Abstract
Leiomodin is an important emerging regulator of thin filaments. As novel molecular, cellular, animal model and human data accumulate, the mechanisms of its action become clearer. Structural studies played a significant part in understanding the functional significance of leiomodin’s interacting partners and functional domains. In this review we present the current state of knowledge on the structural and cellular properties of leiomodin which has led to two proposed mechanisms of its function. Although it is known that leiomodin is essential for life, numerous domains within leiomodin remain unstudied and as such, we outline future directions for investigations that we predict will provide evidence that leiomodin is a multi-functional protein.
In this review, we present the current knowledge on the role of leiomodin in thin-filament length regulation. The latest structural and biochemical findings are discussed within the context of two existing models for the mechanism of leiomodin function. We also outline future directions for research to reconcile the available experimental data in the literature.
Keywords: leiomodin, tropomodulin, actin, sarcomeres, tropomyosin
Graphical Abstract
Introduction
Sarcomeres, the repeating contractile units of striated muscle tissue, are impressively well organized. Despite the seemingly unchanging highly regular sarcomere structure, they are maintained by tightly regulated continuous dynamic processes ensuring the integrity of its protein components [1].
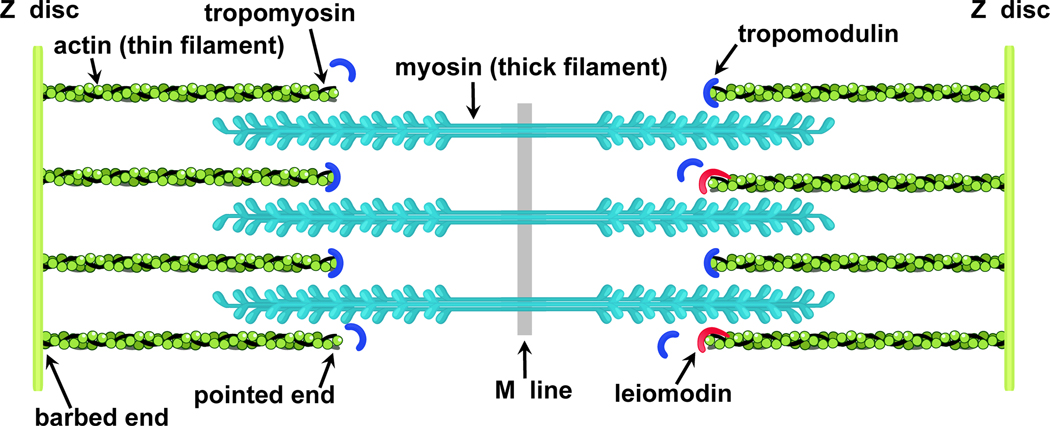
Thin and thick filaments, made predominantly from actin and myosin, respectively, are the main functional elements of sarcomeres. The filaments are overlapped (Fig. 1), and since the length of thick filaments is considered constant and measure at ~1.6 μm in both skeletal and cardiac muscles from different vertebrates [2], thin filament length, which varies in different muscle types and in different species, determines the effective overlap. According to the sliding filament theory [3], contraction is produced by relative sliding of the thin and thick filaments. Optimal overlap is required for efficient force generation, and dysregulation of the thin filament length is associated with severe cardiac and skeletal myopathies [4–8].
Figure 1.
A schematic view of the sarcomere showing actin (green), myosin (cyan), tropomysin (black), tropomodulin (blue) and leiomodin (red) (modified from [51]).
Understanding of the mechanisms that regulate thin filament length is undergoing rapid evolution [2]. New factors are continuously being identified, and roles of the previously known factors are being reassessed. Leiomodin (Lmod), which is a regulatory actin-binding protein involved in sarcomere assembly and organization [9], is one of such proteins. This is an exciting time for the field since new functions of this protein, that is essential for life, are emerging. In this review we will focus on the current state of knowledge on the functional roles of Lmod derived from a plethora of structural and biochemical investigations, as well as its involvement in thin filament assembly and maintenance.
Lmods are members of the leiomodin/tropomodulin protein family. They are represented by three isoforms, Lmod1-Lmod3, expressed mainly in muscle [10–12]. Lmod1 is found mostly in smooth muscle but also in cardiac muscle [11–13], and, surprisingly, in the central nervous system [14, 15], although its function there is unclear. Lmod2 is considered a predominantly cardiac protein [6, 9, 16], but it is also present in skeletal muscle [11, 17, 18]. Recently, Lmod2 has been found to be a critical component of a dual nebulin/Lmod2 thin-filament length-regulation mechanism in some skeletal muscle types [18]. Lmod3 is expressed primarily in skeletal and to some extent in cardiac muscle [4, 19–21]. Several recent reports showed that Lmod dysfunctions result in debilitating myopathies. More specifically, a homozygous premature termination codon in Lmod1 was demonstrated to lead to a visceral myopathy consistent with the megacystis microcolon intestinal hypoperistalsis syndrome [13]. Several Lmod3 mutations were linked to severe and often fatal nemaline myopathy [4, 22], whereas an Lmod2 biallelic mutation has been recently demonstrated to cause neonatal dilated cardiomyopathy [7].
The structural organization of Lmods is modular (Fig. 2), which plays an important part in how they perform their function realized through physical interaction with the thin filament. The modular architecture of Lmods and its relation to Lmod’s function is best visualized and understood when compared with that of a smaller homologous protein, tropomodulin (Tmod).
Figure 2.
A: A schematic view of Tmod and Lmod molecules B: A Clustal Omega multiple sequence alignment [74] of human Tmod1 (NP_001159588), Lmod1 (NP_036266.2), Lmod2 (NP_997046.1), and Lmod3 (NP_938012.2) showing in color TpmBS1 (cyan), ABS1 (green), TpmBS2 (gray), ABS2 (yellow), polyP (magenta), and WH2 (red).
Architecture and Function of Tropomodulins
Unlike Lmod, Tmod is an ubiquitous protein; its isoforms are expressed in both muscle and non-muscle cells [23–27]. Its main identified molecular function is to regulate actin filament length via capping the pointed end of the actin-thin filament (Fig. 1); that is, it prevents actin filament depolymerization and elongation [28, 29]. Tight interaction of Tmod with the actin-thin filament is achieved by simultaneous binding of four binding sites within Tmod to two actin plus two tropomyosin (Tpm) protomers at the pointed end [30–32]. The two Tpm- and two actin-binding sites (TpmBS and ABS, respectively) are localized to sequentially non-overlapping regions of Tmod, and they are numbered according to their position in the Tmod amino acid sequence as TpmBS1, ABS1, TpmBS2, and ABS2 (Fig. 2). Covering such an extensive interaction surface with a relatively small (~350 aa) Tmod molecule is only possible because the N-terminal half of Tmod containing TpmBS1, ABS1, and TpmBS2 is intrinsically disordered [33]. Only one binding site of Tmod, i.e. ABS2, is a folded globular domain [34] representing topologically a consensus leucine-rich repeat (LRR) [35]. The flexible nature of the N-terminal region allows Tmod to wrap around and effectively cover the relatively large surface of the pointed end of the actin filament [32, 36, 37].
Four Tmod isoforms (Tmod1-Tmod4) are present in vertebrates [23, 26, 38, 39]. The diversity of Tmod and Tpm isoforms (~40 in mammals) [40] combined with the wide range of isoform-specific Tmod affinities for Tpm [41] gave rise to a hypothesis that the variability in Tmod/Tpm binding strength is the basis of tissue-specific regulation of pointed-end capping [42, 43].
Architecture and Functions of Leiomodins
Compared to widespread Tmods, Lmods are more specialized and function as muscle proteins. While Lmod is located at the pointed end of thin filaments, unlike Tmod, localization of Lmod is not restricted to the pointed end. Lmod’s staining pattern suggests that it may also bind along thin filaments, next to the pointed end. Lmod is a potent nucleator of actin in biochemical assays and Lmod knockdown results in disrupted sarcomere assembly in cardiomyocytes [4, 9]. Therefore, it has been proposed that the main function of Lmod is that of a thin filament nucleator [9, 44–46]. On the other hand, the influence of Lmod expression level on pointed-end elongation [16, 47] and Lmod localization at or close to the pointed end [9, 16] in isolated myocytes and in hearts in vivo suggested that Lmod acts as a pointed-end regulator (elongator) of thin filament length [16, 18, 47–49]. Additionally, recent findings suggested that Lmod can also have a role as a regulator of myosin activity [50], but this possibility has not been extensively studied. Although these three functions are not mutually exclusive, there is no consensus on which of these play essential roles in vivo.
Clarifying the true place of Lmod in the complex process of muscle actin dynamics starts with understanding of its modular molecular structure and its interaction with binding partners, which is discussed below. Similarly to Tmod, Lmod is an intrinsically disordered modular protein built from distinct functional segments [51]. However, the homology between Lmods and Tmods is shared for some regions but not others (Fig. 2, Table 1), which makes drawing conclusions on Lmod’s function, based solely on homology, unreliable. For example, Lmods bind to Tpm via a TpmBS1 site but they do not have a TpmBS2 site [52, 53], whereas both TpmBS1 and TpmBS2 sites are important for Tmod’s capping function [31, 32]. Additionally, the assignment of binding sites and identification of interacting partners of Lmod and Tmod is incomplete which has resulted in disputes about how, where, when and for what purpose Lmods interact with the thin filament [44, 46, 51, 53].
Table 1.
Pairwise sequential homology percentages for TpmBS1, ABS1, and ABS2 regions of human Tmod1 (NP_001159588), Lmod1 (NP_036266.2), Lmod2 (NP_997046.1), and Lmod3 (NP_938012.2). Sequence alignment and calculations of homology percentages were performed using an online pairwise sequence alignment tool EMBOSS-Needle [74]. The percentages are shown for identical/similar residues.
| Lmod1 TpmBS1 | Lmod2 TpmBS1 | Lmod3 TpmBS1 | |
|---|---|---|---|
| Tmod1 TpmBS1 * | 38/55% | 46/68% | 33/58% |
| Lmod1 TpmBS1 | 48/67% | 30/56% | |
| Lmod2 TpmBS1 | 37/54% | ||
| Lmod1 ABS1 | Lmod2 ABS1 | Lmod3 ABS1 | |
| Tmod1 ABS1 * | 38/58% | 42/58% | 41/59% |
| Lmod1 ABS1 | 45/76% | 28/66% | |
| Lmod2 ABS1 | 46/70% | ||
| Lmod1 ABS2 | Lmod2 ABS2 | Lmod3 ABS2 | |
| Tmod1 ABS2 * | 44/64% | 49/69% | 43/66% |
| Lmod1 ABS2 | 67/83% | 60/77% | |
| Lmod2 ABS2 | 62/77% |
For calculations, we used Tmod1 amino acids 1–38 (TpmBS1), 43–88 (ABS1), and 179–349 (ABS2).
Tropomyosin-binding site.
Lmods have only one of the Tpm-binding sites (TpmBS1) present in Tmods [52, 53] (Fig. 2A). The existence of this Tpm-binding site was first assumed based on sequence homology with Tmods and later confirmed in binding assays [10, 52]. The affinities of peptides corresponding to TpmBS1 in Lmods and Tmods for long Tpm isoforms are comparable [4, 41, 52, 54].
Sequences of TpmBS1 in Tmods and Lmods are highly conserved and are expected to share the same mode of binding to Tpm across different Tmod and Lmod isoforms (Fig. 2B). Until very recently, that mode of binding remained largely unknown. The first model of TpmBS1/Tpm complex was proposed as early as 2007 by comparing the [15N-1H]- heteronuclear single quantum coherence (HSQC) nuclear magnetic resonance (NMR) spectra of an 15N-labeled coiled-coil model Tpm peptide in the presence and absence of TpmBS1 from Tmod1 [32]. The model Tpm peptide contained 19 N-terminal residues of a non-muscle Tpm fused at the C-terminus with a part of the 33-residue leucine-zipper coiled-coil domain of the yeast transcription factor GCN4 [55]. The GCN4 fragment sequence used in the study promoted formation of a stable coiled coil. Fusing GCN4 and Tpm fragment sequences is a typical approach to recreate a coiled-coil Tpm conformation in shorter Tpm fragments (e.g. [56, 57]).
The observed spectral changes indicated that a 38-residue TpmBS1 peptide was binding to the 19 N-terminal residues of Tpm, while the following GCN4 residues were not affected. Since no non-specific binding between TpmBS1 and GCN4 residues was detected in the complex, it was proposed that the mode of TpmBS1 binding represented a hairpin-like structure, with the middle of the TpmBS1 being on top of the Tpm N-terminus [32]. In the absence of more detailed structural information, another mode of interaction was proposed where the orientation of the TpmBS1 “hairpin” was inverted, with the tip of the clamp pointing towards the C-terminus of Tpm [36].
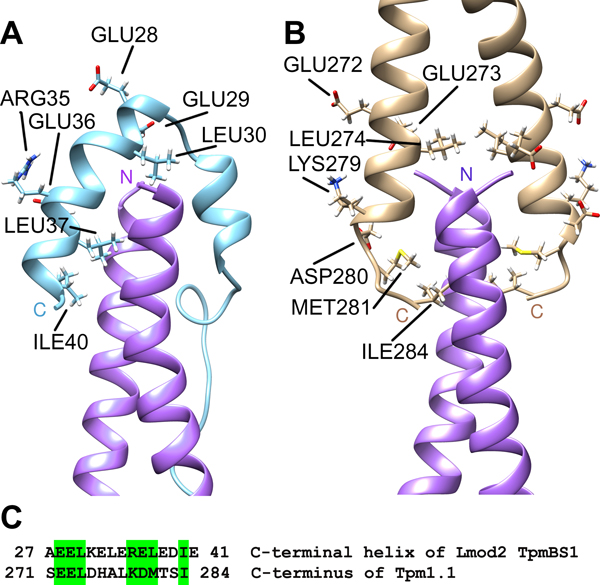
The question about the mode of binding between Tpm and Lmod TpmBS1 has been recently resolved [48]. This was accomplished by utilizing three-dimensional (3D) NMR spectral analysis and NMR-guided molecular dynamics simulations to generate a 3D model of a complex between Lmod2 TpmBS1 and the N-terminus of striated muscle Tpm. The model was validated (1) by residual dipolar coupling data; this approach reports information on internuclei vector orientations [58], and (2) by spectral effects of amino acid substitutions in Tpm. The Lmod2 TpmBS1 folds into an α-helical hairpin, which is positioned over the N-terminal side of the Tpm coiled coil to form a four-helix bundle (Fig. 3). In the bundle, two helices of Lmod occupy opposite corners of the bundle thus generating a crisscross topology similar to that of the head-to-tail (N-terminus to C-terminus) Tpm overlap complex [56, 59, 60] (Fig. 3).
Figure 3.
Comparison of the complex of Lmod2 TpmBS1 with the N-terminus of striated muscle tropomyosin Tpm1.1 (PDB ID 6UT2) (A) and the head-to-tail Tpm1.1 overlap junction (PDB ID 2G9J) (B). The sequence alignment of the structurally homologous parts of the C-terminal helix of Lmod2 TpmBS1 and the C-terminus of Tpm 1.1 (C). Similar residues are marked in green on the sequence and their side chains are dispayed on the 3D structures. Helices of the N-terminal Tpm1.1 peptides in (A) and (B) are shown in magenta. Modified from [48].
The immediate implication of the obtained structure is that the Lmod TpmBS1 can only be bound to an unobstructed N-terminus of Tpm, which is only located near the pointed end of the thin filament, near the M-line [48]. The N-termini of other actin-bound Tpm molecules form the head-to-tail overlap complexes and are therefore blocked by the C-termini of adjacent Tpm molecules inside the continuous helical Tpm cable spanning the entire filament [61]. The broader meaning of these and other structural results will be discussed below in a separate section summarizing proposed Lmod functions and mechanisms of action.
The importance of TpmBS1 in thin filament nucleation remains unclear. Direct quantitative evaluation of the effects of TpmBS1 on the nucleation is challenging because Tpm inhibits actin nucleation in a dose-dependent manner [45, 62]. Skwarek-Maruszewska and co-authors compared rates of actin nucleation induced by either full-length Lmod or by truncated Lmod lacking the TpmBS1 region [45]. At 1 μM Tpm (which approximately corresponds to a dissociation constant, Kd, value for the TpmBS1/Tpm complex [52, 63]) the presence of TpmBS1 was shown to improve nucleation by 30–40 %. However, at slightly higher Tpm concentrations, Tpm inhibits nucleation to the point where experimental errors make it hard to discriminate between nucleation rates in the presence of full-length and truncated Lmod [45]. The effects of TpmBS1 truncation were also small at Tpm concentrations below 1 μM, which is not surprising because if the Tpm concentration is lower than the Kd value, then the population of seeds bound to Tpm via TpmBS1 is smaller than the population of those without Tpm. Therefore,Lmod TpmBS1 appears to promote Tpm attachment only moderately and only in a very narrow range of Tpm concentrations. Additionally, only one Tpm molecule can be recruited by Lmod vs two Tpm molecules required for two sides of a mature thin filament. To summarize, Tpm binding to Lmod is not likely to be essential for Lmod-induced thin-filament nucleation. On the other hand, a single-point mutation reducing TpmBS1 binding to Tpm virtually destroyed the ability of exogenous Lmod2 to make thin filaments longer in cardiomyocytes [48]. This result confirmed that Lmod is part of another regulatory mechanism for which TpmBS1 binding to Tpm is critical.
Actin-binding site 1.
Attempts to map the ABS1 site of Lmod based on its sequential homology with that of Tmod led to conflicting results. The Lmod and Tmod regions of ~55–60 residues immediately following the TpmBS1 show a considerable degree of similarity (Fig. 2, Table 1), suggesting that both of them (residues Pro42–Lys96 of human Lmod2 and residues Pro39–Lys99 of human Tmod1) might bind actin, with the majority of these homologous residues directly involved in the interaction. Nevertheless, a crystal structure of a complex between G-actin and the human Tmod1 region encompassing residues Gln50–Lys101 fused with human gelsolin segment 1 (GS1, residues 52–176) showed that only residues Pro58–Lys99 form contacts with actin [36]. The residues Gln50–Ala57 in the studied peptide were not visible, and therefore it was concluded that the entire N-terminal end of the putative Tmod1 ABS1 region (Pro39–Ala57) does not interact with actin. Residues Arg64–Glu77 formed a helix important for binding. A secondary-structure prediction matched this experimental result reasonably well by identifying residues Arg64–Ala75 as α-helical residues [51].
The dissociation constant (Kd) of the complex between G-actin and the fragment Gln50–Lys101 of Tmod1 was in the low μM range (~8 and ~9–12 μM for ATP-G-actin and ADP-G-actin, respectively) as detected by isothermal titration calorimetry (ITC) [46]. Since the identification of the structure of a complex between G-actin and Lmod ABS1 has not been done so far, the structural results obtained for Tmod ABS1 were extended to Lmod. The Lmod residues corresponding to the Tmod1 residues Pro39–Ala57 were considered irrelevant for actin binding and excluded from the Lmod fragment tested by ITC for actin interactions [46]. Rather surprisingly, unlike the corresponding Tmod region, the Lmod ABS1 region lacking the N-terminal end did not interact with actin at a detectable level. As a result, a conclusion was made that the ABS1 site might not exist in Lmod altogether [44, 46].
Contrary to these conclusions described in the paragraph above, when the N-terminal end of the Lmod ABS1 homology region was included in the analyses, the binding of Lmod ABS1 to actin was detected by three independent techniques. More specifically, N-terminal fragments of Lmod2 containing ABS1 (but not other ABSs) were shown to inhibit actin pointed-end polymerization in a pyrene-actin polymerization assay [53]. In the same work, an Lmod2 fragment corresponding to ABS1 (Asn45–Glu94) was shown to bind to G-actin in an NMR titration assay. In another study, interactions of Lmod2 with G-actin were quantified by atomic force microscopy [64]. The distribution of an unbinding force measured when the full-length Lmod2 was pulled away from G-actin revealed three distinct unbinding events – manifested as three well-resolved force peaks. Each of the three unbinding force peaks was assigned to a particular Lmod2 region by testing interactions of actin with a set of truncated Lmod2. If an interacting binding site was removed by truncation, the corresponding unbinding force peak was not observed. Using this approach, the N-terminal Lmod2 fragment 1–201 was shown to be responsible for one of the force peaks, consistent with the existence of ABS1 within this region.
To summarize, these studies suggest that ABS1 is present in Lmods, and it is homologous to that found in Tmods. However, there are differences between the way Lmod and Tmod ABS1s bind to actin. In Tmods, ABS1 binding is dominated by interactions of the central α-helix (Arg64–Glu77 helix in human Tmod1), whereas in Lmods the N-terminal end of the ABS1 (corresponding to the residues N45LPVGQR51 of chicken Lmod2) region plays an important role adding to the strength of interactions between ABS1 and actin. Consistently, this region is more conserved among both Tmod and Lmod isoforms than the part corresponding to the central helix [46, 53]. Weaker interactions of the central helix with actin in Lmods might be important for a “swinging gate” mode of action of Lmod, allowing it to function as a “leaky cap” at the pointed end. The detailed description of the “swinging gate” mechanism is provided below in the Conclusions section.
The inability to detect an interaction of the N-terminal part of the Tmod ABS1 fragment with actin in the crystal structure can be explained by the fact that, for crystallization, the chosen fragment(1) lacked residues 43– 49, and (2) was fused at the N-terminus with a gelsolin fragment via a 9-aa flexible linker to ensure binding to actin. The gelsolin fragment binds at the opposite side of actin monomer, keeping the linker and residues 50–57 of Tmod1 in a fully extended conformation preventing any possible interactions with actin via this region (for more details see [51]).
Linker between actin-binding site 1 and actin-binding site 2.
ABS1 in Lmod is followed by a long stretch of amino acids connecting ABS1 with ABS2 (Fig. 2B). The linker is likely to be disordered because of the high content of negatively charged amino acid residues. In Tmods, this region houses TpmBS2, which is absent in Lmods [53]. Currently, the function of this region, other than being a linker between ABS1 and ABS2, is unknown. It is predicted, however, to play an as yet unknown auxiliary role because its length significantly exceeds the length required to connect the two ABSs [48].
Actin-binding site 2 (LRR domain).
The crystal structure of Lmod ABS2, alone and bound to actin [46, 65, 66], is structurally homologous to that of Tmod ABS2 [34, 36]. Like the ABS2 in Tmods, it is also folded as a typical LRR, but it misses an N-terminal segment that in Tmods interacts with the DNase I-binding loop in actin [36]. The N-terminal segment of Tmod ABS2 is conserved in Tmods, but not in Lmods [44]. Also, residues from this region in Lmod are not structurally defined in the X-ray structure of the complex between Lmod ABS2 and G-actin [65, 66], suggesting that they are not constrained by direct interactions with actin. In Tmods, this segment is essential for Tmod capping function because it blocks intrastrand actin-actin interactions. Therefore, Lmod ABS2, unlike a Tmod ABS2, is predicted to not compete with another actin subunit for binding at the pointed end. In other words, intrastrand interactions between actin subunits should not interfere with the binding of Lmod ABS2 to actin.
The first in vitro evidence suggesting that ABS2 can bind to the side of an actin filament came from sedimentation data. In sedimentation assays, Lmod and F-actin cosedimented at Lmod/actin ratios considerably exceeding the ratio that can be explained by binding of Lmod only at the pointed ends of the filaments [45, 50, 67]. Since ABS1 is considered specific for the pointed end, while ABS3 (which will be discussed later) is considered specific for the barbed end of F-actin, the only candidate domain left to mediate this binding is seemingly ABS2.
Surprisingly, binding of a truncated version of Lmod lacking the entire C-terminal extension to F-actin was much weaker than binding of full length Lmod to F-actin in cosedimentation assays [45]. The C-terminal-truncated version of Lmod used in the assay included ABS1 and ABS2, but not the C-terminal extension missing in Tmods. From this experiment, it can be ascertained that the presence of the C-terminal extension is required for formation of a sufficiently long-lived Lmod/F-actin complex that can be readily observed in cosedimentation assays. Moreover, truncated Lmod localized in cells similar to Tmod1 as a sharp band exclusively at the pointed end, suggesting that no side-binding was occuring [16].
Interestingly, low-speed sedimentation experiments and visualization of actin filaments by transmission electron microscopy demonstrated that Lmod side-binding to F-actin induces bundling of actin filaments [67]. Centrifugation at low speed sedimented filament bundles more readily than isolated filaments, and the amount of actin that was spun down was highly dependent on the presence of Lmod. These experiments not only confirmed the in vitro side binding of Lmod to F-actin, but also suggested that more than one Lmod ABS is involved in the interactions, providing non-covalent cross-linking of individual actin filaments.
Lmod-dependent actin filament bundling in a test tube might not be physiologically relevant, but may be a manifestation of the di- or polyvalent nature of Lmod binding to the sides of F-actin. Although ABS2 is capable of actin nucleation [46] and could, in principle, link two filaments, results of the cosedimentation assays with the truncated Lmod [45] showed that ABS2 alone does not bind to the filament strongly enough for this binding to be detected in the assays. It is reasonable to assume that simultaneous binding of ABS2 and at least one other actin-binding site located in the C-terminal part of Lmod enhances Lmod interactions with the sides of F-actin through a so-called avidity effect. The biochemical results on side binding of Lmod are consistent with the diffuse localization of Lmod near the pointed end in sarcomeres [45]. However, as discussed above, binding of Lmod to Tpm via TpmBS1 can only happen at the pointed end.
Linker between ABS2 and ABS3.
ABS2 Lmods have an ~20 kDa extension with the actin-binding site 3 (ABS3) located at the C-terminus of the extension (Fig. 2). The extension is not present in Tmods in which ABS2 is the last C-terminal domain. The linker between ABS2 and ABS3 is not conserved between different Lmod isoforms and it is most likely disordered, since no electron density was seen for this region in a crystal structure of Lmod2 that revealed the positions of both ABS2 and ABS3 [65, 66]. Currently, nothing has been reported about the function of the linker between ABS2 and ABS3.
The linker contains a polyproline region (polyP) which varies in length depending on the Lmod isoform and the studied species (from 12–18 consecutive Pro residues in Lmod2 from different species to ~7 Pro residues in Lmod1). It is known that profilin, another actin-binding protein, can bind to polyP regions in other proteins. A polyP region should contain a minimum of 5 consecutive Pro residues and no bulky side chains of neighboring residues to bind profiin [68–70]. It was therefore tempting to hypothesize that profilin binds to Lmod and affects its function. However, based on conclusions reported by Chereau and co-authors, profilin does not bind to Lmod because it had no effect on nucleation by Lmod2 [9]. In these experiments, profilin was added to a mixture of G-actin and Lmod2 fragment (res. 162–495), containing polyP or with polyP replaced with Gly-Ser repeats (named polyP-GS). For both fragments, profilin caused a decrease in Lmod’s nucleation due to its actin-sequestering ability. The authors concluded that the decrease in nucleation was the same for the fragment containing the polyP region and for the fragment polyP-GS, and therefore, neither of the Lmod2 fragments binds profilin. Although not discussed, it is surprising that polyP-GS had a very low nucleation effect compared to the wild-type fragment [9]. In addition, the Lmod2 version used in this study contained only 8 Pro (res 440–449 of 495), while the longest uninterrupted polyP sequences in Lmod2 from different sources vary between 12 and 18 residues. As such, it is possible that this polyP region was not long enough to bind profilin.
Experiments studying the effects of profilin on nucleation by Lmod1 were also performed [46]. In these studies, nucleation by full-length Lmod1 and the ABS2 fragment lacking both the N-terminal and the C-terminal regions were compared in the presence of profilin causing a concentration-dependent decrease in nucleation. The authors concluded that profilin does not bind to Lmod1; however, the concentration effects of profilin were not linear for both proteins rendering the conclusion unreliable.
Based on the data from both manuscripts, it cannot be concluded with certainty that profilin does not bind to Lmod’s polyP region. More experiments including direct studies of binding between Lmod and profilin are necessary to clarify the role of this region.
Actin-binding site 3 (WASP-Homology 2 domain).
The third known actin-binding site in Lmods (ABS3) is located in the C-terminal extension, which is absent in Tmods (Fig. 2). ABS3 represents a WASP-Homology 2 (WH2) domain which often functions as an actin-recruiting domain within actin nucleators [71, 72]. The Lmod2 WH2 domain binds between subdomains 1 and 3 of an actin subunit, in the barbed-end groove [65]. The Lmod2 WH2 domain binding to actin creates steric hindrance for the adjacent intrastrand actin molecule. The WH2 domain/actin complex is not likely to be stable in the middle of the F-actin filament and therefore is predicted to be observed at the barbed end of the filament [72].
Initially, on the basis of its ability to bind actin, the Lmod WH2 domain was assigned a critical role in actin nucleation and formation of an F-actin seed [9]. A recombinant C-terminal-truncated version of Lmod devoid of the WH2 domain is a markedly less potent nucleator than the full-length Lmod. It inhibits pointed-end polymerization of actin in a Tpm-dependent manner [16], much like Tmod does, but with one important difference. The inhibition by the Lmod fragment is only partial; in the presence of saturating concentrations of Lmod elongation of the pointed end is still allowed although at a slower rate. Once the ability of ABS2 to promote nucleation by itself was established, the role of the WH2 domain in nucleation was reassigned to auxiliary [46]. Moreover, further studies of truncated Lmod versions in actin nucleation assays led to the conclusion that the influence of the whole C-terminal extension on nucleation is larger than that provided by the WH2 domain alone [65].
Conclusions: Physiological Roles of Lmod and Future Directions
Currently, there is no consensus on what Lmod does in cells and what it does not do. The available data suggest that it serves as a powerful actin nucleator and a specific pointed-end regulator (elongator) of thin-filament length. As Lmod’s biochemical and in-cell properties are better characterized, additional functionalities of Lmods are predicted to emerge. The existence of many structural similarities between functionally different Tmods and Lmods adds to the confusion of deciphering their physiological roles. Available biological, biochemical and structural information about Tmods is consistent and defines it as a pointed-end cap protein. With Lmods, the picture is not nearly as clear. In fact, it is complex.
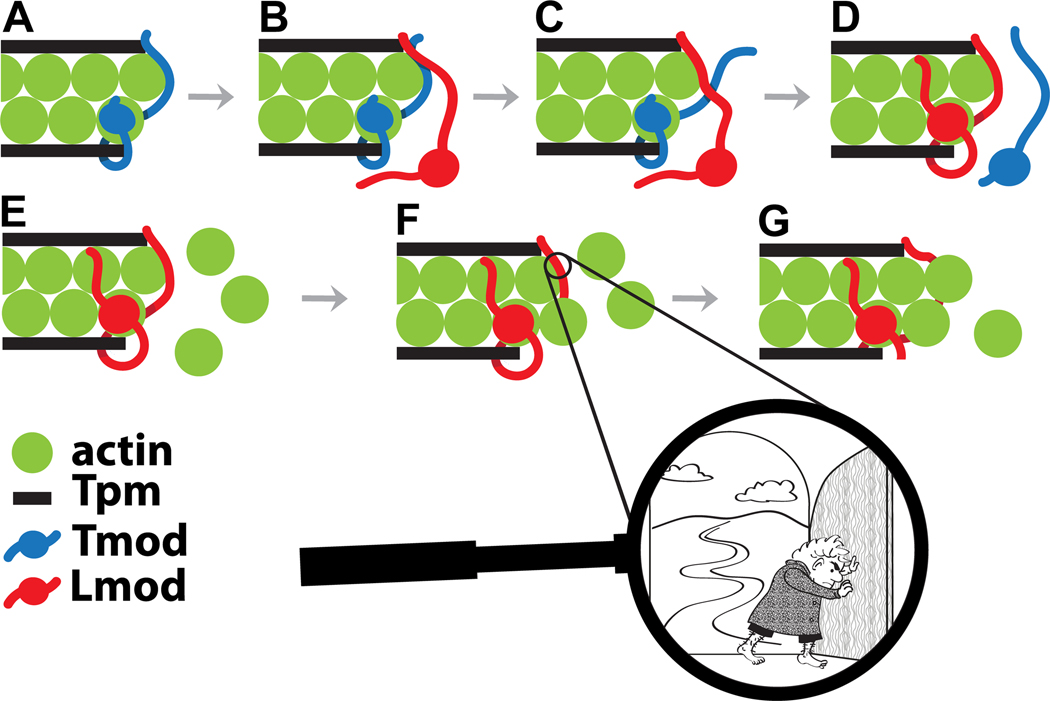
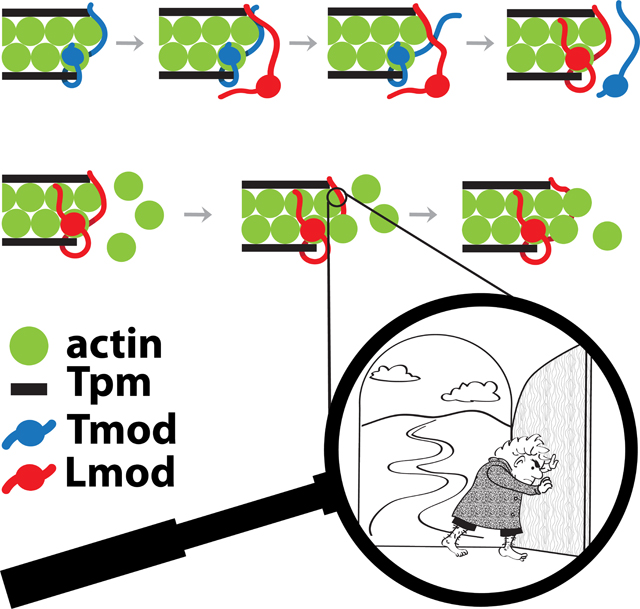
Binding of Lmod to the pointed ends of mature thin filaments and its competitive displacement of Tmod is one of the theories proposed for the mechanism of pointed end thin filament length regulation. The ability of Lmod to displace Tmod from the pointed end of actin filaments and the effect of Tpm on this process was shown in co-sedimetation experiments [54] and by high-resolution immunofluorescence imaging [16]. According to the competition model, Lmod binds to the pointed end via three of its domains: TpmBS1, ABS1 and ABS2; these interactions displace Tmod from the pointed end, allowing thin-filament elongation [16] (Fig. 4). The kinetic rate of Lmod/Tmod exchange can be quite fast due to the instrinsically disordered nature and multi-site structural organization of Tmods and Lmods. Indeed, the mechanisms of dissociation of polyvalent and monovalent non-covalent complexes are different [73]. For a polyvalent intrinsically disordered protein, each binding site might bind/unbind comparatively independently. Then, in the presence of a competitor, stepwise “Velcro-like” unbinding events accelerate the kinetics of dissociation of a multivalent complex considerably [73] – this is what we predict is the mechanism of thin filament regulation by Lmod and Tmod.
Figure 4.
Putative schematic steps of competitive displacement of Tmod by Lmod followed by Lmod-controlled elongation at the pointed-end. The displacement of Tmod (shown in blue) is initiated by a Lmod molecule (shown in red) approaching the Tmod-capped pointed end (A). Sequential binding of Lmod’s TpmBS1 (B), ABS1 (C) and ABS2 (D) removes Tmod from the pointed end. The pointed end occupied by Lmod (E) can now bind G-actin molecules (shown in green) in the process of slow pointed-end elongation (F, G). In the first step of Lmod-dependent elongation, an actin monomer attaches to the filament strand interacting with the Lmod ABS2 (F). This allows a conformational change of the Lmod ABS1 and opens up a “gate” for further elongation (G). Magnified: what might be a molecular mechanism allowing this elongation?
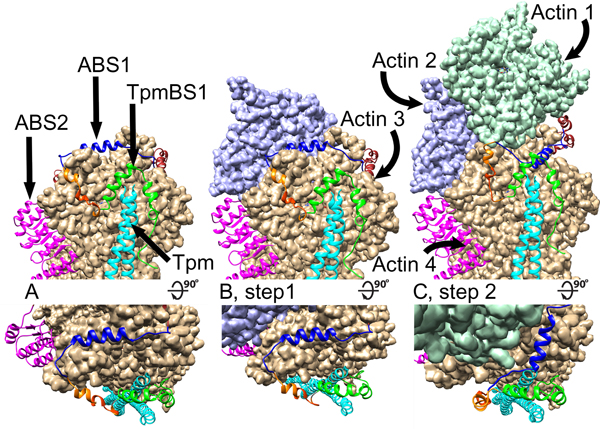
A recent publication sheds more light on the specific mechanism of thin filament elongation upon Lmod binding in the competition model [48]. We can envision that intrinsically disordered regions of Lmod anchors to the pointed end of the thin filament via TpmBS1 and displaces Tmod by stepwise engaging ABS1 and ABS2 in the interactions (Fig. 4). In this scenario, the ABS1 and TpmBS1 subdomains of Lmod serve as specificity determinants for Tmod displacement at the pointed end. Once Lmod removes Tmod and settles on the pointed end, the suggested ability of Lmod ABS2 to bind to the side of thin filament without obstructing the pointed end, as well as weaker interactions of the Lmod ABS1 central helix with the pointed end makes the central helix act as a “swinging gate” [48]. This allows limited actin polymerization to occur from the actin filament pointed end (Fig. 4). The “swinging gate” molecular mechanism of the competition model is depicted in more details in Figure 5. The pointed end with bound Lmod is shown in Figure 5A. Next, the actin subunit 2 attaches to the unobstructed pointed-end side of actin subunit 4 (Fig. 5B). At this step ABS1 is in a “closed gate” position over the pointed-end side of actin subunit 3. As the next step (Fig. 5C)), since the ABS1 helix is not forming very efficient contacts for binding, it swings away, now adopting an “open gate” conformation to allow actin subunit 1 to bind to the pointed-end side of actin subunit 3. Interstrand interactions between actin subunits 1 and 2 facilitate the transition from the “closed gate” to “open gate” conformations of Lmod. In the following step, six more subunits of actin attach to each strand of the filament. This step is concluded upon binding of two protomers of Tpm making the pointed end available for association with either Lmod or Tmod. This mechanism accounts for the observed diffuse nature of Lmod localization at the pointed end because Lmod might remain attached to the filament via side binding.
Figure 5.
A “swinging gate” 3D model for the leaky cap created by Lmod2 binding at the pointed end of the thin filament (modified from [48]). For each pointed-end elongation step (labeled as A, B and C) shown are a side view (top panel) and a view from the top (bottom panel) of the Lmod/pointed end assembly. (A) The initial position of Lmod2 assembled at the pointed end. ABS1 is in a “closed gate” conformation with the blue helix of ABS1 obstructing the attachment of an actin molecules to actin 3. Tpm is shown in cyan, ABS2 is shown in magenta, and Lmod2 TpmBS1 is shown in green. (B) Step 1, an actin molecule 2 (shown in purple) attaches to actin 4. Actin molecule 2 does not interfere with the Lmod2 binding, and the ABS1 of Lmod2 remains in the “closed gate” conformation. (C) An actin molecule 1 (shown in turquoise) attaches to actin 3. Interstrand interactions between actin molecules 1 and 2 facilitate displacement of Lmod2 ABS1 helix, and ABS1 adopts an “open gate” conformation (the blue helix moves away from the point of intrachain actin-actin interaction interface). This allows further actin polymerization until new tropomyosin N-termini are available for Lmod or Tmod attachment.
The roles of Lmod as an actin nucleator and the pointed-end competitor are linked to specific domains within the Lmod molecule; however, functions of some Lmod regions remain unexplained. For example, the linker between ABS1 and ABS2 is much longer than what is required for unobstructed arrangement of Lmod binding sites at the pointed end of the mature thin filament or during nucleation. The role of the C-terminal extension that includes the polyP region and the WH2 domain is also not clear. In cells, while the full length Lmod2 promotes thin filament elongation, the Lmod2 fragment devoid of the WH2 domain shortens thin filaments [16]. This observation suggests a critical role of WH2 in thin-filament length regulation. However, it is not understood yet how WH2 interaction with the barbed-end groove fits into the pointed-end binding of Lmod. The answer to these apparent inconsistencies suggest that Lmod is a multifunctional protein with some of its domains serving distinct functions. For example, Lmod can be involved in regulatory processes that are not yet sufficiently studied, such as Lmod-dependent regulation of myosin activity during the cross-bridge cycle [50] or regulation of thin filament maintenance [48]. Exploring these potential functional roles are some of the promising directions for future investigations
A deeper understanding of Lmod functions can be achieved by gaining structural knowledge on the interactions of full-length Lmod with thin filaments. All of the currently known structural data have been obtained by X-ray and NMR using, due to the methodological limitations, complexes between Lmod fragments and monomeric actin or Tpm fragments [48, 65] rather than complexes between full-length proteins and F-actin. Currently, the field demands engaging other structural methods, such as super-resolution cryo-EM, to fully understand modes of Lmod binding to actin filaments and molecular mechanisms of its roles in the sarcomere. Lastly, the recent identifications of devastating disease-associated mutations in human Lmod1, Lmod2 and Lmod3 outlined in the Introduction section [4, 7, 13, 22] provide a new avenue for modeling human disease and the functional properties of the Lmods therein.
Acknowledgements.
These studies were supported by RO1 grants from the National Institutes of Health GM120137 and HL123078. Authors thank Julia Bolchakova for help with illustrations.
Abbreviations:
- Lmod
leiomodin
- Tmod
tropomodulin
- Tpm
tropomyosin
- LRR
leucin-rich repeat
- ABS
actin-binding site
- TpmBS
tropomyosin-binding site
- NMR
nuclear magnetic resonance
Footnotes
Conflicts of interest: none
References
- 1.Martin TG & Kirk JA (2020) Under construction: The dynamic assembly, maintenance, and degradation of the cardiac sarcomere, J Mol Cell Cardiol. 148, 89–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tskhovrebova L. & Trinick J. (2017) Titin and Nebulin in Thick and Thin Filament Length Regulation, Subcell Biochem. 82, 285–318. [DOI] [PubMed] [Google Scholar]
- 3.Huxley HE (2004) Fifty years of muscle and the sliding filament hypothesis, Eur J Biochem. 271, 1403–1415. [DOI] [PubMed] [Google Scholar]
- 4.Yuen M, Sandaradura SA, Dowling JJ, Kostyukova AS, Moroz N, Quinlan KG, Lehtokari VL, Ravenscroft G, Todd EJ, Ceyhan-Birsoy O, Gokhin DS, Maluenda J, Lek M, Nolent F, Pappas CT, Novak SM, D’Amico A, Malfatti E, Thomas BP, Gabriel SB, Gupta N, Daly MJ, Ilkovski B, Houweling PJ, Davidson AE, Swanson LC, Brownstein CA, Gupta VA, Medne L, Shannon P, Martin N, Bick DP, Flisberg A, Holmberg E, Van den Bergh P, Lapunzina P, Waddell LB, Sloboda DD, Bertini E, Chitayat D, Telfer WR, Laquerriere A, Gregorio CC, Ottenheijm CA, Bonnemann CG, Pelin K, Beggs AH, Hayashi YK, Romero NB, Laing NG, Nishino I, Wallgren-Pettersson C, Melki J, Fowler VM, MacArthur DG, North KN & Clarke NF (2014) Leiomodin-3 dysfunction results in thin filament disorganization and nemaline myopathy, The Journal of clinical investigation. 124, 4693–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Winter JM, Joureau B, Lee EJ, Kiss B, Yuen M, Gupta VA, Pappas CT, Gregorio CC, Stienen GJ, Edvardson S, Wallgren-Pettersson C, Lehtokari VL, Pelin K, Malfatti E, Romero NB, Engelen BG, Voermans NC, Donkervoort S, Bonnemann CG, Clarke NF, Beggs AH, Granzier H. & Ottenheijm CA (2016) Mutation-specific effects on thin filament length in thin filament myopathy, Annals of neurology. 79, 959–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pappas CT, Mayfield RM, Henderson C, Jamilpour N, Cover C, Hernandez Z, Hutchinson KR, Chu M, Nam KH, Valdez JM, Wong PK, Granzier HL & Gregorio CC (2015) Knockout of Lmod2 results in shorter thin filaments followed by dilated cardiomyopathy and juvenile lethality, Proceedings of the National Academy of Sciences of the United States of America. 112, 13573–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ahrens-Nicklas RC, Pappas CT, Farman GP, Mayfield RM, Larrinaga TM, Medne L, Ritter A, Krantz ID, Murali C, Lin KY, Berger JH, Yum SW, Carreon CK & Gregorio CC (2019) Disruption of cardiac thin filament assembly arising from a mutation in LMOD2: A novel mechanism of neonatal dilated cardiomyopathy, Science advances. 5, eaax2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ottenheijm CA, Witt CC, Stienen GJ, Labeit S, Beggs AH & Granzier H. (2009) Thin filament length dysregulation contributes to muscle weakness in nemaline myopathy patients with nebulin deficiency, Hum Mol Genet. 18, 2359–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chereau D, Boczkowska M, Skwarek-Maruszewska A, Fujiwara I, Hayes DB, Rebowski G, Lappalainen P, Pollard TD & Dominguez R. (2008) Leiomodin is an actin filament nucleator in muscle cells, Science. 320, 239–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Conley CA (2001) Leiomodin and tropomodulin in smooth muscle, Am J Physiol Cell Physiol. 280, C1645–56. [DOI] [PubMed] [Google Scholar]
- 11.Conley CA, Fritz-Six KL, Almenar-Queralt A. & Fowler VM (2001) Leiomodins: larger members of the tropomodulin (Tmod) gene family, Genomics. 73, 127–39. [DOI] [PubMed] [Google Scholar]
- 12.Nanda V. & Miano JM (2012) Leiomodin 1, a New Serum Response Factor-dependent Target Gene Expressed Preferentially in Differentiated Smooth Muscle Cells, Journal of Biological Chemistry. 287, 2459–2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Halim D, Wilson MP, Oliver D, Brosens E, Verheij JB, Han Y, Nanda V, Lyu Q, Doukas M, Stoop H, Brouwer RW, van IWF, Slivano OJ, Burns AJ, Christie CK, de Mesy Bentley KL, Brooks AS, Tibboel D, Xu S, Jin ZG, Djuwantono T, Yan W, Alves MM, Hofstra RM & Miano JM (2017) Loss of LMOD1 impairs smooth muscle cytocontractility and causes megacystis microcolon intestinal hypoperistalsis syndrome in humans and mice, Proceedings of the National Academy of Sciences of the United States of America. 114, E2739–E2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnson TP, Tyagi R, Lee PR, Lee M-H, Johnson KR, Kowalak J, Elkahloun A, Medynets M, Hategan A, Kubofcik J, Sejvar J, Ratto J, Bunga S, Makumbi I, Aceng JR, Nutman TB, Dowell SF & Nath A. (2017) Nodding syndrome may be an autoimmune reaction to the parasitic worm Onchocerca volvulus, Science Translational Medicine. 9, eaaf6953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nauen DW, Haffner MC, Kim J, Zheng Q, Yin H, DeMarzo AM, Mahairaki V, Colantuoni C, Pickering JG & Johnson TP (2020) Putative Autoantigen Leiomodin-1 Is Expressed in the Human Brain and in the Membrane Fraction of Newly Formed Neurons, Pathogens. 9, 1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tsukada T, Pappas CT, Moroz N, Antin PB, Kostyukova AS & Gregorio CC (2010) Leiomodin-2 is an antagonist of tropomodulin-1 at the pointed end of the thin filaments in cardiac muscle, Journal of cell science. 123, 3136–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gokhin DS, Ochala J, Domenighetti AA & Fowler VM (2015) Tropomodulin 1 directly controls thin filament length in both wild-type and tropomodulin 4-deficient skeletal muscle, Development. 142, 4351–4362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kiss B, Gohlke J, Tonino P, Hourani Z, Kolb J, Strom J, Alekhina O, Smith JE, Ottenheijm C, Gregorio C. & Granzier H. (2020) Nebulin and Lmod2 are critical for specifying thin-filament length in skeletal muscle, Science advances. 6, eabc1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cenik BK, Garg A, McAnally JR, Shelton JM, Richardson JA, Bassel-Duby R, Olson EN & Liu N. (2015) Severe myopathy in mice lacking the MEF2/SRF-dependent gene leiomodin-3, Journal of Clinical Investigation. 125, 1569–1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tian L, Ding S, You Y, Li T. r., Liu Y, Wu X, Sun L. & Xu T. (2015) Leiomodin-3-deficient mice display nemaline myopathy with fast-myofiber atrophy, Disease Models & Mechanisms. 8, 635–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nworu CU, Kraft R, Schnurr DC, Gregorio CC & Krieg PA (2014) Leiomodin 3 and tropomodulin 4 have overlapping functions during skeletal myofibrillogenesis, Journal of cell science. 128, 239–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Michael E, Hedberg-Oldfors C, Wilmar P, Visuttijai K, Oldfors A. & Darin N. (2019) Long-term follow-up and characteristic pathological findings in severe nemaline myopathy due to LMOD3 mutations, Neuromuscul Disord. 29, 108–113. [DOI] [PubMed] [Google Scholar]
- 23.Almenar-Queralt A, Lee A, Conley CA, de Pouplana LR & Fowler VM (1999) Identification of a novel tropomodulin isoform, skeletal tropomodulin, that caps actin filament pointed ends in fast skeletal muscle, Journal of Biological Chemistry. 274, 28466–28475. [DOI] [PubMed] [Google Scholar]
- 24.Fowler VM (1990) Tropomodulin: a cytoskeletal protein that binds to the end of erythrocyte tropomyosin and inhibits tropomyosin binding to actin, J Cell Biol. 111, 471–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fowler VM, Sussmann MA, Miller PG, Flucher BE & Daniels MP (1993) Tropomodulin is associated with the free (pointed) ends of the thin filaments in rat skeletal muscle, J Cell Biol. 120, 411–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Watakabe A, Kobayashi R. & Helfman DM (1996) N-tropomodulin: a novel isoform of tropomodulin identified as the major binding protein to brain tropomyosin, Journal of cell science. 109 ( Pt 9), 2299–310. [DOI] [PubMed] [Google Scholar]
- 27.Cox PR & Zoghbi HY (2000) Sequencing, expression analysis, and mapping of three unique human tropomodulin genes and their mouse orthologs, Genomics. 63, 97–107. [DOI] [PubMed] [Google Scholar]
- 28.Weber A, Pennise CR, Babcock GG & Fowler VM (1994) Tropomodulin caps the pointed ends of actin filaments, The Journal of cell biology. 127, 1627–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gregorio CC, Weber A, Bondad M, Pennise CR & Fowler VM (1995) Requirement of pointed-end capping by tropomodulin to maintain actin filament length in embryonic chick cardiac myocytes, Nature. 377, 83–6. [DOI] [PubMed] [Google Scholar]
- 30.Fowler VM, Greenfield NJ & Moyer J. (2003) Tropomodulin Contains Two Actin Filament Pointed End-capping Domains, Journal of Biological Chemistry. 278, 40000–40009. [DOI] [PubMed] [Google Scholar]
- 31.Kostyukova AS, Choy A & Rapp BA (2006) Tropomodulin binds two tropomyosins: a novel model for actin filament capping, Biochemistry. 45, 12068–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kostyukova AS, Hitchcock-Degregori SE & Greenfield NJ (2007) Molecular basis of tropomyosin binding to tropomodulin, an actin-capping protein, Journal of molecular biology. 372, 608–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kostyukova A, Maeda K, Yamauchi E, Krieger I. & Maeda Y. (2000) Domain structure of tropomodulin: distinct properties of the N-terminal and C-terminal halves, Eur J Biochem. 267, 6470–5. [DOI] [PubMed] [Google Scholar]
- 34.Krieger I, Kostyukova A, Yamashita A, Nitanai Y. & Maeda Y. (2002) Crystal structure of the C-terminal half of tropomodulin and structural basis of actin filament pointed-end capping, Biophysical journal. 83, 2716–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kobe B. (2001) The leucine-rich repeat as a protein recognition motif, Current Opinion in Structural Biology. 11, 725–732. [DOI] [PubMed] [Google Scholar]
- 36.Rao JN, Madasu Y. & Dominguez R. (2014) Mechanism of actin filament pointed-end capping by tropomodulin, Science. 345, 463–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tolkatchev D, Kuruba B, Smith GE, Swain KD, Smith KA, Moroz N, Williams TJ & Kostyukova AS (2020) Structural insights into the tropomodulin assembly at the pointed ends of actin filaments, Protein Science. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sung LA, Fowler VM, Lambert K, Sussman MA, Karr D. & Chien S. (1992) Molecular cloning and characterization of human fetal liver tropomodulin. A tropomyosin-binding protein, Journal of Biological Chemistry. 267, 2616–2621. [PubMed] [Google Scholar]
- 39.Fischer RS, Fritz-Six KL & Fowler VM (2003) Pointed-end capping by tropomodulin3 negatively regulates endothelial cell motility, The Journal of cell biology. 161, 371–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Geeves MA, Hitchcock-DeGregori SE & Gunning PW (2015) A systematic nomenclature for mammalian tropomyosin isoforms, J Muscle Res Cell Motil. 36, 147–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Uversky VN, Shah SP, Gritsyna Y, Hitchcock-DeGregori SE & Kostyukova AS (2011) Systematic analysis of tropomodulin/tropomyosin interactions uncovers fine-tuned binding specificity of intrinsically disordered proteins, Journal of molecular recognition : JMR. 24, 647–55. [DOI] [PubMed] [Google Scholar]
- 42.Colpan M, Moroz NA & Kostyukova AS (2013) Tropomodulins and tropomyosins: working as a team, J Muscle Res Cell Motil. 34, 247–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gray KT, Kostyukova AS & Fath T. (2017) Actin regulation by tropomodulin and tropomyosin in neuronal morphogenesis and function, Molecular and cellular neurosciences. 84, 48–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fowler VM & Dominguez R (2017) Tropomodulins and Leiomodins: Actin Pointed End Caps and Nucleators in Muscles, Biophysical journal. 112, 1742–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Skwarek-Maruszewska A, Boczkowska M, Zajac AL, Kremneva E, Svitkina T, Dominguez R. & Lappalainen P. (2010) Different localizations and cellular behaviors of leiomodin and tropomodulin in mature cardiomyocyte sarcomeres, Molecular biology of the cell. 21, 3352–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Boczkowska M, Rebowski G, Kremneva E, Lappalainen P. & Dominguez R. (2015) How Leiomodin and Tropomodulin use a common fold for different actin assembly functions, Nature communications. 6, 8314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pappas CT, Farman GP, Mayfield RM, Konhilas JP & Gregorio CC (2018) Cardiac-specific knockout of Lmod2 results in a severe reduction in myofilament force production and rapid cardiac failure, J Mol Cell Cardiol. 122, 88–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tolkatchev D, Smith GE Jr., Schultz LE, Colpan M, Helms GL, Cort JR, Gregorio CC & Kostyukova AS (2020) Leiomodin creates a leaky cap at the pointed end of actin-thin filaments, PLOS Biology. 18, e3000848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mi-Mi L, Farman GP, Mayfield RM, Strom J, Chu M, Pappas CT & Gregorio CC (2020) In vivo elongation of thin filaments results in heart failure, PLoS One. 15, e0226138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Szatmari D, Bugyi B, Ujfalusi Z, Grama L, Dudas R. & Nyitrai M. (2017) Cardiac leiomodin2 binds to the sides of actin filaments and regulates the ATPase activity of myosin, PLOS ONE. 12, e0186288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tolkatchev D, Smith GE Jr. & Kostyukova AS (2019) Role of intrinsic disorder in muscle sarcomeres, Progress in Molecular Biology and Translational Science. 166, 311–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kostyukova AS (2007) Leiomodin/tropomyosin interactions are isoform specific, Archives of biochemistry and biophysics. 465, 227–30. [DOI] [PubMed] [Google Scholar]
- 53.Ly T, Moroz N, Pappas CT, Novak SM, Tolkatchev D, Wooldridge D, Mayfield RM, Helms G, Gregorio CC & Kostyukova AS (2016) The N-terminal tropomyosin- and actin-binding sites are important for leiomodin 2’s function, Molecular biology of the cell. 27, 2565–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Colpan M, Ly T, Grover S, Tolkatchev D. & Kostyukova AS (2017) The cardiomyopathy-associated K15N mutation in tropomyosin alters actin filament pointed end dynamics, Archives of biochemistry and biophysics. 630, 18–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.O’Shea EK, Klemm JD, Kim PS & Alber T. (1991) X-ray structure of the GCN4 leucine zipper, a two-stranded, parallel coiled coil, Science. 254, 539–44. [DOI] [PubMed] [Google Scholar]
- 56.Greenfield NJ, Huang YJ, Swapna GV, Bhattacharya A, Rapp B, Singh A, Montelione GT & Hitchcock-DeGregori SE (2006) Solution NMR structure of the junction between tropomyosin molecules: implications for actin binding and regulation, Journal of molecular biology. 364, 80–96. [DOI] [PubMed] [Google Scholar]
- 57.Nitanai Y, Minakata S, Maeda K, Oda N. & Maeda Y. (2007) Crystal structures of tropomyosin: flexible coiled-coil, Advances in experimental medicine and biology. 592, 137–51. [DOI] [PubMed] [Google Scholar]
- 58.Prestegard JH, Bougault CM & Kishore AI (2004) Residual dipolar couplings in structure determination of biomolecules, Chemical reviews. 104, 3519–40. [DOI] [PubMed] [Google Scholar]
- 59.Greenfield NJ, Kotlyanskaya L. & Hitchcock-DeGregori SE (2009) Structure of the N terminus of a nonmuscle alpha-tropomyosin in complex with the C terminus: implications for actin binding, Biochemistry. 48, 1272–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Frye J, Klenchin VA & Rayment I. (2010) Structure of the tropomyosin overlap complex from chicken smooth muscle: insight into the diversity of N-terminal recognition, Biochemistry. 49, 4908–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lehman W. & Craig R. (2008) Tropomyosin and the Steric Mechanism of Muscle Regulation, Advances in experimental medicine and biology. 644, 95–109. [DOI] [PubMed] [Google Scholar]
- 62.Hitchcock-DeGregori SE, Sampath P. & Pollard TD (1988) Tropomyosin inhibits the rate of actin polymerization by stabilizing actin filaments, Biochemistry. 27, 9182–5. [DOI] [PubMed] [Google Scholar]
- 63.Colpan M, Tolkatchev D, Grover S, Helms GL, Cort JR, Moroz N. & Kostyukova AS (2016) Localization of the binding interface between leiomodin-2 and alpha-tropomyosin, Biochim Biophys Acta. 1864, 523–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Arslan B, Colpan M, Gray KT, Abu-Lail NI & Kostyukova AS (2018) Characterizing interaction forces between actin and proteins of the tropomodulin family reveals the presence of the N-terminal actin-binding site in leiomodin, Archives of biochemistry and biophysics. 638, 18–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Boczkowska M, Yurtsever Z, Rebowski G, Eck MJ & Dominguez R. (2017) Crystal Structure of Leiomodin 2 in Complex with Actin: A Structural and Functional Reexamination, Biophysical journal. 113, 889–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chen X, Ni F, Kondrashkina E, Ma J. & Wang Q. (2015) Mechanisms of leiomodin 2-mediated regulation of actin filament in muscle cells, Proceedings of the National Academy of Sciences of the United States of America. 112, 12687–12692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ly T, Pappas CT, Johnson D, Schlecht W, Colpan M, Galkin VE, Gregorio CC, Dong WJ & Kostyukova AS (2019) Effects of cardiomyopathy-linked mutations K15N and R21H in tropomyosin on thin-filament regulation and pointed-end dynamics, Molecular biology of the cell. 30, 268–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Reinhard M, Giehl K, Abel K, Haffner C, Jarchau T, Hoppe V, Jockusch BM & Walter U. (1995) The proline-rich focal adhesion and microfilament protein VASP is a ligand for profilins, EMBO J. 14, 1583–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Purich DL & Southwick FS (1997) ABM-1 and ABM-2 homology sequences: consensus docking sites for actin-based motility defined by oligoproline regions in Listeria ActA surface protein and human VASP, Biochem Biophys Res Commun. 231, 686–91. [DOI] [PubMed] [Google Scholar]
- 70.Boukhelifa M, Moza M, Johansson T, Rachlin A, Parast M, Huttelmaier S, Roy P, Jockusch BM, Carpen O, Karlsson R. & Otey CA (2006) The proline-rich protein palladin is a binding partner for profilin, FEBS J. 273, 26–33. [DOI] [PubMed] [Google Scholar]
- 71.Qualmann B. & Kessels MM (2009) New players in actin polymerization – WH2-domain-containing actin nucleators, Trends in Cell Biology. 19, 276–285. [DOI] [PubMed] [Google Scholar]
- 72.Dominguez R. (2016) The WH2 Domain and Actin Nucleation: Necessary but Insufficient, Trends in Biochemical Sciences. 41, 478–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rao J, Lahiri J, Isaacs L, Weis RM & Whitesides GM (1998) A trivalent system from vancomycin.D-ala-D-Ala with higher affinity than avidin.biotin, Science. 280, 708–11. [DOI] [PubMed] [Google Scholar]
- 74.Madeira F, Park YM, Lee J, Buso N, Gur T, Madhusoodanan N, Basutkar P, Tivey ARN, Potter SC, Finn RD & Lopez R. (2019) The EMBL-EBI search and sequence analysis tools APIs in 2019, Nucleic Acids Res. 47, W636–W641. [DOI] [PMC free article] [PubMed] [Google Scholar]