Abstract
Typing of gonococcal strains is a valuable tool for the biological confirmation of sexual contacts. We have developed a typing method based on DNA sequencing of two overlapping por gene fragments generated by a heminested PCR. We compared sequencing of the por gene (POR sequencing) and typing of the opa gene (OPA typing) for the characterization of strains from 17 sexual partnerships. Both methods were highly discriminatory. A different genotype was detected in 15 of the 17 epidemiologically unconnected couples by POR sequencing and in 16 of the 17 couples by OPA typing with restriction enzyme HpaII. Within partnerships, identical genotypes were obtained from 16 of the 17 known sex contacts by POR sequencing and from 15 of the 17 by OPA typing. Compared to OPA typing, which relies on interpretation of bands in a gel, DNA sequence data offer the advantage of being objective and portable. As costs for sequencing decline, the method should become affordable for most laboratory personnel who wish to type gonococcal strains.
Neisseria gonorrhoeae infection is one of the most common sexually transmitted diseases worldwide. There is no effective vaccine for N. gonorrhoeae; therefore, control of gonococcal infections depends on identification and treatment of infected individuals. The development of effective public health measures can be aided by an understanding of the molecular epidemiology of gonorrhea. Information on gonococcal strains circulating in a community and temporal changes in prevalent strains can be used to identify patterns of transmission of gonorrhea.
Various typing methods have been developed to characterize N. gonorrhoeae strains. The most widely used method is based on auxotyping and serological characterization (12, 32). Auxotyping, which is based on the nutrient growth requirements of strains, is complicated, labor-intensive, and time-consuming. Serotyping requires well-characterized monoclonal antibodies, which are in limited supply and subject to interlot variation. Furthermore, auxotyping and serological characterization are often unable to discriminate among strains from epidemiologically unrelated individuals (3–5, 14, 16, 22–24, 26). A number of rapid DNA-based typing methods have been developed for the characterization of gonococcal strains. The genetic relatedness of strains has been compared by restriction fragment length polymorphism analysis of whole genomic DNA or individual genes and by amplification of polymorphic DNA fragments with arbitrary or repetitive motif-based primers (4, 13, 18, 22, 25, 26, 35). The most extensively tested and discriminatory of the molecular genetic techniques is the opa gene typing method (OPA typing) of O'Rourke et al. (19). For this method, the 11 opa genes are amplified with a single pair of primers, the products are digested with frequently cutting restriction enzymes, the restriction fragments are separated on polyacrylamide gels, and the patterns of bands produced by different strains are compared. The principal limitation of this method, as well as the other genetic typing methods based on the analysis of band patterns, is that results from different laboratories are difficult to compare.
Nucleotide sequencing is the most definitive form of typing and is the approach that we have taken to the typing of N. gonorrhoeae. Sequence data are objective and can be compared readily between laboratories. The data can be stored in a single central database and shared electronically to provide a resource for epidemiological studies. The use of PCR amplification and direct PCR product sequencing with cycle labeling of oligonucleotides has greatly streamlined the process. Another advantage of the availability of nucleotide sequence data is the ability to perform evolutionary genetic analyses.
With current techniques, only a small portion of a bacterial genome can be sequenced in a cost-efficient manner. The most informative regions have to be determined empirically, and the choice of which region to sequence depends on the goals of a study. We have developed a typing system based on nucleotide sequencing of a large segment of the por locus (∼1 kb) (POR sequencing). This locus offers several advantages as a genetic marker for strain typing. The gene is expected to show high levels of genetic variation, since it encodes the serotyping antigen, which is known to be polymorphic (11). Individual N. gonorrhoeae strains carry a single copy of the gene; thus, comparisons between strains involve homologous alleles, a prerequisite for evolutionary genetic analyses. The molecular epidemiology and genealogy of the por gene are also of interest because the Por protein is a potential vaccine candidate (2, 21).
For short-term transmission studies, a typing method has to be sufficiently discriminating to distinguish among epidemiologically unrelated strains; at the same time, the genetic variation observed has to be sufficiently stable that identical types are recovered from known sexual contacts. In order to establish the utility of POR sequencing as a typing tool for transmission studies and to compare POR sequencing with OPA typing, we characterized gonococcal isolates from 17 sex partners by both methods. The sex partners were recruited from the same clinic over a 2-year period. The temporal and geographic proximity of the sampled population was expected to provide a reasonably stringent test of the efficiency of the different typing methods for short-term transmission studies.
MATERIALS AND METHODS
N. gonorrhoeae strains.
Thirty-four gonococcal isolates from 17 reported sexual contact pairs from Baltimore, Md., were examined. The sex partners were recruited from the Baltimore City Sexually Transmitted Disease (STD) Clinic over a 2-year period from 1991 to 1992 (36). To ensure as accurate an identification of sex partners as possible, persons trained in sexual contact tracing conducted the interviews (6). N. gonorrhoeae strains were single colonies picked from a primary isolation plate, subcultured once or twice, and frozen. The strains had not been passaged prior to this study. Auxotyping and serotyping were not done on the strains. For DNA analysis, strains were cultivated from frozen stocks that had been stored at −70°C in Greaves solution (5% bovine serum albumin, 0.5% monosodium glutamate, 10% glycerol). Cells were grown overnight at 37°C in 5% (vol/vol) CO2 on GC medium base agar (Difco) supplemented with the following components (per liter): 3.0 g of glucose, 0.083 g of Fe(NO3)3, 0.1 g of l-glutamine, 0.1 ml of cocarboxylase (2%), and 40 mg of l-cysteine hydrochloride.
Preparation of genomic DNA.
Genomic DNA was isolated from N. gonorrhoeae cells scraped off an agar culture plate. The bacterial cell pellet was resuspended in 600 μl of Tris-EDTA buffer (pH 7.4). Sodium dodecyl sulfate and proteinase K were added to final volumes of 0.5% and 100 μg/ml, respectively. The suspension was mixed by repeated inversion and then incubated for 1 h at 37°C. The lysate was adjusted to 0.7 M NaCl and 1% cetyltrimethylammonium bromide (CTAB), mixed thoroughly, and incubated for 10 min at 65°C in order to precipitate cell wall debris, denatured proteins, and polysaccharides. The supernatant was extracted once with chloroform-isoamyl alcohol (24:1), once with phenol-chloroform-isoamyl alcohol (25:24:1), and a second time with chloroform-isoamyl alcohol. Chromosomal DNA was precipitated with isopropanol, washed with 70% ethanol, and resuspended in 20 μl of distilled water.
Heminested por PCR.
A heminested PCR was performed to amplify two overlapping fragments of the por gene. The primers were complementary to conserved regions in the por gene, as determined by sequence comparisons of PIA and PIB homology group por genes from GenBank. PIA and PIB are structural variants of the POR protein. In the first round, a 2-μl aliquot of gonococcal chromosomal DNA was amplified in a 50-μl reaction volume containing 200 μM each deoxynucleoside triphosphate (dNTP), 0.5 μM each primer (POR-01, 5′-CTGACTTTGGCAGCCCTTCCTGTTG-3′, and POR-14, 5′-CAGATTAGAATTTGTGGCGC-3′), 2.1 U of Expand High Fidelity (Boehringer Mannheim Biochemicals, Indianapolis, Ind.), and the buffer provided by the manufacturer with 1.5 mM MgCl2. A hot-start PCR protocol was performed with wax beads from PE Applied Biosystems, Inc., Foster City, Calif. Cycle conditions were set at 94°C for 2 min to melt the beads; 30 cycles of 94°C for 40 s, 65°C for 40 s, and 72°C for 1 min; and a final extension reaction for 10 min at 72°C. The PCR products were diluted 1:100 in distilled water, and two nested reactions were performed with primers containing 5′ extensions encoding the M13 forward or M13 reverse sequencing primers. A 2-μl aliquot was amplified in a 100-μl reaction volume containing 200 μM each dNTP, 0.5 μM each primer (M13F-POR-01, 5′-GTCACGACGTTGTAAAACGACGGCCAGTCTGACTTTGGCAGCCCTT-3′ [the M13F sequence is underlined], and M13R-POR-08, 5′-CACACAGGAAACAGCTATGACCGTATTGTGCGAAGAAGC-3′ [the M13R sequence is underlined]; or M13F-POR-11, 5′-GTCACGACGTTGTAAAAC GACGGCCAGTCTGTCCGTACGCTACG-3′, and M13R-POR14, 5′-CACACAGGAAACAGCTATGACCAGATTAGAATTTGTGGCGC-3′), 1.4 U of Expand High Fidelity, and the manufacturer's recommended buffer with 1.5 mM MgCl2. A hot-start PCR protocol was used with cycle conditions set at 94°C for 2 min; 35 cycles of 94°C for 40 s, 60°C for 20 s, and 68°C for 40 s; and a final extension reaction for 10 min at 72°C. First- and second-round reactions were performed with a PE Applied Biosystems 9700 thermal cycler.
PCR products were separated by electrophoresis through a 2% agarose gel prepared with Tris-acetate-EDTA buffer. The gel was stained with Ciber Green dye, and bands were visualized under UV illumination with a Fluorimager.
PCR product sequencing.
The PCR products were passed through a GeneClean spin column (Bio 101, Inc., Vista, Calif.) and eluted with 40 μl of distilled H2O, and the recovered DNA was measured by UV spectrometry. For dideoxy sequencing reactions, 60 to 80 ng of PCR product was added to a final reaction volume of 5 μl containing 2 μl of Big Dye terminator RR mix (PE Applied Biosystems) and 1 μM primer (M13 forward or M13 reverse sequencing primer). Cycle conditions were set at 95°C for 15 s, 50°C for 15 s, and 60°C for 4 min. After 25 cycles, reaction products were denatured by being heated to 95°C for 30 s. The reaction volume was diluted with 15 μl of distilled water and passed through a minicolumn (Spin-50; BioMax, Inc., Odenton, Md.) equilibrated with distilled water. The labeled nucleic acids were dried in a Speed Vac concentrator (Savant, Farmingdale, N.Y.), and loaded into the lanes of an ABI 377 automated DNA sequencer (Synthesis and Sequencing Facility, Department of Biological Chemistry, Johns Hopkins University School of Medicine). Trace data were edited and nucleotide sequences were assembled with the SeqMan software program (DNASTAR, Inc., Madison, Wis.). The edited sequences corresponded approximately to the region from amino acid 18 to amino acid 346 of strain MS11 (accession number M21289).
OPA typing.
OPA typing was done by a modification of the method of O'Rourke et al. (19). The principal modifications were the use of longer segments of the opa genes and nonisotopic visualization of restriction fragments by silver staining. Briefly, opa genes were amplified from gonococcal chromosomal DNA by PCR. A 2-μl aliquot of DNA was amplified in a 100-μl reaction volume containing 200 μM each dNTP, 0.5 μM each primer (OPA-01, 5′-ATGTGCAGGCGGATTTAGCC-3′, and OPA-04, 5′-AATGAGGCTTCGTGGGTTTTG-3′), 2.1 U of Expand High Fidelity, and the buffer provided by the manufacturer with 1.5 mM MgCl2. The primers flank those of O'Rourke et al. (19) and generate a product, on average, about 120 bp longer. A hot-start PCR protocol was performed with wax beads from PE Applied Biosystems. Cycle conditions were set at 94°C for 2 min to melt the beads; 25 cycles of 94°C for 20 s, 65°C for 20 s, and 72°C for 40 s; and a final reaction for 10 min at 72°C. PCR was performed with a PE Applied Biosystems 9700 thermal cycler.
PCR products were precipitated with ethanol, washed with 70% ethanol, and resuspended in 50 μl of distilled water. The opa gene fragments (1.5 μg) were digested overnight at 65°C with 3 U of TaqI in a volume of 11 μl with REact 2 buffer (Life Technologies, Gaithersburg, Md.). An equivalent aliquot was digested overnight at 37°C with 3 U of HpaII. In REact 8 buffer, approximately 750 ng of the resulting restriction fragments was fractionated on a precast 12.5% polyacrylamide gel (GeneGel Excell 12.5/24 kit; Amersham Pharmacia Biotech, Uppsala, Sweden) with a GenePhor electrophoresis unit (Amersham Pharmacia Biotech) according to the recommendations of the manufacturer. Bands were visualized by silver staining (1, 17).
DNA sequence analysis.
Sequences were aligned using the program CLUSTAL X and edited manually. The best-fit model of DNA substitution was chosen by performing hierarchical likelihood ratio tests using PAUP* beta 4 and Modeltest 1.05 (27, 31). A neighbor-joining tree was generated by the PHYLIP program, version 3.5c, using parameter estimates from Modeltest (8). The distance matrix was generated by DNADIST, maximum-likelihood option, using a Kimura two-parameter model with a ratio of transitions to transversions of 2.2405 and unequal base frequencies (A, 0.2810; C, 0.2375; G, 0.2375; and T, 0.2054).
Some of the strains that we sequenced were genotyped by hybridization to oligonucleotide probes for PIB por gene variable regions (33). Isolates 409, 407, 510, 419, 582, 804, 7, 5, 133, 223, 144, 243, 783, 950, 692, 559, 453, 451, 731, 909, 633, 432, 1406, 1331, 1389, and 1540 in Thompson et al. (33) correspond to strains 1(F), 1(M), 2(F), 2(M), 3(F), 3(M), 4(F), 4(M), 5(F), 5(M), 6(F), 6(M), 8(F), 8(M), 11(F), 111(M), 12(F), 12(M), 13(F), 13(M), 14(F), 14(M), 15(F), 15(M), 16(F), and 16(M), respectively, in our study.
Nucleotide sequence accession numbers.
Novel sequences determined in this study were deposited in GenBank under accession numbers AF304390 to AF304405.
RESULTS
OPA typing of isolates from 34 sex partners.
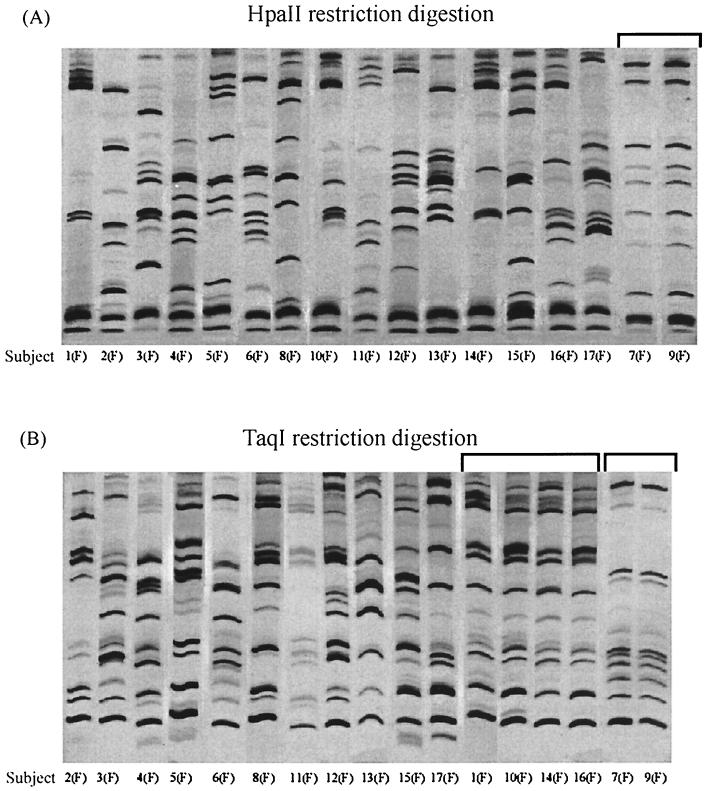
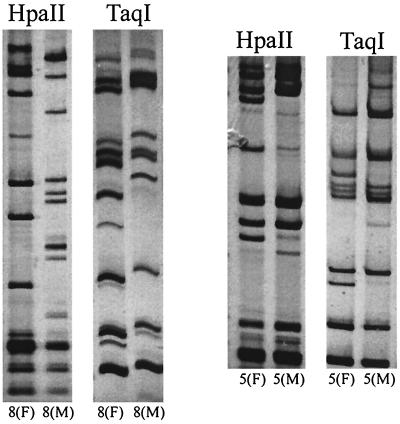
A critical test of the discriminatory power of a typing method for N. gonorrhoeae is the ability to distinguish gonococcal isolates that are epidemiologically unrelated while scoring as identical those from known sexual contacts. To perform such a test, we subjected 34 gonococcal isolates from 17 heterosexual partnerships to OPA typing and POR sequencing. For OPA typing, the corresponding ∼670-bp fragments of the 11 opa genes were amplified and the PCR products were digested with restriction enzymes HpaII and TaqI. The HpaII and TaqI OPA types of the isolates from the female members of the 17 sexual partnerships are shown in Fig. 1. Sixteen of the 17 isolates gave a unique OPA type with HpaII, while the isolates from subjects 7(F) and 9(F) produced an identical band pattern. OPA typing with TaqI was less discriminatory than typing with HpaII. Only 13 of the 17 isolates gave a unique OPA type with TaqI. Isolates from subjects 7(F) and 9(F), which had identical OPA types with HpaII, also had the same OPA type with TaqI. In addition, isolates from four subjects, 1(F), 10(F), 14(F), and 16(F), despite having different HpaII OPA types, had the same TaqI OPA type. With two exceptions (Fig. 2), the OPA types of isolates from sex partners were indistinguishable. Isolates from sex partners 8(F) and 8(M) were markedly different by both HpaII and TaqI OPA typing, with no bands in common between the two isolates. The HpaII and TaqI OPA types of isolates from partners 5(F) and 5(M) had several bands in common but also differed by the presence of two additional DNA fragments in the TaqI digest and two in the HpaII digest of the isolate from subject 5(F). The small differences between the isolates from partners 5(M) and 5(F) are unlikely to be due to a PCR artifact, because the products of a repeat amplification reaction gave the same results.
FIG. 1.
OPA typing of strains from the female members of 17 sex partnerships. Gonococcal chromosomal DNA was amplified by PCR, and the resulting fragments were digested with HpaII (A) or TaqI (B), fractionated on polyacrylamide, and stained with silver nitrate. Brackets mark identical OPA types from different partnerships. The samples were electrophoresed on the same polyacrylamide gel, but lanes were repositioned for purposes of illustration.
FIG. 2.
OPA types of sexual contacts with nonidentical genotypes. Paired samples were electrophoresed on the same polyacrylamide gel.
POR sequencing of isolates from 34 sex partners.
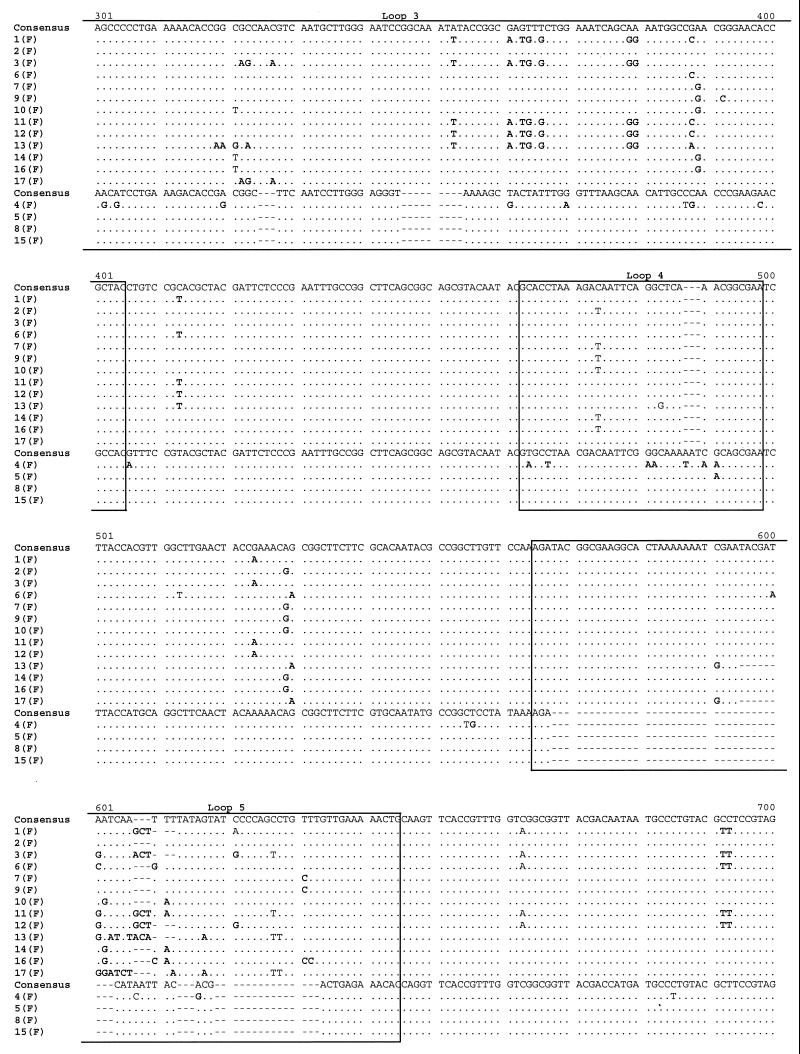
POR sequencing was also performed on the 34 isolates. An alignment of the por sequences of the isolates from the female members of the 17 sexual partnerships is shown in Fig. 3. By comparison to reference strains, 13 sequences were assigned to the B homology group, which encodes PIB Por proteins, and 4 were assigned to the A homology group, which encodes PIA Por proteins. The isolates from 15 of the 17 female subjects had unique por sequences. However, an identical por sequence was recovered from subjects 8(F) and 15(F); similarly, subjects 10(F) and 14(F) harbored an identical, although unique, por sequence. Of note, the isolates from subjects 7(F) and 9(F), which were identical by OPA typing, had por sequences that differed by a single base substitution. In confirmation of the findings, the same single base difference was detected in the por genes of the isolates from the male partners of these women. Inspection of the alignment indicates that nucleotide substitutions were distributed along the entire length of the open reading frame and concentrated in regions predicted to be surface-exposed loops. The majority of substitutions encoded amino acid replacements. These observations are consistent with previous analyses of por nucleotide sequences by others and us (28, 30).
FIG. 3.
Alignment of por nucleotide sequences from female members of 17 sex partnerships. Nucleotides that are identical to the consensus sequence are represented by dots. Dashes indicate alignment gaps. Boxes mark the locations of surface-exposed loops, as predicted by the structural model of Derrick et al. (5a). The por A and B homology group sequences are indicated. Nucleotide substitutions resulting in amino acid replacements are shown in bold type.
A BLAST search of GenBank showed that 2 of the 16 unique por sequences identified in this study matched perfectly sequences previously submitted to the database. One sequence (8F-15F) was identical to six entries, representing por genes from temporally and geographically diverse gonococcal strains, including sequences that we had previously obtained from clients attending the Baltimore City STD Clinic in 1997. A second sequence from this study (8M) was identical to the sequence of a por gene isolated in the 1980s from a person from the United Kingdom. The remaining 14 por sequences were unique but differed in most cases by only one or a few nucleotide differences from the closest match.
Cluster analysis of 34 por sequences from 17 sex partners.
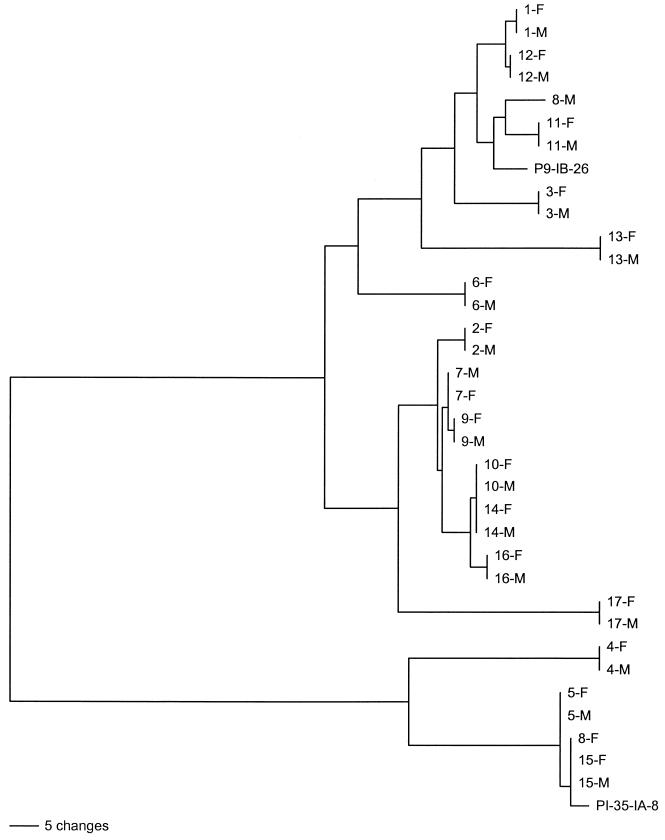
A dendrogram of the por sequences from the 17 sex partners is shown in Fig. 4. Horizontal branches delineate unique sequences and clusters of sequences. The vertical bars at the tips of the branches indicate identical sequences. The two major clusters correspond to por B and A homology group sequences. With a single exception, the por sequences from sexual contacts were identical. The por sequences of isolates from sex partners 8(F) and 8(M) differed by multiple nucleotide substitutions and were assigned by cluster analysis to different homology groups. The isolates from these presumed sex partners were also different by OPA typing, as described above. Compared to the differences in their OPA types, the isolates from sex partners 5(F) and 5(M) had identical por sequences. The interpartnership similarities described above are also illustrated on the dendogram. The por gene of 8(F) was identical to that recovered from sex partners 15(M) and 15(F), although there was no known contact between 8(F) and the couple. The por gene of the isolate from 8(M), the partner of 8(F), was unrelated to other por genes sequenced in this study. The isolates from sex partners 10(M) and 10(F) and partners 14(M) and 14(F) had identical por sequences, although interviews of the couples did not reveal an epidemiological link between couples 10 and 14. Dendrograms were also constructed with sliding windows of different lengths to determine the minimal gene fragment that provided the same discriminatory power as the full-length DNA sequence. The same topographical separation among the 34 strains was obtained using a 220-bp fragment that spans the region from the beginning of loop 3 to the end of loop 5 (data not shown).
FIG. 4.
Neighbor-joining tree of 34 por sequences from 17 sex partnerships. PI-35-IA and P9-IB-26 are reference strains for A and B homology group sequences, respectively. Branch lengths shown are proportional to the amount of genetic change.
DISCUSSION
The ability to identify accurately the strains of infectious agents that cause disease is central to epidemiological surveillance and public health decisions. The ideal bacterial typing system for epidemiological studies should be highly discriminatory, so that infected individuals can be distinguished from each other by the genetic fingerprint of the strain they harbor. However, in order to trace the spread of organisms through a host community and over time, different strains that are linked by a chain of transmission must bear a predictable genetic evolutionary relationship to each other. If they do, reconstruction of evolutionary relationships by phylogenetic analysis will reveal transmission pathways. The ability to carry out this process for any bacterial species and especially for N. gonorrhoeae is problematic. The principal reason is that recombination plays a major role in the evolution of bacteria. Through the process of transformation, Neisseria species in particular have the ability to undergo extensive recombination (7, 20). As a result of the exchange of large segments of DNA during recombinational events, ancestrally closely related strains may acquire multiple nucleotide differences. Because molecular phylogenetic methods assume that genetic variation is the result of the orderly accumulation of nucleotide substitutions and that the number and type of differences between two sequences are proportional to the time of divergence of the sequences, recombination negates the ability of phylogenetic analysis to infer true genealogical relationships (15). However, the practical limitations of phylogenetic analysis for a particular bacterial species depend on the relative rates of nucleotide substitutions and recombination occurring in nature. For most species, there are insufficient empirical data to establish these rates. One goal of our future research is to obtain DNA sequence data from epidemiologically well-characterized strains from individuals with gonorrhea in order to determine the rates of nucleotide substitutions and recombination for N. gonorrhoeae.
If phylogenetic reconstructions are problematic, it is still possible to use genetic characteristics of strains to identify sex partners and short chains of transmission. For this purpose, the genetic marker should be sufficiently polymorphic that strains from epidemiologically unrelated individuals can be distinguished from each other but stable enough so that recently transmitted strains are identical. With this goal in mind, we compared the ability of two typing methods, a newly developed POR sequencing method and the OPA typing method of O'Rourke et al. (19), to discriminate among different sex partnerships and to confirm self-reported sexual contacts.
Both typing methods were highly discriminatory. Different genotypes were detected in 15 of 17 partnerships by POR sequencing and in 16 of 17 partnerships by OPA typing. When results from the two typing methods were combined, all 17 partnerships were found to be associated with infection with different strains of N. gonorrhoeae. Strains with different opa genes may have had the same por gene, or vice versa, as a result of different rates of genetic change at the two loci due to recombination, mutation, or selection pressure. In the only other published study comparing POR sequencing and OPA typing, the latter method proved somewhat more discriminatory (10). Among 33 isolates from 15 patients with 17 repeat infections, there were 19 separate OPA types and 15 different por sequences. An identical OPA or POR result for strains from unrelated partnerships is not necessarily epidemiologically uninformative. In some cases, an identical OPA or POR genotype in different partnerships may indicate an undisclosed sexual linkage between the individuals. However, to determine whether OPA typing or POR sequencing can provide this kind of information will require genotyping of strains collected in studies of sex networks where short chains of transmission can be identified (9).
By both typing methods, different strains were detected for couple 8. Assuming that there was no misclassification of specimens in the laboratory, possible explanations for the discrepancy include a mixed infection, infection from another contact, or short-term genetic instability. A mixed infection seems unlikely because in our experience with direct PCR product sequencing of por genes from urine pellets, we have not seen evidence in the trace data for the presence of multiple allelic variants (unpublished data). The sensitivity of automated DNA sequencing for detection of mutant alleles has been demonstrated in other systems. In mixing experiments, Wei et al. were able to detect single base substitutions in mutant human immunodeficiency virus sequences that comprised as little as 10% of the total population (34). In conflict with our findings, studies conducted in the 1970s using auxotyping suggested that coinfection with multiple strains of N. gonorrhoeae is common (29). However, because gonococci can transform each other in cell culture, auxotyping, which is an in vitro technique, does not establish that there is a mixed infection in vivo. The magnitude of the genetic differences between the strains from couple 8, with multiple different bands by OPA typing and different homology groups by POR sequencing, makes genetic instability unlikely. Thus, the most likely explanation is that the couple did not report their sexual behavior fully or truthfully. For couple 5, whose strains differed only by OPA typing and whose typing profiles differed by just two bands, short-term genetic instability is a likely explanation for the differences between the strains. Consistent with this interpretation, the interval between sampling events for this couple was approximately 50 days, whereas all the other couples were sampled on the same day or within 5 days of each other. Discrepancies in OPA types of sexual contacts have been reported previously. Among 21 pairs of isolates or larger groups of isolates from known sexual contacts, O'Rourke et al. found 1 isolate that differed from the isolates from the sexual contacts (19). Although anecdotal, these findings suggest there is greater genetic instability of opa genes than of por genes and that OPA typing may be less useful than POR sequencing for tracking transmission beyond very recent sexual contacts.
Our study demonstrates that POR sequencing is comparable to the OPA typing method of O'Rourke et al. (19) as a technique for characterizing gonococcal strains from sexual contacts. Compared to the latter method, which relies on interpretation of bands in a gel, DNA sequence data offer the advantages of being objective and portable. As costs for sequencing decline, the method should become affordable for most laboratory personnel who wish to type gonococcal strains. Although we sequenced an approximately 1-kb DNA fragment in two segments, a single, shorter, 220-bp fragment would have provided the same epidemiological information and cut the sequencing costs in half. Similar to other PCR-based methods, typing by POR sequencing is rapid and, as we have previously shown, can be done with urine specimens (28). The ability of POR sequencing to identify sexual contacts should make the method very useful for differentiating between endemic infection, where strains would be expected to exhibit high levels of genetic variation, and an epidemic introduction, characterized by the detection of an identical genotype in multiple individuals.
ACKNOWLEDGMENTS
This project was supported by NIH grant RO1-HD33982 (to R.P.V.).
We thank Jeffrey Yuenger for strain preparation.
REFERENCES
- 1.Blum H, Beier H, Gross H J. Improved silver staining of plant proteins, RNA and DNA in polyacrylamide gels. Electrophoresis. 1987;8:93–99. [Google Scholar]
- 2.Buchanan T M, Eschenbach D A, Knapp J S, Holmes K K. Gonococcal salpingitis is less likely to recur with Neisseria gonorrhoeae of the same principal outer membrane protein antigenic type. Am J Obstet Gynecol. 1980;138:978–980. doi: 10.1016/0002-9378(80)91091-1. [DOI] [PubMed] [Google Scholar]
- 3.Butt N J, Virji M, Vayreda F, Lambden P R, Heckels J E. Gonococcal outer-membrane protein PIB: comparative sequence analysis and localization of epitopes which are recognized by type-specific and cross-reacting monoclonal antibodies. J Gen Microbiol. 1990;136:2165–2172. doi: 10.1099/00221287-136-11-2165. [DOI] [PubMed] [Google Scholar]
- 4.Camarena J J, Nogueira J M, Dasi M A, Moreno F, Garcia R, Ledesma E, Llorca J, Hernandez J. DNA amplification fingerprinting for subtyping Neisseria gonorrhoeae strains. Sex Transm Dis. 1995;22:128–136. doi: 10.1097/00007435-199503000-00009. [DOI] [PubMed] [Google Scholar]
- 5.Cooke S J, de la Paz H, La Poh C, Ison C A, Heckels J E. Variation within serovars of Neisseria gonorrhoeae detected by structural analysis of outer-membrane protein PIB and by pulsed-field gel electrophoresis. Microbiology. 1997;143:1415–1422. doi: 10.1099/00221287-143-4-1415. [DOI] [PubMed] [Google Scholar]
- 5a.Derrick J P, Urwin R, Suker J, Feavers I M, Maiden M C. Structural and evolutionary inferences from molecular variation in Neisseria porins. Infect Immun. 1999;67:2406–2413. doi: 10.1128/iai.67.5.2406-2413.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ellish N J, Weisman C S, Celentano D, Zenilman J M. Reliability of partner reports of sexual history in a heterosexual population at a sexually transmitted diseases clinic. Sex Transm Dis. 1996;23:446–452. doi: 10.1097/00007435-199611000-00002. [DOI] [PubMed] [Google Scholar]
- 7.Feil E J, Maiden M C, Achtman M, Spratt B G. The relative contributions of recombination and mutation to the divergence of clones of Neisseria meningitidis. Mol Biol Evol. 1999;16:1496–1502. doi: 10.1093/oxfordjournals.molbev.a026061. [DOI] [PubMed] [Google Scholar]
- 8.Felsenstein J. PHYLIP phylogeny inference package (version 3.5c). Seattle: University of Washington; 1993. [Google Scholar]
- 9.Ghani A C, Ison C A, Ward H, Garnett G P, Bell G, Kinghorn G R, Weber J, Day S. Sexual partner networks in the transmission of sexually transmitted diseases. An analysis of gonorrhea cases in Sheffield, UK. Sex Transm Dis. 1996;23:498–503. doi: 10.1097/00007435-199611000-00011. [DOI] [PubMed] [Google Scholar]
- 10.Hobbs M M, Alcorn T M, Davis R H, Fischer W, Thomas J C, Martin I, Ison C, Sparling P F, Cohen M S. Molecular typing of Neiserria gonorrhoeae causing repeated infections: evolution of porin during passage within a community. J Infect Dis. 1999;179:371–381. doi: 10.1086/314608. [DOI] [PubMed] [Google Scholar]
- 11.Johnston K H, Holmes K K, Gotschlich E C. The serological classification of Neisseria gonorrhoeae. I. Isolation of the outer membrane complex responsible for serotypic specificity. J Exp Med. 1976;143:741–758. doi: 10.1084/jem.143.4.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Knapp J S, Tam M R, Nowinski R C, Holmes K K, Sandstrom E G. Serological classification of Neisseria gonorrhoeae with use of monoclonal antibodies to gonococcal outer membrane protein I. J Infect Dis. 1984;150:44–48. doi: 10.1093/infdis/150.1.44. [DOI] [PubMed] [Google Scholar]
- 13.Lau Q C, Chow V T, Poh C L. Differentiation of Neisseria gonorrhoeae strains by polymerase chain reaction and restriction fragment length polymorphism of outer membrane protein IB genes. Genitourin Med. 1995;71:363–366. doi: 10.1136/sti.71.6.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li H, Dillon J A. Utility of ribotyping, restriction endonuclease analysis and pulsed-field gel electrophoresis to discriminate between isolates of Neisseria gonorrhoeae of serovar IA-2 which require arginine, hypoxanthine or uracil for growth. J Med Microbiol. 1995;43:208–215. doi: 10.1099/00222615-43-3-208. [DOI] [PubMed] [Google Scholar]
- 15.Li W -H. Molecular evolution. Sunderland, Mass: Sinauer Asociates, Inc.; 1997. [Google Scholar]
- 16.Mee B J, Thomas H, Cooke S J, Lambden P R, Heckels J E. Structural comparison and epitope analysis of outer-membrane protein PIA from strains of Neisseria gonorrhoeae with differing serovar specificities. J Gen Microbiol. 1993;139:2613–2620. doi: 10.1099/00221287-139-11-2613. [DOI] [PubMed] [Google Scholar]
- 17.Merril C R. Silver staining of proteins and DNA. Nature. 1990;343:779–780. doi: 10.1038/343779a0. [DOI] [PubMed] [Google Scholar]
- 18.Ng L K, Carballo M, Dillon J A. Differentiation of Neisseria gonorrhoeae isolates requiring proline, citrulline, and uracil by plasmid content, serotyping, and pulsed-field gel electrophoresis. J Clin Microbiol. 1995;33:1039–1041. doi: 10.1128/jcm.33.4.1039-1041.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.O'Rourke M, Ison C A, Renton A M, Spratt B G. Opa-typing: a high-resolution tool for studying the epidemiology of gonorrhoea. Mol Microbiol. 1995;17:865–875. doi: 10.1111/j.1365-2958.1995.mmi_17050865.x. [DOI] [PubMed] [Google Scholar]
- 20.O'Rourke M, Stevens E. Genetic structure of Neisseria gonorrhoeae populations: a non-clonal pathogen. J Gen Microbiol. 1993;139:2603–2611. doi: 10.1099/00221287-139-11-2603. [DOI] [PubMed] [Google Scholar]
- 21.Plummer F A, Simonsen J N, Chubb H, Slaney L, Kimata J, Bosire M, Ndinya-Achola J O, Ngugi E N. Epidemiologic evidence for the development of serovar-specific immunity after gonococcal infection. J Clin Investig. 1989;83:1472–1476. doi: 10.1172/JCI114040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Poh C L, Lau Q C. Subtyping of Neisseria gonorrhoeae auxotype-serovar groups by pulsed-field gel electrophoresis. J Med Microbiol. 1993;38:366–370. doi: 10.1099/00222615-38-5-366. [DOI] [PubMed] [Google Scholar]
- 23.Poh C L, Lau Q C, Chow V T. Differentiation of Neisseria gonorrhoeae IB-3 and IB-7 serovars by direct sequencing of protein IB gene and pulsed-field gel electrophoresis. J Med Microbiol. 1995;43:201–207. doi: 10.1099/00222615-43-3-201. [DOI] [PubMed] [Google Scholar]
- 24.Poh C L, Loh G K, Tapsall J W. Resolution of clonal subgroups among Neisseria gonorrhoeae IB-2 and IB-6 serovars by pulsed-field gel electrophoresis. Genitourin Med. 1995;71:145–149. doi: 10.1136/sti.71.3.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Poh C L, Ocampo J C, Sng E H, Bygdeman S M. Rapid in situ generation of DNA restriction endonuclease patterns for Neisseria gonorrhoeae. J Clin Microbiol. 1989;27:2784–2788. doi: 10.1128/jcm.27.12.2784-2788.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Poh C L, Ramachandran V, Tapsall J W. Genetic diversity of Neisseria gonorrhoeae IB-2 and IB-6 isolates revealed by whole-cell repetitive element sequence-based PCR. J Clin Microbiol. 1996;34:292–295. doi: 10.1128/jcm.34.2.292-295.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Posada D, Crandall K A. MODELTEST: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- 28.Posada D, Crandall K A, Nguyen M, Demma J C, Viscidi R P. Population genetics of the porB gene of Neisseria gonorrhoeae: different dynamics in different homology groups. Mol Biol Evol. 2000;17:423–436. doi: 10.1093/oxfordjournals.molbev.a026322. [DOI] [PubMed] [Google Scholar]
- 29.Short H B, Ploscowe V B, Weiss J A, Young F E. Rapid method for auxotyping multiple strains of Neisseria gonorrhoeae. J Clin Microbiol. 1977;6:244–248. doi: 10.1128/jcm.6.3.244-248.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smith N H, Maynard S J, Spratt B G. Sequence evolution of the porB gene of Neisseria gonorrhoeae and Neisseria meningitidis: evidence of positive Darwinian selection. Mol Biol Evol. 1995;12:363–370. doi: 10.1093/oxfordjournals.molbev.a040212. [DOI] [PubMed] [Google Scholar]
- 31.Swofford D. PAUP*: phylogenetic analysis using parsimony and other methods. Beta 4. Sunderland, Mass: Sinauer Associates, Inc.; 1998. [Google Scholar]
- 32.Tam M R, Buchanan T M, Sandstrom E G, Holmes K K, Knapp J S, Siadak A W, Nowinski R C. Serological classification of Neisseria gonorrhoeae with monoclonal antibodies. Infect Immun. 1982;36:1042–1053. doi: 10.1128/iai.36.3.1042-1053.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thompson D K, Deal C D, Ison C A, Zenilman J M, Bash M C. A typing system for Neisseria gonorrhoeae based on biotinylated oligonucleotide probes to PIB gene variable regions. J Infect Dis. 2000;181:1652–1660. doi: 10.1086/315464. [DOI] [PubMed] [Google Scholar]
- 34.Wei X, Ghosh S K, Taylor M E, Johnson V A, Emini E A, Deutsch P, Lifson J D, Bonhoeffer S, Nowak M A, Hahn B H. Viral dynamics in human immunodeficiency virus type 1 infection. Nature. 1995;373:117–122. doi: 10.1038/373117a0. [DOI] [PubMed] [Google Scholar]
- 35.Xia M, Whittington W L, Holmes K K, Plummer F A, Roberts M C. Pulsed-field gel electrophoresis for genomic analysis of Neisseria gonorrhoeae. J Infect Dis. 1995;171:455–458. doi: 10.1093/infdis/171.2.455. [DOI] [PubMed] [Google Scholar]
- 36.Zenilman J M, Ellish N, Fresia A, Glass G E. The geography of sexual partnerships in Baltimore—applications of core theory dynamics using a geographic information system. Sex Transm Dis. 1999;26:75–81. doi: 10.1097/00007435-199902000-00002. [DOI] [PubMed] [Google Scholar]