Abstract
Objective
A detailed understanding of the molecular alterations in different forms of cholangiocarcinogenesis is crucial for a better understanding of cholangiocarcinoma (CCA) and may pave the way to early diagnosis and better treatment options.
Design
We analysed a clinicopathologically well-characterised patient cohort (n=54) with high-grade intraductal papillary (IPNB) or tubulopapillary (ITPN) neoplastic precursor lesions of the biliary tract and correlated the results with an independent non-IPNB/ITPN associated CCA cohort (n=294). The triplet sample set of non-neoplastic biliary epithelium, precursor and invasive CCA was analysed by next generation sequencing, DNA copy number and genome-wide methylation profiling.
Results
Patients with invasive CCA arising from IPNB/ITPN had better prognosis than patients with CCA not associated with IPNB/ITPN. ITPN was localised mostly intrahepatic, whereas IPNB was mostly of extrahepatic origin. IPNB/ITPN were equally associated with small-duct and large-duct type intrahepatic CCA. IPNB exhibited mutational profiles of extrahepatic CCA, while ITPN had significantly fewer mutations. Most mutations were shared between precursor lesions and corresponding invasive CCA but ROBO2 mutations occurred exclusively in invasive CCA and CTNNB1 mutations were mainly present in precursor lesions. In addition, IPNB and ITPN differed in their DNA methylation profiles and analyses of latent methylation components suggested that IPNB and ITPN may have different cells-of-origin.
Conclusion
Integrative analysis revealed that IPNB and ITPN harbour distinct early genetic alterations, IPNB are enriched in mutations typical for extrahepatic CCA, whereas ITPN exhibited few genetic alterations and showed distinct epigenetic profiles. In conclusion, IPNB/ITPN may represent a distinctive, intermediate form of intrahepatic and extrahepatic cholangiocarcinogenesis.
Keywords: cholangiocarcinoma, bilary duct carcinoma, gene mutation
Significance of this study.
What is already known on this subject?
Cholangiocarcinoma (CCA) is a highly aggressive disease with poor prognosis due to late diagnosis. Intraductal papillary (IPNB) and tubulopapillary (ITPN) neoplasms of the bile duct have been described as precursor lesions of CCA.
The genetic and epigenetic alterations involved in the stepwise progression from non-neoplastic biliary epithelium via precursor lesion to invasive CCA are poorly understood.
What are the new findings?
Patients with invasive CCA arising from IPNB/ITPN had better prognosis than patients with non-IPNB/ITPN-CCA.
IPNB and ITPN harbour distinct early genetic alterations.
IPNB are enriched in mutations typical for extrahepatic CCA, whereas ITPN exhibited few genetic alterations.
IPNB and ITPN showed distinct epigenetic profiles and they may represent a distinctive, intermediate form of intrahepatic and extrahepatic cholangiocarcinogenesis.
Certain intrahepatic IPNB and ITPN may represent a to-date not recognised precursor lesion for intrahepatic CCA of the small-duct type.
How might it impact on clinical practice in the foreseeable future?
These newly identified alterations corroborate IPNB and ITPN as unique forms of cholangiocarcinogenesis.
The presence of IPNB or ITPN may serve as an indicator for better patient survival.
The here identified molecular alterations may facilitate early diagnosis and improve treatment options for patients with CCA.
Introduction
Cholangiocarcinoma (CCA) is a highly aggressive disease with poor prognosis due to late diagnosis and chemotherapy resistance.1–3 Based on the anatomic location, CCA is classified as intrahepatic (iCCA), perihilar (pCCA) or distal CCA (dCCA). Irrespective of the localisation, most CCA are unresectable at diagnosis and thus, early detection prior to malignant transformation as non-invasive precursor lesion may dramatically improve patient outcome. Two main subtypes of biliary precursor lesions that precede invasive CCA are nowadays recognised and described in detail in the current, 5th WHO classification of Digestive System Tumours: the microscopic biliary intraepithelial neoplasm (BilIN) and the macroscopically visible intraductal papillary or tubulopapillary neoplasms of the bile duct, IPNB and ITPN, respectively.4 5 While BilIN are frequently encountered in resection specimens of the biliary tract, IPNB and in particular ITPN are less frequent. However, the exact frequencies of IPNB and ITPN are unknown, and misclassification is still an issue. In addition, frequency of intraductal papillary and tubulopapillary biliary precursors seem to depend on geographical and aetiological factors.6–8 Moreover, there might be an underestimation of IPNB/ITPN frequency, because some cases may be misdiagnosed as papillary CCA. A more detailed understanding of cholangiocarcinogenesis may allow standardised screening approaches of high-risk patients and may lead to targeted therapies for unresectable patients. While IPNB has been regarded as the biliary counterpart of intraductal papillary mucinous neoplasm (IPMN) of the pancreas based on their histological similarities,9–11 there is rising evidence that IPMN and IPNB are not identical, in particular regarding morphology, molecular biology and clinical course.12 A recent Taiwanese study of 37 IPNB identified frequent mutations of KRAS (49%), GNAS (32%), RNF43 (24%), APC (24%), TP53 (24%) and CTNNB1 (11%) suggesting that IPNB is a heterogeneous disease involving multiple mutations leading to activation of MAP kinase, Wnt/β-catenin and cAMP signalling.13 However, it appears that mutation frequencies vary between geographic locations as GNAS mutation was a rare event affecting only 2.5% of IPNB in a European study.14 Furthermore, PRKACA and PRKACB gene fusions were exclusively found in intraductal oncocytic papillary neoplasms of the pancreas and bile duct suggesting that oncocytic papillary neoplasms may represent a molecularly distinct subtype.15
DNA methylation signatures have recently gained attention due to their capability of differentiating tumour and tissue types. This resulted in the recent development of promising algorithms to characterise cancers of unknown primary, brain tumours and sarcomas according to their epigenetic signatures.16–18 In addition, integrative genetic and epigenetic analyses may reveal certain subgroups in tumours that are morphologically identical but display distinct molecular patterns including targetable options. Taken together, only few genetic changes have been described in previous studies. Moreover, changes of protein expression of some candidate genes have been described in immunohistochemical studies. However, until now, a systematic and integrative approach in characterising this branch of cholangiocarcinogenesis is lacking. Here, we present a comprehensive and well-characterised single-centre cohort of solely IPNB or ITPN-associated CCA, which we analysed using an integrative histomorphological, genetic and epigenetic approach. We identified genetic differences of IPNB and ITPN, demonstrated that IPNB and ITPN have both similar and distinct mutational profiles compared to the corresponding invasive CCA and correlated these results with clinicopathological data. In addition, we analysed the epigenetic profiles of IPNB and ITPN using DNA methylation profiling indicating different cells-of-origin of IPNB and ITPN.
Materials and methods
Study population and histomorphological subclassification
This study included 54 patients with high-grade intraductal neoplasms of the bile duct and 9 patients with ITPN of the pancreas (ITPN-P; table 1, online supplemental table S1). In detail, 44 patients with IPNB and 10 patients with ITPN were included. IPNB was diagnosed applying the current histomorphological and immunohistochemical criteria provided by WHO5 and ITPN was diagnosed using the criteria of AFIP and keynote publications.19 20 In brief, the definition of ITPN as biliary intraductal neoplasms that are predominantly tubular/trabecular, non-mucinous and negative for mucin 5AC (MUC5AC) was used. Corresponding normal bile duct samples of 41 cases with intraductal neoplasm of the bile duct and of all 9 ITPN-P cases were available. In addition, corresponding invasive CCA tumour tissues of 34 patients with IPNB or ITPN were available. CCA were diagnosed and subgrouped using the current criteria provided by WHO and AFIP.19 For iCCA subgrouping (small-duct versus large-duct) WHO criteria were used (see also online supplemental table S2). All tissue samples were provided by the Tissue Bank of the National Center for Tumor Diseases (NCT, Heidelberg, Germany) in accordance with the regulations of the NCT Tissue Bank.
Table 1.
Clinicopathological characteristics of patients with intraductal neoplasms of the biliary tree and associated invasive CCA (n=54)
| Number (per cent) | Total | IPNB | ITPN | P value |
| Parameter | 54 (100.0) | 44 (81.5) | 10 (18.5) | |
| Age | ||||
| Median years (range) | 66 (39–81) | 68 (39–81) | 57 (40–75) | |
| Mean years (range) | 64.3 (39–81) | 65.2 (39–81) | 57.6 (40–75) | 0.038* |
| Sex | ||||
| Male | 40 (74.1) | 34 (77.3) | 6 (60.0) | |
| Female | 14 (25.9) | 10 (22.7) | 4 (40.0) | 0.424** |
| Location | ||||
| Intrahepatic | 16 (29.6) | 8 (18.2) | 8 (80.0) | |
| Perihilar | 12 (22.2) | 10 (22.7) | 2 (20.0) | |
| Distal | 26 (48.1) | 26 (59.1) | 0 (0.0) | <0.001*** |
| Histology precursor | ||||
| Pancreatobiliary | 38 (70.4) | 29 (65.9) | 9 (90.0) | |
| Gastric | 4 (7.4) | 4 (9.1) | 0 (0.0) | |
| Intestinal | 11 (20.4) | 11 (25.0) | 0 (0.0) | |
| Oncocytic | 1 (1.9) | 0 (0.0) | 1 (10.0) | 0.037*** |
| UICC† | ||||
| UICC0 | 4 (7.4) | 3 (6.8) | 1 (10.0) | |
| UICC1 | 14 (25.9) | 12 (27.3) | 2 (20.0) | |
| UICC2 | 20 (37.0) | 19 (43.2) | 1 (10.0) | |
| UICC3 | 5 (9.3) | 3 (6.8) | 2 (20.0) | |
| UICC4 | 5 (9.3) | 3 (6.8) | 2 (20.0) | |
| NA | 6 (11.1) | 4 (9.1) | 2 (20.0) | 0.189*** |
| pT | ||||
| Tis | 4 (7.4) | 3 (6.8) | 1 (10.0) | |
| T1 | 19 (35.2) | 14 (31.8) | 5 (50.0) | |
| T2 | 23 (42.6) | 19 (43.2) | 4 (40.0) | |
| T3 | 7 (13.0) | 7 (15.9) | 0 (0.0) | |
| T4 | 1 (1.9) | 1 (2.3) | 0 (0.0) | 0.607*** |
| pN | ||||
| N0 | 34 (63.0) | 30 (68.2) | 4 (40.0) | |
| N1 | 12 (22.2) | 9 (20.5) | 3 (30.0) | |
| NA | 8 (14.8) | 5 (11.4) | 3 (30.0) | 0.355** |
| M | ||||
| M0 | 49 (90.7) | 41 (93.2) | 8 (80.0) | |
| M1 | 5 (9.3) | 3 (6.8) | 2 (20.0) | 0.227** |
| G | ||||
| G1 | 2 (3.7) | 2 (4.5) | 0 (0.0) | |
| G2 | 38 (70.4) | 31 (70.5) | 7 (70.0) | |
| G3 | 10 (18.5) | 8 (18.2) | 2 (20.0) | |
| NA (Tis) | 4 (7.4) | 3 (6.8) | 1 (10.0) | 0.790*** |
| Invasive component | ||||
| Yes | 50 (92.6) | 41 (93.2) | 9 (90.0) | |
| No | 4 (7.4) | 3 (6.8) | 1 (10.0) | 0.571** |
Significant p-values (p<0.005) are shown in bold.
*Mann Whitney U-test; **Fisher’s exact test; ***χ² test.
†Cases with pNx had no lymph nodes resected; therefore, UICC status could not be assessed.
CCA, cholangiocarcinoma; IPNB, intraductal papillary neoplasm of the bile duct; ITPN, intraductal tubulopapillary neoplasm of the bile duct; NA, not available; UICC, Union for International Cancer Control.
gutjnl-2020-322983supp001.pdf (2.1MB, pdf)
For clinical analysis, the study population of patients with intraductal neoplasms was compared with an independent set of patients with CCA without intraductal neoplasms. All patients included underwent bile duct or liver surgery at the University Hospital Heidelberg between 1995 and 2016. In total, this cohort included 402 patients and for 294 of these staging and survival data were available.
Each non-neoplastic, precursor and invasive tumour sample was histologically confirmed by at least two experienced consultant pathologists.
Patients or the public were not involved in the design, or conduct, or reporting, or dissemination plans of our research.
Tissue microarrays and immunohistochemical analysis
A tissue microarray (TMA) was fabricated for all IPNB and ITPN samples. The morphological classification into IPNB and ITPN and histomorphological subtyping was supported by immunohistochemical analyses of MUC1, MUC2, MUC5AC, MUC6 and CDX2. In addition, the differential diagnosis of neuroendocrine neoplasia was excluded by performing immunohistochemistry of synaptophysin and chromogranin A for all cases. Technical details of TMA fabrication and immunohistochemistry are provided in the online supplemental material.
Statistical analysis
Statistical analysis and visualisation were performed using the computing environment R (http://www.R-project.org/) and GraphPad Prism 6. Median survival and corresponding 95% confidence interval (CI) were calculated by Kaplan-Meier survival analysis, and the survival distributions for each category were compared using the log-rank test. All reported p values were two-sided and p<0.05 was considered statistically significant. Enrichment analysis was performed using Fisher’s exact or χ² test as appropriate.
Graphical representations of mutational differences by oncoplots and circos plots were created by the publicly available R-packages ComplexHeatmap, circlize and factoextra (http://www.R-project.org/; https://bioconductor.org/).21 Due to limitations in visualising multidimensional data, only pairwise co-occurrences are represented in the circos plots. Frequencies shown by the oncoplots are on a sample per gene basis and do not consider that some genes may contain more than one mutation in the same tumour sample.
Data availability
The Infinium MethylationEPIC BeadChip data generated in this study are available at the Gene Expression Omnibus (GEO) repository under accession GSE156299. All relevant data are available from the authors on request.
Online supplemental methods including TMA fabrication, diagnostic criteria, immunohistochemistry, genomic profiling and DNA methylation profiling are available online.
Results
Clinicopathological characteristics of the study cohort
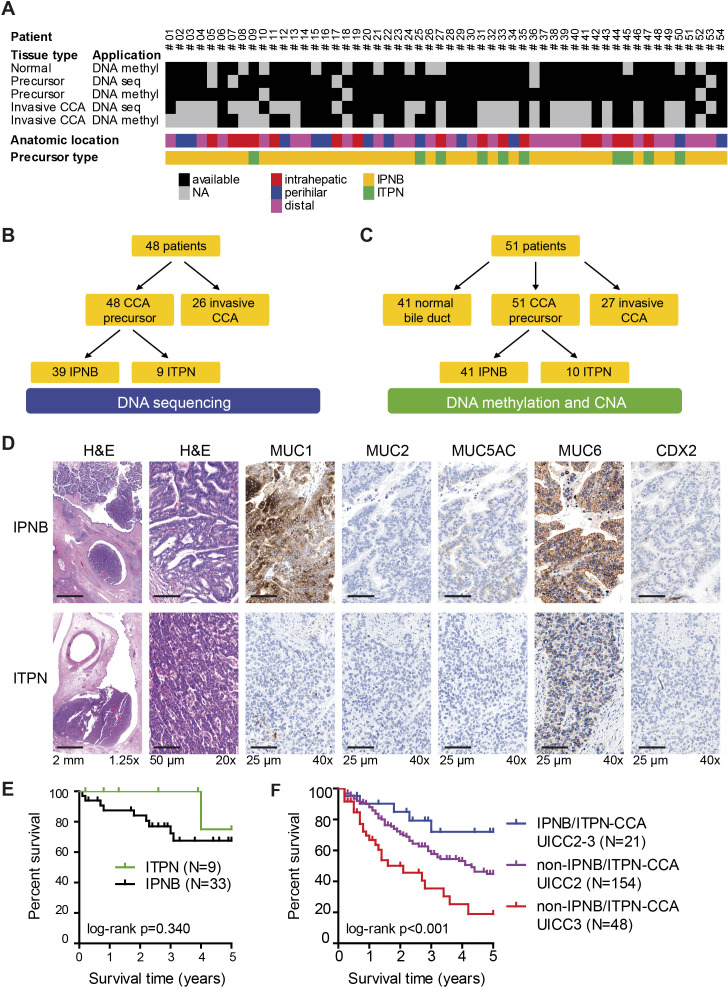
This study included 54 patients with high-grade intraductal neoplasms of the bile duct with intrahepatic, perihilar and distal anatomical location (table 1 and figure 1A). DNA sequencing, methylation and copy number alteration (CNA) analyses were performed for these precursor lesions, the corresponding non-neoplastic bile duct and invasive CCA (figure 1A–C). The intraductal neoplasms of the bile duct were further subdivided based on histomorphological and immunohistochemical characterisation into 44 cases of high-grade IPNB and 10 cases of high-grade ITPN (figure 1D). In addition, it was confirmed that neuroendocrine neoplasias have not been included in this study (online supplemental figure S1). Concomitant invasive CCA was overall present in 92.6% (50/54) of patients, that is, in 93.2% (41/44) of IPNB and 90% (9/10) of ITPN, respectively, and follow-up data were available for 42 patients (table 1). Consistent with other European studies, underlying hepatobiliary diseases associated with the development of CCA included unspecific chronic cholecystitis, cholangitis and pancreatitis. In addition, specific underlying hepatobiliary diseases, such as primary sclerosing cholangitis, IgG4-associated cholangitis, viral hepatitis B, hemochromatosis or Caroli syndrome, were detected overall in 16.7% of patients with IPNB/ITPN (online supplemental table S3).
Figure 1.
Study design, sample and patient characterisation. (A) A total of 54 patients with IPNB or ITPN neoplasms of the biliary tree were included in this study. The heatmap indicates the available DNA sequencing and DNA methylation/CNA data for IPNB/ITPN precursor lesions, invasive CCA and normal tissue. For 45 precursors lesions all three analyses, DNA sequencing, DNA methylation and CNA data were obtained. (B) For DNA sequencing, precursor lesions of 48 patients and 26 corresponding invasive CCA samples were analysed. (C) Genome-wide DNA methylation and genome-wide CNA profiling was performed for precursor lesions of 51 patients and compared with 41 corresponding non-neoplastic bile duct (normal) and 27 corresponding invasive CCA samples. (D) Immunohistochemical and histomorphological subtyping was performed for the entire cohort; immunohistochemical characterisation of one representative IPNB (top row) and one representative ITPN (bottom row) case is depicted. (E) Kaplan-Meier survival curves of IPNB and ITPN cases. (F) Kaplan-Meier survival curves of IPNB/ITPN-CCA cases with UICC2 or UICC3 compared with non-IPNB/ITPN-CCA cases of an independent cohort with matched numbers of UICC2 or UICC3, respectively. CCA, cholangiocarcinoma; CNA, copy number alteration; IPNB, intraductal papillary neoplasm of the bile duct; ITPN, intraductal tubulopapillary neoplasm of the bile duct; NA, not available.
Survival analysis showed that the patients with IPNB-associated and ITPN-associated CCA of our cohort had high 5-year survival rates of 67.6% and 75%, respectively (figure 1E). To test if the favourable outcome of IPNB/ITPN-associated CCA was due to early detection, we compared the survival of patients with or without IPNB/ITPN-associated CCA and drew on our CCA cohort of 294 patients without intraductal neoplasms of the bile duct (non-IPNB/ITPN-CCA) for which staging and survival data were available. These 294 patients with CCA but without intraductal neoplasms of the bile duct had mainly UICC2 (n=154) or UICC3 (n=81) stage (online supplemental figure S2A). As UICC staging is strongly associated with patient outcome, we compared the UICC stages of patients with IPNB/ITPN and non-IPNB/ITPN-associated CCA. This revealed that UICC staging was associated with patient outcome in CCA without IPNB/ITPN but not in IPNB/ITPN-associated CCA (online supplemental figure S2A, B). In addition, we combined patients into low-stage (UICC 0–2) and high-stage (UICC 3–4) patient groups to increase case numbers in the IPNB/ITPN-associated CCA group. This revealed that UICC staging was significantly associated with overall survival in non-IPNB/ITPN-CCA but not in patients with IPNB/ITPN-CCA (online supplemental figure S2C, D). Last, we compared the overall survival of patients with IPNB/ITPN-CCA with patients without IPNB/ITPN-CCA by combining the largest UICC staging group of patients (UICC 2–3; intermediate group) and by using proportional numbers of UICC 2 and UICC 3 for each cohort. This comparative retrospective analysis showed that patients with IPNB/ITPN-CCA have significantly better outcome than patients without IPNB/ITPN-CCA (log-rank p<0.001; figure 1F).
Next, correlation analyses of clinicopathological data revealed that patients with IPNB were older than patients with ITPN (p=0.038) but no significant differences in sex or tumour staging were detected. Only 18.2% (8/44) of IPNB but 80.0% (8/10) of ITPN had an intrahepatic localisation (Fisher’s exact p<0.001, table 1). For 16 patients with intrahepatic intraductal high-grade precursor lesion, 11 corresponding invasive iCCA were available and subtyping according to the current WHO classification5 showed that 45.5% (5/11) were small-duct and 54.5% (6/11) were large-duct iCCA. Of these 11 cases, 4 cases had IPNB with large-duct iCCA, 2 cases had IPNB with small-duct iCCA, 2 cases had ITPN with large-duct iCCA and 3 cases had ITPN with small-duct iCCA (Fisher’s exact p=0.567, online supplemental figures S3–S5). Histomorphology of all iCCA with corresponding IPNB or ITPN is depicted in online supplemental figure S4 and online supplemental figure S5 for large-duct and small-duct type, respectively. Histologically, IPNB had a high frequency of pancreatobiliary subtype (65.9%, 29/44), followed by intestinal (25%, 11/44) and gastric histology (9.1%, 4/44). Oncocytic differentiation was not observed in our IPNB cohort. Interestingly, only one ITPN was oncocytic (10%, 1/10), while the remaining ITPN (90%, 9/10) were not otherwise specified and intestinal or gastric histology was not seen in our ITPN cohort (Fisher’s exact test, p=0.037; table 1). In addition, IPNB subgrouping according to the Japan-Korea Cooperative Study Group22 showed no significant correlation to other clinicopathological or molecular variables (online supplemental table S4). In conclusion, ITPN was significantly more frequent intrahepatic compared with IPNB and for both precursor types an association with iCCA of the small-duct subtype was seen (online supplemental figure S5).
Mutational profiles of intraductal papillary and tubulopapillary neoplasms of the bile duct
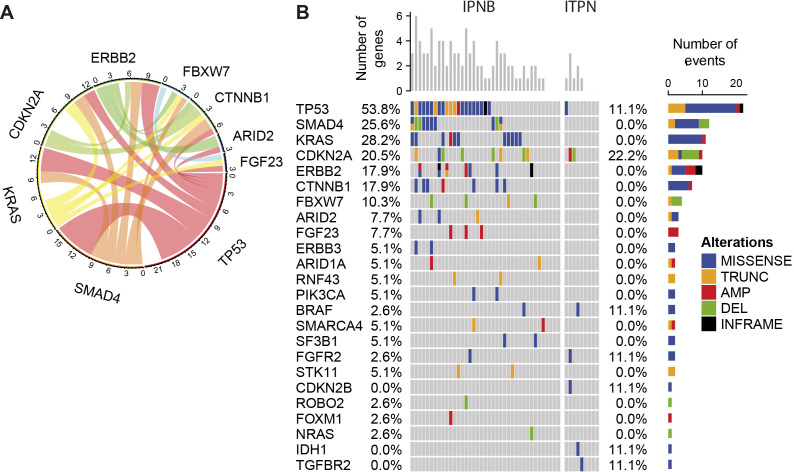
The 54 precursor lesions were analysed by massive parallel next generation sequencing and 48 samples (39 IPNB and 9 ITPN) passed the DNA quality control (figure 1A). In 83.3% (40/48) of precursor lesions, at least one genomic alteration was detected. A total of 137 non-synonymous mutations in 24 genes and 39 CNAs in 13 of the 40 targeted genes were identified. Mutations with the highest frequency were TP53 (45.8%), KRAS (22.9%), SMAD4 (20.8%) and CDKN2A (20.8%; figure 2). The inter-relationship analysis by circos plot revealed that TP53 mutations are linked to KRAS and SMAD4 mutations (figure 2A). Next, the comparison of the 39 IPNB and 9 ITPN revealed that no mutation was detected in 44.4% (4/9) of ITPN but only in 10.3% (4/39) of IPNB (figure 2B). Furthermore, the number of mutations per sample was significantly higher in IPNB with a mean of 2.39 mutations per sample (95% CI 1.91 to 2.86) compared to ITPN with a mean of 0.89 mutations per sample (95% CI 0.08 to 1.70; Mann Whitney U-test, p=0.006).
Figure 2.
Massive parallel sequencing reveals frequent genetic alterations in intraductal neoplasms of the bile duct. (A) Circos plot representing co-occurrence of mutations in all 48 non-invasive IPNB and ITPN samples. A band connecting genes represents co-occurring mutations in a given patient. The width of the band represents the frequency of this mutation pair within the dataset. (B) Oncoplot of genomic alterations in IPNB (n=39) and ITPN (n=9) samples. Missense mutations, in-frame mutations, truncations, amplificationsand deletions of 48 patients with intraductal neoplasms are shown. AMP, amplification; DEL, deletion; INFRAME, in-frame mutation; IPNB, intraductal papillary neoplasm of the bile duct; ITPN, intraductal tubulopapillary neoplasm of the bile duct; TRUNC, truncation.
Next, we compared the mutational profiles of our IPNB/ITPN cases with publicly available data of these precursor lesions and found similar mutation frequencies for TP53, SMAD4, FBXW7 and BRAF (online supplemental table S5).13 23–25 Compared to a large cohort of patients with CCA, the here observed mutation frequencies of IPNB and ITPN mirrored the ones of invasive dCCA and iCCA, respectively.26 Consistent with the low frequency of intrahepatic IPNB and high frequency of intrahepatic ITPN, no IDH1 mutation was observed in IPNB but one ITPN case with associated small-duct type iCCA (online supplemental figure S5E) harboured a hotspot IDH1:p.Arg132Leu mutation which typically occurs in iCCA of the small-duct type (figure 2B, online supplemental table S6). In addition, one intrahepatic ITPN and its associated small-duct iCCA (online supplemental figure S5C) both contained a FGFR2:p.Tyr286Cys mutation and one intrahepatic IPNB and the associated small-duct iCCA showed both the identical ERBB2:p.Ala771_Met774dup mutation (online supplemental figure S5A).
One CTNNB1 amplification and six missense mutations were identified in IPNB exclusively (figure 2B, online supplemental table S6). Thus, ITPN exhibit distinct mutation profiles and harbour overall significantly fewer mutations than IPNB suggesting distinct oncogenic mechanisms in ITPN-derived cholangiocarcinogenesis.
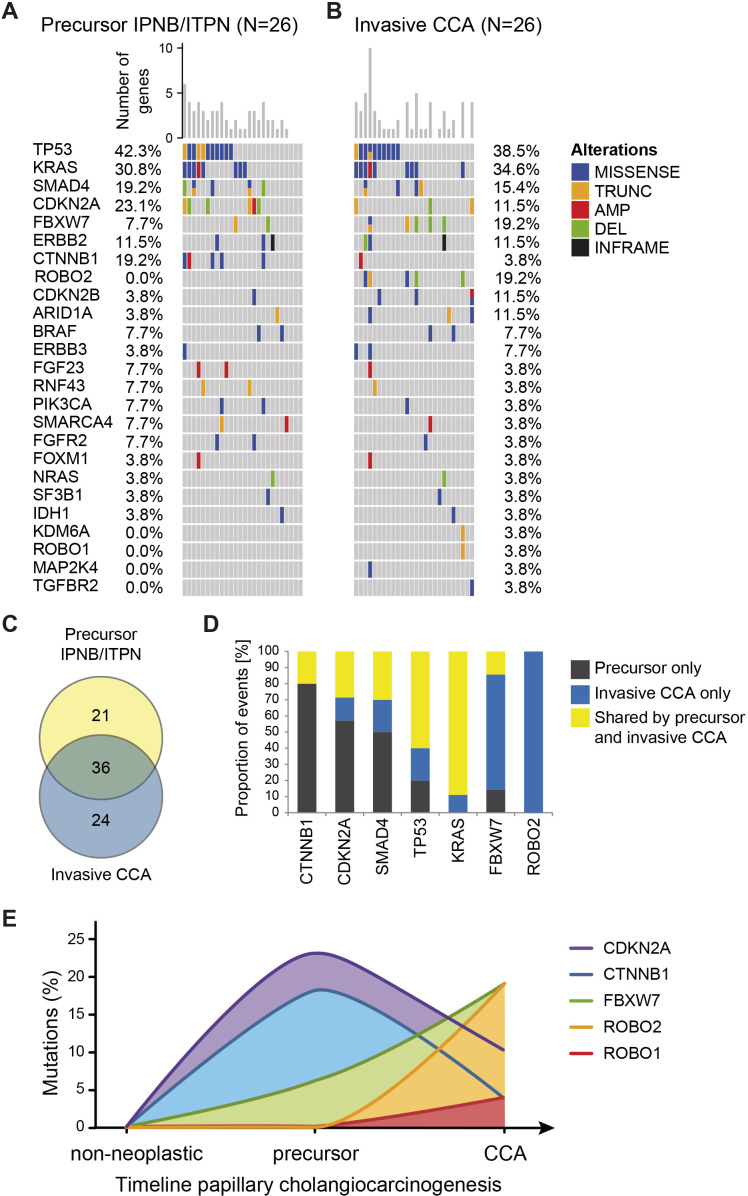
Invasive CCA evolve from IPNB or ITPN and distinct mutational changes occur during tumour evolution
The mutational profiles of corresponding invasive CCA were available for 26 cases. Overall, most mutations were shared between the corresponding precursor lesion and invasive CCA samples (figure 3). Mutation frequencies of TP53 (42.3% vs 38.5%), KRAS (30.8% vs 34.6%) and SMAD4 (19.2% vs 15.4%) were almost identical in precursor lesions and invasive CCA, whereas CTNNB1 (19.2% vs 3.8%), CDKN2A (23.1% vs 11.5%), ROBO2 (0% vs 19.2%) and FBXW7 (7.7% vs 19.2%) differed between precursor lesions and corresponding invasive CCA (online supplemental table S7). Of the 60 distinct mutations observed in invasive CCA and the 57 distinct mutations observed in the precursor lesions, 36 mutations were common for both (figure 3C), supporting the hypothesis that invasive CCA evolved from the corresponding precursor lesion. Furthermore, we performed case-by-case comparison of each alteration distinguishing mutations observed in the precursor lesion only, in the corresponding invasive CCA only or shared by the precursor lesion and corresponding invasive CCA. Among the genes with mutations shared between precursor lesion and invasive CCA were TP53 and KRAS (figure 3D). Mutations in several genes decreased during cholangiocarcinogenesis, including CTNNB1, CDKN2A and SMAD4 which were predominantly observed in the precursor lesion only. In contrast, alterations of other genes increased in CCA development via IPNB or ITPN, in particular ROBO2 and FBXW7 (figure 3D). Thus, CTNNB1 and CDKN2A may be involved in the development of IPNB/ITPN, whereas ROBO2, FBXW7 and CDKN2B may drive late cholangiocarcinogenesis and transformation to invasive CCA.
Figure 3.
IPNB/ITPN and corresponding invasive CCA tissue samples share genetic molecular profiles. (A) Oncoplot of genomic alterations in IPNB/ITPN (n=26) and (B) in the corresponding invasive CCA samples (n=26). Missense mutations, in-frame mutations, truncations, amplifications and deletions are shown. Genes (rows) of both oncoplots (A, B) were sorted by the total number of mutations in all samples. In addition, the order of samples (columns) in both oncoplots is based on the number of alterations in the precursor samples; thus, invasive CCA are sorted identical to the corresponding precursor lesions. (C) Overlap of specific mutations, that is, mutation of a certain amino acid in the respective gene, present in IPNB/ITPN precursor lesions and corresponding invasive CCA. (D) Shown are the proportions of mutations observed exclusively in the precursor lesions, exclusively in the invasive CCA or shared by the precursor lesion and corresponding invasive CCA in the indicated genes. (E) Diagram depicting the mutational alterations during progression from non-neoplastic normal bile to precursor and invasive CCA. AMP, amplification; DEL, deletion; INFRAME, in-frame mutation; IPNB, intraductal papillary neoplasm of the bile duct; ITPN, intraductal tubulopapillary neoplasm of the bile duct; ITPN, intraductal tubulopapillary neoplasm of the bile duct; TRUNC, truncation.
Genome-wide copy number alterations
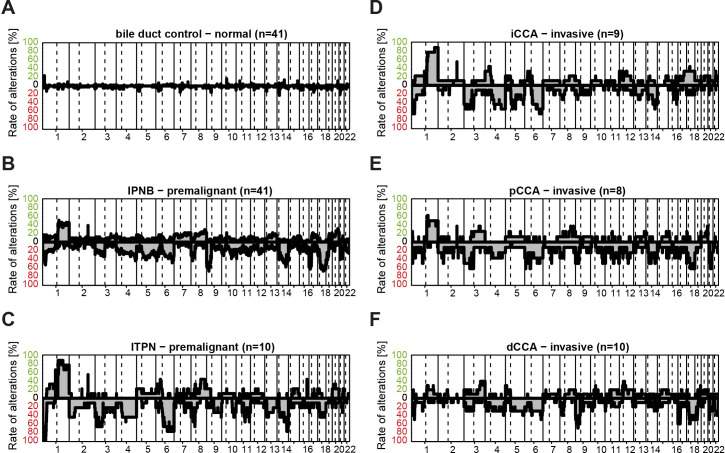
Genomic chromosomal imbalances have been observed in CCA by multiple studies.27–30 Using DNA methylation microarrays, we analysed the genomic profiles of normal bile duct (n=41), IPNB (n=41), ITPN (n=10) and the associated invasive CCA (iCCA: n=9; pCCA: n=8; dCCA: n=10) obtained by 850k DNA methylation analyses (figures 1 and 4). As expected, no significant CNA were observed in the normal bile duct controls (figure 4A). IPNB exhibited frequent deletions on chromosome 9p, 17p and 18q, whereas ITPN showed a high frequency of 1q gain and 6q loss (figure 4B, C). Interestingly, these genomic imbalances reflected the clinicopathologically observed differences in the anatomic localisation of IPNB/ITPN. Consistent with the notion that 80.0% of ITPN were associated with iCCA or had intrahepatic localisation, invasive iCCA genomes exhibited a high frequency of 1q amplification and 6q deletion (figure 4D). However, deletion of chromosome 18q, which is frequently detected in IPNB but not in ITPN, was observed in pCCA but not in iCCA (figure 4D, E). In addition, deletions of 9p and 17p, frequently detected in IPNB, were also frequent in the corresponding invasive dCCA tumour samples (figure 4F). Thus, IPNB/ITPN exhibit frequent CNA which appear to be stable during the transition from precursor to invasive carcinoma and specific to the anatomical location of the invasive carcinoma.
Figure 4.
Copy number alteration profiles of normal, IPNB or ITPN and invasive CCA samples. (A) Copy number alterations of normal bile duct controls, (B) IPNB, (C) ITPN, (D) iCCA, (E) pCCA and (F) dCCA samples are shown along the chromosomes on the x-axes. The relative frequency of observed gains (green) and losses (red) are depicted above and below the horizontal line, respectively. CCA, cholangiocarcinoma; dCCA, distal CCA; iCCA, intrahepatic CCA; IPNB, intraductal papillary neoplasm of the bile duct; ITPN, intraductal tubulopapillary neoplasm of the bile duct; pCCA, perihilar CCA.
DNA methylation profiles reveal distinct epigenetic profiles in IPNB and ITPN
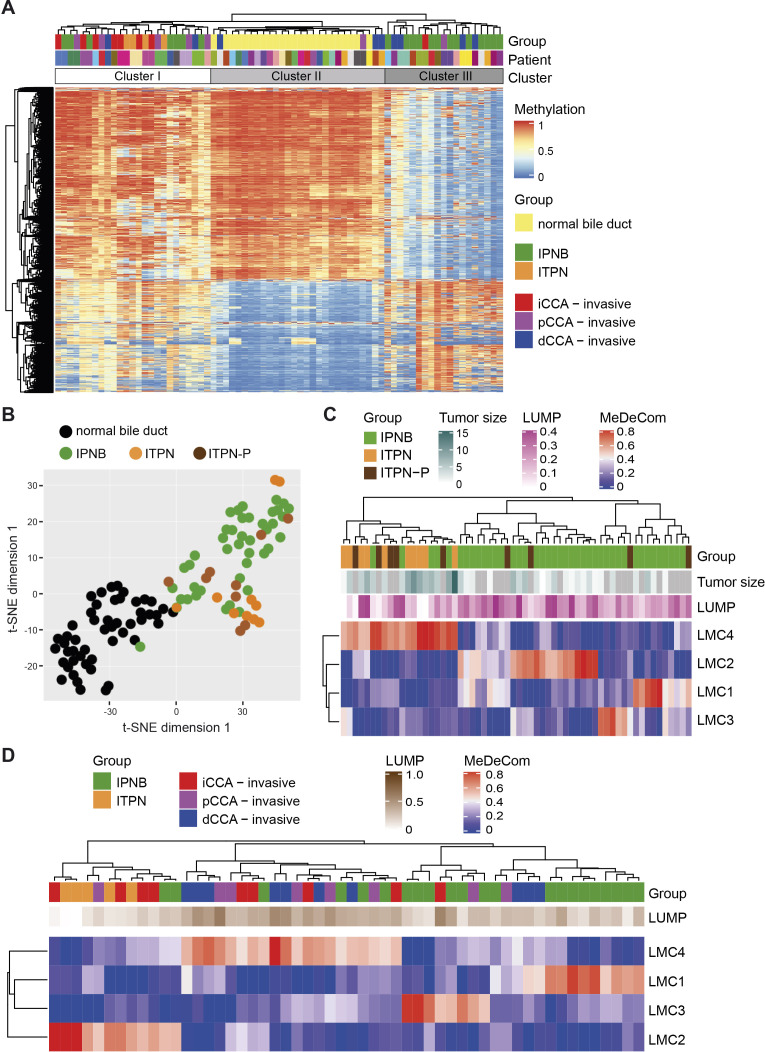
Epigenetic differences between IPNB and ITPN were detected in terms of divergent DNA methylation profiles. In detail, we compared DNA methylation patterns of non-neoplastic (normal) bile duct epithelia to corresponding biliary precursor lesions and invasive CCA. In addition, we included ITPN of the pancreas (ITPN-P) for comparison in the DNA methylation analysis (online supplemental table S1). Unsupervised clustering of all samples (n=137) resulted in two main clusters which included the non-neoplastic normal bile duct samples and the precursors with corresponding invasive CCA samples, respectively (online supplemental figure S6A). To directly compare the triplet samples of normal bile duct, precursor and invasive CCA, we restricted the clustering analysis to patients for whom all three samples were available (n=24). This resulted in separation into three clusters containing mainly the normal bile duct samples (cluster II), ITPN, iCCA and pCCA samples (cluster I) or IPNB and dCCA samples suggesting that the main difference could be observed between normal and precursor or invasive CCA samples (figure 5A). Furthermore, we observed that 14/27 precursor lesions and corresponding invasive CCA clustered directly next to each other proposing that paired precursor lesions and corresponding invasive CCA exhibit similar DNA methylation profiles (online supplemental figure S6B). Visualisation of DNA methylation patterns by two-dimensional t-SNE plot showed that non-neoplastic bile duct samples separated well from IPNB/ITPN samples (figure 5B). IPNB and ITPN did not appear as distinct groups in the t-SNE plot but subtle differences were detectable (figure 5B). To assess the cell type heterogeneity and to trace the cell‐of‐origin in the precursor samples, we used MeDeCom to decompose methylation data into latent methylation components (LMCs) and analysed the proportions of the resulting LMCs in each sample.31 Following this strategy, we found that IPNB, ITPN and ITPN-P samples separated into two main clusters of which one cluster contained all ITPN and 5/41 IPNB cases, whereas the second cluster contained the remaining 36 IPNB cases and no ITPN case (Fisher’s exact test p<0.001; figure 5C). Interestingly, ITPN-P cases were distributed equally in both clusters suggesting that the cells-of-origin of IPNB, ITPN and ITPN-P differ. Furthermore, it appeared that ITPN had high proportions of LMC1, whereas IPNB samples separated further into three groups with high LMC2, LMC3 or LMC4, respectively. To further dissect the similarities between precursor lesions and corresponding invasive CCA, we performed MeDeCom analysis of the 27 paired precursor and invasive CCA samples and found that all ITPN fell into the same cluster (figure 5D). Thus, all ITPN may have the same cell-of-origin but IPNB may arise from at least three different cell-of-origin types.
Figure 5.
DNA methylation profiles of IPNB and ITPN precursor lesions. (A) Unsupervised hierarchical clustering analysis of the triplet normal bile duct, precursor and corresponding invasive CCA of 24 patients. The total number of samples is 72. (B) t-SNE plot of the DNA methylation profiles of bile duct controls of the corresponding IPNB, ITPN and ITPN-P cases (n=50), IPNB (n=41), ITPN (n=10) and ITPN-P (n=9). (C) MeDeCom analysis of IPNB (n=41), ITPN (n=10) and ITPN-P (n=9) samples and (D) of 27 IPNB and ITPN samples with corresponding invasive CCA. CCA, cholangiocarcinoma; IPNB, intraductal papillary neoplasm of the bile duct; ITPN, intraductal tubulopapillary neoplasm of the bile duct; ITPN-P, ITPN of the pancreas.
Discussion
In this study, we characterised a comprehensive cohort of patients with IPNB or ITPN precursor lesions and associated invasive CCA and analysed the genetic and epigenetic alterations. By this integrative analysis and by using intraindividual triplet specimens of non-neoplastic tissue, precursor lesions and invasive CCA, we could delineate distinct molecular patterns in IPNB/ITPN-associated cholangiocarcinogenesis over time and correlate the findings with morphological subtyping and clinicopathological data. Consistent with previous studies of a cohort from Thailand and a mixed American, Asian and European cohort, we demonstrated that the simultaneous presence of IPNB or ITPN precursor lesions is a significant favourable prognostic factor regarding patient survival.25 32
We found that genome-wide DNA methylation profiles of ITPN differ from IPNB profiles, suggesting different epigenetic mechanisms in ITPN and IPNB. In addition, we performed MeDeCom analysis to dissect methylation patterns into LMCs, thereby gaining information on possible cell‐type composition differences. In conclusion, we could reveal (1) that overall DNA methylation patterns between IPNB and ITPN show only modest separation, (2) that IPNB and ITPN may have different cells-of-origin and (3) that IPNB show a higher inter-tumour heterogeneity compared with ITPN.
Panel sequencing data of our cohort confirmed well-known candidate genes that were already discovered in CCA before, such as TP53, KRAS, SMAD4, CDKN2A and ERBB2, to also play a pivotal role in cholangiocarcinogenesis via IPNB.26 33–35 Furthermore, we could deduce a genetic evolution plot presenting distinct mutations that emerge in IPNB/ITPN and vanish during transformation to invasive CCA. In particular, CTNNB1 and CDKN2A mutations occurred in a significant number of the precursor lesions and were then mostly lost in invasive CCA. In contrast, ROBO1, ROBO2 and FBXW7 gene alterations were increasingly detected in late cholangiocarcinogenesis, that is, invasive CCA, and were mostly absent in IPNB/ITPN. Noteworthy, the tumour suppressor gene FBXW7 was previously identified as an epigenetically regulated gene in CCA, and it could be shown that FBXW7 is a direct target for downregulation by miR-200a and miR-429, respectively.36 In addition to epigenetic silencing, genomic loss through chromosome 4q deletion may be an alternative mechanism for shutting down FBXW7.30 The tumour suppressive function of the E3-ubiquitin ligase FBXW7 has been suggested to depend on MYC activation, as a dominant negative form of MYC efficiently inhibited AKT/FBXW7-driven tumourigenesis in a CCA mouse model.37 Furthermore, ROBO1 and ROBO2 mutations were exclusively detected in invasive carcinoma. Recurrent ROBO receptor mutations have been also identified by other studies on CCA but ROBO receptor function has been mainly studied in neuronal development implicating a critical repulsive cue for axon path finding on binding of the Slit ligands.38–40 However, SLIT-ROBO signalling has more recently also been implicated in cancer cell migration, invasion and tumour angiogenesis.41–45 Thereby, ROBO proteins may either inhibit or promote tumourigenesis depending on the tumour entity and subtype. Our data suggest that ROBO1/2 and FBXW7 gene function may play a role in late tumour development and tumour progression of CCA.
Interestingly, all four IPNB cases with CTNNB1 mutation lost the CTNNB1 mutation during cholangiocarcinogenesis, which indicates that either CTNNB1 gene function is not necessary in late cholangiocarcinogenesis or other components of the ß-catenin signalling pathway are functionally crucial at this stage of CCA development. In line with our observation, nuclear ß-catenin accumulation was previously detected in IPNB lesions but not in invasive CCA of the same patients.14 46 Consistently, loss of IPNB-derived CTNNB1 and APC mutations during full malignant transformation has been observed by sequencing approaches.14 46 In contrast to IPNB lesions, neither ITPN nor CCA specimens of our cohort exhibited CTNNB1 mutations. Moreover, we have found in a previous study that epigenetic mechanisms, in particular DNA methylation patterns, play a pivotal role in Wnt pathway regulation of non-IPNB/ITPN associated pCCA and iCCA.47 Earlier, it could be shown that altered expression of CTNNB1 in iCCA may not be caused by genetic mutation.48 Our here presented data confirm these results and expand this concept to IPNB-derived cholangiocarcinogenesis. Similar to CTNNB1, CDKN2A alterations may be involved in early IPNB development but may not be required in invasive CCA. Thus, CTNNB1 may be involved in the development of IPNB and CDKN2A may be involved in the development of IPNB and ITPN, but both are possibly not needed for further progression to invasive CCA, whereas ROBO2, FBXW7 and CDKN2B may exert functional roles in late cholangiocarcinogenesis.
Although our sequencing results are overall consistent with previous studies, we did not find any GNAS mutation in our cohort (online supplemental table S5).13 49 Whether this finding reflects geographic, ethnic or etiological differences cannot be solved here. ITPN harboured significantly fewer mutations than IPNB suggesting distinct oncogenic mechanisms specific to each of these distinct precursor subtypes. This is in line with the observation that ITPN were more often localised intrahepatic in our cohort. Noteworthy, one intrahepatic ITPN was the precursor of a small-duct iCCA with IDH1 mutation (online supplemental table S5 and figure S5E), one intrahepatic ITPN and the associated small-duct iCCA (online supplemental figure S5C) both had the identical FGFR2 mutation and one intrahepatic IPNB and the associated small-duct iCCA showed both the identical ERBB2 mutation (online supplemental figure S5A). Interestingly, in one ITPN of oncocytic histomorphological subtype, no mutation was found in our sequencing approach, suggesting epigenetic instead of genetic alterations or alternative molecular pathways particularly involved in this form of cholangiocarcinogenesis. Indeed, PRKACA and PRKACB gene fusions have been found, for example, in pancreatic and biliary oncocytic papillary neoplasms and may be involved in ITPN development as well.15
According to the current WHO classification, iCCA is subdivided into the small-duct and large-duct type. Since large-duct iCCA are frequently accompanied by high-grade precursor lesions, most often BilIN, we were expecting that IPNB/ITPN would be more frequent in large-duct iCCA.5 Surprisingly, we detected a high proportion of small-duct iCCA in IPNB (33%, 2/6) and ITPN (60%, 3/5) cases. This finding is of particular importance as precursor lesions could not be identified for small-duct iCCA to date.
Molecular data on ITPN of the bile duct are scarce. This is the first comprehensive study identifying distinct genomic and epigenomic alterations in a Western-world cohort including ITPN precursor lesions. In conclusion, our integrative morphomolecular analyses define the molecular landscape of IPNB/ITPN-associated CCA development and propose a putative precursor lesion for small-duct iCCA.
Acknowledgments
We are grateful to Veronika Geißler, Nina Wilhelm and Carolin Kerber (Tissue Bank of the National Center for Tumor Diseases (NCT) Heidelberg, Germany and Institute of Pathology Heidelberg) and Angelika Fraas (Institute of Pathology Heidelberg) for excellent technical assistance. Tissue samples were provided by the tissue bank of the National Center for Tumor Diseases (NCT; Heidelberg, Germany) in accordance with the regulations of the tissue bank and with approval of the Ethics Committee of Heidelberg University (S-206/2005; S-207/2005; S-519/2019).
Footnotes
Twitter: @IEspositoPATH
Contributors: BG initiated the study. BG and SR designed the research project. BG, AM, KH and AMS provided tissue samples and clinicopathological data. BG, DS, SF, ON and SR performed research experiments. BG, DS, SF, MAL, RT, YA, MNV, ON, DW, CP, BK, CS, IE, AVD and SR analysed data or tissue samples. BG and SR wrote the manuscript. All authors read, commented on and approved the manuscript.
Funding: This work was supported by Deutsche Forschungsgemeinschaft (DFG, German Research Foundation)—Project-ID 314905040—SFB/TR 209 Liver Cancer (B01 to SR and Z01, INF to PS) and the European Union’s Horizon 2020 research and innovation programme under grant agreement No 667273 (HEP-CAR) to PS and SR. This study was in part supported by funds from German Cancer Aid (Deutsche Krebshilfe, project no. 70113922) to SR and from the Helmholtz Foundation to CP. We thank the Genomics and Proteomics Core Facility (GPCF) of DKFZ for performing the DNA methylation profiling.
Competing interests: PS: Grant, boards and presentations from Novartis, and boards from Incyte. CP: Advisory board honoraria from BioMedX. BG: Advisory board from Novartis.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Data availability statement
The Infinium MethylationEPIC BeadChip data generated in this study are available at the Gene Expression Omnibus (GEO) repository under accession GSE156299 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE156299). Data are publicly available without any restrictions.
Ethics statements
Patient consent for publication
Not required.
Ethics approval
The study protocol conformed to the ethical guidelines of the 1975 Declaration of Helsinki and was approved by the ethics committee of the University of Heidelberg (S-206/2005, S-207/2005 and S-519/2019).
References
- 1. Banales JM, Cardinale V, Carpino G, et al. Expert consensus document: cholangiocarcinoma: current knowledge and future perspectives consensus statement from the European network for the study of cholangiocarcinoma (ENS-CCA). Nat Rev Gastroenterol Hepatol 2016;13:261–80. 10.1038/nrgastro.2016.51 [DOI] [PubMed] [Google Scholar]
- 2. Valle JW, Borbath I, Khan SA, et al. Biliary cancer: ESMO clinical practice guidelines for diagnosis, treatment and follow-up. Ann Oncol 2016;27:v28–37. 10.1093/annonc/mdw324 [DOI] [PubMed] [Google Scholar]
- 3. Rizvi S, Khan SA, Hallemeier CL, et al. Cholangiocarcinoma - evolving concepts and therapeutic strategies. Nat Rev Clin Oncol 2018;15:95–111. 10.1038/nrclinonc.2017.157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Nagtegaal ID, Odze RD, Klimstra D, et al. The 2019 who classification of tumours of the digestive system. Histopathology 2020;76:182–8. 10.1111/his.13975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. WHO Classification of Tumours Editorial Board . Who classification of tumours: digestive system tumours. 5th Edn. World Health Organization, 2019. [Google Scholar]
- 6. Wan X-S, Xu Y-Y, Qian J-Y, et al. Intraductal papillary neoplasm of the bile duct. World J Gastroenterol 2013;19:8595–604. 10.3748/wjg.v19.i46.8595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rocha FG, Lee H, Katabi N, et al. Intraductal papillary neoplasm of the bile duct: a biliary equivalent to intraductal papillary mucinous neoplasm of the pancreas? Hepatology 2012;56:1352–60. 10.1002/hep.25786 [DOI] [PubMed] [Google Scholar]
- 8. Yeh T-S, Tseng J-H, Chen T-C, et al. Characterization of intrahepatic cholangiocarcinoma of the intraductal growth-type and its precursor lesions. Hepatology 2005;42:657–64. 10.1002/hep.20837 [DOI] [PubMed] [Google Scholar]
- 9. Zen Y, Fujii T, Itatsu K, et al. Biliary papillary tumors share pathological features with intraductal papillary mucinous neoplasm of the pancreas. Hepatology 2006;44:1333–43. 10.1002/hep.21387 [DOI] [PubMed] [Google Scholar]
- 10. Zen Y, Sasaki M, Fujii T, et al. Different expression patterns of mucin core proteins and cytokeratins during intrahepatic cholangiocarcinogenesis from biliary intraepithelial neoplasia and intraductal papillary neoplasm of the bile duct--an immunohistochemical study of 110 cases of hepatolithiasis. J Hepatol 2006;44:350–8. 10.1016/j.jhep.2005.09.025 [DOI] [PubMed] [Google Scholar]
- 11. Klöppel G, Kosmahl M. Is the intraductal papillary mucinous neoplasia of the biliary tract a counterpart of pancreatic papillary mucinous neoplasm? J Hepatol 2006;44:249–50. 10.1016/j.jhep.2005.11.035 [DOI] [PubMed] [Google Scholar]
- 12. Nakanuma Y, Kakuda Y, Uesaka K. Characterization of intraductal papillary neoplasm of the bile duct with respect to the histopathologic similarities to pancreatic intraductal papillary mucinous neoplasm. Gut Liver 2019;13:617–27. 10.5009/gnl18476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yang C-Y, Huang W-J, Tsai J-H, et al. Targeted next-generation sequencing identifies distinct clinicopathologic and molecular entities of intraductal papillary neoplasms of the bile duct. Mod Pathol 2019;32:1637–45. 10.1038/s41379-019-0306-9 [DOI] [PubMed] [Google Scholar]
- 14. Schlitter AM, Born D, Bettstetter M, et al. Intraductal papillary neoplasms of the bile duct: stepwise progression to carcinoma involves common molecular pathways. Mod Pathol 2014;27:73–86. 10.1038/modpathol.2013.112 [DOI] [PubMed] [Google Scholar]
- 15. Singhi AD, Wood LD, Parks E, et al. Recurrent Rearrangements in PRKACA and PRKACB in Intraductal Oncocytic Papillary Neoplasms of the Pancreas and Bile Duct. Gastroenterology 2020;158:573–82. 10.1053/j.gastro.2019.10.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Moran S, Martínez-Cardús A, Sayols S, et al. Epigenetic profiling to classify cancer of unknown primary: a multicentre, retrospective analysis. Lancet Oncol 2016;17:1386–95. 10.1016/S1470-2045(16)30297-2 [DOI] [PubMed] [Google Scholar]
- 17. Capper D, Jones DTW, Sill M, et al. Dna methylation-based classification of central nervous system tumours. Nature 2018;555:469–74. 10.1038/nature26000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Koelsche C, Hartmann W, Schrimpf D, et al. Array-Based DNA-methylation profiling in sarcomas with small blue round cell histology provides valuable diagnostic information. Mod Pathol 2018;31:1246–56. 10.1038/s41379-018-0045-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Torbenson M, Zen Y, Yeh MM. AFIP atlas of tumor pathology. Tumors of the Liver: ARP Press, 2018. [Google Scholar]
- 20. Zen Y, Amarapurkar AD, Portmann BC. Intraductal tubulopapillary neoplasm of the bile duct: potential origin from peribiliary cysts. Hum Pathol 2012;43:440–5. 10.1016/j.humpath.2011.03.013 [DOI] [PubMed] [Google Scholar]
- 21. Gu Z, Eils R, Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 2016;32:2847–9. 10.1093/bioinformatics/btw313 [DOI] [PubMed] [Google Scholar]
- 22. Nakanuma Y, Jang K-T, Fukushima N, et al. A statement by the Japan-Korea expert pathologists for future clinicopathological and molecular analyses toward consensus building of intraductal papillary neoplasm of the bile duct through several opinions at the present stage. J Hepatobiliary Pancreat Sci 2018;25:181–7. 10.1002/jhbp.532 [DOI] [PubMed] [Google Scholar]
- 23. Nakanuma Y, Kakuda Y, Fukumura Y, et al. The pathologic and genetic characteristics of the intestinal subtype of intraductal papillary neoplasms of the bile duct. Am J Surg Pathol 2019;43:1212–20. 10.1097/PAS.0000000000001295 [DOI] [PubMed] [Google Scholar]
- 24. Aoki Y, Mizuma M, Hata T, et al. Intraductal papillary neoplasms of the bile duct consist of two distinct types specifically associated with clinicopathological features and molecular phenotypes. J Pathol 2020;251:38–48. 10.1002/path.5398 [DOI] [PubMed] [Google Scholar]
- 25. Schlitter AM, Jang K-T, Klöppel G, et al. Intraductal tubulopapillary neoplasms of the bile ducts: clinicopathologic, immunohistochemical, and molecular analysis of 20 cases. Mod Pathol 2016;29:93. 10.1038/modpathol.2015.106 [DOI] [PubMed] [Google Scholar]
- 26. Wardell CP, Fujita M, Yamada T, et al. Genomic characterization of biliary tract cancers identifies driver genes and predisposing mutations. J Hepatol 2018;68:959–69. 10.1016/j.jhep.2018.01.009 [DOI] [PubMed] [Google Scholar]
- 27. Chaisaingmongkol J, Budhu A, Dang H, et al. Common molecular subtypes among Asian hepatocellular carcinoma and cholangiocarcinoma. Cancer Cell 2017;32:57–70. 10.1016/j.ccell.2017.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Farshidfar F, Zheng S, Gingras M-C, et al. Integrative genomic analysis of cholangiocarcinoma identifies distinct IDH-Mutant molecular profiles. Cell Rep 2017;18:2780–94. 10.1016/j.celrep.2017.02.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jusakul A, Cutcutache I, Yong CH, et al. Whole-Genome and epigenomic landscapes of etiologically distinct subtypes of cholangiocarcinoma. Cancer Discov 2017;7:1116–35. 10.1158/2159-8290.CD-17-0368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Goeppert B, Toth R, Singer S, et al. Integrative analysis defines distinct prognostic subgroups of intrahepatic cholangiocarcinoma. Hepatology 2019;69:2091–106. 10.1002/hep.30493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lutsik P, Slawski M, Gasparoni G, et al. MeDeCom: discovery and quantification of latent components of heterogeneous methylomes. Genome Biol 2017;18:55. 10.1186/s13059-017-1182-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Luvira V, Pugkhem A, Bhudhisawasdi V, et al. Long-Term outcome of surgical resection for intraductal papillary neoplasm of the bile duct. J Gastroenterol Hepatol 2017;32:527–33. 10.1111/jgh.13481 [DOI] [PubMed] [Google Scholar]
- 33. Nakamura H, Arai Y, Totoki Y, et al. Genomic spectra of biliary tract cancer. Nat Genet 2015;47:1003–10. 10.1038/ng.3375 [DOI] [PubMed] [Google Scholar]
- 34. Goeppert B, Folseraas T, Roessler S, et al. Genomic characterization of cholangiocarcinoma in primary sclerosing cholangitis reveals therapeutic opportunities. Hepatology 2020;72:1253–66. 10.1002/hep.31110 [DOI] [PubMed] [Google Scholar]
- 35. Albrecht T, Rausch M, Rössler S, et al. Her2 gene (ErbB2) amplification is a rare event in non-liver-fluke associated cholangiocarcinogenesis. BMC Cancer 2019;19:1191. 10.1186/s12885-019-6320-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Goeppert B, Ernst C, Baer C, et al. Cadherin-6 is a putative tumor suppressor and target of epigenetically dysregulated miR-429 in cholangiocarcinoma. Epigenetics 2016;11:780–90. 10.1080/15592294.2016.1227899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wang J, Wang H, Peters M, et al. Loss of FBXW7 synergizes with activated Akt signaling to promote c-myc dependent cholangiocarcinogenesis. J Hepatol 2019;71:742–52. 10.1016/j.jhep.2019.05.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Dickson BJ, Gilestro GF. Regulation of commissural axon pathfinding by slit and its Robo receptors. Annu Rev Cell Dev Biol 2006;22:651–75. 10.1146/annurev.cellbio.21.090704.151234 [DOI] [PubMed] [Google Scholar]
- 39. Ong CK, Subimerb C, Pairojkul C, et al. Exome sequencing of liver fluke-associated cholangiocarcinoma. Nat Genet 2012;44:690–3. 10.1038/ng.2273 [DOI] [PubMed] [Google Scholar]
- 40. Kim Y-H, Hong E-K, Kong S-Y, et al. Two classes of intrahepatic cholangiocarcinoma defined by relative abundance of mutations and copy number alterations. Oncotarget 2016;7:23825–36. 10.18632/oncotarget.8183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zhao S-J, Shen Y-F, Li Q, et al. Slit2/Robo1 axis contributes to the Warburg effect in osteosarcoma through activation of SRC/ERK/c-MYC/PFKFB2 pathway. Cell Death Dis 2018;9:390. 10.1038/s41419-018-0419-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Dallol A, Morton D, Maher ER, et al. Slit2 axon guidance molecule is frequently inactivated in colorectal cancer and suppresses growth of colorectal carcinoma cells. Cancer Res 2003;63:1054–8. [PubMed] [Google Scholar]
- 43. Huang Z, Wen P, Kong R, et al. Usp33 mediates slit-robo signaling in inhibiting colorectal cancer cell migration. Int J Cancer 2015;136:1792–802. 10.1002/ijc.29226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Tseng R-C, Lee S-H, Hsu H-S, et al. Slit2 attenuation during lung cancer progression deregulates beta-catenin and E-cadherin and associates with poor prognosis. Cancer Res 2010;70:543–51. 10.1158/0008-5472.CAN-09-2084 [DOI] [PubMed] [Google Scholar]
- 45. Vaughen J, Igaki T. Slit-Robo repulsive signaling Extrudes tumorigenic cells from epithelia. Dev Cell 2016;39:683–95. 10.1016/j.devcel.2016.11.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Fujikura K, Akita M, Ajiki T, et al. Recurrent mutations in APC and CTNNB1 and activated Wnt/β-catenin signaling in intraductal papillary neoplasms of the bile duct: a whole exome sequencing study. Am J Surg Pathol 2018;42:1674–85. 10.1097/PAS.0000000000001155 [DOI] [PubMed] [Google Scholar]
- 47. Goeppert B, Konermann C, Schmidt CR, et al. Global alterations of DNA methylation in cholangiocarcinoma target the Wnt signaling pathway. Hepatology 2014;59:544–54. 10.1002/hep.26721 [DOI] [PubMed] [Google Scholar]
- 48. Sugimachi K, Taguchi K, Aishima S, et al. Altered expression of beta-catenin without genetic mutation in intrahepatic cholangiocarcinoma. Mod Pathol 2001;14:900–5. 10.1038/modpathol.3880409 [DOI] [PubMed] [Google Scholar]
- 49. Sasaki M, Matsubara T, Nitta T, et al. Gnas and KRAS mutations are common in intraductal papillary neoplasms of the bile duct. PLoS One 2013;8:e81706. 10.1371/journal.pone.0081706 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
gutjnl-2020-322983supp001.pdf (2.1MB, pdf)
Data Availability Statement
The Infinium MethylationEPIC BeadChip data generated in this study are available at the Gene Expression Omnibus (GEO) repository under accession GSE156299. All relevant data are available from the authors on request.
Online supplemental methods including TMA fabrication, diagnostic criteria, immunohistochemistry, genomic profiling and DNA methylation profiling are available online.
The Infinium MethylationEPIC BeadChip data generated in this study are available at the Gene Expression Omnibus (GEO) repository under accession GSE156299 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE156299). Data are publicly available without any restrictions.