Abstract
Objective
Therapeutic strategies silencing and reducing the hepatitis B virus (HBV) reservoir, the covalently closed circular DNA (cccDNA), have the potential to cure chronic HBV infection. We aimed to investigate the impact of small interferring RNA (siRNA) targeting all HBV transcripts or pegylated interferon-α (peg-IFNα) on the viral regulatory HBx protein and the structural maintenance of chromosome 5/6 complex (SMC5/6), a host factor suppressing cccDNA transcription. In particular, we assessed whether interventions lowering HBV transcripts can achieve and maintain silencing of cccDNA transcription in vivo.
Design
HBV-infected human liver chimeric mice were treated with siRNA or peg-IFNα. Virological and host changes were analysed at the end of treatment and during the rebound phase by qualitative PCR, ELISA, immunoblotting and chromatin immunoprecipitation. RNA in situ hybridisation was combined with immunofluorescence to detect SMC6 and HBV RNAs at single cell level. The entry inhibitor myrcludex-B was used during the rebound phase to avoid new infection events.
Results
Both siRNA and peg-IFNα strongly reduced all HBV markers, including HBx levels, thus enabling the reappearance of SMC5/6 in hepatocytes that achieved HBV-RNA negativisation and SMC5/6 association with the cccDNA. Only IFN reduced cccDNA loads and enhanced IFN-stimulated genes. However, the antiviral effects did not persist off treatment and SMC5/6 was again degraded. Remarkably, the blockade of viral entry that started at the end of treatment hindered renewed degradation of SMC5/6.
Conclusion
These results reveal that therapeutics abrogating all HBV transcripts including HBx promote epigenetic suppression of the HBV minichromosome, whereas strategies protecting the human hepatocytes from reinfection are needed to maintain cccDNA silencing.
Keywords: liver, hepatitis B, interferon-α, antiviral therapy
Significance of this study.
What is already known on this subject?
There is an urgent need for therapeutic approaches repressing covalently closed circular DNA (cccDNA) activity because silencing of the viral reservoir may promote functional HBV cure. The host restriction factor structural maintenance of chromosome 5/6 (SMC5/6) hinders transcription from the HBV minichromosome, but the viral protein HBx triggers its degradation. The impact of treatments lowering HBV transcripts on HBx, SMC5/6 and cccDNA activity in vivo is unknown.
What are the new findings?
RNA interference targeting all HBV transcripts and pegylated interferon-α abrogate HBx production in a substantial amount of hepatocytes, leading to the prompt reappearance of SMC5/6 and cccDNA silencing in vivo. Viral entry inhibition appears required to prevent cccDNA reactivation in cells that achieved silencing.
How might it impact on clinical practice in the foreseeable future?
Sustained silencing of cccDNA transcription appears achievable by combining interventions both abolishing HBx and shielding the hepatocytes from new infections. These findings can assist in the design of future clinical trials aiming to achieve a functional cure.
Introduction
HBV is a small enveloped DNA virus that causes acute and chronic infections of the liver. Chronic hepatitis B (CHB) affects approximately 250 million individuals worldwide, causing more than 800 000 deaths every year due to complications of liver disease such as cirrhosis and hepatocellular carcinoma.1 Both the lack of robust antiviral immunity and the persistence of an episomal HBV DNA genome, the so-called covalently closed circular DNA (cccDNA), are held responsible for the failure of viral clearance in CHB.2
The viral genome consists of a partially double-stranded relaxed circular DNA that is transferred to the hepatocyte nucleus and converted into a stable cccDNA minichromosome, associated with viral and host proteins similar to host chromatin.3 The cellular transcriptional machinery generates all viral transcripts needed for viral replication. An overlength pregenomic RNA (pgRNA) is reverse transcribed by the viral polymerase within the nucleocapsids before being enveloped and secreted as progeny virus. The infected hepatocytes also secrete the E antigen (HBeAg) and high amounts of subviral particles containing the envelope proteins (HBsAg). High levels of circulating antigens are thought to contribute to immune tolerance and HBV persistence.4 In addition to the capsid protein (HBcAg) and the viral polymerase, HBV produces the regulatory X protein (HBx), which can interfere with several cellular pathways and transcription factors, and is recruited onto the cccDNA.5 6 Studies with HBV X-minus mutant viruses demonstrated that HBx is essential for cccDNA-driven viral transcription.7 8 Moreover, HBx was shown to mediate the degradation of the structural maintenance of chromosome 5/6 complex (SMC5/6),9 10 a multifunctional DNA-binding complex required for chromosome dynamics and stability.11 By binding to the damaged DNA-binding protein 1 (DDB1), an adaptor protein for the E3 ubiquitin ligase complex,12 HBx promotes the interaction of SMC5/6 with the E3 ubiquitin ligase proteasome system, triggering SMC5/6 degradation.13 Various viruses can hijack the DDB1–E3 ubiquitin ligase complex to promote the degradation of host proteins that would otherwise restrict viral replication.14 The SMC5/6 antagonism by HBx seems to be an evolutionarily conserved mechanism among mammalian hepadnaviruses.15 SMC5/6 colocalises with the nuclear domain 10,16 a dynamic nuclear organelle also implicated in the restriction of DNA viruses.17 It is thus not surprising that the SMC5/6 complex can act as a host restriction factor in HBV infection. Despite the increasing awareness of the role of SMC5/6 in counteracting cccDNA activity, data on how antiviral treatments affect the interaction between HBx and SMC5/6 in vivo are absent.
Potent nucleos(t)ide analogues efficiently suppress viral replication but do not target the cccDNA, and CHB is rarely cured.18 19 Thus, interventions silencing and reducing the HBV minichromosome appear essential to cure chronic HBV infection. Small interfering RNAs (siRNAs) targeting the overlapping pregenomic and subgenomic viral RNAs represent a promising therapeutic approach currently under investigation for anti-HBV therapy20–24 because it bears the potential to reduce both viraemia and antigen levels. Interferon-α (IFNα) has immunomodulatory and direct antiviral effects,25–27 including the ability to repress and destabilise the cccDNA.28 29
In this study, we aimed to investigate the impact of antiviral strategies able to reduce levels of HBV transcripts, like siRNA targeting all viral RNAs or pegylated IFNα (peg-IFNα), on HBx and SMC5/6 levels and assessed whether these interventions can induce cccDNA silencing in vivo using HBV-infected human liver chimeric mice. Furthermore, we monitored the fate of SMC5/6 and cccDNA activity after treatment withdrawal, either in the presence or in the absence of the entry inhibitor myrcludex-B (MyrB), now bulevirtide (Hepcludex).30–32 These studies confirm the key role of SMC5/6 in HBV restriction and reveal that shielding the hepatocytes from sodium taurocholate cotransporting polypeptide (NTCP)-dependent infection events enables maintenance of cccDNA silencing.
Materials and methods
Generation, infection and treatment of humanised USG mice
The generation of USG (uPA/SCID/beige/IL2RG–/–) mice reconstituted with human hepatocytes and infected with HBV was conducted as previously reported27 and is briefly described in the online supplemental material. Antiviral treatment was started when mice had reached stable HBV infection levels (>10 weeks after virus inoculation). The siRNA targets all HBV transcripts and was described earlier20 (referred to as W7.5 in the original publication). Mice received one intravenous injection of 1 mg/kg per week. The siRNA was obtained from Dharmacon (Lafayette, Colorado, USA) and formulated in lipid nanoparticles (LNPs) at Precision Nanosystems (Vancouver, British Columbia, Canada). Peg-IFNα (Pegasys; Hoffmann-La Roche, Basel, Switzerland) was administered two times per week intraperitoneally (25 ng/g body weight) for 6 weeks.27 Mice were either sacrificed 24 hours after the last injection or left untreated for follow-up. At the end of siRNA or peg-IFNα treatment, some mice received subcutaneous injections of MyrB (MyrB/bulevirtide; MYR Pharmaceuticals, Bad Homburg, Germany) at 2 µg/g body weight daily for additional 3–8 weeks.33 Additional uninfected mice, either untreated or treated with peg-IFNα for 4 weeks,27 served as controls for SMC6 immunofluorescence.
gutjnl-2020-322571supp001.pdf (8.8MB, pdf)
Immunoprecipitation/western blot
Proteins were extracted from liver tissues with T-Per Tissue Protein Extraction Reagent (Pierce, Rockford, Illinois, USA) supplemented with Complete Protease Inhibitor Cocktail (Roche) and Pefabloc SC-Protease-Inhibitor (Carl Roth, Karlsruhe, Germany). For immunoprecipitation, dynabeads protein G (Thermo Fisher) were incubated with 10 µg of monoclonal rabbit anti-HBx antibody34 for 10 min at room temperature. Protein concentrations were determined by BCA Protein Assay Kit (Pierce), and 1 mg of protein lysate was added to the dynabeads antibody complex and incubated overnight at 4°C. Washing, denaturing and elution were performed according to the manufacturer’s instructions. Proteins were resolved on 12% Mini-PROTEAN TGX Precast Gels (Bio-Rad, Feldkirchen, Germany) and blotted on nitrocellulose membranes (0.2 µm pore size; GE Healthcare, Buckinghamshire, UK) for immunodetection with rabbit anti-HBx (final concentration 40 µg/mL) and VeriBlot for IP Detection Reagent (Abcam, Cambridge, UK) as secondary antibody (1:200). To normalise HBx protein levels to different repopulation levels, 2 µg of the same protein lysates used for immunoprecipitation was loaded on a western blot and detected with a monoclonal mouse antibody (dilution 1:1000) specific for human albumin (#A6684; Sigma-Aldrich, St. Louis, Missouri, USA) and horseradish peroxidase-conjugated AffiniPure goat anti-rabbit IgG (Jackson ImmunoResearch, Suffolk, UK) as secondary antibody (dilution 1:10 000). Signals were visualised with Pierce ECL Western Blotting Substrate (Thermo Fisher) in the Fusion FX Imager (Vilber), and the band volume of the HBx signal was normalised to the band volume of human albumin.
RNA in situ hybridisation plus immunofluorescence
Total HBV RNAs were detected by RNA in situ hybridisation (RNA ISH) using the RNAscope Fluorescent Multiplex kit (Advanced Cell Diagnostics, Newark, California, USA) as published previously.35 The assay was coupled to immunofluorescence staining for the simultaneous detection of SMC6 protein. Briefly, liver sections were first pretreated following the instructions for fresh frozen sample preparation in the RNAscope manual including 4% paraformaldehyde fixation, ethanol dehydration and protease digestion, and then the SMC6 protein was stained as described in the online supplemental material. To increase and stabilise the signal for SMC6, the TSA Fluorescein System (Perkin Elmer, Waltham, Massachusetts, USA) was used. An RNAse inhibitor (RiboGuard RNase Inhibitor; Biozym Scientific, Hessisch Oldendorf, Germany) was added to all buffers during pretreatment and immunofluorescence. Afterwards, RNA ISH was continued according to the manufacturer’s instruction. Briefly, sections were submitted to hybridisation with probes recognising total HBV RNA (assay ID #560071) and beta-2 microglobulin (B2M #484661) as a marker for human hepatocytes for 2 hours at 40°C. The specificity of B2M was confirmed by costaining with murine Gapdh (#442871). This was followed by amplification steps and fluorescent labelling at 40°C and a nuclear counterstain using 4′,6-diamidino-2-phenylindole according to the manufacturer’s instructions. Maximal projections of z-stack images were captured with conventional fluorescence microscopy (BZ-X710; Keyence) using a 60×/1.40 numerical aperture oil objective and identical settings within one experiment.
Measurements of virological parameters and host gene expression, cccDNA quantification, chromatin immunoprecipitation (ChIP) assays and immunofluorescence staining were performed as described in detail in the online supplemental material.
Results
Anti-HBV siRNA treatment potently reduces all HBV markers except cccDNA
To suppress HBV RNA levels, stably HBV-infected human liver chimeric USG mice (median baseline titre of 1.6×109 HBV DNA copies/mL of serum) received weekly intravenous injections of LNP-formulated siRNA that targets the 3′ region of HBV transcripts within the HBV-X open reading frame, thus targeting all HBV RNAs for degradation20 (figure 1A). Four weeks of HBV siRNA treatment decreased HBV DNA viraemia by median of 1.6log (p=0.0003), serum HBsAg levels by 1.3log (p=0.0003) and HBeAg levels by 1.2log (p=0.0003) compared with baseline levels (figure 1B–D). After treatment was stopped, mice were either sacrificed (n=5) or monitored for viral rebound during an additional 4-week period (n=6). During the follow-up phase, mice were left untreated or received the entry inhibitor MyrB to assess the kinetics of viral rebound with and without the interference of new infection events. One additional mouse received MyrB for 8 weeks following treatment. HBV serological rebound occurred within 4 weeks because viraemia and antigenaemia increased up to levels similar to baseline (figure 1B–D). Blocking viral entry with MyrB did not prevent viral rebound in the serum, although these mice reached slightly lower viraemia, HBsAg and HBeAg levels than before treatment (median of 69%, 42% and 35% of their baseline levels, respectively (see online supplemental figure 1A–C)).
Figure 1.

Anti-HBV siRNA treatment potently reduces all HBV markers except cccDNA. (A) Experimental setup. HBV-infected USG mice were treated as indicated and blood was drawn every other week. (B–D) Viral titres were determined by qPCR (B), and serum HBsAg (C) and HBeAg (D) were determined by ELISA (Abbot Architect). Every line represents one mouse. (E, F) Mice were sacrificed at the indicated time points, and cccDNA levels were determined by qPCR in Epicentre-based DNA extracts without proteinase K after PSD digestion (E). cccDNA copies were normalised to the human mitochondrial gene ND2. Each dot represents a single mouse; horizontal lines depict the median. (F) Liver DNA extracts (Epicentre-based extraction without proteinase K) from D were subjected to Southern blot. DNA amounts were normalised to human mitochondrial DNA and digested with PSD before loading. The bar graph below shows the densitometry analysis of the cccDNA band. The blot depicts one representative experiment. cccDNA, covalently closed circular DNA; MyrB, myrcludex-B; NT, not treated; pf-rcDNA, protein-free relaxed circular DNA; qPCR, quantitative PCR; SB, Southern blot; siRNA, small interferring RNA; USG, uPA/SCID/beige/IL2RG–/–. †=this mouse died prematurely at the 2-week blood drawl.
To analyse the intrahepatic impact of siRNA treatment, we euthanised mice at the end of the treatment or after the follow-up phase. Quantitative PCR (qPCR) analyses showed that intrahepatic pgRNA and total HBV RNA levels substantially declined by 1.0log and 1.1log, respectively, in comparison with untreated controls after 4 weeks of treatment (online supplemental figure 1D, E). Consistent with the serological data, average intrahepatic viral RNA levels also increased during follow-up, reaching levels comparable to untreated control mice, although mice receiving the entry inhibitor appeared to reach slightly lower transcript levels. Intrahepatic cccDNA was analysed by qPCR and Southern blot (figure 1E, F). In line with the mode of action of siRNAs targeting HBV transcripts, cccDNA levels did not differ between groups. Longitudinal measurements of human serum albumin levels in mouse serum (online supplemental figure 1F) and the absence of caspase 3 activation (data not shown) indicated that the treatment did not affect hepatocyte viability.
siRNA treatment leads to the reappearance of SMC6 in hepatocytes negative for signs of active HBV replication
To assess the impact of siRNA-mediated HBV suppression at the single cell level and on the SMC5/6 complex, we costained HBcAg and SMC6 protein in mouse liver sections (figure 2A). The SMC6 protein can be used as surrogate to visualise the entire SMC5/6 complex due to its interdependent stability and was detectable in the nuclei of all human hepatocytes of uninfected mice using a human-specific antibody that does not cross-react with murine Smc6 (online supplemental figure 2A). In line with previous results,9 SMC6 staining was lost in stably HBV-infected mice where basically all human hepatocytes stain positive for HBcAg.
Figure 2.
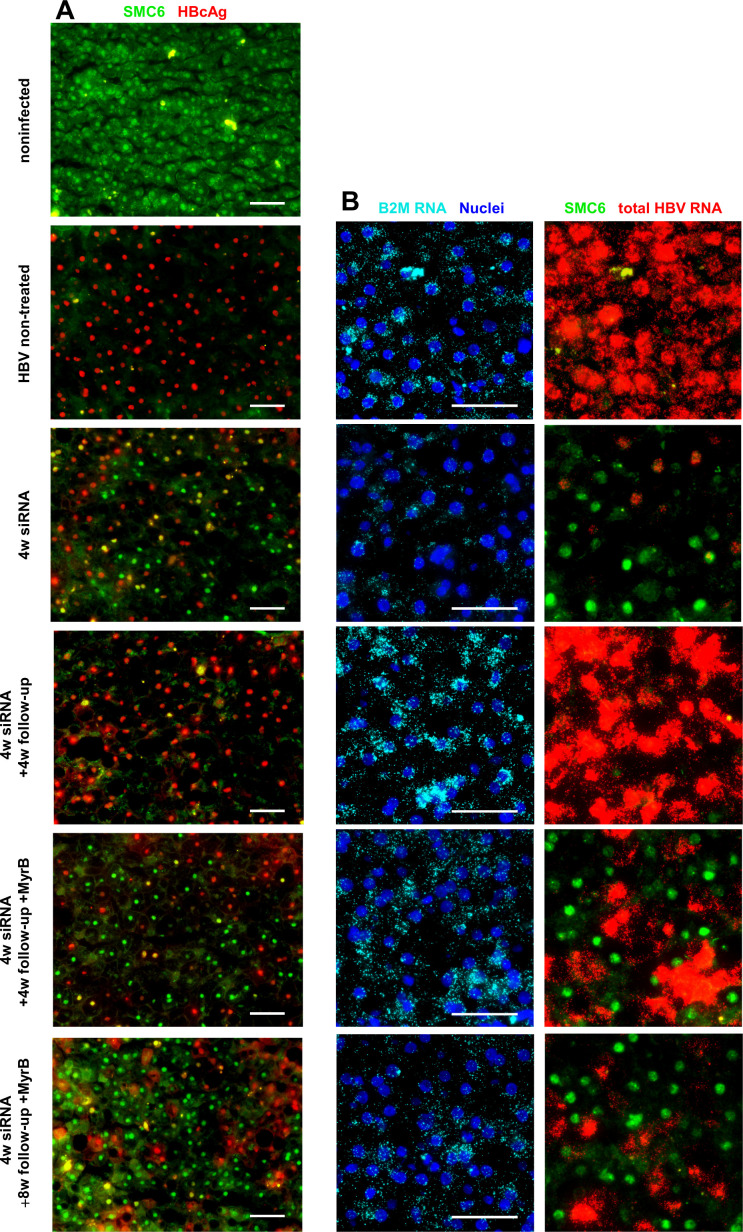
siRNA treatment leads to the reappearance of SMC6 in hepatocytes negative for signs of active HBV replication. (A) Immunofluorescence costaining for HBcAg (red) and SMC6 (green) in cryopreserved liver sections. Shown are representative pictures of one mouse from every treatment group as indicated on the left-hand side. (B) Representative pictures of RNA in situ hybridisation for total HBV RNA combined with immunofluorescence staining for SMC6 protein. Merged pictures of nuclei stained with 4′,6-diamidino-2-phenylindole (blue) and B2M RNA as a marker for human hepatocytes (aqua) are shown in the left column; HBV RNA (red) and SMC6 (green) are shown in the right column. Scale bar 50 µm. MyrB, myrcludex-B; siRNA, small interferring RNA.
At the end of the siRNA treatment, the amount of HBcAg-positive human hepatocytes was strongly reduced and SMC6 reappeared in the nuclei of HBcAg-negative cells (approximately 40% of the human hepatocytes). Four weeks after the treatment was stopped, HBcAg rebounded to levels observed in HBV-infected controls, while SMC6 appeared to be degraded again. Intriguingly, when viral entry was inhibited by MyrB administration, a large proportion of human hepatocytes remained SMC6-positive (approximately 40%), thus resembling the situation determined at the end of the siRNA treatment.
To further investigate the interrelation between SMC6 protein and HBV transcription at the single cell level, we combined the SMC6 staining with an RNA ISH assay detecting all HBV RNAs, as well as human hepatocytes using a species-specific B2M probe (online supplemental figure 2B). Figure 2B depicts merged images, and online supplemental figure 3 depicts all individual channels. We observed a strong reduction in HBV RNA-positive cells after 4 weeks of HBV siRNA treatment. In line with the mode of action of the drug that results in RNA-induced silencing complex-mediated degradation of cytosolic RNAs, HBV RNAs were undetectable in the cytoplasm but remained still detectable in the nuclei of single infected hepatocytes. In a great proportion of human hepatocytes, however, viral RNAs were also undetectable in the nuclei, indicating that viral transcription had been disabled altogether. Of note, the SMC6 protein reappeared exclusively in these hepatocytes, pointing out the possibility that SMC5/6 reappearance was followed by complete abrogation of viral transcription in these cells. SMC5 and SMC6 messenger RNA (mRNA) levels and interferon-stimulated genes (ISGs) did not change over the course of the experiment (online supplemental figure 4A, B).
HBV RNA production was re-established during viral rebound, and conversely SMC6 was degraded again. However, the SMC6 protein was still detected in a substantial proportion of human hepatocytes in mice receiving MyrB, and they were followed up for 4 weeks and even 8 weeks. In contrast, viral transcripts rebounded in the remaining hepatocytes, thus leading to a mixed pattern of SMC6-negative and fully HBV RNA-positive cells and HBV RNA-negative and SMC6-positive cells. Taken together, these analyses indicate that in the absence of de novo infection, HBV rebound could be prevented in SMC6-positive hepatocytes where viral RNA transcription was absent. The reactivation of viral transcription in cells that had been SMC6-negative, on the contrary, explains the overall serological and partial intrahepatic rebound determined in MyrB-treated mice, despite the maintenance of SMC5/6-mediated cccDNA suppression in a significant fraction of cells.
Viral markers including cccDNA are reduced on peg-IFNα treatment and rebound after treatment cessation
IFNα is a cytokine with multiple ascribed direct anti-HBV effects, such as cccDNA epigenetic suppression, destabilisation and post-transcriptional pgRNA suppression.36 To assess the impact of peg-IFNα in reducing both viral RNA and proteins, we performed two independent experiments using two groups of stably HBV-infected, titre-matched mice (median baseline HBV DNA viraemia in experiment 1=1.1×1010; experiment 2=8.5×108 HBV DNA copies/mL) (figure 3A). Mice received two peg-IFNα injections of the human equivalent dose per week, ensuring a constant effect of the drug as measured by the upregulation of ISGs in human hepatocytes (online supplemental figure 4C–F). Similar to previous studies,27 peg-IFNα led to a substantial and continuous reduction in viraemia in all mice (median log reduction, 1.7log in experiment 1 (online supplemental figure 5A) and 1.1log in experiment 2 (p=0.0008 for both experiments) (figure 3B)). After 6 weeks, peg-IFNα was withdrawn and mice were either sacrificed (n=5) or monitored for an additional 3 weeks (experiment 1; n=4) or 6 weeks (experiment 2; n=4) in the absence or presence of MyrB (figure 3A). On drug withdrawal, viraemia rebounded (online supplemental figure 5A; figure 3B). However, mice receiving the entry inhibitor for 6 weeks reached slightly lower viraemia levels than before treatment. Viraemia changes were mirrored by a reduction in HBsAg and HBeAg levels in the mouse serum, although these changes did not reach statistical significance (online supplemental figure 5B, C). At the end of the experiments, mice were sacrificed and intrahepatic markers were analysed collectively. After 6 weeks of treatment, we observed a clear reduction in intrahepatic pgRNA (median fourfold, online supplemental figure 5D) compared with untreated control mice, which increased again on drug withdrawal. However, entry inhibition clearly hindered intrahepatic viral rebound (online supplemental figure 5D).
Figure 3.
Viral markers including cccDNA are reduced on peg-IFNα treatment and rebound after treatment cessation. (A) Experimental setup. HBV-infected USG mice were treated with peg-IFNα as indicated in two independent experiments. (B) Blood was drawn every other week and viral titres were determined by qPCR. The line graph shows mice from experiment 2. Lines depict the median and error bars the range. (C, D) Mice were sacrificed at the indicated time points, and liver DNA extracts (Epicentre-based extraction without proteinase K) were subjected to SB (C). DNA amounts were normalised to human mitochondrial DNA and digested with PSD before loading. The bar graph below shows the densitometry analysis of the cccDNA band. The blot shows mice from experiment 2. (D) cccDNA levels were determined by qPCR in Epicentre-based DNA extracts without proteinase K after PSD digestion in all mice (same extracts as in B). cccDNA copies were normalised to the human mitochondrial gene ND2. Each dot represents a single mouse; horizontal lines depict the median. cccDNA, covalently closed circular DNA; MyrB, myrcludex-B; NT, not treated; IFNα, interferon-α; MyrB, myrcludex-B; NT, not treated; peg-IFNα, pegylated interferon-α; pf-rcDNA, protein-free relaxed circular DNA; qPCR, quantitative PCR; SB, Southern blot; USG, uPA/SCID/beige/IL2RG–/–.
Analysis of cccDNA by Southern blot showed a decrease in cccDNA levels in IFN-treated mice compared with untreated controls, which accounted to a median 3-fold reduction in experiment 1 and 4.5-fold in experiment 2, as determined by densitometry analysis of the 2.1-kb cccDNA band (figure 3C; online supplemental figure 5E). To confirm results obtained by Southern blot with an independent method, qPCR was run in parallel, indicating a median fivefold reduction after 6 weeks of peg-IFNα treatment in both experiments (figure 3D). Remarkably, cccDNA levels increased in the weeks following drug withdrawal but remained low in the groups treated with the entry inhibitor, suggesting that new infection events were necessary to replenish intrahepatic cccDNA loads during viral rebound.
Human albumin levels in mouse serum remained similar during the course of the experiment in both treated and untreated mice (online supplemental figure 5F), and no increase in active caspase 3 as a sign of hepatocyte apoptosis was detected in liver sections of the treated mice (data not shown). In summary, we confirmed the reduction of viral DNA and RNA levels in serum and liver, and observed reduced cccDNA levels on peg-IFNα treatment, whereas no evidence of cytotoxicity in human hepatocytes was observed.
Peg-IFNα treatment leads to the reappearance of the SMC5/6 complex in hepatocytes negative for signs of active replication
We next analysed the impact of peg-IFNα-mediated HBV suppression on the SMC5/6 complex. Costaining of SMC6 protein and HBcAg in liver sections revealed a reduction in HBcAg-positive cells and a concomitant increase in SMC6-positive hepatocyte nuclei (approximately 30%) (figure 4A). During follow-up, HBcAg rebounded to levels observed in HBV-infected controls, whereas SMC6 disappeared again. When viral entry was blocked, a substantial proportion of cells (approximately 30%) remained SMC6-positive both at 3 weeks (data not shown) and at 6 weeks after the end of treatment, resembling the situation determined at the end of peg-IFNα treatment. Only peg-IFNα, but not siRNA treatment, induced the expression of ISGs as measured by qPCR for a set of classical human ISGs (online supplemental figure 4C–F). To assess a possible direct effect of peg-IFNα treatment on the SMC5/6 complex, we analysed SMC6 protein levels by immunofluorescence also in uninfected USG mice that received peg-IFNα for 4 weeks (online supplemental figure 6). Treatment did not change the levels or distribution of SMC6 proteins, indicating that peg-IFNα does not directly regulate this component of the complex. SMC5 and SMC6 mRNA levels did not change over the course of the experiment (online supplemental figure 4A, B).
Figure 4.
Peg-IFNα treatment leads to the reappearance of the SMC5/6 complex in hepatocytes negative for signs of active replication. (A) Immunofluorescence costaining for HBcAg (red) and SMC6 (green) in cryopreserved liver sections. Shown are representative pictures of one mouse from every treatment group as indicated on the left-hand side. (B) Representative pictures of RNA in situ hybridisation for total HBV RNA combined with immunofluorescence staining for SMC6 protein. Merged pictures of nuclei stained with 4′,6-diamidino-2-phenylindole (blue) and B2M RNA as a marker for human hepatocytes (aqua) are shown in the left column; HBV RNA (red) and SMC6 (green) are shown in the right column. Scale bar 50 µm. Peg-IFNα, pegylated interferon-α; SMC5/6, structural maintenance of chromosome 5/6 complex.
Using the combined RNA ISH assay for total HBV RNA and SMC6 immunofluorescence staining, we observed a strong reduction in both HBV RNA-positive cells and HBV transcripts within the infected cells in treated animals (figure 4B and online supplemental figure 7 showing the individual channels). SMC6, on the contrary, was exclusively detected in cells negative for HBV RNA, indicating that the absence of the SMC6 protein in hepatocytes is a reliable marker for HBV activity. Off treatment, HBV RNA production was re-established, and conversely SMC6 was degraded again in all human hepatocytes of mice without entry inhibition. A partial viral rebound was also observed in the serum and liver of MyrB-treated mice. However, a fraction of SMC6-positive hepatocytes comparable to that determined at the end of treatment persisted in these mice. These intrahepatic analyses show that SMC6 persisted in hepatocytes negative for signs of active replication when new infections were excluded. On the contrary, a strong increase in viral RNA levels was observed in the remaining hepatocytes of the same mice, suggesting that cells which had not been efficiently suppressed during treatment served as the source of renewed viral production during rebound. Taken together, peg-IFNα treatment induced the reappearance of the SMC5/6 complex in hepatocytes that had either lost cccDNA or that harboured efficiently suppressed cccDNA molecules.
Both siRNA and peg-IFNα treatment reduce HBx and recruit the SMC5/6 complex to the cccDNA
Previous studies have shown that HBx targets the SMC5/6 complex for proteasomal degradation, and therefore we aimed to analyse HBx protein levels under siRNA and peg-IFNα treatment. Using a novel monoclonal rabbit anti-HBx antibody34 for immunoprecipitation and western blot, we were able to detect the physiological levels of HBx protein in the liver of HBV-infected humanised mice. Alongside the reduction in other viral proteins, HBx decreased to undetectable levels by siRNA treatment (figure 5A) and was strongly reduced by peg-IFNα treatment (figure 5B). However, HBx rebounded after treatment cessation. Of note, intrahepatic HBx protein levels remained lower in mice treated with the entry inhibitor MyrB compared with levels in untreated mice, suggesting that renewed HBx production was limited to a fraction of human hepatocytes.
Figure 5.
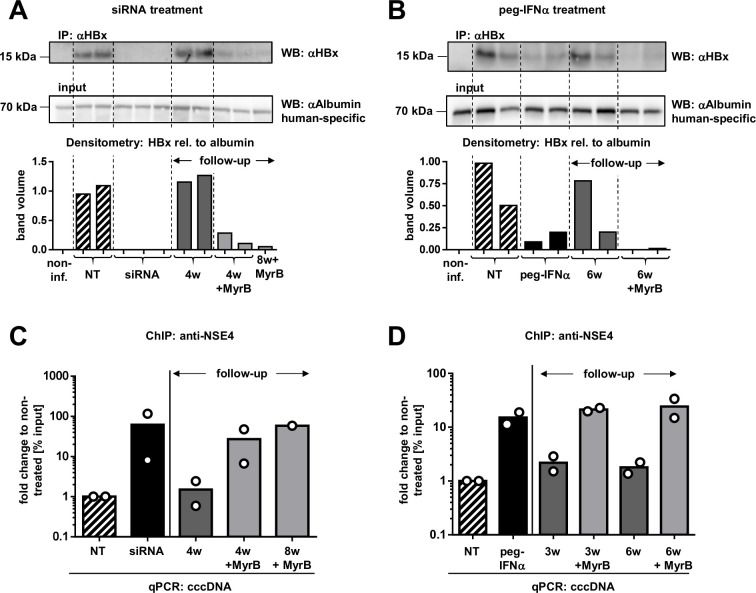
Both siRNA and peg-IFNα treatment reduce HBx and recruit the SMC5/6 complex to the cccDNA. (A, B) HBx levels were measured in liver protein lysates after siRNA (A) and peg-IFNα (B) treatment by IP and WB using the same anti-HBx antibody (upper panel). For normalisation to human hepatocyte content, protein lysates were subjected in parallel to WB using a human-specific antibody recognising albumin (lower panel). The bar graph below shows the densitometry analysis of the HBx signal relative to human albumin. One representative picture from a total of three experiments is shown. (C, D) ChIP assays were performed in two mice from every treatment group (siRNA (C) and peg-IFNα (D)), including untreated controls and follow-up arms. Chromatin from liver tissue was precipitated with anti-NSE4 antibody and analysed by cccDNA-selective qPCR. ChIP assays were performed in triplicates for one of the mouse of every treatment group to confirm accuracy (see online supplemental figure 8). cccDNA, covalently closed circular DNA; ChIP, chromatin immunoprecipitation; IP, immunoprecipitation; MyrB, myrcludex-B; NT, not treated; peg-IFNα, pegylated interferon-α; qPCR, quantitative PCR; WB, western blot; siRNA, small interferring RNA; SMC5/6, structural maintenance of chromosome 5/6 complex.
To assess whether the SMC5/6 complex could associate with the cccDNA during and after antiviral treatment, we performed ChIP-qPCR experiments using antibodies for NSE4, a component of the SMC5/6 complex.9 NSE4 strongly associated with cccDNA on siRNA (figure 5C) and peg-IFNα treatment (figure 5D) but was barely detectable (similar to IgG controls) in untreated HBV-infected control mice. Of note, NSE4 occupancy on the cccDNA diminished to baseline levels during viral rebound, but was still increased in mice receiving MyrB. These data show that the association of this host restriction factor with the cccDNA was maintained until the end of our observation time (8 weeks after stopping siRNA administration) in mice where new infection events were blocked.
Given that cccDNA levels were lower in peg-IFNα-treated mice, the increased occupancy by NSE4 on the remaining cccDNA suggests that IFN treatment induced both silencing and reduction of the intrahepatic cccDNA.
Discussion
In this study, we investigated the impact of two therapeutic approaches, siRNA and peg-IFNα, on the viral regulatory protein HBx and on the HBV restriction factor SMC5/6 in HBV-infected mice with humanised livers. This chromosomal maintenance complex has gained interest in HBV research as it was discovered that HBx, in complex with DDB1, can target this complex for proteasomal degradation. Vice versa, SMC5/6 prevents transcription from the viral episome in HBV mutants lacking HBx expression.9 10 Here, we show for the first time that siRNA-mediated silencing of all HBV transcripts and treatment with peg-IFNα can achieve substantial decrease in HBx protein levels and reappearance of SMC6 in primary human hepatocytes in vivo. In a combined analysis of SMC6 protein and HBV RNAs, we demonstrate that SMC6 is exclusively detected in hepatocytes without active HBV transcription. This is in line with a previous in vitro study showing that treatment with siRNAs that targeted either HBx or DDB1 mRNA, the host factor binding to HBx, led to an increase in SMC6-positive hepatocytes.34
Our in vivo study shows that targeting HBV replication with siRNAs not only enables the reduction of viraemia and antigenaemia but has also the capacity to shut down HBx production and hence suppress cccDNA transcription in HBV-infected human hepatocytes (figure 6). This is particularly apparent in the combined immunofluorescence and RNA ISH assay for SMC6 protein and viral RNAs, where we observed a substantial fraction of human hepatocytes (around 40%) that were SMC6-positive and HBV RNA-negative both in the cytoplasm and in the nucleus. In contrast, HBV RNA production was still detected in the nucleus of some human hepatocytes, and SMC6 negativity was maintained, indicating that the treatment did not achieve full suppression of viral transcription in all hepatocytes. Of note, siRNA treatment neither changed cccDNA levels nor induced the expression of human ISGs, effects that were induced by treatment with peg-IFNα. Nevertheless, ChIP-qPCR assays could confirm an increased recruitment of the SMC5/6 complex onto the cccDNA, thus providing evidence that siRNA treatment targeting the HBx region can promote epigenetic suppression of cccDNA transcription in vivo. Previous cccDNA-ChIP analyses showed that administration of IFNα induces hypoacetylation of the cccDNA-bound histones, as well as active recruitment of transcriptional corepressors, leading to reduced cccDNA transcription both in vitro and in the liver of humanised mice.27 28 Here, we show that 6 weeks of treatment with peg-IFNα substantially reduced HBx protein levels, thereby enabling binding of the SMC5/6 complex to cccDNA.
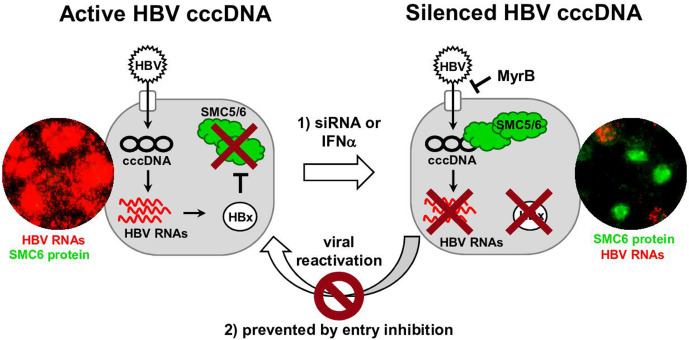
Figure 6.
Graphical summary of the main findings. Schematic representation showing cccDNA-driven production of HBV RNAs and HBx in infected human hepatocytes. Through HBx, the SMC5/6 complex is degraded, thus preventing silencing of the cccDNA. Treatment with siRNA or IFNα lowers HBV RNAs and depletes HBx in vivo, enabling reappearance of the SMC5/6 complex, its recruitment onto the cccDNA and silencing of cccDNA transcription. By combining such interventions with entry/spreading inhibition strategies, long-term suppression of the HBV minichromosome can be maintained. RNA ISH assays combined with SMC6 staining show reappearance of the host restriction factor SMC5/6 in HBV RNA-negative human hepatocytes in chimeric mice. cccDNA, covalently closed circular DNA; MyrB, myrcludex-B; IFNα, interferon-α; ISH, in situ hybridisation; siRNA, small interferring RNA; SMC5/6, structural maintenance of chromosome 5/6.
It must be noted that both antiviral treatments achieved substantial, but not complete, suppression of cccDNA activity, as documented by the persistence of some HBcAg and HBV RNA-positive hepatocytes in the liver of chronically infected humanised mice. Whether this phenomenon reflects intrinsic differences among the hepatocytes to respond to drugs is currently unknown. Moreover, the inability to achieve more profound HBV suppression may also be due to the very high levels of infection present in these immune-deficient chimeric mice. The persistence of such HBV-producing hepatocytes was likely responsible for the rapid serological rebound determined after stopping siRNA or peg-IFNα treatment—both in mice treated with the entry inhibitor or in those without.
Strikingly, the silencing of cccDNA achieved either on peg-IFNα or on siRNA treatment was maintained during follow-up only when new infection events were prevented by administering the entry inhibitor. This long-lasting epigenetic effect (up to 8 weeks) indicates that SMC5/6-mediated silencing is a potent and persisting mechanism that may have key effects for the outcome of anti-HBV therapy. In line with our findings, the absence of HBV RNA transcripts in subpopulations of cccDNA-positive hepatocytes of patients with CHB has been reported using single cell analysis approaches.37 38 It is thus plausible that cells with inactive viral transcription act as a ‘latent reservoir’ that can serve as a source of HBV reactivation in patients interrupting antiviral treatment or receiving immune suppression. A limitation of the USG mouse model used here is the lack of adaptive immune responses. It is tempting to speculate that adaptive responses would contribute to achieve stronger and sustained suppression of viral parameters if reactivated by the reduction in viral load and antigenaemia.
It is important to note that new infection events appeared crucial for the intrahepatic rebound or reactivation of HBV in all human hepatocytes, regardless of whether the cells were uninfected, had cleared cccDNA during IFN treatment or harboured silenced cccDNA molecules. Our cccDNA measurements showed a decrease in the level of intrahepatic cccDNA (threefold to fivefold compared with untreated controls) in mice that received peg-IFNα, thus indicating that several weeks of IFN treatment promoted both transcriptional silencing and partial loss of the HBV minichromosome. This is in line with some previous observations27 and with the notion that IFNα administration can promote partial cccDNA degradation.29 On the contrary, siRNA therapy is not expected to affect cccDNA loads directly, and indeed cccDNA levels, which are generally quite stable in this model in the absence of cell killing or proliferation,35 did not decrease at the end of siRNA treatment. A substantial increase in the cccDNA pool off treatment was also not expected because cccDNA amounts per cell remain low even after long-term infection and unrestricted infection of every hepatocyte.33 35
We cannot prove or unequivocally disprove whether new infections with HBV DNA-containing particles are able to reactivate silenced cccDNA or whether they merely introduce and build new transcriptionally active cccDNA molecules. We cannot exclude either that the NTCP-mediated entry of enveloped and encapsidated HBV RNAs, rather than HBV DNA, may be involved in the reactivation of already established but silenced cccDNA molecules.39 At any rate, our study provides evidence that the blockade of NTCP-mediated pathways determining HBV cell entry plays a key role not only in preventing intrahepatic HBV spreading but also in maintaining transcriptional silencing of the viral minichromosome in those cells that achieved therapeutic HBV silencing. Entry inhibition can be efficiently achieved with MyrB treatment as shown in human liver chimeric mice and HBV/hepatitis D virus coinfected patients,32 33 35 but it remains to be investigated to which extent other antivirals lowering HBV DNA viraemia might prevent de novo infection of hepatocytes and reactivation of persisting cccDNA molecules with comparable efficiency. Finally, degradation or impairment of the human SMC5/6 complex has been shown to be accompanied by an accumulation of DNA damage, p53 activation and mitotic aberrations.40 After reconstitution of the USG mouse livers, human hepatocytes are not subjected to mitotic stimuli any more; thus it is not surprising that a few months of HBV infection and degradation of SMC5/6 did not lead to obvious effects regarding cell viability. It is plausible, however, that in patients with CHB, therapeutic strategies targeting HBx and aiming at restoring SMC5/6 function in infected hepatocytes may reduce progression of liver disease and HBV-related carcinogenesis.
In sum, while elimination of the cccDNA from the infected liver remains challenging, this study demonstrates how distinct treatment interventions can have a profound impact on the transcriptional activity of the cccDNA, and hence contribute to the design of improved therapies aiming to cure chronic HBV infection.
Acknowledgments
We are grateful to R. Reusch and N. Jaeger for their excellent assistance with the mouse colony and C. Dettmer and C. Eggers for their great technical help.
Footnotes
LA and KG contributed equally.
Correction notice: This article has been corrected since it published Online First. The first sentence of the abstract has been updated for clarity.
Contributors: MD initiated and supervised the study. ML, MD, LA, KG, RCM, RKB, SU, HJ and SPF designed the experiments and discussed data. TV and LA generated and treated chimeric mice. LA, KG, and AP performed the experiments and analysed the data. LA, KG and MD wrote the manuscript. All authors read and corrected the manuscript.
Funding: The study was supported by the German Research Foundation (DFG) by a grant to MD (SFB 841 A5) and by the State of Hamburg with the Research Program (LFF-FV44: EPILOG). MD and SU also received funding from the German Center for Infection Research (DZIF-BMBF; TTU-hepatitis 05.815 and 05.704). All funding sources supporting the work are acknowledged.
Competing interests: RCM, RKB, HJ and SPF are employees of Gilead Sciences. SU is co-applicant and co-inventor on patents protecting myrcludex-B (bulevirtide). The other authors have nothing to disclose.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Data availability statement
All data relevant to the study are included in the article or uploaded as supplementary information. Additional data (eg, detailed protocols) are available upon request to m.dandri@uke.de.
Ethics statements
Patient consent for publication
Not required.
References
- 1. Polaris Observatory Collaborators . Global prevalence, treatment, and prevention of hepatitis B virus infection in 2016: a modelling study. Lancet Gastroenterol Hepatol 2018;3:383–403. 10.1016/S2468-1253(18)30056-6 [DOI] [PubMed] [Google Scholar]
- 2. Allweiss L, Dandri M. The role of cccDNA in HBV maintenance. Viruses 2017;9. 10.3390/v9060156. [Epub ahead of print: 21 06 2017]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Schreiner S, Nassal M. A role for the host DNA damage response in hepatitis B virus cccDNA Formation-and beyond? Viruses 2017;9. 10.3390/v9050125. [Epub ahead of print: 22 05 2017]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dembek C, Protzer U, Roggendorf M. Overcoming immune tolerance in chronic hepatitis B by therapeutic vaccination. Curr Opin Virol 2018;30:58–67. 10.1016/j.coviro.2018.04.003 [DOI] [PubMed] [Google Scholar]
- 5. Belloni L, Pollicino T, De Nicola F, et al. Nuclear HBx binds the HBV minichromosome and modifies the epigenetic regulation of cccDNA function. Proc Natl Acad Sci U S A 2009;106:19975–9. 10.1073/pnas.0908365106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dandri M. Epigenetic modulation in chronic hepatitis B virus infection. Semin Immunopathol 2020;42:173–85. 10.1007/s00281-020-00780-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lucifora J, Arzberger S, Durantel D, et al. Hepatitis B virus X protein is essential to initiate and maintain virus replication after infection. J Hepatol 2011;55:996–1003. 10.1016/j.jhep.2011.02.015 [DOI] [PubMed] [Google Scholar]
- 8. Tsuge M, Hiraga N, Akiyama R, et al. HBx protein is indispensable for development of viraemia in human hepatocyte chimeric mice. J Gen Virol 2010;91:1854–64. 10.1099/vir.0.019224-0 [DOI] [PubMed] [Google Scholar]
- 9. Decorsière A, Mueller H, van Breugel PC, et al. Hepatitis B virus X protein identifies the SMC5/6 complex as a host restriction factor. Nature 2016;531:386–9. 10.1038/nature17170 [DOI] [PubMed] [Google Scholar]
- 10. Murphy CM, Xu Y, Li F, et al. Hepatitis B virus X protein promotes degradation of Smc5/6 to enhance HBV replication. Cell Rep 2016;16:2846–54. 10.1016/j.celrep.2016.08.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jeppsson K, Kanno T, Shirahige K, et al. The maintenance of chromosome structure: positioning and functioning of SMC complexes. Nat Rev Mol Cell Biol 2014;15:601–14. 10.1038/nrm3857 [DOI] [PubMed] [Google Scholar]
- 12. Hodgson AJ, Hyser JM, Keasler VV, et al. Hepatitis B virus regulatory HBx protein binding to DDB1 is required but is not sufficient for maximal HBV replication. Virology 2012;426:73–82. 10.1016/j.virol.2012.01.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Minor MM, Hollinger FB, McNees AL, et al. Hepatitis B virus HBx protein mediates the degradation of host restriction factors through the cullin 4 DDB1 E3 ubiquitin ligase complex. Cells 2020;9. 10.3390/cells9040834. [Epub ahead of print: 30 03 2020]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Livingston CM, Ramakrishnan D, Strubin M, et al. Identifying and characterizing interplay between hepatitis B virus X protein and Smc5/6. Viruses 2017;9. 10.3390/v9040069. [Epub ahead of print: 03 04 2017]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Abdul F, Filleton F, Gerossier L, et al. Smc5/6 antagonism by HBx is an evolutionarily conserved function of hepatitis B virus infection in mammals. J Virol 2018;92. 10.1128/JVI.00769-18. [Epub ahead of print: 15 08 2018]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Niu C, Livingston CM, Li L, et al. The SMC5/6 complex restricts HBV when localized to ND10 without inducing an innate immune response and is counteracted by the HBV X protein shortly after infection. PLoS One 2017;12:e0169648. 10.1371/journal.pone.0169648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Scherer M, Stamminger T. Emerging role of PML nuclear bodies in innate immune signaling. J Virol 2016;90:5850–4. 10.1128/JVI.01979-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Revill PA, Penicaud C, Brechot C, et al. Meeting the challenge of eliminating chronic hepatitis B infection. Genes 2019;10. 10.3390/genes10040260. [Epub ahead of print: 01 04 2019]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fanning GC, Zoulim F, Hou J, et al. Therapeutic strategies for hepatitis B virus infection: towards a cure. Nat Rev Drug Discov 2019;18:827–44. 10.1038/s41573-019-0037-0 [DOI] [PubMed] [Google Scholar]
- 20. Thi EP, Dhillon AP, Ardzinski A, et al. ARB-1740, a RNA interference therapeutic for chronic hepatitis B infection. ACS Infect Dis 2019;5:725–37. 10.1021/acsinfecdis.8b00191 [DOI] [PubMed] [Google Scholar]
- 21. Wooddell CI, Yuen M-F, Chan HL-Y, et al. Rnai-Based treatment of chronically infected patients and chimpanzees reveals that integrated hepatitis B virus DNA is a source of HBsAg. Sci Transl Med 2017;9. 10.1126/scitranslmed.aan0241. [Epub ahead of print: 27 Sep 2017]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Shih Y-M, Sun C-P, Chou H-H, et al. Combinatorial RNA interference therapy prevents selection of pre-existing HBV variants in human liver chimeric mice. Sci Rep 2015;5:15259. 10.1038/srep15259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yamamoto N, Sato Y, Munakata T, et al. Novel pH-sensitive multifunctional envelope-type nanodevice for siRNA-based treatments for chronic HBV infection. J Hepatol 2016;64:547–55. 10.1016/j.jhep.2015.10.014 [DOI] [PubMed] [Google Scholar]
- 24. Sato Y, Matsui H, Yamamoto N, et al. Highly specific delivery of siRNA to hepatocytes circumvents endothelial cell-mediated lipid nanoparticle-associated toxicity leading to the safe and efficacious decrease in the hepatitis B virus. J Control Release 2017;266:216–25. 10.1016/j.jconrel.2017.09.044 [DOI] [PubMed] [Google Scholar]
- 25. Wieland SF, Eustaquio A, Whitten-Bauer C, et al. Interferon prevents formation of replication-competent hepatitis B virus RNA-containing nucleocapsids. Proc Natl Acad Sci U S A 2005;102:9913–7. 10.1073/pnas.0504273102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Xu C, Guo H, Pan X-B, et al. Interferons accelerate decay of replication-competent nucleocapsids of hepatitis B virus. J Virol 2010;84:9332–40. 10.1128/JVI.00918-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Allweiss L, Volz T, Lütgehetmann M, et al. Immune cell responses are not required to induce substantial hepatitis B virus antigen decline during PEGylated interferon-alpha administration. J Hepatol 2014;60:500–7. 10.1016/j.jhep.2013.10.021 [DOI] [PubMed] [Google Scholar]
- 28. Belloni L, Allweiss L, Guerrieri F, et al. IFN-α inhibits HBV transcription and replication in cell culture and in humanized mice by targeting the epigenetic regulation of the nuclear cccDNA minichromosome. J Clin Invest 2012;122:529–37. 10.1172/JCI58847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lucifora J, Xia Y, Reisinger F, et al. Specific and nonhepatotoxic degradation of nuclear hepatitis B virus cccDNA. Science 2014;343:1221–8. 10.1126/science.1243462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Schulze A, Schieck A, Ni Y, et al. Fine mapping of pre-S sequence requirements for hepatitis B virus large envelope protein-mediated receptor interaction. J Virol 2010;84:1989–2000. 10.1128/JVI.01902-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Petersen J, Dandri M, Mier W, et al. Prevention of hepatitis B virus infection in vivo by entry inhibitors derived from the large envelope protein. Nat Biotechnol 2008;26:335–41. 10.1038/nbt1389 [DOI] [PubMed] [Google Scholar]
- 32. Bogomolov P, Alexandrov A, Voronkova N, et al. Treatment of chronic hepatitis D with the entry inhibitor myrcludex B: first results of a phase Ib/IIa study. J Hepatol 2016;65:490–8. 10.1016/j.jhep.2016.04.016 [DOI] [PubMed] [Google Scholar]
- 33. Volz T, Allweiss L, Ben MBarek M, et al. The entry inhibitor Myrcludex-B efficiently blocks intrahepatic virus spreading in humanized mice previously infected with hepatitis B virus. J Hepatol 2013;58:861–7. 10.1016/j.jhep.2012.12.008 [DOI] [PubMed] [Google Scholar]
- 34. Kornyeyev D, Ramakrishnan D, Voitenleitner C, et al. Spatiotemporal analysis of hepatitis B virus X protein in primary human hepatocytes. J Virol 2019;93. 10.1128/JVI.00248-19. [Epub ahead of print: 15 08 2019]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Allweiss L, Volz T, Giersch K, et al. Proliferation of primary human hepatocytes and prevention of hepatitis B virus reinfection efficiently deplete nuclear cccDNA in vivo. Gut 2018;67:542–52. 10.1136/gutjnl-2016-312162 [DOI] [PubMed] [Google Scholar]
- 36. Xia Y, Protzer U. Control of hepatitis B virus by cytokines. Viruses 2017;9:18. 10.3390/v9010018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhang X, Lu W, Zheng Y, et al. In situ analysis of intrahepatic virological events in chronic hepatitis B virus infection. J Clin Invest 2016;126:1079–92. 10.1172/JCI83339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Balagopal A, Hwang HS, Grudda T, et al. Single hepatocyte hepatitis B virus transcriptional landscape in HIV coinfection. J Infect Dis 2020;221:1462–9. 10.1093/infdis/jiz607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Stadelmayer B, Diederichs A, Chapus F, et al. Full-Length 5'RACE identifies all major HBV transcripts in HBV-infected hepatocytes and patient serum. J Hepatol 2020;73:40–51. 10.1016/j.jhep.2020.01.028 [DOI] [PubMed] [Google Scholar]
- 40. Venegas AB, Natsume T, Kanemaki M, et al. Inducible degradation of the human Smc5/6 complex reveals an essential role only during interphase. Cell Rep 2020;31:107533. 10.1016/j.celrep.2020.107533 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
gutjnl-2020-322571supp001.pdf (8.8MB, pdf)
Data Availability Statement
All data relevant to the study are included in the article or uploaded as supplementary information. Additional data (eg, detailed protocols) are available upon request to m.dandri@uke.de.