Abstract
Background
With the development of medical imaging and processing tools, accurate diagnosis of diseases has been made possible by intelligent systems. Owing to the remarkable ability of support vector machines (SVMs) for diseases diagnosis, extensive research has been conducted using the SVM algorithm for the classification of Alzheimer's disease (AD) and mild cognitive impairment (MCI).
Objectives
In this study, we applied an automated method to classify patients with AD and MCI and healthy control (HC) subjects based on the diffusion tensor imaging (DTI) features in the superficial white matter (SWM).
Participants
For this purpose, DTI data were downloaded from the Alzheimer's Disease Neuroimaging Initiative (ADNI). This method employed DTI data from 72 subjects: 24 subjects as HC, 24 subjects with MCI, and 24 subjects with AD.
Measure
ments: DTI processing was performed using DSI Studio software and all machine learning analyses were performed using MATLAB software.
Results
The linear kernel of SVM was the best classifier, with an accuracy of 95.8% between the AD and HC groups, followed by the quadratic kernel of SVM with an accuracy of 83.3% between the MCI and HC groups and the Gaussian kernel of SVM with an accuracy of 83.3% between the AD and MCI groups.
Conclusions
Given the importance of diagnosing AD and MCI as well as the role of superficial white matter in the diagnosis of neurodegenerative diseases, in this study, the features of different DTI methods of the SWM are discussed, which could be a useful tool to assist in the diagnosis of AD and MCI.
Keywords: Support vector machine, Diffusion tensor imaging, Alzheimer's disease, Mild cognitive impairment, Superficial white matter
Support vector machine, Diffusion tensor imaging, Alzheimer's disease, Mild cognitive impairment, Superficial white matter.
1. Introduction
Owing to the increase in the aging population, accurate and effective detection of Alzheimer's disease (AD) has become an important issue in the society [1]. Mild cognitive impairment (MCI) is a condition between normal aging-related cognitive decline and the more severe decline of dementia. Since there is no specific cure for MCI and there is a high risk of its progression to dementia, the diagnosis and prevention of the disease is very important [2].
The neurodegenerative disease often affects certain regions of the brain. Superficial white matter (SWM) is one of the regions that has been found to be highly vulnerable to many diseases, according to the retrogenesis model [3, 4]. In this study, we developed a support vector machine (SVM) model that classifies HC, MCI, and AD subjects based on the features of SWM.
Due to the development of automatic systems technology in various fields of medical sciences, machine learning systems help physicians to automatically diagnose diseases [5]. SVM, as a supervised machine learning technique, is a very powerful tool in big data analytics [1].
Brain imaging techniques are widely considered to have the potential for the diagnosis of brain disease. Using these techniques, problems in the human brain can be identified, without the need for invasive neurosurgery. Currently, several accepted safe imaging techniques are being used in research centers and hospitals around the world [6]. Diffusion tensor imaging (DTI) is a novel MRI-based neuroimaging technique that allows the assessment of neuronal fiber tract integrity [7].
DTI data reconstruction can be divided into two categories: model-free and model-based methods. Model-based methods such as DTI reconstruction assume that the shape of water diffusion follows a 3D Gaussian pattern, but there is no assumption on the distribution in the model-free method such as the q-space diffeomorphic reconstruction (QSDR) method. The QSDR method reconstructs data in the Montreal Neurological Institute (MNI) space [8]. DSI Studio software (www.dsi-studio.labsolver.org) supports both model-based and model-free reconstruction methods.
Different measurements are derived from the DTI reconstruction method including mean diffusivity (MD), fractional anisotropy (FA), axial diffusivity (AxD) and radial diffusivity (RD). Density-based measurements derived from the QSDR reconstruction method include quantitative anisotropy (QA), the isotropic value (ISO), restricted diffusion imaging (RDI), and so on.
QA is a metric to quantify the spin population in a specific direction and resolved the fiber population (specifically crossing fibers). The normalized QA (nQA) scale is calculated by normalizing the maximum QA value to one so that QA may be more comparable across the subject [9].
RDI is a method to quantify the density of restricted diffusion with respect to the diffusion displacement range (e.g. 10 microns) [10].
The ability to estimate the main direction of diffusion using the tensor has also yielded the tractography technique, which has been applied to calculate the connectivity matrix and network measures [11].
The brain is represented as a complex network consisting of neurons and brain regions that are structurally and functionally related. A brain network (or graph) consists of nodes (representing neurons or brain regions) connected by lines (connectivity between brain regions) [12].
Network science can help in reducing the analytical brain architecture complexity and understanding brain connectivity patterns and can provide information about clinical disorders [13].
Network measurements include assortativity, efficiency, PageRank, betweenness, small-world network, and so on; they are used to better understand the structure and function of the human brain as a network [14].
Only a few studies have used different analysis approaches such as region of interest (ROI), tractography, and connectivity and network in both DTI and QSDR reconstruction. We applied the SVM technique based on features extracted from the superficial white matter by the above-mentioned analysis methods for the automated binary detection of AD and MCI, AD and HC, and MCI and HC.
2. Materials and methods
The data presented in this article is extracted from a M.Sc. thesis and was reviewed and approved by the Ethical Committee of Mashhad University of Medical Sciences (Ethical number: IR.MUMS.MEDICAL.REC.1397.320).
The steps are as follows:
2.1. Data acquisition
Data of the 72 participants of the three groups were downloaded from the Alzheimer's Disease Neuroimaging Initiative (ADNI) database (http://www.loni.ucla.edu/ADNI/). Subjects of this study including subjects from the AD (n = 24), MCI (n = 24), and control (n = 24) groups were recruited from the ADNI2 project.
A whole-brain DTI of the subjects were generated from the ADNI2 project with the following scanning parameters: Manufacturer = GE MEDICAL SYSTEMS; Matrix X = 256.0 pixels; Matrix Y = 256.0 pixels; Matrix Z = 2714.0; Pixel Size X = 1.4 mm; Pixel Size Y = 1.4 mm; Pulse Sequence = EP/SE; repetition time (TR) of 13000 ms, echo time (TE) of 68.3 ms, flip angle of 90°, field strength of 3.0, slice thickness of 2.7 mm, 41 non-collinear directions with a b-value of 1000 s/mm2, and 5 images with no diffusion weighting. In addition to the images, clinical and neuropsychological data of subjects were also downloaded.
2.2. DTI processing
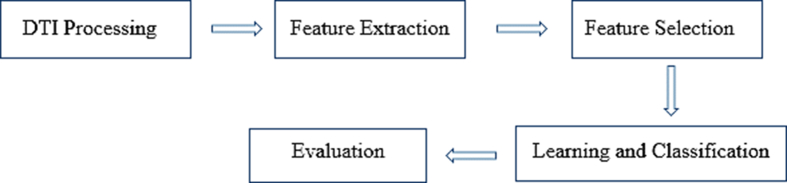
For each raw data, the following main steps were performed to extract the features of the DTI techniques in the SWM (are shown in Figure 1). All these processes were performed using DSI Studio software (developed by Fang-Cheng Yeh from the Advanced Biomedical MRI Lab, National Taiwan University Hospital, Taiwan, supported by Fiber Tractography Lab, University of Pittsburgh, and made available at http://dsi-studio.labsolver.org/Download/).
Figure 1.
Steps to extract the parameters from the DTI data.
2.2.1. Preprocessing and reconstruction step
Before DTI parameter measurement, correction of head motion and eddy-current and skull stripping were done. For skull stripping and filtering the background region, we used the masks provided by DSI-Studio. Then in the next step, we used two different reconstruction methods include model base (DTI) and free model (QSDR) option in DSI Studio; with two different attitudes to process the diffusion images.
2.2.2. ROI approach
After the reconstruction step, different DTI parameters were obtained from ROI, tractography and connectivity, and network methods. The ROI is the identity for a particular purpose, the SWM region in the current study. The mask (in the MNI space) of this region was obtained from a similar study by Arash Nazeri et al [15, 16].
According to the division of brain regions in the Terminologia Anatomica 1998 [17] and Terminologia Neuroanatomica 2017 (FIPAT. Terminologia Neuroanatomica. FIPAT.library.dal.ca. Federative International Programme for Anatomical Terminology, February 2017), we divided the SWM region into 12 anatomical regions including frontal, parietal, temporal, occipital, limbic, and insular lobes (on each side) by the Talairach Atlas [18] (Figure 2). These regions were confirmed by two experienced radiologists. Finally, we added sub-regions SWM atlas for better and easier access to DSI-Studio software atlases. In total, 12 SWM regions and average DTI and QSDR values were calculated for each region.
Figure 2.
Overview of the division of the SWM of the brain into the frontal (green), insular (orange), limbic (purple), parietal (pink), temporal (blue), and occipital (yellow) lobes: a) 3D axial view and b) 3D sagittal view.
2.2.3. Tractography approach
In order to extract the tractography parameters, FA and QA indexes were used for DTI and QSDR reconstruction to determine the fiber tracking threshold, respectively. Initially, SWM regions (as ROI) were placed and tractography was performed separately from the regions. The tractography of each SWM regions was performed with 100000 seeds, randomly generated at the subvoxel positions, and the seeds were placed across all the SWM regions, with a step size of 0 (0.5 voxel to 1.5 voxel distance) and a smoothing value of 1. The tracking from the primary fiber of a seeding point was set to streamline (Euler), and the direction interpolation was set to trilinear. The fiber length range was set between 30 and 300 mm.
2.2.4. Connectivity and network analysis
After performing tractography, the structural connectivity between the brain SWM regions and the brain network measures was obtained by the QSDR reconstruction method. To do this, the "Connectivity matrix" option was used to extract the connectivity and network parameters. So that after performing a tractography of the whole brain, interconnection measurements between the regions of the SWM were evaluated based on the count of connections. Also, the measured information from the network (such as efficiency, assortativity, betweenness, etc.) was extracted from different SWM regions.
2.3. Classification methods
We included the DTI parameters of both reconstruction and measured parameters from the ROI, tractography and connectivity and network methods (i.e., FA, MD, RD, AxD, and QA, nQA, iso, RDI, network values and number of connections between the brain regions). For each group was converted to CSV files to enter MATLAB software for the classification. Features extracted from each consisted of 504 features of the ROI method, 576 features of the tractography method, and 702 features of the connectivity and network method. After sorting the features, the vector of features for each subject was estimated from the 1782 features. In order to prepare the feature matrices as input to the SVM classification model, intergroup matrices (HC-AD, HC-MCI, and MCI-AD) with specific labels for each group were created.
All machine learning analyses were performed using MATLAB software (R2014a). The steps can be divided into the following flow chart (as shown in Figure 3).
Figure 3.
The process flow chart in our study.
After DTI data processing, feature extraction, and feature vector creation, feature selection was performed.
2.3.1. Feature selection
In this neuroimaging research, the number of features per subject was very high. So for identify the most relevant features (or parameters) for the classification, we used a method based on fast correlation-based filter (FCBF), which is a feature selection method for high-dimensional data [19].
After DTI processing, feature selection is performed using FCBF methods and used in SVM for binary classification.
2.3.2. Learning and classification
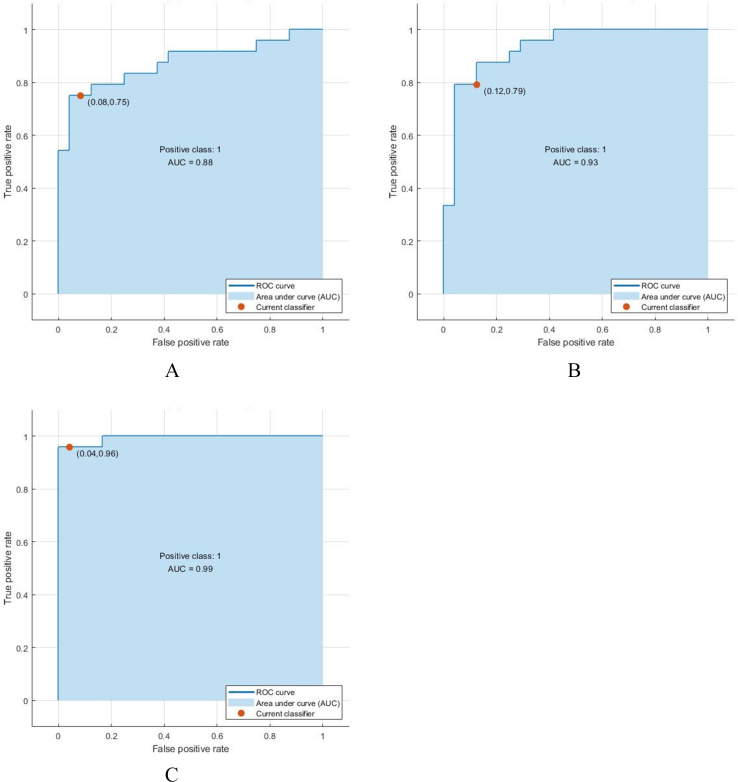
After feature selection, we performed an internal 5-fold cross-validation for the training data and applied the SVM algorithm using binary classification between the three groups. Cross-validation is a model validation technique used to ensure performance generalization; it is also a re-sampling method used to assess a model if we have limited data [20]. In total, we evaluated the Linear, Quadratic, Cubic, and Gaussian kernels (fine, medium, coarse). Due to the better results in Linear, Quadratic and Gaussian kernels and to reduce the complexity of the study, we reported the results of these three kernels. Finally, we show the Receiver operating characteristic (ROC) curve and the area under the curve (AUC) for the best kernel in each classification (as shown in Figure 4).
Figure 4.
A) The ROC curve followed by the quadratic kernel of SVM for HC-MCI classification. B) The ROC curve followed by the Gaussian kernel of SVM for AD-MCI classification. C) The ROC curve followed by the linear kernel of SVM for AD-HC classification.
2.3.3. Evaluation
Once the SVM algorithm has been trained, the results including accuracy, specificity, and sensitivity, which are defined as follows, are used to evaluate the classification performance.
Generally in these equations, true positive (TP) refers to the number of patients predicted correctly, false positive (FP) refers to the number of healthy controls predicted incorrectly as patients, true negative (TN) refers to the number of healthy controls predicted correctly, and false negative (FN) refers to the number of patients predicted incorrectly as healthy [21].
3. Result
3.1. Demographic and clinical characteristics
Demographics and the clinical scores of the participants are shown in Table 1. There were no significant differences (P > 0.05) between the three groups with respect to age and sex (see Table 1). Mini-mental state examination (MMSE), Global clinical dementia rating (CDR), and Functional Activities Questionnaire (FAQ) scores were significantly different among the three groups. Statistical analysis of the basic information was performed using SPSS 24.
Table 1.
Demographics and clinical scores of the participants.
| HC (n = 24) |
MCI (n = 24) |
AD (n = 24) |
p-value | |
|---|---|---|---|---|
| Mean (SD) | Mean (SD) | Mean (SD) | ||
| Age | 75.3 (8.3) | 76 (8.6) | 76.4 (8.2) | 0.89 |
| Sex | 11 M/13 F | 12 M/12 F | 16 M/8 F | 0.3 |
| Global CDR | 0.021 (0.1) | 0.58 (0.19) | 1.1 (0.116) | <0.001 |
| FAQ Total Score | 0.08 (0.4) | 4.9 (6.9) | 19.7 (6.2) | <0.001 |
| MMSE | 29 (1.2) | 26.7 (2) | 20.1 (4.9) | <0.001 |
Note. CDR: Commission on Dietetic Registration, FAQ: Functional Activities Questionnaire, MMSE: minimal-mental simple examination, HC: Healthy control, MCI: Mild cognitive impairment, AD: Alzheimer's disease, M: Male, F: Female.
P < 0.05 was considered statistically significant and the bold font indicates statistical significance.
3.2. The selective features of superficial white matter
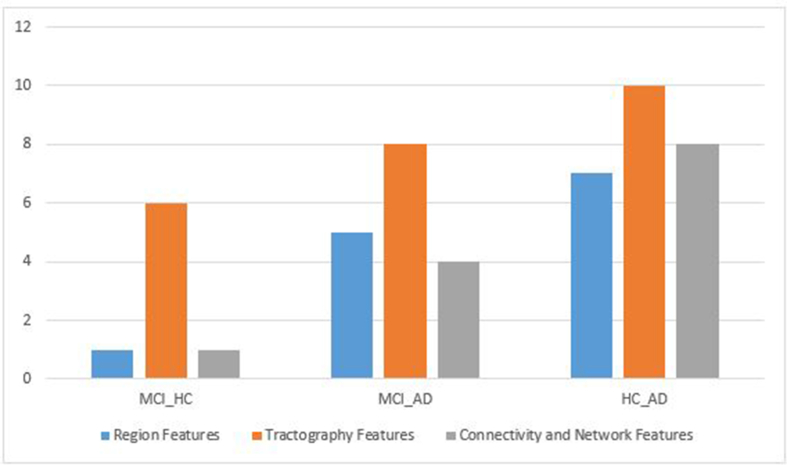
The FCBF method of feature selection showed 8 features for the classification of MCI and HC, 25 features for the classification of AD and HC, and 17 features for the classification of AD and MCI (Table 2). Figure 5 shows the number of selective features of the different methods of DTI.
Table 2.
The selective features for the SVM classifier.
| MCI versus HC |
AD versus HC |
||
|---|---|---|---|
| SWM Regions | Metrics | SWM Regions | Metrics |
| Occipital (L) | PageRank-network | Total | assortativity-network |
| Temporal (L) | rdi02L | Frontal (L) | efficiency-network |
| Insula (L) | Txy mean | Occipital (R) | betweenness-network |
| Occipital (L) | Txz mean | Frontal (R) | eigenvector-network |
| Limbic (R) | Tyz mean | Frontal (R) | PageRank-network |
| Occipital (R) | RD | Parietal (R) | PageRank-network |
| Limbic (R) | iso | Frontal (R) | eccentricity-network |
| Frontal (R) | rdi02L | Parietal(R) | efficiency-network |
| MCI versus AD | Insula (L) | Total-Connect | |
| SWM Regions | Metrics | Occipital-Limbic (L) | Connectivity |
| Total | Small-worldness-network | Temporal-Insula (L) | Connectivity |
| Frontal (R) | Cluster coefficient network | Limbic (L) | Txy mean |
| Occipital-Limbic (L) | Connectivity | Limbic (L) | Tyz mean |
| Parietal-Temporal (R) | Connectivity | Temporal (L) | RD |
| Insula (R) | Region-FA | Occipital (L) | rdi08L |
| Temporal (L) | Tyy mean | Parietal (R) | Tract length |
| Insula (R) | Tyz mean | Parietal (R) | Txx mean |
| Temporal (L) | Tzz mean | Insula (L) | Txy mean |
| Insula (L) | nQA | Limbic (L) | Txy mean |
| Parietal (L) | Tract length | Insula (L) | Tzz mean |
| Parietal (L) | Tract number | Occipital (L) | Tzz mean |
| Insula (R) | Tract-FA | Limbic (L) | AxD |
| Temporal (R) | Txx mean | Frontal (R) | RD |
| Occipital (L) | Txz mean | Insula (R) | iso |
| Insula (L) | Tyz mean | Limbic (L) | iso |
| Insula (L) | AxD | ||
| Occipital (L) | Tract length | ||
Note: FA: Fractional anisotropy; RD: Radial diffusivity; AD: Axial diffusivity; Txx, Txy, Txz, Tyy, Tyz, Tzz: The main values of the diffusion matrix; nQA: Normalized quantitative anisotropy; rdi: Restricted diffusion imaging; HC: Healthy control; MCI: mild cognitive impairment; AD: Alzheimer's disease; L: left; R: right.
Figure 5.
The number of selective features of the different methods.
3.3. The classification performance
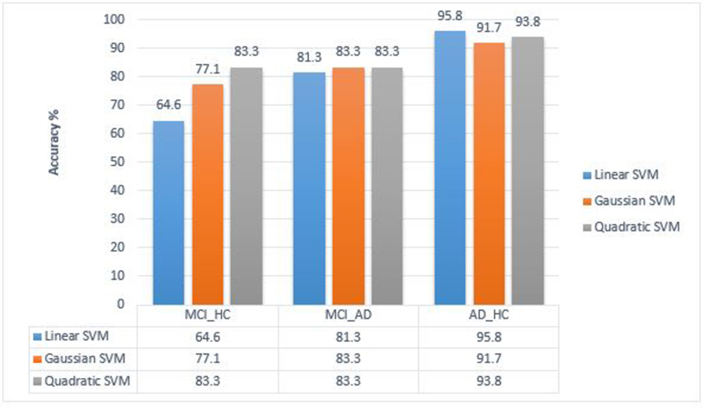
The mean accuracy, sensitivity, and specificity were reported as the results of this study. The linear kernel of SVM was the best classifier, with an accuracy of 95.8%, a sensitivity of 95.8%, and a specificity of 95.8% between the AD and HC groups, followed by the quadratic kernel of SVM with an accuracy of 83.3%, a sensitivity of 94.4%, and a specificity of 76.6% between the MCI and HC groups and the Gaussian kernel of SVM with an accuracy of 83.3%, a sensitivity of 80.7%, and a specificity of 86.3% between the AD and MCI groups) as shown in Table 1). Figure 6 shows the comparison between the three kernels and finds the best kernel in any pair classification (see Table 3).
Figure 6.
Comparison between the three kernels to find the best kernel in any pair classification.
Table 3.
Classification performance for each pair group.
| Pair classifier | Correction | Accuracy | Sensitivity | Specificity | AUC |
|---|---|---|---|---|---|
| MCI-HC | 70.3 | 83.3 | 94.4 | 76.6 | .88 |
| MCI-AD | 87.5 | 83.3 | 80.7 | 86.3 | .93 |
| HC-AD | 95.8 | 95.8 | 95.8 | 95.8 | .99 |
Note. HC: Healthy control, MCI: mild cognitive impairment, AD: Alzheimer's disease, AUC: Area Under the Curve.
4. Discussion
There are various methods for analyzing DTI data, and each method has its strengths and weaknesses. In this study, we used three methods, namely ROI, tractography, connectivity and network analysis, to obtain the characteristics of the superficial white matter [22].
The superficial white matter is one of the regions that has been found to be highly vulnerable to many diseases, according to the retrogenesis model [23]. For this reason, this area has been investigated in this study.
Several studies have been conducted to differentiate between individuals with AD and MCI and healthy individuals and many researchers are interested in finding methods to separate these three groups. In the current study, we applied an automated method to classify subjects with AD and MCI and HC subjects based on DTI features in the SWM.
It is important to note that the fiber architecture of the SWM (containing multiple fiber populations called "crossing fibers") shows a more complex order than the deep white matter (Figure 7). For this reason, it seems necessary to use the QSDR reconstruction technique, because model-free methods are more accurate in voxels containing multiple fiber populations compared with model-based methods [24]. For this purpose, we used the QSDR technique along with the DTI reconstruction technique.
Figure 7.
Example of the complex architecture of the SWM and crossing fiber (The SWM mask is shown in a white background).
Due to the development of intelligent systems in different sciences, a machine learning system was developed for pair classification between the three groups. To the best of our knowledge, this is the first study to use a support vector machine to identify the features of the DTI techniques in the SWM.
Our results from control versus MCI classification showed that the quadratic kernel was the best kernel for this classification, with an accuracy of 83.3%. The discrimination between AD patients and elderly controls showed 95.8% accuracy by the linear kernel. Connectivity parameters showed great importance on the selected features. An example of these connections is shown in Figure 8.
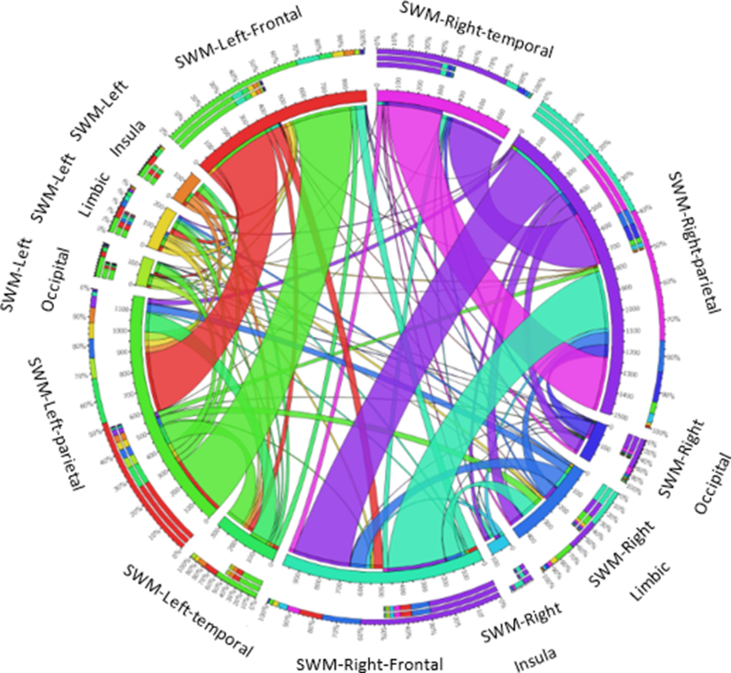
Figure 8.
An example of the connections between the superficial white matter regions provided by http://mkweb.bcgsc.ca/tableviewer/visualize/.
Generally, investigation of brain network features can provide researchers with information on most neurodegenerative diseases, including AD and MCI. Recently, the study of network properties in Alzheimer's disease has attracted the attention of several researchers. Among these researchers are Daianu et al. [25], Seo et al. [26], Jalili et al. [27], Sheng et al. [28] and Sulaimany et al. [29]. They believe that brain network connectivity analysis provides a significant understanding of how neural pathways break down in Alzheimer's disease. For example, as reported in the study of Yongxia Zhou et al., the feature of the small-world network in the brain cortex was able to distinguish between Alzheimer's patients and MCI patients. In this study, the small-world network, as one of the selective features in the superficial white matter, showed the ability to differentiate AD from MCI, and it could be used to explain the decline of memory and cognitive functions, consistent with the findings of previous studies that loss of small-world network characteristics are altered in AD patients [30, 31, 32].
Also, the PageRank measurement can highlight the brain regions with a higher number of external links [33]. In our study, the PageRank in the frontal and parietal regions was one of the top identified features for AD diagnosis and it could play a more important role in the brain.
One of the most important findings of the study is the length and number of the left parietal lobe tracts in the separation of AD from MCI. Desikan et al.’s study mentioned the importance of parietal lobe injury as a predictor of progression from MCI to AD [34]. The tractography results of this study can confirm the conclusion of the previous study in the SWM region. Because MCIs have a higher risk of developing AD than controls, examining of parietal lobe may be a helpful indicator.
Also addition to tractography findings in AD versus MCI comparisons, the disruption in connectivity between the temporal and parietal lobes and also limbic and occipital lobes was observed in ADs. In other word, short-range fibers connections in temporo-parieto can be an essential finding in the separation of these two diseases. The temporo-parieto plays a vital role in high-level human neural functions [35] that may be damaged in Alzheimer's disease. Desikan et al. examined the atrophy of this region in AD [34]. The findings of this study with a new technique and using the features of the DTI method can be effective in separating MCI from AD.A functional magnetic resonance imaging (fMRI) study showed that insula is the key region of the human brain networks and the most vulnerable region of AD [36]; the present study can confirm these results in the superficial white matter because the features of insula such as connectivity, iso, Tzz, and Txy are among the selected and main features of Alzheimer's patients.
Most studies have reported that DTI values change between the AD and MCI groups. Classification accuracy in this study between AD and MCI was 83.3% by the Gaussian kernel, which was the best kernel for this classification.
As a suggestion for future research, it will be interesting to include other modalities and biomarkers in the multimodal study such as fMRI and electroencephalogram (EEG), Positron emission tomography (PET), and CSF proteins data with DTI data and can also be one of our future goals.
5. Conclusion
In conclusion, we performed a method to automatically discriminate between patients with AD and MCI and healthy controls. In this study, we demonstrated that AD or MCI could be distinguished from HC using SWM region features through DTI. Thus, features obtained from the ROI, tractography, and connectivity and network methods could help assist in the diagnosis of AD and MCI. Finally, this study provides a background to evaluate the other automated classification methods in this region. s.
5.1. Limitations
The sample size in machine learning is a crucial factor that impacts the model performance. The study limitation is the small sample size of the included subjects.
Declarations
Author contribution statement
Bahare Bigham: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Seyed Amir Zamanpour: Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Hoda Zare: Conceived and designed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Funding statement
This work was supported by the Office of Vice President for Research Affairs of Mashhad University of Medical Sciences (MUMS), Mashhad, Iran.
Data availability statement
Data included in article/supplementary material/referenced in article.
Declaration of interests statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Acknowledgements
Data used in the preparation of this article were obtained from the Alzheimer's Disease Neuroimaging Initiative (ADNI) database (https://ida.loni.usc.edu/collaboration/access/adni.loni.usc.edu). The ADNI was launched in 2003 as a public–private partnership, led by Principal Investigator Michael W. Weiner, MD. The primary goal of ADNI has been to test whether serial magnetic resonance imaging (MRI), positron emission tomography (PET), other biological markers, and clinical and neuropsychological assessment can be combined to measure the progression of mild cognitive impairment (MCI) and early Alzheimer's disease (AD). For up-to-date information, see https://ida.loni.usc.edu/collaboration/access/www.adni-info.org.
The authors would like to thank Dr. Arash Nazari (arash.nazeri@mail.utoronto.ca) for sharing the superficial white matter (SWM) mask with us.
References
- 1.Lilia M., Marie S., Valerie H.-B., Bruno D., Patrick G., Serge K. DTI and structural MRI classification in Alzheimer’s disease. Adv. Mol. Imag. 2012;2012 [Google Scholar]
- 2.Eshkoor S.A., Hamid T.A., Mun C.Y., Ng C.K. Mild cognitive impairment and its management in older people. Clin. Interv. Aging. 2015;10:687–693. doi: 10.2147/CIA.S73922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Phillips O.R., Joshi S.H., Piras F., Orfei M.D., Iorio M., Narr K.L., et al. The superficial white matter in Alzheimer's disease. Hum. Brain Mapp. 2016;37(4):1321–1334. doi: 10.1002/hbm.23105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reginold W., Luedke A.C., Itorralba J., Fernandez-Ruiz J., Islam O., Garcia A. Altered superficial white matter on tractography MRI in Alzheimer's disease. Dementia Geriatr. Cognit. Disord. Extra. 2016;6(2):233–241. doi: 10.1159/000446770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guo J., Li B. The application of medical artificial intelligence technology in rural areas of developing countries. Health Equity. 2018;2(1):174–181. doi: 10.1089/heq.2018.0037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davis K.D., Flor H., Greely H.T., Iannetti G.D., Mackey S., Ploner M., et al. Brain imaging tests for chronic pain: medical, legal and ethical issues and recommendations. Nat. Rev. Neurol. 2017;13:624. doi: 10.1038/nrneurol.2017.122. [DOI] [PubMed] [Google Scholar]
- 7.Dyrba M., Ewers M., Wegrzyn M., Kilimann I., Plant C., Oswald A., et al. International Workshop on Multimodal Brain Image Analysis. Springer; 2012. Combining DTI and MRI for the automated detection of Alzheimer’s disease using a large European multicenter dataset; pp. 18–28. [Google Scholar]
- 8.Jin Z., Bao Y., Wang Y., Li Z., Zheng X., Long S., et al. Differences between generalized Q-sampling imaging and diffusion tensor imaging in visualization of crossing neural fibers in the brain. Surg. Radiol. Anat. 2019;41(9):1019–1028. doi: 10.1007/s00276-019-02264-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lim S.Y., Tyan Y.-S., Chao Y.-P., Nien F.-Y., Weng J.-C. New insights into the developing rabbit brain using diffusion tensor tractography and generalized q-sampling MRI. PLoS One. 2015;10(3) doi: 10.1371/journal.pone.0119932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yeh F.C., Liu L., Hitchens T.K., Wu Y.L. Mapping immune cell infiltration using restricted diffusion MRI. Magn. Reson. Med. 2017;77(2):603–612. doi: 10.1002/mrm.26143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Prasad G., Nir T.M., Toga A.W., Thompson P.M. 2013 IEEE 10th International Symposium on Biomedical Imaging. IEEE; 2013. Tractography density and network measures in Alzheimer's disease; pp. 692–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mheich A., Wendling F., Hassan M. 2019. Brain Network Similarity: Methods and Applications. arXiv preprint arXiv:190810592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sporns O. Graph theory methods: applications in brain networks. Dialogues Clin. Neurosci. 2018;20(2):111. doi: 10.31887/DCNS.2018.20.2/osporns. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sporns O., Zwi J.D. The small world of the cerebral cortex. Neuroinformatics. 2004;2(2):145–162. doi: 10.1385/NI:2:2:145. [DOI] [PubMed] [Google Scholar]
- 15.Nazeri A., Chakravarty M.M., Rajji T.K., Felsky D., Rotenberg D.J., Mason M., et al. Superficial white matter as a novel substrate of age-related cognitive decline. Neurobiol. Aging. 2015;36(6):2094–2106. doi: 10.1016/j.neurobiolaging.2015.02.022. [DOI] [PubMed] [Google Scholar]
- 16.Nazeri A., Chakravarty M.M., Felsky D., Lobaugh N.J., Rajji T.K., Mulsant B.H., et al. Alterations of superficial white matter in schizophrenia and relationship to cognitive performance. Neuropsychopharmacology: Off. Publ. Am. College Neuropsychopharmacol. 2013;38(10):1954. doi: 10.1038/npp.2013.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ribas G.C. The cerebral sulci and gyri. Neurosurg. Focus. 2010;28(2):E2. doi: 10.3171/2009.11.FOCUS09245. [DOI] [PubMed] [Google Scholar]
- 18.Talairach J. 1988. Co-planar Stereotaxic Atlas of the Human Brain-3-Dimensional Proportional System. An Approach to Cerebral Imaging. [Google Scholar]
- 19.Yu L., Liu H. Proceedings of the 20th International Conference on Machine Learning (ICML-03) 2003. Feature selection for high-dimensional data: a fast correlation-based filter solution; pp. 856–863. [Google Scholar]
- 20.Browne M.W. Cross-validation methods. J. Math. Psychol. 2000;44(1):108–132. doi: 10.1006/jmps.1999.1279. [DOI] [PubMed] [Google Scholar]
- 21.Zhu W., Zeng N., Wang N. Sensitivity, specificity, accuracy, associated confidence interval and ROC analysis with practical SAS implementations. NESUG Proceed.: Health Life Sci. (Baltimore, Maryland) 2010;19:67. [Google Scholar]
- 22.Van Hecke W., Emsell L., Sunaert S. Springer; 2015. Diffusion Tensor Imaging: a Practical Handbook. [Google Scholar]
- 23.Bigham B., Zamanpour S.A., Zemorshidi F., Boroumand F., Zare H. Alzheimer’s Disease Neuroimaging Initiative. Identification of superficial white matter abnormalities in Alzheimer’s disease and mild cognitive impairment using diffusion tensor imaging. J. Alzheimer’s Dis. Rep. 2020;4(1):49–59. doi: 10.3233/ADR-190149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang H., Wang Y., Lu T., Qiu B., Tang Y., Ou S., et al. Differences between generalized q-sampling imaging and diffusion tensor imaging in the preoperative visualization of the nerve fiber tracts within peritumoral edema in brain. Neurosurgery. 2013;73(6):1044–1053. doi: 10.1227/NEU.0000000000000146. [DOI] [PubMed] [Google Scholar]
- 25.Daianu M., Dennis E.L., Jahanshad N., Nir T.M., Toga A.W., Jack C.R., et al. 2013 IEEE 10th International Symposium on Biomedical Imaging. IEEE; 2013. Alzheimer's disease disrupts rich club organization in brain connectivity networks; pp. 266–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seo E.H., Lee D.Y., Lee J.-M., Park J.-S., Sohn B.K., Lee D.S., et al. Whole-brain functional networks in cognitively normal, mild cognitive impairment, and Alzheimer’s disease. PLoS One. 2013;8(1) doi: 10.1371/journal.pone.0053922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jalili M. Graph theoretical analysis of Alzheimer's disease: discrimination of AD patients from healthy subjects. Inf. Sci. 2017;384:145–156. [Google Scholar]
- 28.Sheng J., Wang B., Zhang Q., Zhou R., Wang L., Xin Y. Identifying and characterizing different stages toward Alzheimer's disease using ordered core features and machine learning. Heliyon. 2021 doi: 10.1016/j.heliyon.2021.e07287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sulaimany S., Khansari M., Zarrineh P., Daianu M., Jahanshad N., Thompson P.M., et al. Predicting brain network changes in Alzheimer's disease with link prediction algorithms. Mol. Biosyst. 2017;13(4):725–735. doi: 10.1039/c6mb00815a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yao Z., Zhang Y., Lin L., Zhou Y., Xu C., Jiang T., et al. Abnormal cortical networks in mild cognitive impairment and Alzheimer's disease. PLoS Comput. Biol. 2010;6(11) doi: 10.1371/journal.pcbi.1001006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.He Y., Chen Z., Evans A. Structural insights into aberrant topological patterns of large-scale cortical networks in Alzheimer's disease. J. Neurosci. 2008;28(18):4756–4766. doi: 10.1523/JNEUROSCI.0141-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.He Y., Chen Z., Gong G., Evans A. Neuronal networks in Alzheimer's disease. Neuroscientist. 2009;15(4):333–350. doi: 10.1177/1073858409334423. [DOI] [PubMed] [Google Scholar]
- 33.Ebadi A., Dalboni da Rocha J.L., Nagaraju D.B., Tovar-Moll F., Bramati I., Coutinho G., et al. Ensemble classification of Alzheimer's disease and mild cognitive impairment based on complex graph measures from diffusion tensor images. Front. Neurosci. 2017;11:56. doi: 10.3389/fnins.2017.00056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Desikan R.S., Cabral H.J., Fischl B., Guttmann C.R., Blacker D., Hyman B.T., et al. Temporoparietal MR imaging measures of atrophy in subjects with mild cognitive impairment that predict subsequent diagnosis of Alzheimer disease. Am. J. Neuroradiol. 2009;30(3):532–538. doi: 10.3174/ajnr.A1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu Y., Sun D., Wang Y., Wang Y., Wang Y. Tracing short connections of the temporo-parieto-occipital region in the human brain using diffusion spectrum imaging and fiber dissection. Brain Res. 2016;1646:152–159. doi: 10.1016/j.brainres.2016.05.046. [DOI] [PubMed] [Google Scholar]
- 36.Liu X., Chen X., Zheng W., Xia M., Han Y., Song H., et al. Altered functional connectivity of insular subregions in Alzheimer’s disease. Front. Aging Neurosci. 2018;10:107. doi: 10.3389/fnagi.2018.00107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data included in article/supplementary material/referenced in article.