Abstract
Abstract
Pseudomonas aeruginosa, an ESKAPE pathogen causes many fatal clinical diseases in humans across the globe. Despite an increase in clinical instances of Pseudomonas infection, there is currently no effective vaccine or treatment available. The major membrane protein candidate of the P. aeruginosa bacterial cell is known to be a critical component for cellular bacterial susceptibility to antimicrobial peptides and survival inside the host organisms. Therefore, the current computational study aims to examine P. aeruginosa’s major membrane protein, OprF, and OprI, in order to design linear B-cell, cytotoxic T-cell, and helper T-cell peptide-based vaccine constructs. Utilizing various immune-informatics tools and databases, a total of two B-cells and twelve T-cells peptides were predicted. The final vaccine design was simulated to generate a high-quality three-dimensional structure, which included epitopes, adjuvant, and linkers. The vaccine was shown to be nonallergenic, antigenic, soluble, and had the best biophysical properties. The vaccine and Toll-like receptor 4 have a strong and stable interaction, according to protein-protein docking and molecular dynamics simulations. Additionally, in silico cloning was employed to see how the developed vaccine expressed in the pET28a (+) vector. Ultimately, an immune simulation was performed to see the vaccine efficacy. In conclusion, the newly developed vaccine appears to be a promising option for a vaccine against P. aeruginosa infection.
Graphical Abstract
Supplementary Information
The online version contains supplementary material available at 10.1007/s10989-021-10356-z.
Keywords: Pseudomonas aeruginosa, Major membrane protein, Immunoinformatics, Multi-peptide vaccine, Epitope
Introduction
Pseudomonas aeruginosa, a common Gram-negative bacillus found in the environment, causes 51,000 infections per year, 6700 of which are caused by multi-drug resistant strains, resulting in 440 deaths [1]. The US Center for Disease Control and Prevention has classified P. aeruginosa as a pathogen that poses a significant hazard [2]. This bacterium is known as a major source of nosocomial infections due to its capacity to form biofilms, which are bacterial aggregates wrapped in a self-produced extracellular matrix that are difficult or impossible to eliminate with antibiotic therapy [3]. It affects humans, animals, and plants, and is frequently associated with sporadic clinical mastitis, hospital-acquired pneumonia, cystic fibrosis, UTIs with skin infections like external otitis and folliculitis, soft tissue infections, joint infections, and a variety of other infections, particularly in patients with chronic wounds, severe burns, or people with cancer or AIDS who are immunocompromised [4].
Pseudomonas aeruginosa is an aerobic, motile, lactose non-fermenter that can thrive in a wide range of environments and hosts [5]. In comparison to other sequenced bacteria, P. aeruginosa has a large genome (5.5–7 Mbp), and it contains a huge number of regulatory enzymes involved in the transportation, metabolism, and expulsion of organic compounds [6]. The P. aeruginosa genome’s increased coding capability provides for more metabolic flexibility and response to environmental changes [7]. The outer membranes of Gram-negative bacteria are made up of three different types of compounds: proteins, lipids, and lipopolysaccharides. A few of the proteins are classified as major membrane proteins because they have a high copy number (5 × 104 to 2 × 105 copies per cell) [8]. The outer membrane of P. aeruginosa is made up of several proteins, including lipoproteins and channels [9, 10]. β-barrel proteins that produce waterfilled diffusion channels, known as porins, control nutrient exchange across the outer membrane [11].
OprF, the major porin protein responsible for forming channels across the outer membrane is an eye-catching target for diagnosis and treatment in P. aeruginosa [12]. OprF protein has been suggested as a possible target for monoclonal antibody-based diagnostic and immunotherapeutic intervention [13]. Immunization with OprF protects against recurrent P. aeruginosa infections, according to research [14]. OprF-deficient variants have a spherical form and can only grow in media with high osmolarity, showing that OprF is involved in cell structure and outer membrane integrity [15]. Despite the fact that OprF is a porin, its structure is likely to be substantially different from the bulk of bacterial porins [16]. Similarly, outer membrane protein I (OprI) is a well-known surface lipoprotein that plays a key role in bacterial antimicrobial peptide sensitivity [17]. According to reports, the most potential P. aeruginosa vaccine (IC43) was tested in phase III clinical research (NCT01563263) and is made up of OprI and a portion of the outer membrane protein OprF [18]. OprI also acts as an adjuvant by activating the TLR2-TLR4 pathway, which boosts immunity against diseases including TB and swine flu fever [19, 20].
Due to reduced outer membrane permeability and drug efflux paired with adaptive mechanisms, P. aeruginosa has significant levels of intrinsic antibiotic resistance [21]. Furthermore, during bacterial growth, the establishment of biofilms and quorum sensing systems causes adaptive resistance, resulting in multidrug-resistant strains [22]. Effective vaccines and novel drug molecules would be critical in reducing the higher mortality rates caused by these infections [23]. Against this backdrop, as compared to conventional approaches, the recent immunoinformatics approach helps to reduce the time required for vaccine development, and there are examples of successful vaccine candidate development using in silico approaches [24]. In this study, a multi-epitope vaccine construct was developed consisting of screened Cytotoxic T-cell (CTL), Helper T-cell (HTL), and linear B-cell (LBL) epitopes with the potential to elicit both humoral and cellular immune response. To increase the immunogenicity of the vaccine, potent adjuvant β-defensins were also used. The vaccine has the appropriate physicochemical qualities, such as room temperature stability, antigenicity, and allergy-free. In addition, to enhance the antigen-presenting process, the epitopes are fused with suitable linkers. To obtain a good quality three-dimensional structure of the vaccine protein sequence, modelling, and validation were carried out. Additionally, molecular docking with the TLR-4 receptor and molecular dynamics simulations confirmed the final vaccine construct’s stability. At last, the codon optimization and in silico cloning were performed. The overall schematic workflow employed for vaccine designing is presented in Fig. 1.
Fig. 1.
Schematic representation of the workflow for the development of multi-epitope vaccine against P. aeruginosa infections
Materials and Methods
Sequence Availability
The FASTA sequences of the targeted outer membrane protein F (OprF; UniPort ID P13794) and outer membrane protein I (OprI; UniPort ID P11221) from P. aeruginosa were retrieved from the UniProt database(https://www.uniprot.org/).
Prediction and Assessment of LBL Epitopes
LBL epitopes were predicted using the ABCpred (http://crdd.osdd.net/raghava/abcpred/) tool with threshold of 0.51 and window length of 10. The antigenic, allergic, and toxic properties of the anticipated LBL epitopes were assessed using the VaxiJen V2.0, AllerTOP V2.0, and ToxinPred servers, respectively.
MHC-I Epitopes Prediction
The majority of cytotoxic T-cells have TCRs that identify a particular antigen [25]. Therefore, CTL epitope prediction is critical for vaccine designing concepts. CTL epitopes were predicted on the NetMHCpan4.1 server (http://www.cbs.dtu.dk/services/NetMHCpan/). The threshold for epitope identification for a strong binder was set at 0.5% while at 2% for a weak binder. Peptide binding to MHC-I, proteasomal C-terminus cleavage score, and the transport efficiency of Transporter Associated with Antigen Processing (TAP) were used to make these predictions [26]. The peptide binding to MHC-I and Proteasomal C-terminal cleavage scores were generated using an artificial neural network, whereas the TAP score was calculated by the weight matrix.
MHC-II Epitopes Prediction
Helper T lymphocytes are a type of adaptive immune cell that may produce humoral and cellular immune responses in reaction to foreign antigens [27]. Hence, HTL epitopes that may bind to MHC-II alleles are critical in vaccine development. HTL epitopes prediction was performed on online server IEDB (Immune Epitope Database) (http://www.iedb.org/) [28, 29] considering human HLAs as a reference set by default. The server assigns IC50 values to the predicted epitopes, which are inversely related to the binding affinity towards the MHC-II. IC50 scores of < 50 nM represent a high binding affinity. The IC50 value < 500 nM corresponds to intermediate binding affinity; however, < 5000 nM is related low binding affinity of epitopes towards MHC-II.
Assessment of Epitopes for Immunogenicity Score
The immunogenic potency of epitopes was assessed utilizing the MHC-I immunogenicity tool of the IEDB server (http://tools.iedb.org/immunogenicity/).
Antigenicity, Allergenicity, and Toxicity Analysis of the Epitopes
The antigenicity of the predicted epitopes was confirmed using the Vaxigen v2.0 (http://www.ddg-pharmfac.net/vaxijen/VaxiJen/VaxiJen.html) tool at 0.4 thresholds. The AllerTOP2.0 server (https://www.ddg-pharmfac.net/AllerTOP/) was used to assess the epitope’s allergenicity. The server is based on the translation of protein sequences into uniform equal-length vectors using auto cross-covariance (ACC). Based on a training set of 2210 known allergens from different species and 2210 non-allergens from the same species, the proteins are categorized using the k-nearest neighbor algorithm (kNN, k = 3) [30].
For checking the toxicity of the predicted epitopes, the Toxinpred server (http://crdd.osdd.net/raghava/toxinpred/) was used with default parameters. Other details i.e., net molecular weight, SVM score, hydrophobicity was also determined using the toxinpred tool.
Vaccine Construction
The multi-peptide vaccine candidate was constructed by adjoining the adjuvant, chosen LBL, CTL, and HTL epitopes with the suitable linkers, namely EAAAK, KK, AAY, and GPGPG, to help the amino acids fold into appropriate conformations with maximum flexibility.
Vaccine Construct’s Antigenicity and Allergenicity Profiling
The vaccine construct’s antigenicity and allergenicity profiles were also assessed. The VaxiJen server [34] was used to analyse the antigenicity of the vaccine design, and the AllerTop server [35] was utilised to assess the vaccine construct’s non-allergenicity.
Population Coverage Analysis
The IEDB’s Population Coverage tool (http://tools.iedb.org/population/) was used to look at how the affinity of CTL and HTL epitopes for HLA alleles varies by race, location, and country, affecting the construction of epitope-based vaccines. By keeping the default parameters, the 16 geographical areas were selected for population coverage. The distribution of human MHC binding alleles is used in this tool to estimate population coverage for the T-cell epitopes in different parts across the globe.
Physicochemical Properties and Secondary Structure Assessment
The Expasy ProtParam (https://web.expasy.org/protparam/) server was used to analyze the vaccine construct, which predicts the theoretical pI, Molecular Weight, solubility, Half-Life (in-vitro), and Grand Average of Hydropathicity Values (GRAVY) for the input vaccine protein sequence. The secondary structure prediction was carried out using the online server PSIPRED (http://bioinf.cs.ucl.ac.uk/psipred/).
Tertiary Structure Prediction and Validation
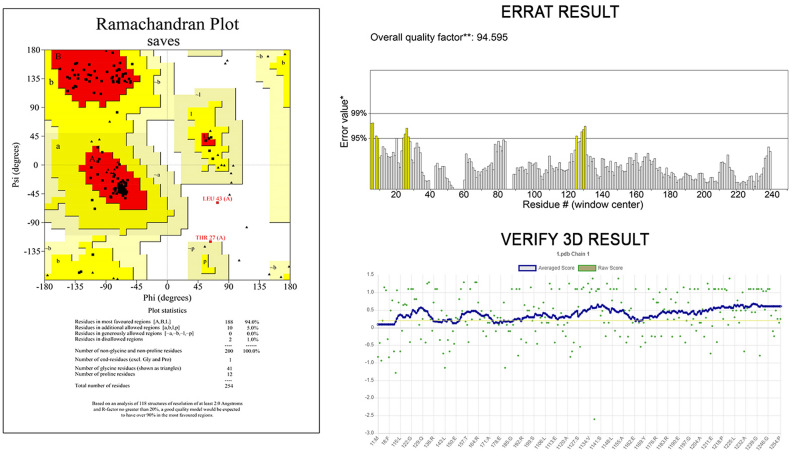
Robetta server (http://robetta.bakerlab.org) was utilized to obtain a tertiary (3D) structure for the vaccine sequence. To predict the 3D model, analyze the query protein sequence into supposed domains. Robetta employs a comparative modelling technique for the structure development if the template structure is identified for the provided amino acid sequence using PSI-BLAST, BLAST, FFAS03, or 3D-Jury. If there is no template is found, then the de novo Rosetta fragment insertion method is employed. The final structure was refined by GalaxyRefine2 (http://galaxy.seoklab.org/cgi-bin/submit.cgi?type=REFINE2) server. The quality of models was assessed by the generation of Ramachandran plot using PROCHECK (https://servicesn.mbi.ucla.edu/PROCHECK/). This server employs the Ramachandran plot to forecast the likelihood of amino acids forming a secondary structure and to display the quality of models based on the fraction of amino acids in the allowed, favored, and outlier regions. The validation was also confirmed using ERRAT (http://services.mbi.ucla.edu/ERRAT/) and Verify 3D (https://saves.mbi.ucla.edu/).
Disulfide Engineering for the Stability of the Vaccine
To obtain stability of the final vaccine construct’s modelled structure, disulfide engineering was performed using Disulfide by Design v2.0 (http://cptweb.cpt.wayne.edu/DbD2/index.php).
Discontinuous B Cell Epitope Prediction
The ElliPro server (http://tools.iedb.org/ellipro/) was used to predict discontinuous B cell epitopes for the final confirmed 3D structure of vaccine with default parameters (Minimum residue score 0.5 and Maximum distance 6 Å). It assigns a score to each epitope, which is the PI (Protrusion Index) value averaged over the epitope’s residues. This approach uses ellipsoids to estimate the 3D structure of a protein. A score of 0.9 PI implies that 90% of the protein’s residues are included, while the remaining 10% are outside of the ellipsoid. This PI number is determined by the centre of mass of each residue, which is located outside of the greatest feasible ellipsoid. Discontinuous epitopes are predicted using the PI value, and their clustering is based on the distance R (distance between the centre of mass of the residue), which is measured in. A higher R value suggests that there are a greater number of expected discontinuous epitopes.
Protein–Protein Docking Studies
Cluspro 2.0 (http://cluspro.bu.edu/login.php) is a rigid-body protein–protein docking service that predicts interactions between two proteins. It is entirely automated and quick. This software employs 3 distinct procedures: the first is a unique fast Fourier transform (FFT) correlation, the second is clustering the best energy conformations, and the third is evaluating cluster stability using brief Monte Carlo simulations. The 3D structure of TLR4 was retrieved from the RCSB PDB database and equipped for docking analysis by utilizing the PyMol visualization software. PyMol was used to depict the protein–peptide interactions of the best-docked postures.
Molecular Dynamics Simulation
The MD simulations were performed for the apo form of the designed vaccine and TLR4, as well as for the complex of TLR4-vaccine construct by the Schrodinger of Desmond package 19Des2. The OPLS3e force field was used to simulate water molecules using a predefined TIP3P water model. To balance the system charge and neutralize the solvated system, sufficient sodium ions were used. After constructing the solvated system with docked complex, the system was reduced and relaxed using the Desmond module’s default protocol and OPLS3e force field parameters. The NPT ensemble was used to run MD simulations with periodic boundary conditions. Simulations were run for 100 ns at a constant temperature of 300 K and a pressure of 1 atm. During the 100 ns MD simulations, the root mean square deviation (RMSD) of α-carbon chains and the root mean square fluctuation (RMSF) of multi-epitope residues were plotted versus time (Ps) to demonstrate the stability of the developed vaccine construct.
Codon Optimization and In Silico Cloning
In silico cloning was employed to better understand the expression of our developed multi epitopic vaccine candidate on E. coli hosts. The vaccine construct was codon-optimized using a web server called Java Codon Adaptation Tool (JCAT) (http://www.jcat.de/) for high-level production of the vaccine sequence in the E. coli K12 strain. The output of the JCat tool includes the codon adaptation index (CAI), the ideal CAI should be greater than 0.8 and scores up to 1.0. The ideal CAI score should be 1.0. The GC content percentage should range between 30 and 70%. In silico cloning of the vaccine sequence was carried out using the pET28b (+) expression vector. Here, the SnapGene 5.2.4 (https://www.snapgene.com/) restriction cloning tool was employed for finalizing in silico cloning.
Immune Simulation for Vaccine Efficacy
To evaluate the immunogenic features of a multiepitope vaccination in real-life conditions, in silico immune simulations were performed using the C-ImmSim server (http://150.146.2.1/CIMMSIM/index.php). C-ImmSim is an agent-based dynamics simulator for immune reactions that uses the PSS matrix and machine learning approaches to predict the peptides and immunological interactions. The duration between doses 1 and 2 for vaccination should be at least 4 weeks. As a result, 3 injections containing one thousand vaccine proteins were administered 4 weeks apart at 1, 84, and 168 time-steps (each time-step equals 8 h in real life, and time-step 1 is injection at time = 0), for a total of 1050 simulation steps (parameters were set in the C-ImmSim immune simulator). All of the remaining simulation settings were left at their default values. In addition, 3 injections of the chosen peptide were given four weeks apart to replicate recurring antigen exposure and investigate clonal selection in a typical endemic region. The Simpson index (D) is a measure of diversity that was calculated using the graph.
Result
Prediction and Assessment of LBL Epitopes
The online server ABCpred was used to predict linear B-cell epitopes of the two protein sequences (Supplementary Table 1). Epitopes selected based on the prediction scores were further screened for antigenicity scores on the Vaxigen server. The allergenicity prediction on the AllerTOP tool showed that the vaccine is not allergenic. Toxinpred server predicted that all the peptides are non-toxic along with their hydrophobicity, hydrophilicity, hydropathicity, and molecular weight. Epitopes that were identified as allergenic for humans and classified as toxic were separated from the predicted list of peptides. Two epitopes were chosen for final vaccine constructions from a total of 18 epitopes, namely GTPGVGLRPY (antigenicity score = 1.8921) and HSKETEARLT (antigenicity score = 2.0946), based on the highest antigenicity score are tabulated in Table 1.
Table 1.
Predicted B-cell epitopes from P. aeruginosa MMP protein and their corresponding immunogenic properties
MHC-I Epitopes Prediction
The major membrane protein of P. aeruginosa was chosen for chimeric vaccine development to find immunogenic factors for the vaccine construct. MHC-I epitopes were identified with the help of the NetMHCpan 4.1server. The length of the produced epitopes was 9mers (Supplementary Table 2). Epitopes were further screened for antigenicity, allergenicity, and toxicity. The top 10 chosen epitopes showed maximum binding to all HLA supertypes (A1, A2, A3, A24, A26, B7, B8, B27, B39, B44, B58, and B62) were selected of each protein are listed in Table 2.
Table 2.
Predicted CTL epitopes from P. aeruginosa proteins to design multi-epitope vaccine construct with their corresponding MHC Class I alleles and their immunogenic properties
| Uniprot_ID | CTL Epitope | Alleles | Position | Score | Antigencity Score | Immunogenicity | Toxicity |
|---|---|---|---|---|---|---|---|
| P13794 | GTYETGNKK | HLA-A*01:01 | 79 | 4.012 | 1.3730 | 0.03779 | Non-toxin |
| DLYGGSIGY | HLA-A*02:01 | 51 | 6.825 | 0.04229 | 0.04229 | Non-toxin | |
| VGFNFGGSK | HLA-A*02:01 | 180 | 1.596 | 1.4581 | 0.10933 | Non-toxin | |
| AGLGVGFNF | HLA-A*24:02 | 176 | 2.33 | 2.0917 | 0.19762 | Non-toxin | |
| NATAEGRAI | HLA-B*07:02 | 329 | 6.37 | 1.2860 | 0.24791 | Non-toxin | |
| EGRAINRRV | HLA-B*08:01 | 333 | 6.283 | 1.6282 | 0.2536 | Non-toxin | |
| GRAINRRVE | HLA-B*27:05 | 334 | 6.149 | 1.4441 | 0.25684 | Non-toxin | |
| YHFGTPGVG | HLA-B*39:01 | 99 | 1.543 | 0.7858 | 0.15218 | Non-toxin | |
| NEYGVEGGR | HLA-B*40:01 | 306 | 4.762 | 2.0516 | 0.21575 | Non-toxin | |
| P11221 | HSKETEARL | HLA-A*01:01 | 23 | 7.395 | 2.1549 | 0.22606 | Non-toxin |
MHC-II Epitopes Prediction
Using the Immune Epitopes Database (IEDB) (https://www.iedb.org/), immunogenic T-Cells (MHC-II) were identified [31]. For the MHC epitope binding study, it uses a consensus prediction method. To improve our confidence in predicting epitopes for MHC Class II alleles, we kept binding affinity below 250 nM. With an IC50 value ranging from 6 to 350 nM, these 15-mer amino acid residues interacted with a variety of different and/or common MHC II alleles (Supplementary Table 3). A good epitope should interact with as many MHC alleles as possible. Epitopes were further screened for antigenicity, allergenicity, and toxicity. The top two most probable epitopic candidates (GVGFNFGGSKAAPAP and LKFSALALAAVLATG) interacting with several MHC class II alleles, as well as their scores, have been shown in Table 3.
Table 3.
Predicted HTL epitopes from P. aeruginosa proteins to design multi-epitope vaccine construct with their corresponding MHC Class II alleles and their immunogenic properties
| Uniprot_ID | MHC II Epitope | Alleles | Pos | IC50 value | Percentile_Rank | Antigencity Score | Toxicity |
|---|---|---|---|---|---|---|---|
| P13794 | GVGFNFGGSKAAPAP | HLA-DPB1*01:01, HLA-DRB1*01:01, HLA-DRB1*09:01,HLA-DRB3*02:02, HLA-DRB1*13:02, HLA-DRB1*11:01, HLA-DRB1*04:01, HLA-DRB1*12:01, HLA-DPA1*03:01, HLA-DPB1*04:02, HLA-DRB1*04:05, HLA-DRB1*15:01, HLA-DQA1*01:01, HLA-DQB1*05:01, HLA-DRB1*08:02, HLA-DPA1*02:01, HLA-DPB1*14:01, HLA-DPA1*01:03, HLA-DPB1*04:01, HLA-DQA1*05:01, HLA-DQB1*03:01, HLA-DQA1*04:01, HLA-DQB1*04:02, HLA-DPA1*02:01, HLA-DPA1*02:01, HLA-DPB1*05:01, HLA-DPA1*01:03, HLA-DPB1*02:01, HLA-DQA1*05:01, HLA-DQB1*02:01, HLA-DQA1*03:01, HLA-DQB1*03:02, HLA-DQA1*01:02, HLA-DQB1*06:02, HLA-DRB3*01:01, HLA-DRB5*01:01, HLA-DRB1*07:01, HLA-DRB4*01:01, HLA-DRB1*03:01 | 179-193 | 7 | 0.48 | 1.1979 | Non-toxin |
| P11221 | LKFSALALAAVLATG | HLA-DRB3*01:01, HLA-DPA1*03:01, HLA-DPB1*04:02, HLA-DPA1*01:03, HLA-DPB1*02:01,HLA-DRB1*01:01, HLA-DRB1*09:01,HLA-DRB3*02:02, HLA-DRB1*13:02, HLA-DRB1*11:01, HLA-DRB1*04:01, HLA-DRB1*12:01,HLA-DRB1*04:05, HLA-DRB1*15:01, HLA-DQA1*01:01, HLA-DQB1*05:01, HLA-DRB1*08:02, HLA-DPA1*02:01, HLA-DPB1*14:01, HLA-DPA1*01:03, HLA-DPB1*04:01, HLA-DQA1*05:01, HLA-DQB1*03:01, HLA-DQA1*04:01, HLA-DQB1*04:02, HLA-DPA1*02:01, HLA-DPB1*01:01, HLA-DPA1*02:01, HLA-DPB1*05:01,HLA-DQA1*05:01, HLA-DQB1*02:01, HLA-DQA1*03:01, HLA-DQB1*03:02, HLA-DQA1*01:02, HLA-DQB1*06:02, HLA-DRB5*01:01, HLA-DRB1*07:01, HLA-DRB4*01:01, HLA-DRB1*03:01 | 5--19 | 6 | 0.38 | 0.5480 | Non-toxin |
Assessment of Epitopes for Immunogenicity Score
IEDB class I immunogenicity tool was employed to find out the immunogenicity score. Epitopes having positive immunogenicity scores were selected for further studies.
Vaccine Construction
The final vaccine construct comprises the adjuvant β-defensins (P81534) fused to the epitope of the B-cell with an EAAAK linker at the N-terminal end. Both the B-cells were separated by KK linkers, subsequently, the B-cell epitopes were fused with CTL peptide with AAY linker. Finally, using the GPGPG linker, HTL epitopes were fused at the C-terminal. Figure 2 shows a schematic diagram of the vaccine construct.
Fig. 2.
The structural arrangement of B and T-cell epitopes along with linkers and adjuvant for the final multi-epitope vaccine construct
Vaccine’s Antigenicity and Allergenicity Profiling
In addition to evaluating the antigenicity and allergenicity of individual epitopes, the final proposed vaccine construct should be highly antigenic in order to generate a stronger immune response. As a result, the antigenicity of the entire vaccine design was found to be high, with a score of 1.0957 predicted by Vaxigen V2.0, indicating that the vaccine is immunogenic and capable of eliciting a robust immune response. AllerTOP V2.0 tool also classified the construct to be non-allergen against humans.
Population Coverage Analysis
Since MHC molecules’ peptide-binding regions are enormously variable, they have a wide range of binding specificity. As a result, population coverage analysis is essential for developing a successful epitope-based subunit vaccine that binds to a wide range of HLA supertype alleles. The predicted epitopic core sequences, as well as the MHC Class I and MHC Class II alleles that bind to them, were submitted to the Immuno Epitope Database’s population coverage analysis tool to calculate the population coverage. The IEDB findings confirmed that 98.62% of the world’s population would be covered if the intended vaccine architecture was administered, which covers 109 countries in 16 different geographical regions. It was highlighted, in particular, that the population coverage rate in practically all countries worldwide exceeded 90%. (Supplementary Table 4).
Physicochemical Properties Assessment and Secondary Structure Analysis
The constructed vaccine was analyzed by the ProtParam server. Supplementary Table 5 shows the physicochemical parameters of the peptides. The molecular weight (MW) was measured at 26726.58 kDa with a theoretical PI of 9.85. The construct was found to be soluble and stable. PSIPRED 4.0 server was utilized to predict the secondary structure. As shown in Fig. 3, the vaccine contains 32.17% α-helices together with 11.417% -strands, and 55.905% random coils. The solubility of the vaccine was found to be 0.63 by SOLpro server.
Fig. 3.
Secondary structure prediction of the final multi-epitope vaccine construct by using PSIPRED tool
Tertiary Structure Prediction and Validation
3D model of the vaccine protein sequence was generated by the Robetta server (Fig. 4). GalaxyRefine2 was used to improve the Robetta server’s predicted tertiary structure. The best model was chosen for further analysis and included the following parameters: RMSD, 0.314; MolProbity, 1.916; Clash score, 13.7; Poor rotamers, 0; Rama preferred, 96.0. When compared to the original structure developed by the Robetta server, analysis of the Ramachandran plot of the refined protein obtained from GalaxyRefine at the GalaxyWEB server revealed better results with two residue in disallowed regions (94.0% of residues in favoured region, 5.0% residues in allowed region, and none in outlier region) (92.5% of residues in favoured region, 6.0% residues in allowed region and 1.5 of residues in outlier region). Similarly, additional structural validation tools such as ERRAT and VERIFY3D revealed that the model derived after GalaxyRefine had higher stereochemical quality (Fig. 5).
Fig. 4.

Homology modeling of the three-dimensional structure of the final multi-epitope vaccine construct
Fig. 5.
Structure validation tool results confirmed the developed multi-epitope vaccine to be reliable and accurate
Disulfide Engineering for the Stability of the Vaccine
There are a total of 24 pairs of residues that can be exploited for disulfide engineering, according to the findings (Supplementary Table 6). However, after considering additional factors such as energy and χ3 value, only two pairs of residues were chosen because their values fall within the acceptable range, i.e., energy must be less than 2.2 and Chi3 must be between − 87 and + 97° [32]. As a result, two mutations were generated on the residue pairs ALA9-ALA232, and VAL210-GLY246 with a χ3 angle of − 86.78° and energy of 1.91 kcal/mol, respectively. The disuphide bonds (red and magenta colour) are shown in the mutant created (Fig. 6).
Fig. 6.

Disulphide engineering of the vaccine protein. Residue pairs showed in red (ALA9 and VAL210) and magenta (ALA232 and GLY246) spheres were mutated to Cysteine residues to form disulphide bridge between them
Defining Discontinuous B-cell Epitopes
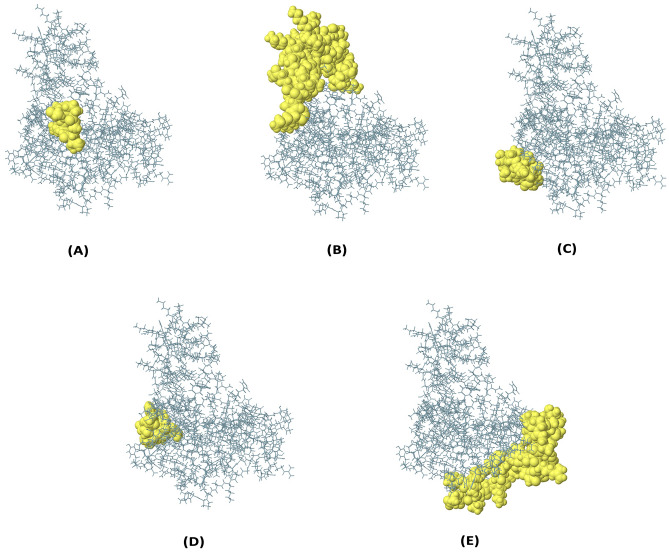
Ellipro Server was used to predict discontinuous B-cell epitopes from the vaccine protein’s 3D structure. Discontinuous B-cell epitopes were predicted with scores ranging from 0.696 to 0.808. Supplementary Table 7 lists the amino acid residues, the number of residues, the sequence location, and their scores. Figure 7 depicts a graphical depiction of the discontinuous epitopes.
Fig. 7.
The conformational B-lymphocyte epitopes present in the vaccine. The yellow spheres showing epitopes containing a 7 residues (AA 85–91) with 0.808; b 49 residues (AA 29–30, AA 37–51, and AA 53–84) with 0.773; c 10 residues (AA 163–172) with 0.748; d 9 residues (AA 110–118) with 0.715; e 45 residues (AA 196–220, AA 235-254) with 0.696 (Color figure online)
Protein–Protein Docking Studies
The ClusPro server was employed for the molecular docking. To evaluate the protein-protein interaction, the vaccine was docked with the immunological receptor TLR-4 (PDB ID:5YJB). For further analysis, the model with the lowest binding energy (-857.9) was chosen. The hydrogen bond interacting residues in the vaccine-TLR4 docked complex were: PRO87-ASN492, SER141-ASP483, GLY20-GLN508, ARG92-SER536, ARG92-ASP515, TYR94-ASP515, TYR94-GLN540, ARG80-GLN540, ALA143-TRP511, GLY22-ASP534, GLY20-GLN508, ARG80-TYR542, GLU76-ARG564, LYS66-THR566, ARG64-ASP563, GLN51-ASP563, GLU49-LYS594, GLY59-THR619, LYS54-ASP586, GLN29-ASP559, THE27-ASP559 (Fig. 8).
Fig. 8.
Molecular interaction of multi-epitope vaccine construct with TLR2
Molecular Dynamics Simulation
The stable interaction between the receptor molecule (TLR 4) and the multi-peptide subunit vaccine was determined using the molecular dynamics simulation. The RMSD graph showed that the proposed vaccine remained stable in combination with TLR 4 throughout the simulation time, especially after 60 nanoseconds (Fig. 9). MD simulation results were also used to determine the most flexible areas of the vaccine molecule. The root-mean-square fluctuations (RMSF) of vaccine-TLR4 complex residues were assessed in this regard (Fig. 9). The small degrees of fluctuations of the vaccine residues corroborate that the modeled vaccine construct has established sustainable interactions with the receptor protein. The majority of the vaccine molecule was found to be stiff and have low RMSF values.
Fig. 9.
Root mean square deviation (RMSD) and root mean square fluctuation (RMSF) analysis of protein backbone and side chain residues of MD simulated vaccine construct
Optimization of the Codon and In Silico Cloning
In silico cloning was employed to better comprehend the expression of our designed multi-peptide vaccine candidate on the E. coli host. The Java Codon Adaptation tool was used to optimize our vaccine design for expression in the E. coli (K12 strain). We also chose three filter parameters to minimize pointless errors: (a) eliminating rho-independent transcription terminators, (b) avoiding prokaryotic ribosome binding sites, and (c) avoiding restriction enzyme cleavage sites. The Codon adaptation index (CAI) value of the optimized vaccine design is 1.0 (Range 0.8 to 1.0), while the ideal GC (guanine–cytosine) content range is 53.28 (Range 30–70%), indicating a high likelihood of protein expression. In the next stage, we added 2 restriction endonucleases (EcoRI and BamHI) to both terminal ends of the vaccine construct to help in cloning. Finally, by using the SnapGene software, the vaccine design was cloned into the pET28b (+) vector (Fig. 10). The clone had a length of 6135 bp.
Fig. 10.
The in silico cloning of the designed vaccine into the pET-28a (+) vector. Herein, black color represents the vector DNA, while the red color indicates the adapted DNA sequence of the designed vaccine
Immune Simulation for Vaccine Efficacy
As shown in Fig. 11, the in silico immune response produced in the C-ImmSim immune simulator was compatible with the real immunological responses. After the first, second, and third doses of the proposed vaccine, antibody titers were significantly higher. The secondary and tertiary responses exceeded the initial response by a wide margin. Immunoglobulin activity (IgG1 + IgG2, IgM, and IgG + IgM antibodies) was high in both secondary and tertiary reactions, with a proportional decrease in antigen concentration. Several B-cell isotypes with long-term activity were also discovered, demonstrating the possibility of isotype switching and memory formation. The TH and TC cell populations showed an identical high reaction, as well as memory acquisition, which is necessary to support the immune response. Whereas the anergic B and T cells were continuously decreasing which conferred a good sign for the vaccine efficacy. Increased macrophage activity was observed throughout the exposure, whereas dendritic cell and natural killer cell activity remained consistent. There was a rise in the levels of IFN-g, TGF-B, IL-10, and IL-12, which are in line with the report of high levels of cytokines in the recovered individuals from Pseudomonas infection. The decrease in Simpson value ‘D’ over time suggests a complex immune response, which is understandable given the chimera vaccine’s several epitopes [33]. This immunogenicity profile shows the emergence of immunological memory and, as a result, higher antigen clearance in subsequent exposures.
Fig. 11.
In silico simulation of immune response using vaccine as antigen: A Antigen and immunoglobulins, B B-cell population, C B-cell population per state, D Helper T-cell population, E helper T-cell population per state, F cytotoxic T-cell population per state, G macrophage population per state, H dendritic cell population per state, and I production of cytokine and interleukins with Simpson index D of immune response
Discussion
Pseudomonas major membrane proteins have a variety of functions during visceral infection. The protein helps in the growth of bacteria in high-osmolarity environments. They contribute to the maintenance of contamination by interfering with macrophage microbicidal capabilities and play a key role in bacterial susceptibility to antimicrobial peptides in a later stage. The identification of P. aeruginosa major membrane proteins (OprF and OprI) immunogenic epitopes and their interactions with host MHC alleles and immunological cells are extremely important in developing new therapeutic strategies for the improvement of immunizations against P. aeruginosa. Immunoinformatics approach employing different bioinformatics tools and databases have recently emerged as the most cost-effective, rapid, and reliable method for the prediction of most antigenic regions of protein as potential targets for the advancement of subunit vaccines [34–36].
Therefore, an attempt has been made in this study to create a novel multi epitope-based vaccine for P. aeruginosa infection targeting the major membrane proteins (OprF and OprI proteins) utilizing the immune-informatics approach. promiscuous highly immunogenic, non-toxic, and non-allergenic B-cell and T-cell (MHC-I and MHC-II) epitopes were consensually predicted through multiple epitope prediction tools. These epitopes also had a high affinity for binding to the highest number of human leukocyte antigen alleles. Consequently, the predicted epitopes along with specific linkers and β-defensin as an adjuvant were used to construct the final vaccine construct. Physiochemical features of vaccines revealed that the vaccine’s molecular weight is 26,726.58 kDa, which is within the acceptable range for a subunit vaccine [37–39]. The vaccine has a theoretical PI score of 9.85, indicating that it is basic in nature. The high aliphatic index indicated high thermostability, while the negative GRAVY value shows its hydrophilic nature. The vaccine protein is classified as stable (Instability index < 40) by ProtParam. PSIPRED V3.3 was used to predict and analyze the vaccine’s secondary structure, which indicated the presence of 32.17% alpha-helix, 11.41% beta-strand, and 55.90% coil. Following that, a homology modelling approach was utilized to construct the tertiary structure of the final designed vaccine [40]. Ramachandran plot results showed that the overall model is reasonable because more than 99% of residues were located in the allowed region. Likewise, other structure validation tools like ERRAT, Verify 3D corroborated the stereochemical accuracy of the developed structure. Furthermore, the vaccine was docked with the TLR-4 receptor to determine the vaccine’s ability to elicit a significant immunological response. Finally, to confirm the stability of the interaction, MD simulation up to 100 ns was performed on the docked vaccine–receptor complex. The RMSD graph showed fluctuation from 40 to 60 ns while RMSF values were observed to be uniform throughout the time period. To ensure the complex’s stability, we performed MD simulations for up to 100 ns. This probably suggests the vaccine construct is robust. In silico cloning was used to analyze the expression of the vaccine construct in the host E. coli K12 strain. According to the findings of the immune simulation, using this multi epitopic vaccine protein as a potential candidate against pseudomonas infection could result in high amounts of antibacterial cytokines, as well as humoral and innate immune responses. In conclusion, this peptide will undoubtedly aid in the development of broad-spectrum peptide vaccines against P. aeruginosa bacteria in the near future.
Conclusions
Defining epitopes possessing the ability to bind with different MHC alleles is the major requirement of cellular immunity. The immunoinformatics tools and databases employed in this study helped to determine immunogenic peptides from the antigenic major membrane proteins of P. aeruginosa. LBL, CTL, and HTL epitopes that are nonallergenic, highly antigenic, and nontoxic, as well as 100% conserved, were considered for final vaccine development, this would elicit a strong immunogenic response while causing no harmful side effects within the body. With high-cost requirements and multiple restrictions for developing the live, attenuated, or inactivated vaccine preparation, these peptide-based vaccine candidates could be useful in further experimental validation to combat pseudomonas infections.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Electronic supplementary material 1 (XLSX 11 kb)
Electronic supplementary material 2 (XLSX 33 kb)
Electronic supplementary material 3 (XLSX 12 kb)
Electronic supplementary material 4 (XLSX 16 kb)
Electronic supplementary material 5 (DOCX 18 kb)
Electronic supplementary material 6 (XLSX 13 kb)
Electronic supplementary material 7 (DOCX 14 kb)
Acknowledgements
We acknowledge infrastructure support available through DBT-BUILDER program (BT/INF/22/SP42155/2021) at KIIT Deemed to Be University, Bhubaneswar. We would like to thank Mr. Krishn Kumar Verma (Associate - Scientific Visualizer, KIIT-TBI) for his contribution in designing graphical representation of figures.
Author Contributions
JD, SRM: Designed and performed the in silico experiments; GSK: performed molecular dynamics simulation study; Sibabratta Patnaik, S Lata, Rakesh Kumar Panda: Data curation; JD, SRM: wrote the paper; Namrata Misra and Mrutyunjay Suar: supervised the research and edited the manuscript. All authors read and approved the manuscript.
Funding
The present study is an in house exploratory research work, authors received no funding support from an external funding source.
Declarations
Conflict of interest
The authors declare no competing interest.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Jyotirmayee Dey and Soumya Ranjan Mahapatra have contributed equally to this work.
Contributor Information
Namrata Misra, Email: namrata@kiitincubator.in.
Mrutyunjay Suar, Email: mrutyunjay@kiitincubator.in, Email: msuar@kiitbiotech.ac.in.
References
- 1.McPhee JB, Tamber S, Bains M, Maier E, Gellatly S, Lo A, Hancock RE. The major outer membrane protein OprG of Pseudomonas aeruginosa contributes to cytotoxicity and forms an anaerobically regulated, cation-selective channel. FEMS Microbiol Lett. 2009;296(2):241–247. doi: 10.1111/j.1574-6968.2009.01651.x. [DOI] [PubMed] [Google Scholar]
- 2.Lister PD, Wolter DJ, Hanson ND. Antibacterial-resistant Pseudomonas aeruginosa: clinical impact and complex regulation of chromosomally encoded resistance mechanisms. Clin Microbiol Rev. 2009;22(4):582–610. doi: 10.1128/CMR.00040-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sharma G, Rao S, Bansal A, Dang S, Gupta S, Gabrani R. Pseudomonas aeruginosa biofilm: potential therapeutic targets. Biologicals. 2014;42(1):1–7. doi: 10.1016/j.biologicals.2013.11.001. [DOI] [PubMed] [Google Scholar]
- 4.Gellatly SL, Hancock RE. Pseudomonas aeruginosa: new insights into pathogenesis and host defenses. Pathog Dis. 2013;67(3):159–173. doi: 10.1111/2049-632X.12033. [DOI] [PubMed] [Google Scholar]
- 5.Campodónico VL, Llosa NJ, Grout M, Döring G, Maira-Litrán T, Pier GB. Evaluation of flagella and flagellin of Pseudomonas aeruginosa as vaccines. Infect Immun. 2010;78(2):746–755. doi: 10.1128/IAI.00806-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pang Z, Raudonis R, Glick BR, Lin TJ, Cheng Z. Antibiotic resistance in Pseudomonas aeruginosa: mechanisms and alternative therapeutic strategies. Biotechnol Adv. 2019;37(1):177–192. doi: 10.1016/j.biotechadv.2018.11.013. [DOI] [PubMed] [Google Scholar]
- 7.Klockgether J, Cramer N, Wiehlmann L, Davenport CF, Tümmler B. Pseudomonas aeruginosa genomic structure and diversity. Front Microbiol. 2011;2:150. doi: 10.3389/fmicb.2011.00150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chevalier S, Bouffartigues E, Bodilis J, Maillot O, Lesouhaitier O, Feuilloley MG, Cornelis P. Structure, function and regulation of Pseudomonas aeruginosa porins. FEMS Microbiol Rev. 2017;41(5):698–722. doi: 10.1093/femsre/fux020. [DOI] [PubMed] [Google Scholar]
- 9.Remans K, Vercammen K, Bodilis J, Cornelis P. Genome-wide analysis and literature-based survey of lipoproteins in Pseudomonas aeruginosa. Microbiology. 2010;156(9):2597–2607. doi: 10.1099/mic.0.040659-0. [DOI] [PubMed] [Google Scholar]
- 10.Woodruff WA, Hancock RE. Pseudomonas aeruginosa outer membrane protein F: structural role and relationship to the Escherichia coli OmpA protein. J Bacteriol. 1989;171(6):3304–3309. doi: 10.1128/jb.171.6.3304-3309.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fernández L, Hancock RE. Adaptive and mutational resistance: role of porins and efflux pumps in drug resistance. Clin Microbiol Rev. 2012;25(4):661–681. doi: 10.1128/CMR.00043-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peluso L, De Luca C, Bozza S, Leonardi A, Giovannini G, Lavorgna A, Rossano F. Protection against Pseudomonas aeruginosa lung infection in mice by recombinant OprF-pulsed dendritic cell immunization. BMC Microbiol. 2010;10(1):1–11. doi: 10.1186/1471-2180-10-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sugawara E, Nestorovich EM, Bezrukov SM, Nikaido H. Pseudomonas aeruginosa porin OprF exists in two different conformations. J Biol Chem. 2006;281(24):16220–16229. doi: 10.1074/jbc.M600680200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hassan R, El-Naggar W, El-Aziz A, Shaaban AM, Kenawy M, Ali YM. Immunization with outer membrane proteins (OprF and OprI) and flagellin B protects mice from pulmonary infection with mucoid and nonmucoid Pseudomonas aeruginosa. J Microbiol Immunol Infect. 2018;51(3):312–320. doi: 10.1016/j.jmii.2016.08.014. [DOI] [PubMed] [Google Scholar]
- 15.Fito-Boncompte L, Chapalain A, Bouffartigues E, Chaker H, Lesouhaitier O, Gicquel G, Chevalier S. Full virulence of Pseudomonas aeruginosa requires OprF. Infect Immun. 2011;79(3):1176–1186. doi: 10.1128/IAI.00850-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sugawara E, Nagano K, Nikaido H. Alternative folding pathways of the major porin OprF of Pseudomonas aeruginosa. FEBS J. 2012;279(6):910–918. doi: 10.1111/j.1742-4658.2012.08481.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang SH, Wang CF, Chang TW, Wang YJ, Liao YD. Oligomerization and insertion of antimicrobial peptide TP4 on bacterial membrane and membrane-mimicking surfactant sarkosyl. PLoS ONE. 2019;14(5):e0216946. doi: 10.1371/journal.pone.0216946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang F, Gu J, Yang L, Gao C, Jing H, Wang Y, Zhang J. Protective efficacy of the trivalent Pseudomonas aeruginosa vaccine candidate PcrV-OprI-Hcp1 in murine pneumonia and burn models. Sci Rep. 2017;7(1):1–13. doi: 10.1038/s41598-017-04029-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gartner T, Baeten M, Otieno S, Revets H, De Baetselier P, Huygen K. Mucosal prime-boost vaccination for tuberculosis based on TLR triggering OprI lipoprotein from Pseudomonas aeruginosa fused to mycolyl-transferase Ag85A. Immunol Lett. 2007;111(1):26–35. doi: 10.1016/j.imlet.2007.04.010. [DOI] [PubMed] [Google Scholar]
- 20.Rau H, Revets H, Cornelis P, Titzmann A, Ruggli N, McCullough KC, Summerfield A. Efficacy and functionality of lipoprotein OprI from Pseudomonas aeruginosa as adjuvant for a subunit vaccine against classical swine fever. Vaccine. 2006;24(22):4757–4768. doi: 10.1016/j.vaccine.2006.03.028. [DOI] [PubMed] [Google Scholar]
- 21.Breidenstein EB, de la Fuente-Núñez C, Hancock RE. Pseudomonas aeruginosa: all roads lead to resistance. Trends Microbiol. 2011;19(8):419–426. doi: 10.1016/j.tim.2011.04.005. [DOI] [PubMed] [Google Scholar]
- 22.Bjarnsholt T, Tolker-Nielsen T, Høiby N, Givskov M. Interference of Pseudomonas aeruginosa signalling and biofilm formation for infection control. Expert Rev Mol Med. 2010 doi: 10.1017/S1462399410001420. [DOI] [PubMed] [Google Scholar]
- 23.Pires J, Siriwardena TN, Stach M, Tinguely R, Kasraian S, Luzzaro F, Endimiani A. In vitro activity of the novel antimicrobial peptide dendrimer G3KL against multidrug-resistant Acinetobacter baumannii and Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2015;59(12):7915–7918. doi: 10.1128/AAC.01853-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bhatnager R, Bhasin M, Arora J, Dang AS. Epitope based peptide vaccine against SARS-COV2: an immune-informatics approach. J Biomol Struct Dyn. 2020 doi: 10.1080/07391102.2020.1787227. [DOI] [PubMed] [Google Scholar]
- 25.Chaudhry A, Rudra D, Treuting P, Samstein RM, Liang Y, Kas A, Rudensky AY. CD4+ regulatory T cells control TH17 responses in a Stat3-dependent manner. Science. 2009;326(5955):986–991. doi: 10.1126/science.1172702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Larsen MV, Lundegaard C, Lamberth K, Buus S, Lund O, Nielsen M. Large-scale validation of methods for cytotoxic T-lymphocyte epitope prediction. BMC Bioinform. 2007;8(1):1–12. doi: 10.1186/1471-2105-8-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nielsen M, Lundegaard C, Lund O. Prediction of MHC class II binding affinity using SMM-align, a novel stabilization matrix alignment method. BMC Bioinform. 2007;8(1):1–12. doi: 10.1186/1471-2105-8-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nielsen PH, Kragelund C, Seviour RJ, Nielsen JL. Identity and ecophysiology of filamentous bacteria in activated sludge. FEMS Microbiol Rev. 2009;33(6):969–998. doi: 10.1111/j.1574-6976.2009.00186.x. [DOI] [PubMed] [Google Scholar]
- 29.Pandey RK, Ojha R, Aathmanathan VS, Krishnan M, Prajapati VK. Immunoinformatics approaches to design a novel multi-epitope subunit vaccine against HIV infection. Vaccine. 2018;36(17):2262–2272. doi: 10.1016/j.vaccine.2018.03.042. [DOI] [PubMed] [Google Scholar]
- 30.Dimitrov I, Flower DR, Doytchinova I. AllerTOP-a server for in silico prediction of allergens. BMC Bioinform. 2013;14(6):1–9. doi: 10.1186/1471-2105-14-S6-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shey RA, Ghogomu SM, Esoh KK, Nebangwa ND, Shintouo CM, Nongley NF, Souopgui J. In-silico design of a multi-epitope vaccine candidate against onchocerciasis and related filarial diseases. Sci Rep. 2019;9(1):1–18. doi: 10.1038/s41598-019-40833-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chatterjee R, Sahoo P, Mahapatra SR, Dey J, Ghosh M, Kushwaha GS, Son YO. Development of a conserved chimeric vaccine for induction of strong immune response against Staphylococcus aureus using immunoinformatics approaches. Vaccines. 2021;9(9):1038. doi: 10.3390/vaccines9091038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mahapatra SR, Dey J, Kushwaha GS, Puhan P, Mohakud NK, Panda SK, Suar M. Immunoinformatic approach employing modeling and simulation to design a novel vaccine construct targeting MDR efflux pumps to confer wide protection against typhoidal Salmonella serovars. J Biomol Struct Dyn. 2021;31:1–13. doi: 10.1080/07391102.2021.1964600. [DOI] [PubMed] [Google Scholar]
- 34.Khan M, Khan S, Ali A, Akbar H, Sayaf AM, Khan A, Wei DQ. Immunoinformatics approaches to explore Helicobacter pylori proteome (Virulence Factors) to design B and T cell multi-epitope subunit vaccine. Sci Rep. 2019;9(1):1–13. doi: 10.1038/s41598-019-49354-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dey J, Mahapatra SR, Singh P, Patro S, Kushwaha GS, Misra N, Suar M. B and T cell epitope-based peptides predicted from clumping factor protein of Staphylococcus aureus as vaccine targets. Microb Pathog. 2021;160:105171. doi: 10.1016/j.micpath.2021.105171. [DOI] [PubMed] [Google Scholar]
- 36.Panda PK, Arul MN, Patel P, Verma SK, Luo W, Rubahn HG, Ahuja R. Structure-based drug designing and immunoinformatics approach for SARS-CoV-2. Sci Adv. 2020;6(28):eabb8097. doi: 10.1126/sciadv.abb8097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ahmad I, Ali SS, Zafar B, Hashmi HF, Shah I, Khan S, Wei DQ. Development of multi-epitope subunit vaccine for protection against the norovirus’ infections based on computational vaccinology. J Biomol Struct Dyn. 2020 doi: 10.1080/07391102.2020.1845799. [DOI] [PubMed] [Google Scholar]
- 38.Mahapatra SR, Sahoo S, Dehury B, Raina V, Patro S, Misra N, Suar M. Designing an efficient multi-epitope vaccine displaying interactions with diverse HLA molecules for an efficient humoral and cellular immune response to prevent COVID-19 infection. Expert Rev Vaccines. 2020;19(9):871–885. doi: 10.1080/14760584.2020.1811091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mahapatra SR, Dey J, Kaur T, Sarangi R, Bajoria AA, Kushwaha GS, Suar M. Immunoinformatics and molecular docking studies reveal a novel multi-epitope peptide vaccine against pneumonia infection. Vaccine. 2021;39(42):6221–6237. doi: 10.1016/j.vaccine.2021.09.025. [DOI] [PubMed] [Google Scholar]
- 40.Narang PK, Dey J, Mahapatra SR, Ghosh M, Misra N, Suar M, Raina V. Functional annotation and sequence-structure characterization of a hypothetical protein putatively involved in carotenoid biosynthesis in microalgae. S Afr J Bot. 2021;141:219–226. doi: 10.1016/j.sajb.2021.04.014. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Electronic supplementary material 1 (XLSX 11 kb)
Electronic supplementary material 2 (XLSX 33 kb)
Electronic supplementary material 3 (XLSX 12 kb)
Electronic supplementary material 4 (XLSX 16 kb)
Electronic supplementary material 5 (DOCX 18 kb)
Electronic supplementary material 6 (XLSX 13 kb)
Electronic supplementary material 7 (DOCX 14 kb)