Abstract
The full potential of diagnostic PCR is limited, in part, by the presence of inhibitors in complex biological samples that reduce the amplification efficiency. Therefore, different pre-PCR treatments are being used to reduce the effects of PCR inhibitors. The aim of the present study was to investigate the effects of 16 amplification facilitators to enhance DNA amplification in the presence of blood, feces, or meat. Different concentrations of amplification facilitators and inhibitory samples were added to PCR mixtures containing rTth or Taq DNA polymerase. The addition of 0.6% (wt/vol) bovine serum albumin to reaction mixtures containing Taq DNA polymerase reduced the inhibitory effect of blood and allowed DNA amplification in the presence of 2% instead of 0.2% (vol/vol) blood. Furthermore, the addition of bovine serum albumin (BSA) to reaction mixtures containing feces or meat enhanced the amplification capacities of both polymerases. Taq DNA polymerase was able to amplify DNA in the presence of 4% instead of 0.4% (vol/vol) feces and 4% instead of 0.2% (vol/vol) meat, and rTth was able to amplify DNA in the presence of 4% instead of 0.4% (vol/vol) feces and 20% instead of 2% (vol/vol) meat. The single-stranded DNA binding T4 gene 32 protein (gp32) had a relieving effect similar to that of BSA, except when it was added to PCR mixtures of rTth containing meat and of Taq DNA polymerase containing feces. The relieving effects of betaine and a cocktail of proteinase inhibitors were more sample specific. The addition of 11.7% (wt/vol) betaine allowed Taq DNA polymerase to amplify DNA in the presence of 2% (vol/vol) blood, while the addition of proteinase inhibitors allowed DNA amplification by both polymerases in the presence of 4% (vol/vol) feces. When various combinations of betaine, BSA, gp32, and proteinase inhibitors were tested, no synergistic or additive effects were observed. The effects of facilitators on real-time DNA synthesis instead of conventional PCR were also studied.
Diagnostic PCR is limited, in part, by the presence of inhibitory substances in complex biological samples, which may interfere with the cell lysis step, inactivate the thermostable DNA polymerase, and/or interfere with nucleic acids (1, 4, 9, 15, 19, 28). Much effort is being devoted to the development of various sample pretreatments to generate PCR-compatible samples (for a review, see reference 11). However, sample preparation techniques are at present complicated, require experience, are difficult to handle for large numbers of samples, and are time-consuming. An alternative strategy that can be used to overcome PCR inhibition is to enhance the efficiency of PCR in the presence of complex biological samples. This can be done by using an alternative thermostable DNA polymerase more resistant to inhibitors (3, 9, 13, 27) and by using amplification facilitators such as bovine serum albumin (BSA), single-stranded DNA binding T4 gene 32 protein (gp32), organic solvents, and proteinase inhibitors (4, 5, 10, 13–15). The addition of amplification facilitators has also been found to improve the specificity of PCR and allow the amplification of GC-rich DNA sequences (8, 18, 22, 26), and/or increase the fidelity of DNA synthesis (29).
The aim of this study was to investigate the abilities of 16 amplification facilitators to enhance the amplification efficiencies of rTth and Taq DNA polymerases in the presence of blood, feces, and meat in conventional PCR. The abilities of the amplification facilitators to mediate DNA synthesis in reaction mixtures containing rTth and inhibitory samples with a single-stranded poly(dA) template with an oligo(dT) primer annealed to the 3′ end were also investigated. The reason for using this simplified system instead of conventional PCR was to avoid the interference of primer dimers and nonspecific amplicons.
MATERIALS AND METHODS
Template DNA.
The DNA of Listeria monocytogenes 167 vet, which was obtained from Swedish Meats R&D (Kävlinge, Sweden), was used as the target DNA in this study. The DNA extraction was performed in accordance with a standard technique described by Sambrook et al. (20), modified by the addition of 30 U of mutanolysin (Sigma Chemical Co., St. Louis, Mo.) per ml to the lysis solution. The concentration of DNA was determined spectrophotometrically (20).
PCR-inhibitory samples.
The blood sample was drawn from a healthy person and placed into 5-ml evacuated blood collection tubes containing 0.1 ml (0.47 mol/liter) of EDTA (Terumo Europe N. V., Leuven, Belgium). A fecal sample (henceforth referred to as feces) was obtained from a healthy person and was diluted 10-fold in physiological saline solution and homogenized for 2 min in a stomacher (Lab-Blender 400; Steward Laboratory, London, United Kingdom). The minced pork meat (henceforth referred to as meat) was diluted 10-fold in physiological saline solution and was homogenized for 2 min in a stomacher. Each PCR-inhibitory sample was poured into sterile 1.5-ml Eppendorf tubes, and the tubes were stored at −20°C. The frozen blood, feces, and meat homogenates were thawed at room temperature, mixed with a vortex mixer, and left for 5 min to allow the large particles to settle before they were diluted and/or added to the PCR mixtures.
PCR assay and incubation conditions.
The total volume of the PCR mixtures was 25 μl. The PCR assay was carried out as previously described by Lantz et al. (12). The PCR mixtures contained each of the primers rU8 and LM2 at a concentration of 0.5 μM (12, 16) and each of the deoxyribonucleoside triphosphates at a concentration of 0.2 mM. Reaction buffers for the DNA polymerases, as specified by the manufacturers, were as follows. The PCR buffer for rTth DNA polymerase (Perkin-Elmer Cetus, Norwalk, Conn.) contained 5% (vol/vol) glycerol, 10 mM Tris-HCl (pH 8.3), 0.1 M KCl, 0.05% (wt/vol) Tween 20, 0.75 mM EGTA [ethylene glycol-bis(β-aminoethyl ether)-N,N,N′,N′-tetraacetic acid], 2.5 mM MgCl2, and 1.25 U of rTth DNA polymerase. The PCR buffer for Taq DNA polymerase (Roche Molecular Biochemicals, Basel, Switzerland) contained 10 mM Tris-HCl, 1.5 mM MgCl2, 50 mM KCl (pH 8.3; 20°C), and 0.75 U of Taq DNA polymerase. The 0.55-kb PCR product was visualized by 1.3% agarose gel electrophoresis containing ethidium bromide (20). The gel was analyzed with a gel documentation system (Bio-Rad Laboratories, Hercules, Calif.). The results were recorded as + (PCR product of high yield), ± (PCR product of low yield), or − (no PCR product).
Real-time DNA synthesis conditions.
The reaction volumes were 25 μl. All mixtures contained 0.2 mM dTTP, a 1:10,000-diluted stock solution of SYBR Green I (Roche Molecular Biochemicals), 4 mM MgCl2, and 10 ng of poly(dA) with oligo(dT)12–18 (Amersham Pharmacia Biotech, Uppsala, Sweden). The reaction mixtures for rTth DNA polymerase contained 1× chelating buffer and 1.25 U of rTth (Perkin-Elmer Cetus). Different concentrations of PCR inhibitors (1, 0.2, and 0.04% [vol/vol] blood; 20, 4, and 2% [vol/vol] feces or meat), with or without PCR facilitators (11.7% [wt/vol] betaine, 0.4% [wt/vol] BSA, 0.01% [wt/vol] gp32, 1× proteinase inhibitor cocktail), were added to glass capillary tubes, and the tubes were incubated at 65°C. The background fluorescence for each sample was measured by measuring the fluorescence of a reaction mixture containing all the sample components except rTth. Ninety fluorescence measurements were taken at 20-s intervals. The fluorescence of the samples was monitored online with a LightCycler Instrument (Roche Molecular Biochemicals). The increase in fluorescence due to DNA synthesis was considered the difference between the sample fluorescence and the background fluorescence. The mean fluorescence level of three independent experiments was calculated.
RESULTS
Abilities of amplification facilitators to relieve inhibition of rTth and Taq DNA polymerases.
The effects of 16 PCR facilitators on the amplification capacities of rTth and Taq DNA polymerases were tested in the presence of different concentrations of blood, feces, and meat (Tables 1 and 2). Except for gp32, three different concentrations that had no positive or negative effect on the detection limit in the absence of added PCR-inhibitory samples were investigated. Among the 16 facilitators tested, only BSA reduced the inhibition of both rTth and Taq DNA polymerases in the presence of all types of inhibitory samples. The addition of 0.4% (wt/vol) BSA allowed DNA amplification by Taq DNA polymerase in the presence of 2% instead of 0.02% (vol/vol) blood, 4% instead of 0.4% (vol/vol) feces, and 4% instead of 0.2% (vol/vol) meat. The corresponding values for rTth were 4% instead of 0.4% (vol/vol) feces and 20% instead of 0.4% (vol/vol) meat. rTth was, as observed earlier (3), able to amplify DNA in the presence of 20% (vol/vol) blood without the addition of any facilitators. When 0.01% (wt/vol) gp32 was added to the PCR mixtures, the inhibitory effects of blood and meat on Taq DNA polymerase were reduced by the same level as the addition of 0.4% (wt/vol) BSA. A similar effect was also observed when gp32 was added to reaction mixtures of rTth containing feces or meat. However, the ability of gp32 to reduce the inhibition of Taq DNA polymerase by feces was not reproducible when different batches of Taq DNA polymerase and buffers were used. For example, in the first run of experiments, Taq DNA polymerase amplified DNA in the presence of 2% (vol/vol) feces. However, in the second run of an experiment with a new Taq DNA polymerase and buffer, the addition of gp32 could not overcome the inhibitory effect of feces. The addition of 11.7% (wt/vol) betaine allowed the Taq DNA polymerase to amplify the specific product in the presence of 2% (vol/vol) blood and 0.4% (vol/vol) meat, in comparison to weak amplification in the presence of 0.2 or 0.04% (vol/vol) blood and 0.2% (vol/vol) meat without any facilitator. Betaine was also found to relieve inhibition of rTth by meat and to allow DNA amplification in the presence of 4% instead of 0.4% (vol/vol) meat. Without proteinase inhibitors, rTth and Taq DNA polymerase were able to amplify DNA in the presence of 0.4% and 0.27% (vol/vol) feces, respectively. The addition of proteinase inhibitors, however, reduced the inhibition of both polymerases by feces and allowed DNA amplification in the presence of 4% (vol/vol) feces. No enhanced efficiency of PCR was observed for the other 12 facilitators.
TABLE 1.
Effects of 16 PCR facilitators on amplification capacity of Taq DNA polymerase in the presence of blood, feces, and meat
| Facilitator and facilitator concnc | PCR resultsa with the following at the indicated concn (% [vol/vol])b:
|
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Blood
|
Feces
|
Meat
|
||||||||||||||
| 20 | 2 | 0.2 | 0.04 | 0.02 | 20 | 4 | 2 | 0.4 | 0.27 | 0.2 | 20 | 4 | 2 | 0.4 | 0.2 | |
| Acetamide | ||||||||||||||||
| 0 | −, − | −, − | +, − | +, + | +, ± | −, − | − | −, − | −, − | + | −, + | −, − | −, − | −, − | −, − | +, + |
| 1 | −, − | +, − | +, − | +, − | +, − | −, − | − | −, − | −, − | + | −, + | −, − | −, − | −, − | −, − | +, ± |
| 2.5 | −, − | −, − | ±, − | +, − | +, − | −, − | − | −, − | −, − | + | −, + | −, − | −, − | −, − | −, − | ±, − |
| 4 | −, − | −, − | −, − | −, − | ±, − | −, − | − | −, − | −, − | + | −, + | −, − | −, − | −, − | −, − | −, − |
| Betaine | ||||||||||||||||
| 0 | −, − | −, − | −, ± | −, ± | −, − | −, − | − | −, − | −, + | + | −, + | −, − | −, − | −, − | −, − | +, ± |
| 5.9 | −, − | +, + | +, + | +, + | +, + | −, − | − | −, − | −, + | + | −, + | −, − | −, − | −, − | ±, ± | +, + |
| 8.8 | −, − | ±, + | +, + | +, + | +, + | −, − | − | −, − | −, + | + | −, + | −, − | −, − | −, − | −, + | +, + |
| 11.7 | −, − | +, + | +, + | +, + | +, + | −, − | − | −, − | −, + | + | −, + | −, − | −, − | −, − | ±, + | +, + |
| BSA | ||||||||||||||||
| 0 | −, − | −, − | −, − | −, − | ±, − | −, − | − | −, − | −, + | + | −, + | −, − | −, − | −, − | −, − | ±, − |
| 0.2 | −, − | −, − | −, ± | ±, + | +, + | −, − | − | +, + | +, + | + | +, + | −, − | ±, + | +, + | +, + | +, + |
| 0.4 | −, − | −, + | −, + | +, + | +, + | −, − | ± | +, + | +, + | + | +, + | −, − | −, + | +, + | +, + | +, + |
| 0.6 | −, − | −, + | ±, + | +, + | +, + | −, − | + | +, + | +, + | + | +, + | −, − | −, + | +, + | +, + | +, + |
| Dextran 40 | ||||||||||||||||
| 0 | −, − | ±, − | ±, − | ±, ± | ±, + | −, − | − | −, − | −, − | + | −, + | −, − | −, − | −, − | ±, − | ±, ± |
| 1 | −, − | ±, − | −, − | −, + | −, + | −, − | − | −, − | −, − | + | −, + | −, − | −, − | −, − | +, − | +, + |
| 2.5 | −, − | −, − | −, ± | −, + | −, + | −, − | − | −, − | −, − | + | −, + | −, − | −, − | −, − | ±, − | +, + |
| 4 | −, − | −, − | −, ± | −, + | −, + | −, − | − | −, − | −, − | + | −, + | −, − | −, − | ±, − | ±, − | +, + |
| Dextran 500 | ||||||||||||||||
| 0 | −, − | −, − | −, ± | −, + | −, ± | −, − | − | −, − | −, − | + | −, + | −, − | −, − | −, − | ±, − | ±, + |
| 1 | −, − | −, − | −, − | −, + | −, + | −, − | − | −, − | −, − | + | −, + | −, − | −, − | −, − | +, + | +, + |
| 2.5 | −, − | −, − | −, − | −, ± | −, ± | −, − | − | −, − | −, ± | + | −, + | −, − | −, − | −, − | +, − | ±, + |
| 4 | −, − | −, ± | −, ± | −, ± | −, ± | −, − | − | −, − | −, + | + | −, + | −, − | +, − | +, − | +, ± | +, ± |
| Dimethyl sulfoxide | ||||||||||||||||
| 0 | −, − | −, − | −, − | −, − | −, − | −, − | − | −, − | −, − | + | −, + | −, − | −, − | −, − | −, − | +, − |
| 2.5 | −, − | −, − | −, − | −, − | −, − | −, − | − | −, − | −, − | + | −, + | −, − | −, − | −, − | −, − | +, − |
| 5 | −, − | −, − | −, − | −, − | −, − | −, − | − | −, − | −, − | + | −, + | −, − | −, − | −, − | −, − | +, − |
| 7.5 | −, − | −, − | −, − | −, − | −, − | −, − | − | −, − | −, − | + | −, + | −, − | −, − | −, − | −, − | +, − |
| Formamide | ||||||||||||||||
| 0 | −, − | −, − | −, − | −, − | −, − | −, − | − | −, − | −, + | + | −, + | −, − | −, − | −, − | ±, − | +, − |
| 0.25 | −, − | −, − | −, − | −, − | −, − | −, − | − | −, − | −, + | + | −, + | −, − | −, − | −, − | ±, − | +, + |
| 0.5 | −, − | −, − | −, − | −, − | −, − | −, − | − | −, − | −, ± | + | −, + | −, − | −, − | −, − | ±, − | +, − |
| 1 | −, − | −, − | −, − | −, − | −, − | −, − | − | −, − | −, − | − | −, − | −, − | −, − | −, − | ±, − | +, − |
| Glycerol | ||||||||||||||||
| 0 | −, − | −, ± | ±, ± | −, − | −, − | −, − | − | −, − | −, − | + | −, + | −, − | −, − | −, − | ±, − | +, ± |
| 5 | −, − | −, ± | −, ± | −, − | −, − | −, − | − | −, − | −, − | + | −, + | −, − | −, − | −, − | −, − | +, − |
| 7.5 | −, − | −, − | −, − | −, − | −, − | −, − | − | −, − | −, − | + | −, + | −, − | −, − | −, − | −, − | ±, − |
| 10 | −, − | −, − | −, − | −, − | −, − | −, − | − | −, − | −, − | + | −, + | −, − | −, − | −, − | −, − | +, − |
| gp32 | ||||||||||||||||
| 0 | −, − | +, + | +, + | −, − | −, − | −, − | −, − | −, − | −, − | ±, − | −, + | −, − | −, − | −, − | −, − | +, ± |
| 0.01 | −, − | +, + | +, + | +, + | +, + | −, − | −, − | −, ± | −, + | −, ± | −, + | −, − | +, + | +, + | +, + | +, + |
| Nonidet P-40 | ||||||||||||||||
| 0 | −, − | −, − | +, − | +, − | +, − | −, − | − | −, − | −, − | + | −, + | −, − | −, − | −, − | ±, − | +, − |
| 0.1 | −, − | −, − | −, − | +, − | +, − | −, − | − | −, − | −, − | + | −, + | −, − | −, − | −, − | −, − | +, ± |
| 0.25 | −, − | −, − | −, − | ±, − | ±, − | −, − | − | −, − | −, − | + | −, + | −, − | −, − | −, − | −, − | +, − |
| 0.5 | −, − | −, − | +, − | ±, − | ±, − | −, − | − | −, − | −, − | + | −, + | −, − | −, − | −, − | −, − | +, − |
| Polyethylene glycol 400 | ||||||||||||||||
| 0 | −, − | −, − | −, − | −, − | −, − | −, − | − | −, − | −, − | + | −, + | −, − | −, − | −, − | −, − | +, − |
| 1 | −, − | −, − | ±, − | −, − | +, − | −, − | − | −, − | −, ± | + | −, + | −, − | −, − | −, − | −, − | +, − |
| 2 | −, − | −, − | ±, − | ±, − | ±, − | −, − | − | −, − | −, + | + | −, + | −, − | −, − | −, − | −, − | +, − |
| 4 | −, − | −, − | ±, − | +, − | +, − | −, − | − | −, − | −, + | + | −, + | −, − | −, − | −, − | −, − | −, − |
| Polyethylene glycol 4000 | ||||||||||||||||
| 0 | −, − | −, − | −, − | −, − | −, − | −, − | − | −, − | −, − | + | −, + | −, − | −, − | −, − | +, − | +, − |
| 1 | −, − | −, − | −, − | −, − | −, − | −, − | − | −, − | −, ± | + | −, + | −, − | −, − | −, − | ±, − | +, − |
| 2.5 | −, − | −, − | −, − | −, − | −, − | −, − | − | −, − | −, ± | + | −, + | −, − | −, − | −, − | −, − | +, − |
| 4 | −, − | −, − | −, − | −, − | −, − | −, − | − | −, − | −, + | + | −, + | −, − | −, − | −, − | −, − | +, − |
| Proteinase inhibitord | ||||||||||||||||
| 0 | −, − | −, − | +, − | +, − | +, − | −, − | − | −, − | −, − | + | −, + | −, − | −, − | −, − | +, − | +, − |
| 0.25× | −, − | −, − | ±, − | +, − | +, − | −, − | − | −, + | +, + | + | +, + | −, − | −, − | −, − | ±, − | +, + |
| 0.5× | −, − | −, − | −, − | ±, − | +, − | −, − | − | +, + | +, + | + | +, + | −, − | −, − | −, − | ±, − | +, − |
| 1× | −, − | −, − | −, − | −, − | ±, − | −, − | + | +, + | +, + | + | +, + | −, − | −, − | −, − | +, − | +, − |
| TMACe | ||||||||||||||||
| 0 | −, − | −, − | −, − | −, − | −, − | −, − | − | −, − | −, ± | + | −, + | −, − | −, − | −, − | +, − | +, − |
| 2.8 × 10−4 | −, − | −, − | −, − | −, − | −, − | −, − | − | −, − | −, ± | + | −, + | −, − | −, − | −, − | +, − | +, − |
| 5.5 × 10−4 | −, − | −, − | −, − | −, − | −, − | −, − | − | −, − | −, ± | + | −, + | −, − | −, − | −, − | +, − | +, − |
| 1.1 × 10−3 | −, − | −, − | −, − | −, − | −, − | −, − | − | −, − | −, ± | + | −, + | −, − | −, − | −, − | +, − | +, − |
| Tween 20 | ||||||||||||||||
| 0 | −, − | −, − | ±, − | +, − | +, − | −, − | − | −, − | −, − | + | −, + | −, − | −, − | −, − | −, − | ±, − |
| 0.1 | −, − | −, − | ±, − | +, − | +, − | −, − | − | −, − | −, − | + | −, + | −, − | −, − | −, − | −, − | ±, − |
| 0.25 | −, − | −, − | −, − | +, − | +, − | −, − | − | −, − | −, + | + | −, + | −, − | −, − | −, − | −, − | ±, − |
| 0.5 | −, − | −, − | −, − | ±, − | +, − | −, − | − | −, − | −, + | + | −, + | −, − | −, − | −, − | −, − | ±, − |
| Tween 80 | ||||||||||||||||
| 0 | −, − | −, − | −, − | −, − | −, ± | −, − | − | −, − | −, + | + | −, + | −, − | −, − | −, − | +, − | +, ± |
| 0.1 | −, − | −, − | −, − | −, ± | −, + | −, − | − | −, − | −, + | + | −, + | −, − | −, − | −, − | ±, − | +, ± |
| 0.2 | −, − | −, − | −, − | −, − | −, ± | −, − | − | −, − | −, + | + | −, + | −, − | −, − | −, − | ±, − | +, ± |
| 0.5 | −, − | −, − | −, − | −, − | −, ± | −, − | − | −, − | −, + | + | −, + | −, − | ±, − | −, − | −, − | +, ± |
Results are of one or two independent PCRs; +, PCR product of high yield; ±, PCR product of low yield; −, no PCR product. The results were recorded in the order in which the PCRs were done.
Percentage (vol/vol) of blood and homogenates of feces and meat in PCR mixture for Taq DNA polymerase containing 1 ng of L. monocytogenes DNA.
Percentage (wt/vol) of 16 amplification facilitators in the reaction mixtures. The PCR sensitivity in the presence of the concentrations of these facilitators was the same as that with a solution of pure water with DNA.
One tablet of Proteinase Inhibitor Complete Mini cocktail (EDTA-free; Roche Molecular Biochemicals) was dissolved in 2 ml of deionized water. This concentration was five times greater than that recommended by the supplier.
Tetramethylammonium chloride.
TABLE 2.
Effects of 16 PCR facilitators on amplification capacity of rTth DNA polymerase in the presence of feces and meat
| Facilitator and facilitator concnc | PCR resultsa with the following at the indicated concn (% [vol/vol])b:
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Feces
|
Meat
|
|||||||||
| 20 | 4 | 2 | 0.4 | 0.2 | 20 | 4 | 2 | 0.4 | 0.2 | |
| Acetamide | ||||||||||
| 0 | −, − | −, − | −, − | +, ± | +, + | −, − | −, − | ±, − | +, ± | +, ± |
| 1 | −, − | −, − | −, − | +, − | +, ± | −, − | −, − | −, − | +, ± | +, + |
| 2.5 | −, − | −, − | −, − | +, − | +, ± | −, − | −, − | −, − | +, − | +, ± |
| 4 | −, − | −, − | −, − | ±, − | +, − | −, − | −, − | −, − | ±, − | ±, − |
| Betaine | ||||||||||
| 0 | −, − | −, − | −, − | +, − | +, + | −, − | −, − | −, − | ±, + | ±, + |
| 5.9 | −, − | −, − | −, − | +, − | +, + | −, − | ±, − | +, − | ±, + | +, + |
| 8.8 | −, − | −, − | −, − | +, ± | +, + | −, − | −, ± | ±, ± | +, + | +, + |
| 11.7 | −, − | −, − | −, − | +, + | +, + | −, − | −, − | +, − | +, + | +, + |
| BSA | ||||||||||
| 0 | −, − | −, − | −, − | −, + | +, + | −, − | −, − | ±, − | +, + | +, + |
| 0.2 | −, − | +, − | ±, ± | −, + | +, + | +, + | +, + | +, + | +, + | +, + |
| 0.4 | −, − | +, ± | +, + | +, + | +, + | +, + | +, + | +, + | +, + | +, + |
| 0.6 | −, − | −, ± | −, + | +, + | +, + | +, + | +, + | +, + | +, + | +, + |
| Dextran 40 | ||||||||||
| 0 | −, − | −, − | −, − | +, + | +, + | −, − | −, − | ±, − | +, + | +, + |
| 1 | −, − | −, − | −, − | +, + | +, + | −, − | +, − | ±, + | +, + | +, + |
| 2.5 | −, − | −, − | +, − | +, + | +, + | −, − | −, − | −, − | +, + | +, + |
| 4 | −, − | −, − | −, − | +, + | +, + | −, − | −, − | +, + | +, + | +, + |
| Dextran 500 | ||||||||||
| 0 | −, − | −, − | −, + | +, + | +, + | −, − | −, − | +, ± | +, + | +, + |
| 1 | −, − | −, − | −, − | +, + | +, + | −, − | −, − | ±, − | +, + | +, + |
| 2.5 | −, − | −, − | −, − | +, + | +, + | −, − | −, − | ±, − | +, + | +, + |
| 4 | −, − | −, − | −, − | +, + | +, + | −, − | −, − | −, − | ±, + | +, + |
| Dimethyl sulfoxide | ||||||||||
| 0 | −, − | −, − | −, − | +, + | +, + | −, − | −, − | ±, − | ±, + | +, + |
| 2.5 | −, − | −, − | −, − | +, + | +, + | −, − | −, − | −, − | ±, + | +, + |
| 5 | −, − | −, − | −, − | +, + | +, + | −, − | −, − | −, − | −, ± | +, + |
| 7.5 | −, − | −, − | −, − | +, − | +, − | −, − | −, − | −, − | −, − | ±, − |
| Formamide | ||||||||||
| 0 | −, − | −, − | −, − | +, + | +, + | −, − | −, − | ±, ± | ±, + | +, + |
| 0.25 | −, − | −, − | −, − | +, − | +, + | −, − | ±, − | ±, − | ±, + | +, + |
| 0.5 | −, − | −, − | −, − | +, − | +, + | −, − | −, − | ±, − | +, − | +, + |
| 1 | −, − | −, − | −, − | +, − | +, − | −, − | −, − | ±, − | ±, − | +, − |
| Glycerol | ||||||||||
| 0 | −, − | −, − | −, − | +, + | +, + | −, − | −, − | −, − | +, + | +, + |
| 5 | −, − | −, − | −, − | +, + | +, + | −, − | −, ± | −, ± | +, + | +, + |
| 7.5 | −, − | −, − | −, − | +, + | +, + | −, − | −, ± | +, ± | +, + | +, + |
| 10 | −, − | −, − | −, − | +, + | +, + | −, − | −, ± | −, + | +, + | +, + |
| gp32 | ||||||||||
| 0 | −, − | −, − | +, + | +, + | +, + | −, − | −, − | −, − | +, + | +, + |
| 0.01 | −, − | +, + | +, + | +, + | +, + | −, − | +, + | +, + | +, + | +, + |
| Nonidet P-40 | ||||||||||
| 0 | −, − | −, − | −, − | +, + | +, + | −, − | −, − | −, ± | +, + | +, + |
| 0.1 | −, − | −, − | −, − | +, + | +, + | −, − | −, − | −, − | +, + | +, + |
| 0.25 | −, − | −, − | −, − | +, + | +, + | −, − | −, − | −, − | +, + | +, + |
| 0.5 | −, − | −, − | −, ± | +, + | +, + | −, − | −, − | −, − | +, + | +, + |
| Polyethylene glycol 400 | ||||||||||
| 0 | −, − | −, − | −, − | +, + | +, + | −, − | −, − | −, − | +, + | +, + |
| 1 | −, − | −, − | −, ± | +, + | +, + | −, − | −, − | −, − | −, + | ±, + |
| 2 | −, − | −, − | −, ± | +, + | +, + | −, − | −, − | −, − | −, + | ±, + |
| 4 | −, − | −, − | −, ± | +, + | +, + | −, − | −, − | −, − | −, ± | −, + |
| Polyethylene glycol 4000 | ||||||||||
| 0 | −, − | −, − | −, − | +, + | +, + | −, − | −, − | −, − | +, + | +, + |
| 1 | −, − | −, − | −, ± | +, + | +, + | −, − | −, − | −, − | +, + | +, + |
| 2.5 | −, − | −, − | −, + | +, + | +, + | −, − | −, − | −, − | −, + | +, + |
| 4 | −, − | −, − | −, ± | +, + | +, + | −, − | −, − | −, − | ±, ± | ±, + |
| Proteinase inhibitord | ||||||||||
| 0 | −, − | −, − | −, − | +, + | +, + | −, − | −, − | ±, − | +, + | +, + |
| 0.25× | −, − | −, − | +, ± | +, + | +, + | −, − | −, − | −, − | +, + | +, + |
| 0.5× | −, − | −, − | +, ± | +, + | +, + | −, − | −, − | ±, − | +, − | +, + |
| 1× | −, − | +, ± | +, ± | +, + | +, + | −, − | −, − | −, − | ±, − | +, + |
| TMAC | ||||||||||
| 0 | −, − | −, − | −, − | +, ± | +, + | −, − | −, − | −, − | ±, + | +, + |
| 2.8 × 10−4 | −, − | −, − | −, − | +, ± | +, ± | −, − | −, − | −, − | +, + | +, + |
| 5.5 × 10−4 | −, − | −, − | −, − | +, ± | +, + | −, − | −, − | −, − | ±, + | +, + |
| 1.1 × 10−3 | −, − | −, − | −, − | +, ± | +, + | −, − | −, − | −, − | ±, + | +, + |
| Tween 20 | ||||||||||
| 0 | −, − | −, − | −, − | +, + | +, + | −, − | −, − | −, − | ±, ± | +, + |
| 0.1 | −, − | −, − | −, − | +, + | +, + | −, − | −, − | ±, − | ±, ± | +, + |
| 0.25 | −, − | −, − | −, − | +, + | +, + | −, − | −, − | −, − | +, ± | +, + |
| 0.5 | −, − | −, − | −, − | +, + | +, + | −, − | −, − | −, − | ±, + | +, + |
| Tween 80 | ||||||||||
| 0 | −, − | −, − | −, − | +, + | +, + | −, − | −, − | −, − | +, + | +, + |
| 0.1 | −, − | −, − | −, − | +, + | +, + | −, − | −, − | ±, − | +, ± | +, + |
| 0.25 | −, − | −, − | −, ± | +, + | +, + | −, − | −, − | ±, − | +, + | +, + |
| 0.5 | −, − | −, − | −, + | +, + | +, + | −, − | −, − | ±, − | +, + | +, + |
Results are of two independent PCR results; +, PCR product of high yield; ±, PCR product of low yield; −, no PCR product. The results were recorded in the order in which the PCRs were done.
Percentage (vol/vol) of homogenates of feces and meat in PCR mixture for rTth containing 1 ng of L. monocytogenes DNA.
Percentage (wt/vol) of 16 amplification facilitators in the reaction mixtures. The PCR sensitivity in the presence of the concentrations of these facilitators was the same as that with a solution of pure water with DNA.
One tablet of Proteinase Inhibitor Complete Mini cocktail (EDTA-free; Roche Molecular Biochemicals) was dissolved in 2 ml of deionized water. This concentration was five times greater than that recommended by the supplier.
Four facilitators (11.7% betaine, 0.4% BSA, 0.01% gp32, and 1× proteinase inhibitor mixture) which had the highest relieving effects were selected to study the effects of their combinations on the amplification capacities of rTth and Taq DNA polymerases in the presence of blood, feces, and meat (Table 3). No synergistic or additive effects were observed by the different combinations of the four facilitators. However, the PCR product yield was increased when betaine was combined with BSA or gp32 and when BSA was combined with gp32 in reaction mixtures of Taq DNA polymerase containing feces.
TABLE 3.
Effects of different combinations of four amplification facilitators on amplification capacities of rTth and Taq DNA polymerases in the presence of blood, feces, and meat
| Polymerase and facilitator (facilitators concnc) | PCR resultsa with the following at the indicated concn (% [vol/vol])b:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Bloodd
|
Feces
|
Meat
|
|||||||
| 20 | 4 | 2 | 20 | 4 | 2 | 20 | 4 | 2 | |
| rTth | |||||||||
| BSA (0.4), gp32 (0.01) | −, − | +, + | +, + | +, + | +, + | +, + | |||
| Betaine (11.7), gp32 (0.01) | −, − | −, + | +, + | −, − | +, + | +, + | |||
| gp32 (0.01), proteinase inhibitor (1×) | −, − | −, − | −, − | −, − | +, + | +, + | |||
| BSA (0.4), betaine (11.7) | −, ± | +, + | +, + | +, + | +, + | +, + | |||
| BSA (0.4), proteinase inhibitor (1×) | −, − | +, + | +, + | −, − | +, + | +, + | |||
| Betaine (11.7), proteinase inhibitor (1×) | −, − | +, + | +, + | −, − | −, − | −, − | |||
| Taq | |||||||||
| BSA (0.4), gp32 (0.01) | −, − | +, + | +, + | −, − | +, + | +, + | +, − | +, + | +, + |
| Betaine (11.7), gp32 (0.01) | −, − | −, − | −, − | −, − | +, + | +, + | −, − | +, + | +, + |
| gp32 (0.01), proteinase inhibitor (1×) | −, − | −, − | −, − | −, − | −, − | −, − | −, − | +, − | +, − |
| BSA (0.4), betaine (11.7) | −, − | +, + | +, + | −, − | +, + | +, + | −, − | +, + | +, + |
| BSA (0.4), proteinase inhibitor (1×) | −, − | +, + | +, + | −, − | +, + | +, + | −, − | +, + | +, + |
| Betaine (11.7), proteinase inhibitor (1×) | −, − | +, + | +, + | −, − | −, − | −, − | −, − | −, + | −, + |
Results are of two independent PCRs; +, PCR product of high yield; ±, PCR product of low yield; −, no PCR product. The results were recorded in the same order in which the PCRs were done.
Percentage (vol/vol) of blood and homogenates of feces and meat in the PCR mixtures for rTth and Taq DNA polymerase containing 1 ng of L. monocytogenes DNA.
Percentage (wt/vol) of amplification facilitators in the reaction mixtures.
The polymerase rTth was able to amplify DNA in the presence of all concentrations of blood tested without the addition of any amplification facilitators.
Quantitative effects of amplification facilitators.
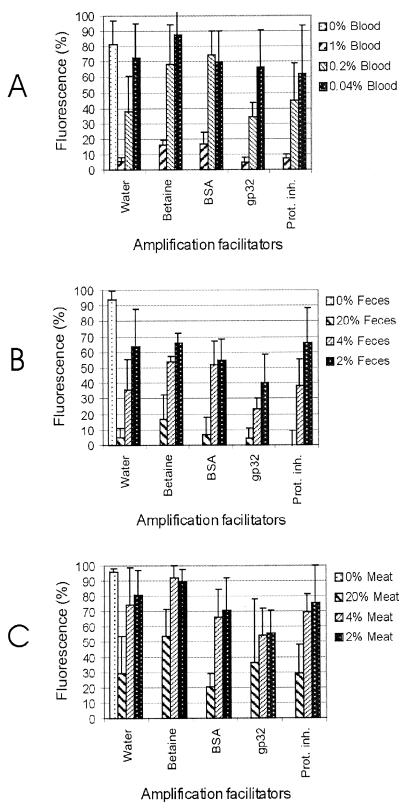
The capacity of rTth to synthesize DNA was monitored by the increase in fluorescence levels as a result of SYBR Green I binding to the double-stranded DNA formed. The effects of betaine, BSA, gp32, and proteinase inhibitors on real-time DNA synthesis of rTth in the presence of blood, feces, and meat are shown in Fig. 1. These fluorescence measurements were taken at the end of the incubation period. The maximum fluorescence was 100, even though the fluorescence for 21 of a total of 144 samples exceeded this value. The background fluorescence of reaction mixtures without inhibitory samples or facilitators was less than 1.5, which excludes the interference of poly(dA) with oligo(dT) on the fluorescence level. The highest background fluorescence (39%) was in the presence of 20% (vol/vol) feces, while the highest background fluorescence signals in the presence of 1% (vol/vol) blood and 20% (vol/vol) meat were 2 and 24%, respectively. The standard deviation values of the mean fluorescence showed large variations between the three runs of experiments and were found to increase as the mean fluorescence signal increased. The mean fluorescence signal was the lowest in the presence of blood and the highest in the presence of meat. Inclusion of higher concentrations of blood was not possible due to the precipitation of blood proteins, which hindered the detection of the fluorescence at the tip of the glass capillary tubes. Despite the limitation of the assay, it was shown that addition of PCR-inhibitory samples reduced the fluorescence signal, and a linear relation was observed between the decrease in the concentration of inhibitor and the increase in the fluorescence signal, mainly when blood and feces were added to the reaction mixture without amplification facilitators (Fig. 1A and B). Addition of 11.7% (wt/vol) betaine was found to enhance the fluorescence signals in the reaction mixtures containing blood, feces, and meat. The maximum enhancement effect was found in the presence of meat, so that the addition of meat homogenate at ≤4% was no longer inhibitory compared to the inhibitory effects of reaction mixtures without inhibitors or facilitators, as determined from the fluorescence signals. In addition, a linear relation was observed between decreasing the concentration of inhibitors and an increase in the fluorescence signal in the presence of betaine. The fluorescence signals were only slightly affected in the presence of BSA, gp32, and proteinase inhibitors.
FIG. 1.
Effects of amplification facilitators on real-time DNA synthesis of rTth in the presence of blood (A), feces (B), and meat (C). prot. inh., proteinase inhibitors.
DISCUSSION
Betaine, BSA, gp32, and proteinase inhibitors improved the amplification capacities of rTth and Taq DNA polymerases in the presence of blood, feces, and meat when the effects of 16 amplification facilitators were investigated. BSA and, to a lesser extent, gp32 were the most efficient facilitators in conventional PCR. The presence of 0.4 to 0.6% (wt/vol) BSA was found to partially relieve the inhibitory effects of blood, feces, and meat when Taq DNA polymerase was used in conventional PCR. In a recent study (3), it was found that rTth was more resistant to biological samples than Taq DNA polymerase. In this study, it was shown that the resistance of rTth to biological samples was improved by including BSA in the reaction mixture. The ability of albumin to relieve inhibition may be related, in part, to its ability to bind to inhibitors such as heme (4). In a study by Tsutsui and Mueller (25), it was found that addition of heme-binding protein from rabbit serum completely restored the activity of Rauscher murine leukemia virus reverse transcriptase in the presence of 10−4 M hemin, whereas addition of ovalbumin, a non-heme-binding protein, had no effect on heme inhibition. In a similar study (23), inhibition of PCR detection of Epstein-Barr virus by paraffin-embedded gastric carcinoma tissue was removed successfully by the addition of BSA and other proteins (plasma α2-macroglobulin, rabbit muscle phosphorylase b, rabbit muscle lactate dehydrogenase, and chicken egg white lysozyme). In a previous study (2), it was found that addition of 0.5 μg of BSA per reaction tube removed the PCR-inhibitory span at a dilution of about 1:500 of the blood culture medium containing 26.8% whole blood, while the undiluted blood culture medium remained PCR inhibitory.
The relieving effect of gp32 was noted previously with DNA polymerases and reverse transcriptases (13, 24). The gp32 protein is a single-stranded DNA-binding protein, which is encoded by gene 32 of bacteriophage T4 (6). The first mechanism by which gp32 may relieve PCR inhibition is through protection of single-stranded DNA from nuclease digestion (29). The second possible mechanism may be by binding to inhibitors, which is similar to the ability of BSA and other proteins to bind to inhibitors. Feces are known to contain different proteinases. BSA, gp32, and proteinase inhibitors reduced the level of PCR inhibition by feces (Tables 1 and 2). The mechanism by which BSA and gp32 remove the effects of proteinases may be by being the main targets for these polymerases. Proteinases have been found to be PCR inhibitory in milk (15) and blood (3), and the addition of BSA or proteinase inhibitors (soybean inhibitor or α2-macroglobulin) was found to overcome the inhibition of Taq DNA polymerase by milk, while the addition of lima bean trypsin inhibitor reduced the inhibition of rTth by blood.
Betaine was found to reduce the inhibition of Taq DNA polymerase by blood. Betaine (N,N,N-trimethylglycine) carries both positive and negative charges at pH close to neutrality (17). Betaine has been used to enhance the yields and specificities of PCR amplifications (7), which has been suggested to be due to its ability to destabilize GC-rich DNA sequences, while AT-rich DNA sequences are destabilized much less (17). Betaine has also been found to increase the thermal unfolding transition temperatures of proteins (21).
When the results of the conventional PCR were compared with the results of the real-time DNA synthesis, it was found that feces and meat were less inhibitory than blood. The relieving effects of BSA, gp32, and proteinase inhibitors were not seen. Among the four amplification facilitators tested, only betaine was found to enhance the fluorescence signal in the presence of blood, feces, and meat. These results showed that the effects of amplification inhibitors and facilitators on DNA synthesis by conventional PCR were different from those on real-time DNA synthesis. This could be related to the difference in the principles of the two assays. The real-time DNA synthesis excludes (i) the effects of amplification inhibitors and facilitators on the efficiency of primer annealing and (ii) the effect of high temperature on the different components of the reaction mixtures. In addition, new factors can interfere with DNA synthesis and/or detection of the double-stranded DNA (dsDNA) formed, for example, (i) the ability of glass capillary tubes to bind to different components of the reaction mixture including inhibitors and facilitators, (ii) the interference of inhibitors with SYBR Green I and/or dsDNA can reduce the number of dye molecules that bind to the dsDNA formed, and (iii) the formation of an opaque precipitate can block fluorescence detection. Therefore, further investigations are needed to overcome these limitations and to enhance the reproducibility. This study demonstrates, however, that the PCR-inhibitory effects of biological samples can be reduced or eliminated by the use of an appropriate combination of thermostable DNA polymerase and PCR facilitator. For example, the combination of rTth and BSA eliminated the inhibition of 20% (vol/vol) meat. In addition, we showed that it is possible to study quantitatively the effects of inhibitors and facilitators by using real-time DNA synthesis instead of conventional PCR.
ACKNOWLEDGMENT
This work was supported by the Swedish National Board for Industrial and Technical Development.
REFERENCES
- 1.Abu Al-Soud W, Jönsson L J, Rådström P. Identification and characterization of immunoglobulin G in blood as a major inhibitor of diagnostic PCR. J Clin Microbiol. 2000;38:345–350. doi: 10.1128/jcm.38.1.345-350.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abu Al-Soud W, Lantz P-G, Bäckman A, Olcén P, Rådström P. A sample preparation method which facilitates detection of bacteria in blood cultures by the polymerase chain reaction. J Microbiol Methods. 1998;32:217–224. [Google Scholar]
- 3.Abu Al-Soud W, Rådström P. Capacity of nine thermostable DNA polymerases to mediate DNA amplification in the presence of PCR-inhibiting samples. Appl Environ Microbiol. 1998;64:3748–3753. doi: 10.1128/aem.64.10.3748-3753.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Akane A, Matsubara K, Nakamura H, Takahashi S, Kimura K. Identification of the heme compound copurified with deoxyribonucleic acid (DNA) from bloodstains, a major inhibitor of polymerase chain reaction (PCR) amplification. J Forensic Sci. 1994;39:362–372. [PubMed] [Google Scholar]
- 5.Chandler D P, Wagnon C A, Bolton H., Jr Reverse transcriptase (RT) inhibition of PCR at low concentrations of template and its implications for quantitative RT-PCR. Appl Environ Microbiol. 1998;64:669–677. doi: 10.1128/aem.64.2.669-677.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chase J W, Williams K R. Single-stranded DNA binding proteins required for DNA replication. Annu Rev Biochem. 1986;55:103–136. doi: 10.1146/annurev.bi.55.070186.000535. [DOI] [PubMed] [Google Scholar]
- 7.Frackman S, Kobs G, Simpson D, Storts D. Betaine and DMSO: enhancing agents for PCR. Promega Notes. 1998;65:27. [Google Scholar]
- 8.Innis M A, Myambo K B, Gelfand D H, Brow M A D. DNA sequencing with Thermus aquaticus DNA polymerase and direct sequencing of polymerase chain reaction-amplified DNA. Proc Natl Acad Sci USA. 1988;85:9436–9440. doi: 10.1073/pnas.85.24.9436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Katcher H L, Schwartz I. A distinctive property of Tth DNA polymerase: enzymatic amplification in the presence of phenol. BioTechniques. 1994;16:84–92. [PubMed] [Google Scholar]
- 10.Kreader C A. Relief of amplification inhibition in PCR with bovine serum albumin or T4 gene 32 protein. Appl Environ Microbiol. 1996;62:1102–1106. doi: 10.1128/aem.62.3.1102-1106.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lantz P-G, Abu Al-Soud W, Knutsson R, Hahn-Hägerdal B, Rådström P. Biotechnical use of the polymerase chain reaction for microbial analysis of biological samples. In: El-Gewely M R, editor. Biotechnology annual review. Vol. 5. Amsterdam, The Netherlands: Elsevier Science B. V.; 2000. pp. 87–130. [DOI] [PubMed] [Google Scholar]
- 12.Lantz P-G, Tjerneld F, Borch E, Hahn-Hg̈erdal B, Rådström P. Enhanced sensitivity in PCR detection of Listeria monocytogenes in soft cheese through use of an aqueous two-phase system as a sample preparation method. Appl Environ Microbiol. 1994;60:3416–3418. doi: 10.1128/aem.60.9.3416-3418.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Panaccio M, Lew A. PCR based diagnosis in the presence of 8% (v/v) blood. Nucleic Acids Res. 1991;19:1151. doi: 10.1093/nar/19.5.1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pomp D, Medrano J F. Organic solvents as facilitators of polymerase chain reaction. BioTechniques. 1991;10:58–59. [PubMed] [Google Scholar]
- 15.Powell H A, Gooding C M, Garrett S D, Lund B M, McKee R A. Proteinase inhibition of the detection of Listeria monocytogenes in milk using the polymerase chain reaction. Lett Appl Microbiol. 1994;18:59–61. [Google Scholar]
- 16.Rådström P, Bäckman A, Qian N, Kragsbjerg P, Påhlson C, Olcén P. Detection of bacterial DNA in cerebrospinal fluid by an assay for simultaneous detection of Neisseria meningitidis, Haemophilus influenzae, and streptococci using a seminested PCR strategy. J Clin Microbiol. 1994;32:2738–2744. doi: 10.1128/jcm.32.11.2738-2744.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rees W A, Yager T D, Korte J, von Hippel P H. Betaine can eliminate the base pair composition dependence of DNA melting. Biochemistry. 1993;32:137–144. doi: 10.1021/bi00052a019. [DOI] [PubMed] [Google Scholar]
- 18.Reysenbach A L, Giver L J, Wickham G S, Pace N R. Differential amplification of rRNA genes by polymerase chain reaction. Appl Environ Microbiol. 1992;58:3417–3418. doi: 10.1128/aem.58.10.3417-3418.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rossen L, Nøskov P, Holmstrøm K, Rasmussen O F. Inhibition of PCR by components of food samples, microbial diagnostic assays and DNA-extraction solution. Int J Food Microbiol. 1992;17:37–45. doi: 10.1016/0168-1605(92)90017-w. [DOI] [PubMed] [Google Scholar]
- 20.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Vol. 3. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 21.Santoro M M, Liu Y, Khan S M, Hou L X, Bolen D W. Increased thermal stability of proteins in the presence of naturally occurring osmolytes. Biochemistry. 1992;31:5278–5283. doi: 10.1021/bi00138a006. [DOI] [PubMed] [Google Scholar]
- 22.Sarkar G, Kapelner S, Sommer S S. Formamide can dramatically improve the specificity of PCR. Nucleic Acids Res. 1990;18:7465. doi: 10.1093/nar/18.24.7465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Satoh Y, Takasaka N, Hoshikawa Y, Osaki M, Ohfuji S, Ito H, Kaibara N, Kurata T, Sairenji T. Pretreatment with restriction enzyme or bovine serum albumin for effective PCR amplification of Epstein-Barr virus DNA in DNA extracted from paraffin-embedded gastric carcinoma tissue. J Clin Microbiol. 1998;36:3423–3425. doi: 10.1128/jcm.36.11.3423-3425.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Topal M D, Sinha N K. Products of bacteriophage T4 genes 32 and 45 improve the accuracy of DNA replication in vitro. J Biol Chem. 1983;258:12274–12279. [PubMed] [Google Scholar]
- 25.Tsutsui K, Mueller G C. Hemin inhibits virion-associated reverse transcriptase of murine leukemia virus. Biochem Biophys Res Commun. 1987;149:628–634. doi: 10.1016/0006-291x(87)90414-1. [DOI] [PubMed] [Google Scholar]
- 26.Varadaraj K, Skinner D M. Denaturants or cosolvents improve the specificity of PCR amplification of a G + C-rich DNA using genetically engineered DNA polymerases. Gene. 1994;140:1–5. doi: 10.1016/0378-1119(94)90723-4. [DOI] [PubMed] [Google Scholar]
- 27.Wiedbrauk D L, Werner J C, Drevon A M. Inhibition of PCR by aqueous and vitreous fluids. J Clin Microbiol. 1995;33:2643–2646. doi: 10.1128/jcm.33.10.2643-2646.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wilson I G. Inhibition and facilitation of nucleic acid amplification. Appl Environ Microbiol. 1997;63:3741–3751. doi: 10.1128/aem.63.10.3741-3751.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu J R, Yeh Y C. Requirement of a functional gene 32 product of bacteriophage T4 in UV. J Virol. 1973;12:758–765. doi: 10.1128/jvi.12.4.758-765.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]