefaciens ExoR controls acid response genes and impacts exopolysaccharide synthesis, horizontal gene transfer, and virulence gene expression. J. Bacteriol. 196, 3221–3233. doi: 10.1128/JB.01751-14, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellweg C., Pühler A., Weidner S. (2009). The time course of the transcriptomic response of Sinorhizobium meliloti 1021 following a shift to acidic pH. BMC Microbiol. 9, 37–53. doi: 10.1186/1471-2180-9-37, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch A. M., Lum M. R., Downie J. A. (2001). What makes the rhizobia-legume symbiosis so special? Plant Physiol. 127, 1484–1492. doi: 10.1104/pp.010866, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horváth B., Domonkos Á., Kereszt A., Szűcs A., Ábrahám E., Ayaydin F., et al. (2015). Loss of the nodule-specific cysteine rich peptide, NCR169, abolishes symbiotic nitrogen fixation in the Medicago truncatula dnf7 mutant. Proc. Natl. Acad. Sci. U. S. A. 112, 15232–15237. doi: 10.1073/pnas.1500777112, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howieson J. G., Ewing M. A., D’Antuono M. F. (1988). Selection for acid tolerance in rhizobium meliloti. Plant Soil 105, 179–188. doi: 10.1007/BF02376781 [DOI] [Google Scholar]
- Hunt S., King B. J., Canvin D. T., Layzell D. B. (1987). Steady and nonsteady state gas exchange characteristics of soybean nodules in relation to the oxygen diffusion barrier. Plant Physiol. 84, 164–172. doi: 10.1104/pp.84.1.164, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt S., Layzell D. B. (1993). Gas exchange of legume nodules and the regulation of nitrogenase activity. Annu. Rev. Plant Physiol. Plant Mol. Biol. 44, 483–511. doi: 10.1146/annurev.pp.44.060193.002411 [DOI] [Google Scholar]
- Jamet A., Mandon K., Puppo A., Hérouart D. (2007). H2O2 is required for optimal establishment of the Medicago sativa/Sinorhizobium meliloti symbiosis. J. Bacteriol. 189, 8741–8745. doi: 10.1128/JB.01130-07, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamet A., Sigaud S., Van de Sype G., Puppo A., Herouart D. (2003). Expression of the bacterial catalase genes during Sinorhizobium meliloti-Medicago sativa symbiosis and their crucial role during the infection process. Mol. Plant-Microbe Interact. 16, 217–225. doi: 10.1094/MPMI.2003.16.3.217, PMID: [DOI] [PubMed] [Google Scholar]
- Jones D. L., Dennis P. G., Owen A. G., Van Hees P. A. W. (2003). Organic acid behavior in soils - misconceptions and knowledge gaps. Plant Soil 248, 31–41. doi: 10.1023/A:1022304332313 [DOI] [Google Scholar]
- Jones K. M., Kobayashi H., Davies B. W., Taga M. E., Walker G. C. (2007). How rhizobial symbionts invade plants: the Sinorhizobium-Medicago model. Nat. Rev. Microbiol. 5, 619–633. doi: 10.1038/nrmicro1705, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jungk A. O. (2002). “Dynamics of nutrient movement at the soil root interface,” in Plant Roots. The Hidden Half. Kafkafi U., Waisel Y., Eshel (Eds.) A. (United States: CRC Press; ), 587–616. [Google Scholar]
- Kannenberg E. L., Brewin N. J. (1989). Expression of a cell surface antigen from rhizobium leguminosarum 3841 is regulated by oxygen and pH. J. Bacteriol. 171, 4543–4548. doi: 10.1128/jb.171.9.4543-4548.1989, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaharada Y., Kelly S., Nielsen M. W., Hjuler C. T., Gysel K., Muszyński A., et al. (2015). Receptor-mediated exopolysaccharide perception controls bacterial infection. Nature 523, 308–312. doi: 10.1038/nature14611, PMID: [DOI] [PubMed] [Google Scholar]
- Keating D. H., Willits M. G., Long S. R. (2002). A Sinorhizobium meliloti lipopolysaccharide mutant altered in cell surface sulfation. J. Bacteriol. 184, 6681–6689. doi: 10.1128/JB.184.23.6681, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Király Z., El-Zahaby H. M., Klement Z. (1997). Role of extracellular polysaccharide (EPS) slime of plant pathogenic bacteria in protecting cells to reactive oxygen species. J. Phytopathol. 145, 59–68. doi: 10.1111/j.1439-0434.1997.tb00365.x [DOI] [Google Scholar]
- Lehman A. P., Long S. R. (2013). Exopolysaccharides from Sinorhizobium meliloti can protect against H2O2-dependent damage. J. Bacteriol. 195, 5362–5369. doi: 10.1128/JB.00681-13, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lelpi L., Dylan T., Ditta G. S., Helinski D. R., Stanfield S. W. (1990). The ndvB locus of rhizobium meliloti encodes a 319-kDa involved in the production of β-(1-2)-glucan. J. Biol. Chem. 265, 2643–2651. [PubMed] [Google Scholar]
- Li L., Jia Y., Hou Q., Charles T. C., Nester E. W., Pan S. Q. (2002). A global pH sensor: agrobacterium sensor protein ChvG regulates acid-inducible genes on its two chromosomes and Ti plasmid. Proc. Natl. Acad. Sci. U. S. A. 99, 12369–12374. doi: 10.1073/pnas.192439499, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y., Cao Y., Tanaka K., Thibivilliers S., Wan J., Choi J., et al. (2013). Nonlegumes respond to Rhizobial nod factors by suppressing the innate immune response. Science 341, 1384–1387. doi: 10.1126/science.1242736, PMID: [DOI] [PubMed] [Google Scholar]
- Lohar D. P., Haridas S., Gantt J. S., VandenBosch K. A. (2007). A transient decrease in reactive oxygen species in roots leads to root hair deformation in the legume-rhizobia symbiosis. New Phytol. 173, 39–49. doi: 10.1111/j.1469-8137.2006.01901.x, PMID: [DOI] [PubMed] [Google Scholar]
- Lohar D. P., Sharopova N., Endre G., Peñuela S., Samac D., Town C., et al. (2006). Transcript analysis of early nodulation events in Medicago truncatula. Plant Physiol. 140, 221–234. doi: 10.1104/pp.105.070326.1, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H. Y., Luo L., Yang M. H., Cheng H.-P. (2012). Sinorhizobium meliloti ExoR is the target of periplasmic proteolysis. J. Bacteriol. 194, 4029–4040. doi: 10.1128/JB.00313-12, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madsen E. B., Madsen L. H., Radutoiu S., Olbryt M., Rakwalska M., Szczyglowski K., et al. (2003). A receptor kinase gene of the LysM type is involved in legume perception of rhizobial signals. Nature 425, 637–640. doi: 10.1038/nature02045, PMID: [DOI] [PubMed] [Google Scholar]
- Maillet F., Fournier J., Mendis H. C., Tadege M., Wen J., Ratet P., et al. (2020). Sinorhizobium meliloti succinylated high-molecular-weight succinoglycan and the Medicago truncatula LysM receptor-like kinase MtLYK10 participate independently in symbiotic infection. Plant J. 102, 311–326. doi: 10.1111/tpj.14625, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maroti G. G., Kereszt A., Kondorosi E., Mergaert P. (2011). Natural roles of antimicrobial peptides in microbes, plants and animals. Res. Microbiol. 162, 363–374. doi: 10.1016/j.resmic.2011.02.005, PMID: [DOI] [PubMed] [Google Scholar]
- Mendis H. C., Madzima T. F., Queiroux C., Jones K. M. (2016). Function of succinoglycan polysaccharide in Sinorhizobium meliloti host plant invasion depends on succinylation, not molecular weight. MBio 7:e00606-16. doi: 10.1128/mBio.00606-16, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mergaert P., Nikovics K., Kelemen Z., Maunoury N., Vaubert D., Kondorosi A., et al. (2003). A novel family in Medicago truncatula consisting of more than 300 nodule-specific genes coding for small, secreted polypeptides with conserved cysteine motifs. Plant Physiol. 132, 161–173. doi: 10.1104/pp.102.018192, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mergaert P., Uchiumi T., Alunni B., Evanno G., Cheron A., Catrice O., et al. (2006). Eukaryotic control on bacterial cell cycle and differentiation in the rhizobium-legume symbiosis. Proc. Natl. Acad. Sci. U. S. A. 103, 5230–5235. doi: 10.1073/pnas.0600912103, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller K. J., Kennedy E. P., Reinhold V. N. (1986). Osmotic adaptation by gram-negative bacteria: possible role for periplasmic oligosaccharides. Science 231, 48–51. doi: 10.1126/science.3941890, PMID: [DOI] [PubMed] [Google Scholar]
- Miller K. J., Wood J. M. (1996). Osmoadatptation by rhizosphere bacteria. Annu. Rev. Microbiol. 6:e23307. doi: 10.1371/journal.pone.0023307, PMID: [DOI] [PubMed] [Google Scholar]
- Miller-Williams M., Loewen P. C., Oresnik I. J. (2006). Isolation of salt-sensitive mutants of Sinorhizobium meliloti strain Rm1021. Microbiology 152, 2049–2059. doi: 10.1099/mic.0.28937-0, PMID: [DOI] [PubMed] [Google Scholar]
- Muglia C. I., Grasso D. H., Aguilar O. M. (2007). Rhizobium tropici response to acidity involves activation of glutathione synthesis. Microbiology 153, 1286–1296. doi: 10.1099/mic.0.2006/003483-0, PMID: [DOI] [PubMed] [Google Scholar]
- Mylona P., Pawlowski K., Bisseling T. (1995). Symbiotic nitrogen fixation. Plant Cell 7, 869–885. doi: 10.1097/00010694-199511000-00009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagpal P., Khanuja S. P., Stanfield S. W. (1992). Suppression of the ndv mutant phenotype of rhizobium meliloti by cloned exo genes. Mol. Microbiol. 6, 479–488. doi: 10.1111/j.1365-2958.1992.tb01492.x, PMID: [DOI] [PubMed] [Google Scholar]
- Ojeda K. J., Simonds L., Noel K. D. (2013). Roles of predicted glycosyltransferases in the biosynthesis of the rhizobium etli CE3 O antigen. J. Bacteriol. 195, 1949–1958. doi: 10.1128/JB.02080-12, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldroyd G. E. D. (2013). Speak, friend, and enter: signalling systems that promote beneficial symbiotic associations in plants. Nat. Rev. Microbiol. 11, 252–263. doi: 10.1038/nrmicro2990, PMID: [DOI] [PubMed] [Google Scholar]
- Oldroyd G. E. D., Downie J. A. (2008). Coordinating nodule morphogenesis with rhizobial infection in legumes. Annu. Rev. Plant Biol. 59, 519–546. doi: 10.1146/annurev.arplant.59.032607.092839, PMID: [DOI] [PubMed] [Google Scholar]
- Oldroyd G. E. D., Murray J. D., Poole P. S., Downie J. A. (2011). The rules of engagement in the legume-rhizobial symbiosis. Annu. Rev. Genet. 45, 119–144. doi: 10.1146/annurev-genet-110410-132549, PMID: [DOI] [PubMed] [Google Scholar]
- Pauly N., Pucciariello C., Mandon K., Innocenti G., Jamet A., Baudouin E., et al. (2006). Reactive oxygen and nitrogen species and glutathione: key players in the legume-rhizobium symbiosis. J. Exp. Bot. 57, 1769–1776. doi: 10.1093/jxb/erj184, PMID: [DOI] [PubMed] [Google Scholar]
- Penterman J., Abo R. P., De Nisco N. J., Arnold M. F. F., Longhi R., Zanda M., et al. (2014). Host plant peptides elicit a transcriptional response to control the Sinorhizobium meliloti cell cycle during symbiosis. Proc. Natl. Acad. Sci. U. S. A. 111, 3561–3566. doi: 10.1073/pnas.1400450111, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Montaño F., del Cerro P., Jiménez-Guerrero I., López-Baena F. J., Cubo M. T., Hungria M., et al. (2016). RNA-seq analysis of the rhizobium tropici CIAT 899 transcriptome shows similarities in the activation patterns of symbiotic genes in the presence of apigenin and salt. BMC Genomics 17, 198–111. doi: 10.1186/s12864-016-2543-3, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierre O., Engler G., Hopkins J., Brau F., Boncompagni E., Hérouart D. (2013). Peribacteroid space acidification: A marker of mature bacteroid functioning in Medicago truncatula nodules. Plant Cell Environ. 36, 2059–2070. doi: 10.1111/pce.12116, PMID: [DOI] [PubMed] [Google Scholar]
- Pueppke S. G., Broughton W. J. (1999). Rhizobium sp. strain NGR234 and R. fredii USDA257 share exceptionally broad, nested host ranges. Mol. Plant-Microbe Interact. 12, 293–318. doi: 10.1094/MPMI.1999.12.4.293, PMID: [DOI] [PubMed] [Google Scholar]
- Puppo A., Pauly N., Boscari A., Mandon K., Brouquisse R. (2013). Hydrogen peroxide and nitric oxide: key regulators of the legume-rhizobium and mycorrhizal symbioses. Antioxid. Redox Signal. 18, 2202–2219. doi: 10.1089/ars.2012.5136, PMID: [DOI] [PubMed] [Google Scholar]
- Pusztahelyi T. (2018). Chitin and chitin-related compounds in plant–fungal interactions. Mycology 9, 189–201. doi: 10.1080/21501203.2018.1473299, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramu S. K., Peng H. M., Cook D. R. (2002). Nod factor induction of reactive oxygen species production is correlated with expression of the early nodulin gene rip1 in Medicago truncatula. Mol. Plant-Microbe Interact. 15, 522–528. doi: 10.1094/MPMI.2002.15.6.522, PMID: [DOI] [PubMed] [Google Scholar]
- Reeve W. G., Dilworth M. J., Tiwari R. P., Glenn A. R. (1997). Regulation of exopolysaccharide production in rhizobium leguminosarum biovar viciae WSM710 involves exoR. Microbiology 143, 1951–1958. doi: 10.1099/00221287-143-6-1951, PMID: [DOI] [PubMed] [Google Scholar]
- Reuber T. L., Urzainqui A., Glazebrook J., Reed J. W., Walker G. C. (1990). Genetic analyses and manipulations of rhizobium meliloti exopolysaccharides. Nov. Biodegrad. Microb. Polym. 12, 285–294. doi: 10.1007/978-94-009-2129-0_24 [DOI] [Google Scholar]
- Reyes-González A., Talbi C., Rodríguez S., Rivera P., Zamorano-Sánchez D., Girard L. (2016). Expanding the regulatory network that controls nitrogen fixation in Sinorhizobium meliloti: elucidating the role of the two-component system hFixL-FxkR. Microbiology 162, 979–988. doi: 10.1099/mic.0.000284, PMID: [DOI] [PubMed] [Google Scholar]
- Riccillo P. M., Muglia C. I., De Bruijn F. J., Roe A. J., Booth I. R., Aguilar O. M. (2000). Glutathione is involved in environmental stress responses in rhizobium tropici, including acid tolerance. J. Bacteriol. 182, 1748–1753. doi: 10.1128/JB.182.6.1748-1753.2000, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutten P. J., Steel H., Hood G. A., Ramachandran V. K., McMurtry L., Geddes B., et al. (2021). Multiple sensors provide spatiotemporal oxygen regulation of gene expression in a rhizobium-legume symbiosis. PLoS Genet. 17:e1009099. doi: 10.1371/journal.pgen.1009099, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos R., Hérouart D., Sigaud S., Touati D., Puppo A. (2001). Oxidative burst in alfalfa- Sinorhizobium meliloti symbiotic interaction. Mol. Plant-Microbe Interact. 14, 86–89. doi: 10.1094/MPMI.2001.14.1.86, PMID: [DOI] [PubMed] [Google Scholar]
- Schopfer C. R. (1999). The male determinant of self-incompatibility in brassica. Science 286, 1697–1700. doi: 10.1126/science.286.5445.1697, PMID: [DOI] [PubMed] [Google Scholar]
- Shaw S. L., Long S. R. (2003). Nod factor inhibition of reactive oxygen efflux in a host legume. Plant Physiol. 132, 2196–2204. doi: 10.1104/pp.103.021113, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith L. T., Allaith A. A., Smith G. M. (1994). Mechanism of osmotically regulated N-acetylglutaminylglutamine amide production in rhizobium meliloti. Plant Soil 161, 103–108. doi: 10.1007/BF02183090 [DOI] [Google Scholar]
- Streng A., Op Den Camp R., Bisseling T., Geurts R., Camp R., Bisseling T., et al. (2011). Evolutionary origin of rhizobium nod factor signaling. Plant Signal. Behav. 6, 1510–1514. doi: 10.4161/psb.6.10.17444, PMID: . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y., Hollingsworth R. I. (1998). Regulation of lipid synthesis in Bradyrhizobium japonicum: low oxygen concentrations trigger phosphatidylinositol biosynthesis. Appl. Environ. Microbiol. 64, 1963–1966. doi: 10.1128/AEM.64.5.1963-1966.1998, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tóth K., Stacey G. (2015). Does plant immunity play a critical role during initiation of the legume-rhizobium symbiosis? Front. Plant Sci. 6, 401–408. doi: 10.3389/fpls.2015.00401, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Velde W., Zehirov G., Szatmari A., Debreczeny M., Ishihara H., Kevei Z., et al. (2010). Plant peptides govern terminal differentiation of bacteria in symbiosis. Science 327, 1122–1126. doi: 10.1126/science.1184057, PMID: [DOI] [PubMed] [Google Scholar]
- Via V. D., Zanetti M. E., Blanco F. (2016). How legumes recognize rhizobia. Plant Signal. Behav. 11:e1120396. doi: 10.1080/15592324.2015.1120396, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virts E. L., Stanfield S. W., Helinski D. R., Ditta G. S. (1988). Common regulatory elements control symbiotic and microaerobic induction of nifA in rhizobium meliloti. Proc. Natl. Acad. Sci. U. S. A. 85, 3062–3065. doi: 10.1073/pnas.85.9.3062, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkel-Shirley B. (2001). Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol. 126, 485–493. doi: 10.1104/pp.126.2.485, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yancey P. H., Clark M. E., Hand S. C., Bowlus R. D., Somero G. N. (1982). Living with water stress: evolution of osmolyte systems. Science 217, 1214–1222. doi: 10.1126/science.7112124, PMID: [DOI] [PubMed] [Google Scholar]
- Yao S. Y., Luo L., Har K. J., Becker A., Rüberg S., Yu G. Q., et al. (2004). Sinorhizobium meliloti ExoR and ExoS proteins regulate both succinoglycan and flagellum production. J. Bacteriol. 186, 6042–6049. doi: 10.1128/JB.186.18.6042-6049.2004, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- York W. S., McNeil M., Darvill A. G., Albersheim P. (1980). Beta-2-linked glucans secreted by fast-growing species of rhizobium. J. Bacteriol. 142, 243–248. doi: 10.1128/jb.142.1.243-248.1980, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- York G. M., Walker G. C. (1997). The rhizobium meliloti exoK gene and prsD/prsE/exsH genes are components of independent degradative pathways which contribute to production of low-molecular-weight succinoglycan. Mol. Microbiol. 25, 117–134. doi: 10.1046/j.1365-2958.1997.4481804.x, PMID: [DOI] [PubMed] [Google Scholar]
- Zamorano-Sánchez D., Girard L. (2015). “FNR-like proteins in rhizobia: past and future,” in Biology Nitrogen Fixation. Brujin (Ed.) F. J. (United States: John Wiley & Sons; ), 155–166. [Google Scholar]
- Zhou P., Silverstein K. A., Gao L., Walton J. D., Nallu S., Guhlin J., et al. (2013). Detecting small plant peptides using SPADA (small peptide alignment discovery application). BMC Bioinfo. 14, 335–351. doi: 10.1186/1471-2105-14-335, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel C., Oldroyd G. E. D. (2017). Plant signalling in symbiosis and immunity. Nature 543, 328–336. doi: 10.1038/nature22009, PMID: [DOI] [PubMed] [Google Scholar]