Abstract
COVID-19 is both a viral illness and a disease of immunopathology. Proximal events within the innate immune system drive the balance between deleterious inflammation and viral clearance. We hypothesize that a divergence between the generation of excessive inflammation through over activation of the TLR associated myeloid differentiation primary response (MyD88) pathway relative to the TIR-domain-containing adaptor-inducing IFN-β (TRIF) pathway plays a key role in COVID-19 severity. Both viral elements and damage associated host molecules act as TLR ligands in this process. In this review, we detail the mechanism for this imbalance in COVID-19 based on available evidence, and we discuss how modulation of critical elements may be important in reducing severity of disease.
Keywords: TLR, COVID-19, innate immunity, interferon, CD14
The innate immune system's antiviral response
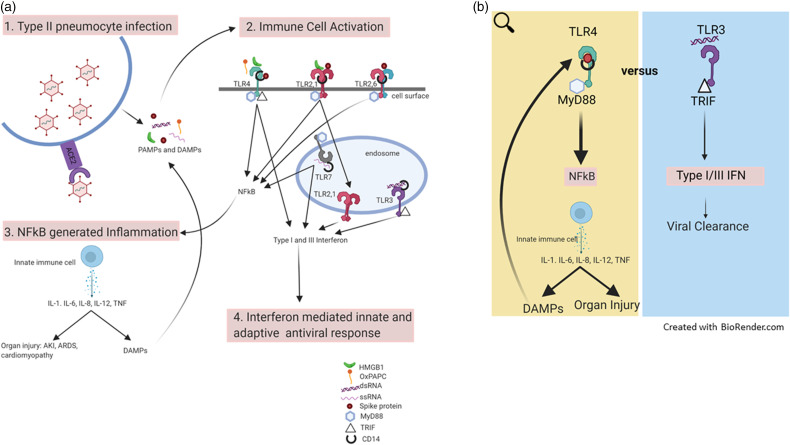
Upon infection with SARS-CoV-2, the virus that causes COVID-19, an innate immune response is activated through the phylogenetically preserved TLR family. TLRs are germline encoded PRRs that are believed to be important for both the protective and maladaptive response to lower respiratory viral infection. 1 Each TLR recognizes ligands comprised of both PAMPs elaborated by the virus and damage associated molecular patterns (DAMPs) released by host injured and dying cells. The leukocytes of the innate immune system, including neutrophils, monocytes, and NK cells, depend on TLR signaling to appropriately localize to the site of infection and target virus and virally infected host cells. 2 This initial response helps control viral infection but also causes cell damage and release of DAMPs, further TLR activation, and more inflammation. This cycle is thought to drive organ injury. TLR binding is necessary for the maturation of APCs through production of pro-inflammatory cytokines (e.g. TNF-α), chemokines, chemokine receptors, and co-stimulatory molecules. 1 These early innate immune responses initiate and direct the subsequent Ag-specific adaptive immune responses necessary for viral clearance. 3 This leads to a race between ongoing damage from direct viral infection and the innate hyperinflammatory response versus the protective innate and adaptive antiviral responses (Figure 1).
Figure 1.
(a) Schematic detailing the innate immune response in COVID-19 beginning with viral infection of type II pneumocytes. Viral elements (PAMPs) and DAMPs are generated and act as ligands for TLR activation. Several cell surface and endosomal TLRs are activated. Through MyD88-dependent and TRIF-dependent pathways, an NF-κB derived inflammatory response and an IFN response are generated. (b) NF-κB derived inflammation generates DAMPs and initiates a cycle of innate immune activation. This inflammation may drive organ injury. The IFN response is critical for viral clearance. As both pathways are active, a race between viral clearance and host injury ensue. In severe disease, the balance is shifted toward injurious NF-κB derived inflammation.
DAMP: damage associated molecular pattern.
In viral infection, it has been demonstrated that the cell surface receptors TLR1/2, TLR2/6, and TLR4 recognize structural components including proteins and glycolipids. TLR3, TLR7, TLR8, and TLR9 are expressed on endosomes and recognize nucleic acids. 4 Each TLR has an extracellular leucine-rich repeat domain and an intracellular Toll/IL-1 receptor (TIR) domain. 5 Upon TLR binding the TIR domain recruits adaptor proteins which leads to an MyD88- or TRIF-dependent signaling cascade. The MyD88-dependent pathway generates pro-inflammatory cytokine (Il-1, Il-6, TNF-α, etc.) production via transcription factor NF-κB. Alternatively, the TRIF-dependent pathway leads to activation of the type 1 and type III IFN critical for viral defense in addition to inflammatory cytokines. Depending on the TLR, cell type and subcellular compartment, downstream signaling through either MyD88 or TRIF can generate both pro-inflammatory cytokines and a type I/ III IFN response.6,7 The heterodimers TLR2/6 and TLR2/1 are MyD88-dependent as cell surface receptors but can traffic to endosomes and generate a type I IFN response. TLR3 recognizes dsRNA and signals solely through the TRIF pathway to generate a type I/III IFN response. TLR4 uses both MyD88- and TRIF-dependent pathways to generate a variable response. The prototypical TLR4 ligand is LPS but other ligands including glycolipids, viral structural proteins, and DAMPs such as HMGB1 have been identified.8,9 TLR7 and TLR8 recognize ssRNA and TLR9 recognizes unmethylated CpG DNA. TLR7 and TLR9 are MyD88-dependent but effectively generate a type I/ III IFN response. 10 Other PRRs including the cytosolic RNA sensing RIG-I-like receptors RIG-I and MDA5 also participate in viral recognition and response through TLR-independent mechanisms. 11 Therefore, upon infection with SARS-CoV-2, viral particles bind different TLRs and other PRRs and activate the innate immune system. This response consists of the generation of inflammation through transcription factor NF-κB and an antiviral response through the TRIF pathway. 12
The antiviral properties of TLR induced type I and type III IFN are critical for coordinated viral control and clearance. 13 Type I IFN consist predominantly of IFN-α and IFN-β and these cytokines signal through a common receptor IFNAR1/2. IFNAR1/2 binding leads to signal transduction via the JAK/STAT pathway and eventual formation of the transcription factor ISGF3 and expression of IFN-stimulated genes (ISGs). Type III IFN consist of the cytokines IFN-λ(1/2/3) which are structurally unique from IFN-α and IFN-β but exhibit similar functions; the main difference is that type III IFN receptors primarily localize to epithelial surfaces, for example airway epithelial cells. When type III IFN bind their receptor, similarly, signal transduction is initiated through the JAK/STAT pathway with eventual formation of ISGs. 14 Protein kinase R (PKR) and 2′-5′oligoadenylate are two well-characterized ISGs that act on translational machinery and nucleic acids directly to impair viral replication. 15 MERS-CoV, another coronavirus with high sequence homology to SARS-CoV-2, produces proteins that antagonized PKR so as to subvert the host antiviral response. 16 , 17 Within the innate immune system IFN production activates DCs, NK cells, and macrophages to directly clear and kill virally infected cells. IFN also play an immunomodulator role on APCs and lymphocytes to generate an effective adaptive immune response. 18 In vitro, SARS-CoV-2 is sensitive to IFN-α pre-treatment; specifically, when Vero and Calu3 cells are pre-treated with IFN-α, SARS-CoV-2 infection is unable to halt phosphorylation of STAT and downstream expression of ISG proteins resulting in resolution of SARS-CoV-2 infection. 19 However, there is limited data about the timing of TLR mediated type I IFN induction in COVID-19.
Concurrently to this TLR-mediated IFN response, TLR activation generates a robust inflammatory response through the MyD88-dependent pathway and transcription factor NF-κB. This pro-inflammatory response leads to an increase in cytokines and chemokines and an influx of activated immune cells (neutrophils, monocytes, NK cells) to the site of infection.2,20 Patients with severe COVID-19 compared to uninfected patients have higher serum levels of pro-inflammatory cytokines and chemokines including IL-2, IL-6, IL-1β, IL-8, IL-17, G-CSF, GM-CSF, IP10, MCP1, MIP1α, and TNF-α.21–23 This pro-inflammatory state and direct infection by virus lead to host cell death and organ injury. Damaged host cells release DAMPS like HMGB1 and oxPAPC that are capable of amplifying inflammation by TLR binding particularly through the MyD88-NF-κB pathway. Pro-inflammatory states are typically accompanied by an increase in anti-inflammatory cytokine production, serving as a negative feedback loop. IL-10 is a negative regulator of the immune response produced down-stream of MyD88. In translational studies, IL-10, like pro-inflammatory cytokines, is elevated in COVID-19, and it is higher in critically ill patients.24,25 Interestingly, when compared to critically ill community acquired pneumonia patients, critically ill COVID-19 patients had higher circulating IL-1β and IL-6 levels but less IL-10, potentially adding to the pro-inflammatory milieu. 26 Severe illness in COVID-19 is most commonly accompanied by lung injury or ARDS. 27 Emerging evidence also reports injury to multiple other organ systems: cardiac, liver, kidney, hematologic, and integumentary.28–31 Therefore, increased TLR activation and signaling through the MyD88 pathway and transcription factor NF-κB may drive the severity of clinical disease.
The immune response to SARS-CoV-2
In COVID-19, severe disease correlates with an immune signature characterized by an early cytokine and chemokine rich inflammatory response likely through NF-κB and very low level of antiviral IFN gene activation. This has been described in patients, in whom a low ISG signature in peripheral blood correlated with disease severity; additionally, in patients with severe disease an increased SARS-CoV-2 viral load was observed suggesting delayed viral clearance. 32 In this same study, elevated NF-κB pathway expression of IL-6 and TNF-α was also observed. 32 Similar findings have been observed across experimental methods including in vitro tissue culture, ex vivo infection of primary cells, and in vivo samples derived from patients and animal studies. 33 However, in contrast to these findings, Broggi et al. found that in COVID-19 disease severity correlated with increased IFN-λ expression within the lungs. Further, in an animal model, they found that exposure of murine lungs to a synthetic RNA virus led to lung DC IFN-λ production. This then caused epithelial barrier damage and an increased susceptibility to bacterial co-infection. 34 One proposed explanation for these discordant findings is that while local type III IFN production in the lungs may contribute to immunopathology, systemic type I and type III IFN production may be protective. IFN is being actively studied as a therapeutic in COVID-19.35–38 Intra-nasal IFN-α prophylaxis in addition to PPE reduced the incidence of SARS-CoV-2 infection in at risk personnel. 35 In severe COVID-19, in a small randomized controlled trial, systemic IFN-β therapy improved discharge rate and reduced mortality. 38 In another randomized control trial, IFN-β in combination with lopinavir-ritonavir and ribavirin compared to lopinavir-ritonavir alone reduced time to negative nasopharyngeal SARS-CoV-2 PCR result. 36 In a non-randomized retrospective cohort study, IFN-α with or without the antiviral arbidol improved time to viral clearance and reduced systemic inflammation. 37 Interestingly, in COVID-19 patients, IFN stimulation appears preserved when exogenously stimulated by other agonists; this raises the question of how upstream proximal factors in TLR activation may impact the adaptive immune response in COVID-19. 39
One possibility is that the virus itself may possess an evasion mechanism to protect against viral dsRNA and ssRNA recognition by TLRs 3 and 7 as was the case with SARS-CoV-1.40,41 Han et al. report that the SARS-CoV-2 accessory protein ORF9b impairs induction of types I and III IFN in part by targeting components of the TLR3-TRIF endosomal RNA sensing pathway in vitro. 42 Yin et al. report in vitro data describing that the PRRs MDA5 and LGP2, as opposed to TLR3, primarily recognize SARS-CoV-2 RNA and regulate a delayed IFN response in airway epithelial cells. 43 TLRs 3 and 7 are critical activators of the type I/III IFN antiviral response, and these data suggest SARS-CoV-2 evasion. Others suggest that DEAD-box RNA helicases in humans may induce a TLR independent anti-viral IFN response to SARS-CoV-2 though evidence is lacking. 44
The notion of a TLR mediated dys-regulated immune response was previously investigated in SARS. In a mouse model of SARS-CoV-1 infection, it was found that MyD88 knockout mice and TRIF knockout mice had similar mass loss and mortality. In this same study, the TRIF knockout mice had a more robust cytokine response and a higher viral load in type II pneumocytes. Additionally, it was found that TLR3 knockout mice, TLR4 knockout mice and TRAM (TLR4-TRAM-TRIF pathway) knockout mice all lost mass but recovered. These findings suggested redundancy in TLR viral recognition. It also suggested impaired IFN generation was associated with a relative increase in inflammation and decrease in viral clearance akin to clinical COVID-19 infection. 45 This suggests that while both the MyD88 pathway and TRIF pathway play a role in viral defense, the intact MyD88 pathway drives a robust inflammatory response, whereas, the TRIF pathway is critical for viral clearance. Similar studies using humanized animal models are needed to better understand the relative activation of MyD88 and TRIF-dependent pathways in response to TLR activation by SARS-CoV-2 infection.
TLR recognition of PAMPs initiates a coordinated adaptive immune response including directing the phenotype of lymphocytes and other white blood cells present. 2 A defective adaptive immune response has been associated with more severe COVID-19; 46 in fact, in one study the presence of a SARS-CoV-2 specific CD4 + T cells and CD8 + T cells was associated with less severe disease independent of a neutralizing Ab titer. 47 Lymphopenia and an increase in neutrophil count in comparison to other white blood cells are associated with disease severity and poor clinical outcome. 48 Type I/ III IFN are necessary for viral Ag presentation by DC cells and therefore a specific antiviral response. 49 Further, TGF-β, which is activated following acute inflammatory responses and down-regulates the immune response, may actually promote a chronic adaptive immune reaction with Ab production lacking specificity to SARS-CoV-2. This may contribute to persistent infection. 50 A poorly coordinated adaptive immune response may start with proximal TLR pathway events.
In COVID-19, the kinetics and details of the innate and adaptive immune responses are under active investigation. There is a need to better understand the role of TLRs in driving both the harmful and beneficial immune response in COVID-19. We suspect that a net increase in TLR-MyD88-NF-κB response and a relative decrease in TLR-TRIF-IFN antiviral response leads to immunopathology and ineffective viral clearance. In time, manipulation of TLRs and pathway components may prove critical to improved outcomes. Here we will review the data on the role of TLRs and associated molecules in COVID-19.
TLRs in COVID-19
TLR4
TLR4 is positioned to respond to both exogenous PAMPs and DAMPs released as part of tissue injury. TLR4 cooperates with MD-2 and CD14 to bind extra-cellular ligands. TLR4 signals both through the MyD88-dependent and TRIF-dependent pathways. LPS, a component of most Gram-negative bacterial cell envelopes, is its classic ligand. TLR4 activation has been implicated in the development of a pro-inflammatory state as in Gram-negative bacterial sepsis. This response is critical to host defense in Gram-negative bacterial infection. For example, mice strains with TLR4 genotypes that lead to LPS hypo-responsiveness have increased susceptibility to Gram-negative infections. 51 However, excessive LPS directed TLR4 stimulation can lead to damaging inflammation. In fact, the administration of lipid A (a component of LPS), is extremely toxic at high doses and leads to a hyper-inflammatory response and fulminant sepsis-like syndrome in animals. This response is thought to be driven primarily through MyD88 activation as opposed to TRIF signaling. 52 In vaccine development, scientists have looked to develop modified lipid A with a bias away from the MyD88 pathway and toward TRIF-IFN pathways so as to harness a protective and not maladaptive immune response. 52
In COVID-19, it is proposed that the viral structural proteins and glycolipids are PAMPS that bind TLR4 initiating an innate immune response. Computer-based modelling has found that the S protein of SARS-CoV-2 is predicted to bind to TLR4. 53 Further, emerging data shows that purified SARS-CoV-2 S protein binds TLR4 with strong affinity in vitro, and that purified S protein induces a TLR4-dependent IL-1β response akin to the response observed with LPS challenge. 9 The balance in downstream activation of MyD88 versus TRIF-dependent pathways is unknown. However, higher MyD88 activation relative to TRIF activation may contribute to excess NF-κB activation and inflammatory cytokine production as TLR4 has been heavily implicated in the injurious inflammation and lung injury seen SARS-CoV-1 and MERS. 54 TLR4 modulation or blockade has been shown to reduce lung injury in animal models by reducing NF-κB pathway activation. 55 Further work is needed to determine the extent by which TLR4 binding of viral PAMPs including the SARS-CoV-2 S protein drives lung injury and viral sepsis in COVID-19.
In addition to recognition of viral proteins, several DAMPS also act as ligands for TLR4. Viral infection and subsequent inflammation lead to the generation of DAMPs. For example, HSP60 and HSP70, released from virally infected and stressed cells are TLR4 agonists. 56 In addition, fibrinogen, an acute phase reactant, binds and activates TLR4. 57 HMGB1 is a DAMP that binds TLR4 and has been studied in the context of influenza-associated ALI. 58 OxPAPC, a heterogeneous group of lipids (oxidized phosphorylcholine derivatives) released from dying virally infected cells, is another DAMP that has been shown to promote inflammatory responses. TLR4 binding of oxPAPC in cooperation with CD14, generates inflammatory cytokines at levels akin to LPS binding. 59 Both the MyD88 and TRIF pathways are activated by DAMPS but there may be a bias toward MyD88 signaling and inflammatory cytokine production. In several viral pneumonia models, TLR4 blockade led to a reduction in inflammation, DAMP accumulation and ALI suggesting that TLR4 activation preferentially generates an injurious pro-inflammatory state. 60
TLR4 binding may impact the characteristics and efficacy of an adaptive immune response. The TLR4 pathway is capable of elaborating type I/III IFN which are necessary for a virus specific adaptive immune response. 2 Past work has shown that the dose of LPS signaling through TLR4 differentially regulates a Th1 versus Th2 response. 61 However if TLR4 binding leads to excess cytokine production, for example il-6 as is seen in COVID-19, cytotoxic CD8 + T cells and cell mediated immunity may be impaired. 62 To date, the effect of IL-6 blockade on clinical outcomes in COVID-19 has been mixed.63–65
TLR7
Functional TLR7 genetic variants have been linked to COVID-19 severity in multiple studies, strongly suggesting a key role for TLR7 in COVID-19 pathogenesis. TLR7 binds ssRNA and imidazoquinolones. While TLR7 signals through the MyD88-dependent pathway, downstream signaling leads to both NF-κB mediated inflammation and a type I/III IFN response; the balance between NF-κB mediated inflammation and the IFN response is critical. 66 Whole exome sequencing in families with multiple cases of COVID-19 identified rare loss of function variants in TLR7. Further, in vitro studies in these patients revealed that type I/III IFN responses to imiquimod, a strong TLR7 agonist, were greatly down-regulated in peripheral blood mononuclear cells. 67 These data suggest aberrant type I/III IFN responses due to a functional genetic mutation are specifically tied to severe COVID-19. Given that the gene for TLR7 is found on the X-chromosome it has also been proposed that TLR7 might play a role in sex-based differences in COVID-19 susceptibility and severity. Men are nearly two times as likely to develop respiratory failure or die from COVID-19. 68 Incomplete X-inactivation in immune cells resulting in an increased gene-dosage of TLR7 in women relative to men could lead to a more effective early antiviral response. 69 Common variants in TLR7 have also been identified in association with severe COVID-19 in an exploratory study using multilevel filtering (e.g. frequency, chromosomal, geographic, and functional filters). 70
Imiquod, a TLR7 an agonist, is currently under investigation as an immune modulator in COVID-19. 71 Imidazoquinolones have well-described antiviral properties through activation of TLR7 and the MyD88-dependent pathway with downstream type I/ III IFN generation. 72 Imiquod has been previously studied for its antiviral effects in anal condylomata in HIV. 73 In addition to its antiviral properties TLR7 activation may decrease inflammation. In an asthma model, TLR7 agonist RSQ reduced allergen induced airway inflammation and decrease reactive oxygen species. Interestingly, inhibition of MyD88 reversed RSQ-mediated effects. 74 It has also been studied as an immune modulator and as a vaccine adjuvant in influenza. 75
Additionally, Rutin, a flavonoid compound, that effectively binds TLR7 (and TLR2/TLR6) has been identified as a potential immune modulator. In addition to binding TLRs, rutin binds and potentially blocks SARS-CoV-2 main protease (Mpro). 76 Mpro is an important enzyme that mediates viral replication and transcription in coronaviruses, and it has been identified as an attractive target in drug development. 77 Rutin may play a role in both immune modulation via TLR agonism and an inhibitor of viral replication via Mpro antagonism. TLR7 is emerging as a proximal element in the immune response to viral infection that when activated may lead to downstream elaboration of a net antiviral response.
TLR3
TLR3 has the potential to improve both innate and adaptive immune responses in COVID-19 through type I/III IFN production. TLR3 agonists include dsRNA and poly IC, a synthetic RNA analogue. It is presumed to be critical in the recognition of respiratory RNA viruses like SARS-CoV-2; during viral replication, dsRNA is generated and stimulates endosomal TLR3 in addition to other intracellular receptors (RIG1/MDA5). 78 Unlike all other TLRs, TLR3 is TRIF dependent and MyD88 independent. Stimulation of TLR3 results in recruitment of the TRIF adaptor protein and down-stream activation of ISGs. TLR3 does not directly activate the pro-inflammatory NF-κB pathway. Because TLR3 activation favors type I/III IFN production, it is thought to be critical in viral defense. In fact, TLR3 mutations and deficiency have been described as monogenic drivers of other viral illnesses. Patients with TLR3 mutations and deficiency are highly susceptible to pediatric herpes virus encephalitis, 79 and patients with TLR3 deficiency are at higher risk of developing ARDS in Influenza A viral infection. 80 Fortunately, in these influenza ARDS cases, in vitro type I IFN therapy, the downstream product of TLR3 activation, reduced the susceptibility of airway epithelial cells and fibroblasts to influenza A viral infection suggesting modulation may be helpful. 80
Antiviral properties of TLR3 and disease modulation with TLR3 agonists have been described in several respiratory viral infections including SARS-CoV. In a SARS-CoV mouse model, TLR3/TRIF signaling played a critical role in the innate immune response through ISG protein production. 45 Additionally, in aged mice, intra-nasal poly IC protected mice from lethal SARS-CoV. 81 Synthetic TLR3 agonists have been studied in the context of numerous other viral infections in animal models. In fact, Poly IC or modified forms has been protected against the following viral infections in animal models: influenza virus, Herpes simplex virus type 2, western equine encephalitis virus, yellow fever virus, RSV, and others. 78 While these data suggests a protective role, the use of Poly IC has been largely limited by systemic toxicity. However, scientists have investigated modifications to limit adverse effects, and modified versions have been safely used as vaccine adjuvants.78,82 The protective role of synthetic Poly IC is currently being investigated in a phase 1 trial in COVID 19 (NCT: 04672291).
TLR2 heterodimers
The TLR1/TLR2 heterodimer binding by SARS-CoV-2 S protein may contribute to the hyper-inflammatory state and lung injury seen in COVID 19. TLR2 forms heterodimers with TLR1 and TLR6, respectively, to create functional cell surface receptors. In vitro and in vivo studies show that TLR2 recognizes the SARS-CoV-2 envelope protein resulting in MyD88-dependent inflammation. 83 In silico studies suggest that TLR1, TLR2, and TLR6 effectively bind the viral S protein in SARS-CoV-2. 53 In particular, specific peptides have been identified within the S protein as TLR2 agonists. It is not clear to what extent S protein binding of these heterodimers impacts the immune response in COVID-19. While these heterodimers typically signal via the MyD88 pathways, in certain immune cells TLR2 may be internalized and localize to endosomes facilitating a type I/III IFN response. 6 These specific peptides and TLR ligands within the S protein are being further investigated as SARS-CoV-2 specific and antigenic molecules that may be used in vaccine development. 84 As noted, TLR1 acts exclusively as a heterodimer with TLR2. In sepsis, TLR1 polymorphisms, in particular TLR1-7202G, may impact patient outcomes including the predisposition to the development of ARDS. Further in trauma related-sepsis, there was a trend towards a higher incidence of ARDS among patients homozygous for TLR1-7202G. 85 Using translation methods, it was shown that a specific TLR1 polymorphism was associated with increased TLR1 expression, TLR1 mediated NF-κB activity, and cytokine production. 86 TLR1 polymorphisms may also impact the regulation of the pro-inflammatory state through their influence on T regulatory cells. 87 This data suggests that functional variants in TLR1 lead to variable cytokine expression and lung injury. The SARS-CoV-2 S protein is predicted to bind TLR1/2; TLR1/2 agonism may contribute to the pathophysiology of cytokine storm and lung injury seen in patients with severe COVID-19.
The TLR immunomodulator PUL-042 is under investigation in COVID-19. PUL-042 is a combination drug comprised of the TLR2/6 ligand Pam2CSK4 and the TLR9 ligand ODN M362. It was developed in order to augment host response to respiratory infections. In fact, in animal models it has been shown to effectively improve survival and reduce pathogen burden in otherwise lethal SARS-CoV and MERS-CoV infections. 88
TLR5
TLR5 is most relevant in viral vaccine development as an adjuvant. TLR5 is activated by bacterial flagellin. It has been studied for immunotherapeutic development in respiratory infections because of its presence on respiratory tract epithelium; for this reason, it has been proposed as a candidate target in COVID-19 vaccine and drug development. 89 Administration of flagellin has been previously studied in influenza virus; in a mouse model, flagellin administration reduced viral replication in an interferon-independent manner. Flagellin administration also enhanced the efficacy of oseltamivir. 90 Interestingly, the protective affects appears to be independent of interferon. Flagellin has been a successful adjuvant in several viral vaccines including IAV flu models 91 although its use has been limited by toxicity. In COVID-19, S protein multi-epitope vaccines have been developed with antigenicity confirmed by the degree of TLR5 docking. 92
CD14, a TLR accessory molecule
As a proximal regulator of multiple TLRs, CD14 modulation may impact inflammation and organ injury in COVID-19. CD14 is a protein that is both membrane-bound on immune cells and soluble. It has a large flexible hydrophobic pocket and several grooves that are accessible for ligand binding. It acts in concert with TLR1/TLR2, TLR2/TLR6, TLR4, and TLR9 by binding a variety of PAMPs and DAMPs. 93 Its role in as a co-receptor for nucleic acids for TLR3 and TLR7 is less certain in humans.
CD14 may facilitate an initial innate immune system response to PAMPS like protein S and other viral structural proteins and glycolipids. However, with disease evolution it also likely recognizes DAMPs in addition to PAMPs leading to a net increase in immunopathology. As noted previously, one such DAMP: oxPAPC, an LPS-like DAMP generated from host cell death facilitates innate immune system hyperactivation via pro-inflammatory signaling pathways. 59
In COVID-19, using a novel ultra-high throughout proteomic approach, up-regulation of CD14 correlated with severity of COVID-19 infection. 94 Further, in COVID-19, soluble CD14 levels were associated with severity of disease. 95 Additionally, an increase in CD14 + CD16 + monocytes were identified in the peripheral blood of critically ill patients with COVID-19. 96 Presepsin, a soluble N-terminal component of CD14, has been identified as a biomarker associated with disease severity in COVID-19. 97 Because of these findings and previous work in sepsis and ARDS, CD14 inhibition has been identified as a potential means to target aberrant innate immune system activation in COVID-19. 98
It is possible that CD14 is a proximal modulator of several TLRs and plays a central role in regulating the inflammatory response observed in severe COVID-19. CD14 blockade may impart a net protective effect through modulation of several TLRs. As noted above, CD14 complexes with TLR1/TLR2, TLR2/TLR6, TLR4, and TLR9 to bind PAMPs and DAMPs. However, CD14 has not been definitively shown to interact with TLR3 and TLR7 in humans. TLR3 and TLR7 seem to be critical for an effective antiviral response through the TRIF-IFN pathway. Therefore, CD14 blockade may not decrease the TLR-TRIF-IFN response. TLR1/TLR2, TLR2/TLR6, TLR4, are predicted to bind PAMPs and DAMPs in COVID-19; this may drive an excessive MyD88-NF-κB-dependent inflammation. Therefore, blockade with an anti-CD14 Ab, may actually lead to reduced inflammation while not interfering with the antiviral response. A phase 2, anti-CD14 Ab clinical trial in COVID-19 is currently underway (NCT04391309).
CD14 has been implicated in several deleterious pro-inflammatory disease states: sepsis and septic shock, 99 InfluenzaA 100 infection and ARDS. 101 Previous work has shown that the CD-14 mAb is safe and that when co-administered, less inflammation is generated by LPS challenge. While sCD14 levels increases with Ab administration, inflammatory cytokine levels decrease. CD14 mAbs were previously studied in a phase 1 severe sepsis clinical trial. While CD14-Abs reduced inflammation, the study was underpowered to detect clinical benefit, and CD14 Abs antibodies are not used routinely in clinical practice. 102
Other TLR accessory molecules in COVID-19
IRAK-1 is a critical proximal component of the TLR MyD88 pathway. It may functionally bridge different pathways including the JAK/STAT- and TRIF-dependent pathways which may be a critical regulator of the pro-inflammatory versus type I/III IFN response. IRAK-1 has been implicated as a potential gene variant critical to the pathogenesis of COVID-19. 70 IRAK-1 has been previously studied in the context of endotoxin shock and autoimmune disease. 103 IRAK-4 also acts within the MyD88 pathway. IRAK-4 mutation has been implicated in recurrent and life threatening bacterial infections in children. 104 Its expression was up-regulated in a small group of patient with COVID-19. 105
CD24Fc is a biologic immunomodulator. CD24 acts as a checkpoint molecule that modulates TLR4 signaling in response to DAMPS, as in viral infection. For example, upon oxPAPC binding CD24 is recruited to the intracellular TLR4 module and ameliorates or blocks pro-inflammatory signaling. 106 CD24Fc has been designed to reduce pathologic activation of TLR4. While previously studied in the context of GVHD after stem cell transplant, CD24Fc has recently been shown to reduce the development of ARDS in viral pneumonia. 107 CD24Fc is being studied as a therapeutic in COVID-19. Initial reports from a phase III randomized control clinical trial suggest CD24Fc may reduce disease progression, shorten length of stay, and may blunt the immune response. Specifically, CD24Fc treated patients had reduced activation of CD8 + and NK cells and reduced cytokine and chemokine levels. 108
Conclusion
Upon infection with SARS-CoV-2, TLRs and their proximal accessory molecules play a critical role in both the protective and maladaptive innate and adaptive immune system responses. We hypothesize that the balance between MyD88-NF-κB-dependent excessive inflammation and TRIF-IFN-dependent antiviral response drives severity of disease in COVID-19. The goal of proximal modulation of the innate immune system should be to shift the balance towards a systemic antiviral TLR-TRIF-IFN response and away from immunopathology related to excessive TLR-MyD88-NF-κB activation. As discussed in detail above, one strategy is to inhibit CD14 which acts as a co-receptor for multiple TLRs and in particular those associated with the generation of excessive inflammation including TLR4 and TLR2.
Further investigation into how each TLR contributes to the net immune response and the pathophysiology of COVID-19 is needed. Future work should also be directed at better understanding how modulation of TLRs and accessory molecules impact each phase of the innate and adaptive immune responses to SARS-CoV-2.
Footnotes
Declaration of conflicting interests: The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: Frances Linzee Mabrey received support through the NIH under the following training grant: T32 HL007287
ORCID iD: Frances L Mabrey https://orcid.org/0000-0003-0263-9601
References
- 1.Akira S, Takeda K, Kaisho T. Toll-like receptors: critical proteins linking innate and acquired immunity. Nat Immunol 2001; 2: 675–680. [DOI] [PubMed] [Google Scholar]
- 2.Iwasaki A, Medzhitov R. Toll-like receptor control of the adaptive immune responses. Nat Immunol 2004; 5: 987–995. [DOI] [PubMed] [Google Scholar]
- 3.Aoshi T, Koyama S, Kobiyama Ket al. et al. Innate and adaptive immune responses to viral infection and vaccination. Curr Opin Virol 2011; 1: 226–232. [DOI] [PubMed] [Google Scholar]
- 4.Lester SN, Li K. Toll-Like receptors in antiviral innate immunity. J Mol Biol 2014; 426: 1246–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Medzhitov R, Preston-Hurlburt P, Janeway CA. A human homologue of the drosophila toll protein signals activation of adaptive immunity. Nature 1997; 388: 394–397. [DOI] [PubMed] [Google Scholar]
- 6.Barbalat R, Lau L, Locksley RMet al. et al. Toll-like receptor 2 on inflammatory monocytes induces type I interferon in response to viral but not bacterial ligands. Nat Immunol 2009; 10: 1200–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kawai T, Akira S. Toll-like receptors and their crosstalk with other innate receptors in infection and immunity. Immunity 2011; 34: 637–650. [DOI] [PubMed] [Google Scholar]
- 8.Lee K-M, Seong S-Y. Partial role of TLR4 as a receptor responding to damage-associated molecular pattern. Immunol Lett 2009; 125: 31–39. [DOI] [PubMed] [Google Scholar]
- 9.Zhao Y, Kuang M, Li J, et al. SARS-CoV-2 spike protein interacts with and activates TLR41. Cell Res 2021; 31: 818–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoshino K, Sugiyama T, Matsumoto M, et al. I[kappa]B kinase-[alpha] is critical for interferon-[alpha] production induced by toll-like receptors 7 and 9. Nat Lond 2006; 440: 949–953. [DOI] [PubMed] [Google Scholar]
- 11.Ramos HJ, Gale M. RIG-I like receptors and their signaling crosstalk in the regulation of antiviral immunity. Curr Opin Virol 2011; 1: 167–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hirano T, Murakami M. COVID-19: a New virus, but a familiar receptor and cytokine release syndrome. Immunity 2020; 52: 731–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu S-Y, Sanchez DJ, Aliyari Ret al. et al. Systematic identification of type I and type II interferon-induced antiviral factors. Proc Natl Acad Sci U S A 2012; 109: 4239–4244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kotenko SV, Gallagher G, Baurin VV, et al. IFN-lambdas mediate antiviral protection through a distinct class II cytokine receptor complex. Nat Immunol 2003; 4: 69–77. [DOI] [PubMed] [Google Scholar]
- 15.Davidson S, Maini MK, Wack A. Disease-Promoting effects of type I interferons in viral, bacterial, and coinfections. J Interferon Cytokine Res 2015; 35: 252–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rabouw HH, Langereis MA, Knaap RCM, et al. Middle East Respiratory coronavirus accessory protein 4a inhibits PKR-mediated antiviral stress responses. PLOS Pathog 2016; 12: e1005982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Comar CE, Goldstein SA, Li Y, Yount B, Baric RS, Weiss SR. Antagonism of dsRNA-Induced Innate Immune Pathways by NS4a and NS4b Accessory Proteins during MERS Coronavirus Infection. mBio. 2019;10(2):e00319-19. Published 2019 Mar 26. doi:10.1128/mBio.00319-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Teijaro JR. Type I interferons in viral control and immune regulation. Curr Opin Virol 2016; 16: 31–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lokugamage KG, Hage A, de Vries M, et al. Type I Interferon Susceptibility Distinguishes SARS-CoV-2 from SARS-CoV. J Virol. 2020;94(23):e01410-20. Published 2020 Nov 9. doi:10.1128/JVI.01410-20 [DOI] [PMC free article] [PubMed]
- 20.Biron CA, Nguyen KB, Pien GCet al. et al. Natural killer cells in antiviral defense: function and regulation by innate cytokines. Annu Rev Immunol 1999; 17: 189–220. [DOI] [PubMed] [Google Scholar]
- 21.Orlov M, Wander PL, Morrell EDet al. et al. A case for targeting Th17 cells and IL-17A in SARS-CoV-2 infections. J Immunol 2020; 205: 892–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. The Lancet 2020; 395: 497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tan M, Liu Y, Zhou R, et al. Immunopathological characteristics of coronavirus disease 2019 cases in Guangzhou, China. Immunology. 2020;160(3):261-268. doi:10.1111/imm.13223 [DOI] [PMC free article] [PubMed]
- 24.Han H, Ma Q, Li C, et al. Profiling serum cytokines in COVID-19 patients reveals IL-6 and IL-10 are disease severity predictors. Emerg Microbes Infect 2020; 9: 1123–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao Y, Qin L, Zhang P, et al. Longitudinal COVID-19 profiling associates IL-1RA and IL-10 with disease severity and RANTES with mild disease. JCI Insight. 2020;5(13):e139834. Published 2020 Jul 9. doi:10.1172/jci.insight.139834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McElvaney OJ, McEvoy NL, McElvaney OF, et al. Characterization of the inflammatory response to severe COVID-19 illness. Am J Respir Crit Care Med 2020; 202: 812–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bhatraju PK, Ghassemieh BJ, Nichols M, et al. Covid-19 in critically Ill patients in the Seattle region — case series. N Engl J Med 2020; 382: 2012–2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou F, Yu T, Du R, et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. The Lancet 2020; 395: 1054–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shi S, Qin M, Shen B, et al. Association of cardiac injury with mortality in hospitalized patients with COVID-19 in Wuhan, China. JAMA Cardiol 2020; 5: 802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guan W, Ni Z, Hu Y, et al. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med 2020; 382: 1708–1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gupta A, Madhavan MV, Sehgal K, et al. Extrapulmonary manifestations of COVID-19. Nat Med 2020; 26: 1017–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hadjadj J, Yatim N, Barnabei L, et al. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science 2020; 369: 718–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Blanco-Melo D, Nilsson-Payant BE, Liu W-C, et al. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell 2020; 181: 1036–1045. e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Broggi A, Ghosh S, Sposito B, et al. Type III interferons disrupt the lung epithelial barrier upon viral recognition. Science 2020; 369: 706–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Acharya D, Liu G, Gack MU. Dysregulation of type I interferon responses in COVID-19. Nat Rev Immunol 2020; 20: 397–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hung IF-N, Lung K-C, Tso EY-K, et al. Triple combination of interferon beta-1b, lopinavir–ritonavir, and ribavirin in the treatment of patients admitted to hospital with COVID-19: an open-label, randomised, phase 2 trial. The Lancet 2020; 395: 1695–1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou: Interferon-α2b Treatment for COVID-19 - Google Scholar [Internet]. [cited 2021 May 13]. Available from: https://scholar.google.com/scholar_lookup?author=Q + Zhou&author=V + Chen&author=CP + Shannon&title=Interferon-%CE%B12b + treatment + for + COVID-19&publication_year=2020&journal=Front + Immunol&volume=11
- 38.Davoudi-Monfared: A randomized clinical trial of… Google Scholar [Internet]. [cited 2021 May 13]. Available from: https://scholar.google.com/scholar_lookup?author=E + Davoudi-Monfared&author=H + Rahmani&author=H + Khalili&title=A + randomized + clinical + trial + of + the + efficacy + and + safety + of + interferon + %CE%B2-1a + in + treatment + of + severe + COVID-19&publication_year=2020&journal=Antimicrob + Agents + Chemother&volume=64
- 39.Zhou Z, Ren L, Zhang L, et al. Heightened innate immune responses in the respiratory tract of COVID-19 patients. Cell Host Microbe 2020; 27: 883–890. e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Knoops K, Kikkert M, Worm Svd, et al. SARS-Coronavirus Replication Is supported by a reticulovesicular network of modified endoplasmic Reticulum. PLOS Biol 2008; 6: e226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bouvet M, Debarnot C, Imbert I, et al. In vitro reconstitution of SARS-coronavirus mRNA Cap methylation. PLOS Pathog 2010; 6: e1000863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Han L, Zhuang M-W, Deng J, et al. SARS-CoV-2 ORF9b antagonizes type I and III interferons by targeting multiple components of the RIG-I/MDA-5-MAVS, TLR3–TRIF, and cGAS-STING signaling pathways. J Med Virol 2021; 93: 5376–5389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yin X, Riva L, Pu Y, et al. MDA5 Governs the innate immune response to SARS-CoV-2 in lung epithelial cells. Cell Rep 2021; 34: 108628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Squeglia F, Romano M, Ruggiero A, Maga G, Berisio R. Host DDX Helicases as Possible SARS-CoV-2 Proviral Factors: a Structural Overview of Their Hijacking Through Multiple Viral Proteins. Front Chem. 2020;8:602162. Published 2020 Dec 10. doi:10.3389/fchem.2020.602162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Totura AL, Whitmore A, Agnihothram S, et al. Toll-Like Receptor 3 Signaling via TRIF Contributes to a Protective Innate Immune Response to Severe Acute Respiratory Syndrome Coronavirus Infection. mBio. 2015;6(3):e00638-15. Published 2015 May 26. doi:10.1128/mBio.00638-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yu K, He J, Wu Y, et al. Dysregulated adaptive immune response contributes to severe COVID-19. Cell Res 2020; 30: 814–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rydyznski Moderbacher C, Ramirez SI, Dan JM, et al. Antigen-Specific Adaptive Immunity to SARS-CoV-2 in Acute COVID-19 and Associations with Age and Disease Severity. Cell. 2020;183(4):996-1012.e19. doi:10.1016/j.cell.2020.09.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang B, Zhou X, Zhu C, et al. Immune Phenotyping Based on the Neutrophil-to-Lymphocyte Ratio and IgG Level Predicts Disease Severity and Outcome for Patients With COVID-19. Front Mol Biosci. 2020;7:157. Published 2020 Jul 3. doi:10.3389/fmolb.2020.00157. [DOI] [PMC free article] [PubMed]
- 49.Le Bon A, Etchart N, Rossmann C, et al. Cross-priming of CD8 + T cells stimulated by virus-induced type I interferon. Nat Immunol 2003; 4: 1009–1015. [DOI] [PubMed] [Google Scholar]
- 50.Ferreira-Gomes M, Kruglov A, Durek P, et al. SARS-CoV-2 in severe COVID-19 induces a TGF-β-dominated chronic immune response that does not target itself. Nat Commun 2021; 12: 1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hagberg L, Hull R, Hull Set al. et al. Difference in susceptibility to gram-negative urinary tract infection between C3H/HeJ and C3H/HeN mice. Infect Immun 1984; 46: 839–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bohannon JK, Hernandez A, Enkhbaatar Pet al. et al. The immunobiology of TLR4 agonists: from endotoxin tolerance to immunoadjuvants. Shock Augusta Ga 2013; 40: 451–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Choudhury A, Mukherjee S. In silico studies on the comparative characterization of the interactions of SARS-CoV-2 spike glycoprotein with ACE-2 receptor homologs and human TLRs. J Med Virol. 2020;92(10):2105-2113. doi:10.1002/jmv.25987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Olejnik J, Hume AJ, Mühlberger E. Toll-like receptor 4 in acute viral infection: too much of a good thing. PLoS Pathog. 2018;14(12):e1007390. Published 2018 Dec 20. doi:10.1371/journal.ppat.1007390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cao C, Yin C, Shou S, et al. Ulinastatin protects against LPS-induced acute lung injury By attenuating TLR4/NF-κB pathway activation and reducing inflammatory mediators. Shock 2018; 50: 595–605. [DOI] [PubMed] [Google Scholar]
- 56.Ohashi K, Burkart V, Flohé Set al. et al. Cutting edge: heat shock protein 60 Is a putative endogenous ligand of the toll-like receptor-4 Complex. J Immunol 2000; 164: 558–561. [DOI] [PubMed] [Google Scholar]
- 57.Smiley ST, King JA, Hancock WW. Fibrinogen stimulates macrophage chemokine secretion through toll-like receptor 4. J Immunol 2001; 167: 2887–2894. [DOI] [PubMed] [Google Scholar]
- 58.Shirey KA, Lai W, Patel MC, et al. Novel strategies for targeting innate immune responses to influenza. Mucosal Immunol 2016; 9: 1173–1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zanoni I, Tan Y, Di Gioia Met al. et al. By capturing inflammatory lipids released from dying cells, the receptor CD14 induces inflammasome-dependent phagocyte hyperactivation. Immunity 2017; 47: 697–709. e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Patel MC, Shirey KA, Boukhvalova MS, Vogel SN, Blanco JCG. Serum High-Mobility-Group Box 1 as a Biomarker and a Therapeutic Target during Respiratory Virus Infections. mBio. 2018;9(2):e00246-18. Published 2018 Mar 13. doi:10.1128/mBio.00246-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Eisenbarth SC, Piggott DA, Huleatt JWet al. et al. Lipopolysaccharide-enhanced, toll-like receptor 4-dependent T helper cell type 2 responses to inhaled antigen. J Exp Med 2002; 196: 1645–1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Velazquez-Salinas L, Verdugo-Rodriguez A, Rodriguez LL, Borca MV. The Role of Interleukin 6 During Viral Infections. Front Microbiol. 2019;10:1057. Published 2019 May 10. doi:10.3389/fmicb.2019.01057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stone JH, Frigault MJ, Serling-Boyd NJ, et al. Efficacy of tocilizumab in patients hospitalized with covid-19. N Engl J Med 2020; 0: null. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hermine O, Mariette X, Tharaux P-L, et al. Effect of tocilizumab vs usual care in adults hospitalized With COVID-19 and moderate or severe pneumonia: a randomized clinical trial. JAMA Intern Med 2021; 181: 32–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Salvarani C, Dolci G, Massari M, et al. Effect of tocilizumab vs standard care on clinical worsening in patients hospitalized With COVID-19 pneumonia: a randomized clinical trial. JAMA Intern Med 2021; 181: 24–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Blasius AL, Beutler B. Intracellular toll-like receptors. Immunity 2010; 32: 305–315. [DOI] [PubMed] [Google Scholar]
- 67.van der Made CI, Simons A, Schuurs-Hoeijmakers J, et al. Presence of Genetic Variants Among Young Men With Severe COVID-19. JAMA. 2020;324(7):663-673. doi:10.1001/jama.2020.13719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Klein SL, Dhakal S, Ursin RLet al. et al. Biological sex impacts COVID-19 outcomes. PLOS Pathog 2020; 16: e1008570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Souyris M, Cenac C, Azar P, et al. TLR7 escapes X chromosome inactivation in immune cells. Sci Immunol. 2018;3(19):eaap8855. doi:10.1126/sciimmunol.aap8855 [DOI] [PubMed] [Google Scholar]
- 70.Allen J. Severe COVID-19 infection via a gene variant [internet]. Rochester, NY: Social Science Research Network, 2020 Jun [cited 2020 Aug 30]. Report No.: ID 3622655. Available from: https://papers.ssrn.com/abstract=3622655 [Google Scholar]
- 71.Angelopoulou A, Alexandris N, Konstantinou E, et al. Imiquimod - A toll like receptor 7 agonist - Is an ideal option for management of COVID 19. Environ Res 2020; 188: 109858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hemmi H, Kaisho T, Takeuchi O, et al. Small anti-viral compounds activate immune cells via the TLR7 MyD88–dependent signaling pathway. Nat Immunol 2002; 3: 196–200. [DOI] [PubMed] [Google Scholar]
- 73.Meier A, Chang JJ, Chan ES, et al. Sex differences in the toll-like receptor–mediated response of plasmacytoid dendritic cells to HIV-1. Nat Med 2009; 15: 955–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nadeem A, Siddiqui N, Al-Harbi NOet al. et al. TLR-7 agonist attenuates airway reactivity and inflammation through Nrf2-mediated antioxidant protection in a murine model of allergic asthma. Int J Biochem Cell Biol 2016; 73: 53–62. [DOI] [PubMed] [Google Scholar]
- 75.To EE, Erlich J, Liong F, et al. Intranasal and epicutaneous administration of toll-like receptor 7 (TLR7) agonists provides protection against influenza A virus-induced morbidity in mice. Sci Rep 2019; 9: 2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hu X, Cai X, Song X, et al. Possible SARS-coronavirus 2 inhibitor revealed by simulated molecular docking to viral main protease and host toll-like receptor. Future Virol. 2020;10.2217/fvl-2020-0099. doi:10.2217/fvl-2020-0099 [Google Scholar]
- 77.Jin Z, Du X, Xu Y, et al. Structure of M pro from SARS-CoV-2 and discovery of its inhibitors. Nature 2020; 582: 289–293. [DOI] [PubMed] [Google Scholar]
- 78.Christopher M, Wong J. Use of toll-like receptor 3 agonists against respiratory viral infections. Anti-Inflamm Anti-Allergy Agents Med Chem 2011; 10: 327–338. [Google Scholar]
- 79.Casanova J-L. Severe infectious diseases of childhood as monogenic inborn errors of immunity. Proc Natl Acad Sci 2015; 112: E7128–E7137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lim HK, Huang SXL, Chen J, et al. Severe influenza pneumonitis in children with inherited TLR3 deficiency. J Exp Med 2019; 216: 2038–2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhao J, Wohlford-Lenane C, Zhao J, et al. Intranasal treatment with poly(I·C) protects aged mice from lethal respiratory virus infections. J Virol 2012; 86: 11416–11424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Martins KA, Bavari S, Salazar AM. Vaccine adjuvant uses of poly-IC and derivatives. Expert Rev Vaccines 2015; 14: 447–459. [DOI] [PubMed] [Google Scholar]
- 83.Zheng M, Karki R, Williams EP, et al. TLR2 Senses the SARS-CoV-2 envelope protein to produce inflammatory cytokines. Nat Immunol 2021; 22: 829–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Das RP, Jagadeb M, Rath SN. Identification of peptide candidate against COVID-19 through reverse vaccinology: an immunoinformatics approach [internet]. Bioinformatics 2020; 1-11. http://biorxiv.org/lookup/doi/10.1101/2020.07.01.150805. cited 2020 Jul 28]. [Google Scholar]
- 85.Thompson CM, Holden TD, Rona G, et al. Toll-like receptor 1 polymorphisms and associated outcomes in sepsis after traumatic injury: a candidate gene association study. Ann Surg. 2014;259(1):179-185. doi:10.1097/SLA.0b013e31828538e8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wurfel MM, Gordon AC, Holden TD, et al. Toll-like receptor 1 polymorphisms affect innate immune responses and outcomes in sepsis. Am J Respir Crit Care Med 2008; 178: 710–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mikacenic C, Schneider A, Radella Fet al. et al. Cutting edge: genetic variation in TLR1 is associated with Pam3CSK4-induced effector T cell resistance to regulatory T cell suppression. J Immunol 2014; 193: 5786–5790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Evans SE, Tseng CK, Scott BL, Höök AM, Dickey BF. Inducible Epithelial Resistance against Coronavirus Pneumonia in Mice. Am J Respir Cell Mol Biol. 2020;63(4):540-541. doi:10.1165/rcmb.2020-0247LE [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chakraborty C, Sharma AR, Bhattacharya M, Sharma G, Lee SS, Agoramoorthy G. Consider TLR5 for new therapeutic development against COVID-19. J Med Virol. 2020;92(11):2314-2315. doi:10.1002/jmv.25997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Georgel A-F, Cayet D, Pizzorno A, et al. Toll-like receptor 5 agonist flagellin reduces influenza A virus replication independently of type I interferon and interleukin 22 and improves antiviral efficacy of oseltamivir. Antiviral Res 2019; 168: 28–35. [DOI] [PubMed] [Google Scholar]
- 91.Song L, Xiong D, Kang X, et al. An avian influenza A (H7N9) virus vaccine candidate based on the fusion protein of hemagglutinin globular head and Salmonella typhimurium flagellin. BMC Biotechnol 2015; 15: 79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bhattacharya M, Sharma AR, Patra P, et al. Development of epitope-based peptide vaccine against novel coronavirus 2019 (SARS-COV-2): immunoinformatics approach. J Med Virol 2020; 92: 618–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Di Gioia M, Zanoni I. Toll-like receptor co-receptors as master regulators of the immune response. Mol Immunol 2015; 63: 143–152. [DOI] [PubMed] [Google Scholar]
- 94.Messner CB, Demichev V, Wendisch D, et al. Ultra-High-Throughput Clinical Proteomics Reveals Classifiers of COVID-19 Infection. Cell Syst. 2020;11(1):11-24.e4. doi:10.1016/j.cels.2020.05.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bowman ER, Cameron CMA, Avery A, et al. Levels of Soluble CD14 and Tumor Necrosis Factor Receptors 1 and 2 May Be Predictive of Death in Severe Coronavirus Disease 2019. J Infect Dis. 2021;223(5):805-810. doi:10.1093/infdis/jiaa744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zhou Y, Fu B, Zheng X, et al. Pathogenic T cells and inflammatory monocytes incite inflammatory storm in severe COVID-19 patients. Natl Sci Rev. 2020;nwaa041. Published 2020 Mar 13. doi:10.1093/nsr/nwaa041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Fukada A, Kitagawa Y, Matsuoka M, et al. Presepsin as a predictive biomarker of severity in COVID-19: A case series. J Med Virol. 2021;93(1):99-101. doi:10.1002/jmv.26164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Martin TR, Wurfel MM, Zanoni Iet al. et al. Targeting innate immunity by blocking CD14: novel approach to control inflammation and organ dysfunction in COVID-19 illness. EBioMedicine 2020; 57: 102836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wright SD. CD14 And innate recognition of bacteria. J Immunol 1995; 155: 6–8. [PubMed] [Google Scholar]
- 100.Pauligk C, Nain M, Reiling Net al. et al. CD14 Is required for influenza A virus-induced cytokine and chemokine production. Immunobiology 2004 Aug 23; 209: 3–10. [DOI] [PubMed] [Google Scholar]
- 101.Martin TR, Rubenfeld GD, Ruzinski JT, et al. Relationship between soluble CD14, lipopolysaccharide binding protein, and the alveolar inflammatory response in patients with acute respiratory distress syndrome. Am J Respir Crit Care Med 1997; 155: 937–944. [DOI] [PubMed] [Google Scholar]
- 102.Axtelle T, Pribble J. An overview of clinical studies in healthy subjects and patients with severe sepsis with IC14, a CD14-specific chimeric monoclonal antibody. J Endotoxin Res 2003; 9: 385–389. [DOI] [PubMed] [Google Scholar]
- 103.An H, Hou J, Zhou J, et al. Phosphatase SHP-1 promotes TLR- and RIG-I-activated production of type I interferon by inhibiting the kinase IRAK1. Nat Immunol 2008; 9: 542–550. [DOI] [PubMed] [Google Scholar]
- 104.Picard C, von Bernuth H, Ghandil P, et al. Clinical features and outcome of patients with IRAK-4 and MyD88 deficiency. Medicine (Baltimore) 2010; 89: 403–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ong EZ, Chan YFZ, Leong WY, et al. A dynamic immune response shapes COVID-19 progression. Cell Host Microbe 2020; 27: 879–882. e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Liu Y, Chen G-Y, Zheng P. CD24-Siglec G/10 discriminates danger- from pathogen-associated molecular patterns. Trends Immunol 2009; 30: 557–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Tian R-R, Zhang M-X, Liu M, et al. CD24Fc Protects against viral pneumonia in simian immunodeficiency virus-infected Chinese rhesus monkeys. Cell Mol Immunol 2020; 17: 887–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Song NJ, Allen C, Vilgelm AE, et al. IMMUNOLOGICAL INSIGHTS INTO THE THERAPEUTIC ROLES OF CD24Fc AGAINST SEVERE COVID-19. Preprint. medRxiv. 2021;2021.08.18.21262258. Published 2021 Aug 23. doi:10.1101/2021.08.18.21262258 [Google Scholar]