Abstract
Mitotic spindle is a self-assembling macromolecular machine responsible for the faithful segregation of chromosomes during cell division. Assembly of the spindle is believed to be governed by the ‘Search & Capture’ (S&C) principle in which dynamic microtubules explore space in search of kinetochores while the latter capture microtubules and thus connect chromosomes to the spindle. Due to the stochastic nature of the encounters between kinetochores and microtubules, the time required for incorporating all chromosomes into the spindle is profoundly affected by geometric constraints, such as the size and shape of kinetochores as well as their distribution in space at the onset of spindle assembly. In recent years, several molecular mechanisms that control these parameters have been discovered. It is now clear that stochastic S&C takes place in structured space, where components are optimally distributed and oriented to minimize steric hindrances. Nucleation of numerous non-centrosomal microtubules near kinetochores accelerates capture, while changes in the kinetochore architecture at various stages of spindle assembly promote proper connection of sister kinetochores to the opposite spindle poles. Here we discuss how the concerted action of multiple facilitating mechanisms ensure that the spindle assembles rapidly yet with a minimal number of errors.
Keywords: Mitosis, Spindle assembly, Microtubules, Chromosome architecture, Chromosome biorientation, Kinetochore
1. Introduction
A healthy cell is ‘euploid’, it contains the entire karyotype of an organism, not a chromosome more, nor a chromosome less. To maintain euploidy in generations, replicated chromosomes must segregate evenly during cell division (mitosis) and consequences of just a single error are dire. Not only the daughter cells are born lacking a chromosome or bearing an extra chromosome (i.e., aneuploid), but their progeny will likely enter a never-ending series of chromosome segregation errors in subsequent divisions [1]. This degradation of control over proper chromosome segregation, known as Chromosomal Instability (CIN), is a hallmark of malignancy [2,3]. Clearly, a robust machinery must be in place to ensure that all chromosomes reach their proper destinations during myriads of mitoses. This machinery, referred to as the ‘mitotic spindle’ due to its characteristically rhomboid shape (Fig. 1A), assembles during each division and the assembly relies on numerous interactions among complexly shaped elements scattered in space. Understanding molecular mechanisms that allow these elements to find one another and establish proper connections while avoiding erroneous ones, is essential to understanding how the high fidelity of chromosome segregation is achieved.
Fig. 1.
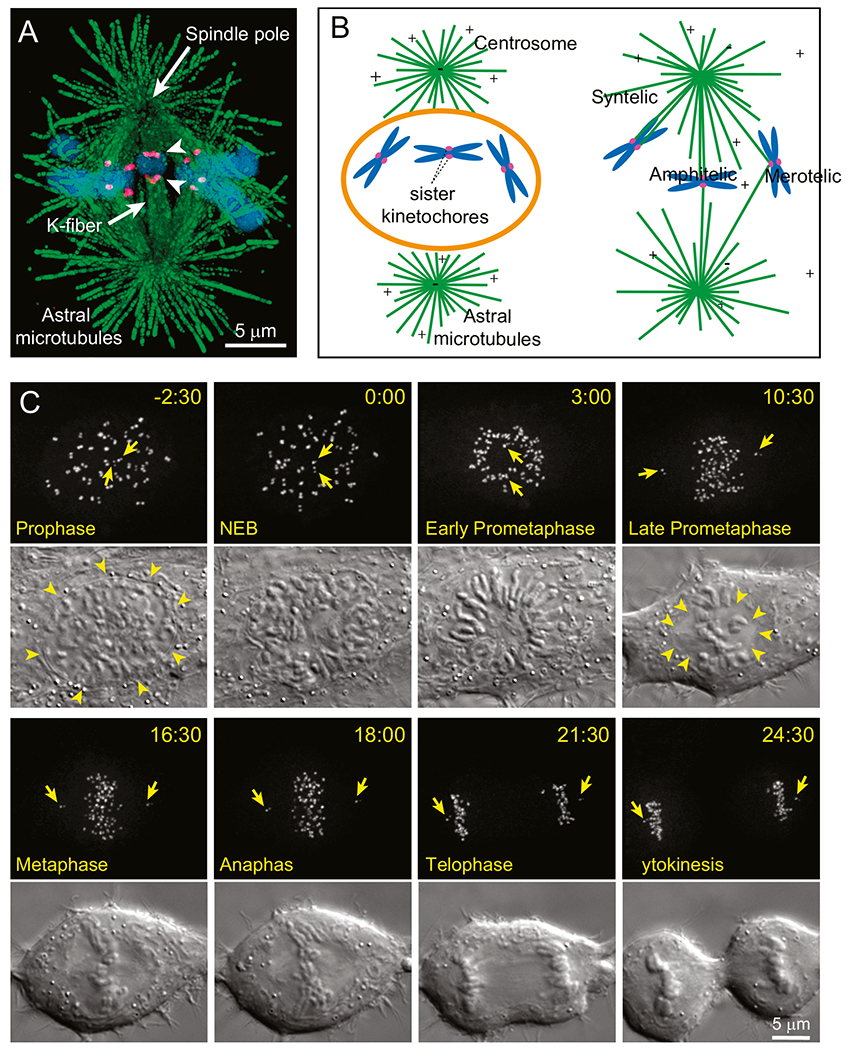
Mitotic machinery and the principle of spindle assembly (A) Architecture of mitotic spindle. Two radial arrays of microtubules (green) emanate from the spindle poles. Bundles of MTs (K-fibers) connect spindle poles and kinetochores (magenta), specialized organelles on chromosomes (blue). Arrowheads denote sister kinetochores on a chromosome that is simultaneously attached to the opposite spindle poles (i.e., ‘amphitelic’). The image depicts a medial slice through the metaphase spindle of Indian muntjac, a species of deer that possesses the largest kinetochores among mammals. (B) Cartoon of the ‘Search and Capture’ model for spindle assembly. Breakdown of the nuclear envelope (orange) at the onset of mitosis allows microtubules (green) that radially emanate from the centrosomes to reach chromosomes (blue). Connection to a spindle pole is achieved via capture of microtubules by the kinetochores (magenta). Due to stochasticity in the distribution of chromosomes and growth of microtubules, capture may lead to a proper amphitelic attachment as well as to erroneous configurations when a single kinetochore simultaneously attaches to both spindle poles (‘merotelic’ attachment) or when sister kinetochores attach to the same spindle pole (‘syntelic’ attachment). For faithful chromosome segregation, amphitelic attachments should be promoted while erroneous attachments – suppressed. (C) Progression through mitosis in a typical human cell. Kinetochores and centrosomes, labelled with CENP-A-GFP and centrin1-GFP respectively, are shown as Maximum Intensity Projections through the entire cell. Arrows denote pairs of centrioles within each centrosome/spindle pole. Changes in cell morphology are shown in Differential Interference Contrast (DIC). Spindle assembly initiates when nuclear envelope breaks down (NEB, arrowheads). Chromosomes arrange in a characteristic ring around the forming spindle during Early Prometaphase and later repopulate the centre of the inner parts of the spindle forming a tight plate at the equator by Metaphase. The spindle assembles within a clear zone devoid of large organelles (Late Prometaphase, arrowheads). Notice rounding of the cell as it progresses through spindle assembly. Chromosome segregation occurs rapidly during Anaphase that initiates less than 20 min after NEB and daughter cells form during Cytokinesis.
In higher eukaryotes, the spindle comprises hundreds of microtubules [4], dynamic filaments composed of αβ-tubulin dimers. The goal of spindle assembly is to connect each chromosome with the opposite poles of the spindle by attaching bundles of microtubules to ‘kinetochores’, a pair of macromolecular complexes that reside at the chromosome’s centromere [5]. Attachment of sister kinetochores to microtubules from the opposite poles, termed ‘amphitelic attachment’, ensures that replicated DNA molecules within the chromosome distribute evenly into the daughter cells (Fig. 1A). A fundamental question that has inspired generations of researchers on mitosis is how amphitelic attachments are formed.
Our current views on the mechanism of spindle assembly are guided by the ‘Search and Capture’ (S&C) hypothesis (Fig. 1B), originally formulated by Kirschner and Mitchison in 1986 [6]. Inspired by their discovery of microtubule ‘dynamic instability’ [7], these researchers proposed that repetitive cycles of growth and shrinkage allow microtubules to search for kinetochores that are scattered in space. In turn, serendipitously discovered kinetochores ‘capture’ (attach to) plus ends of microtubules and thus connect to the spindle poles where the minus ends of microtubules reside [6]. Capture of individual microtubules by kinetochores was subsequently observed in live vertebrate cells [8–10] as well as in yeast [11] and several molecules have been implicated in this process [12–15]. Further, theoretical analyses support the notion that dynamic instability of the microtubule plus end is uniquely advantageous for space exploration over other types of filament behaviour [16]. However, computer simulations based on real-life parameters of microtubule dynamics [17] suggest that unfacilitated search for 46 chromosomes would take hours, which is much longer than duration of mitosis in a typical human cell (Fig. 1C). Moreover, the stochastic nature of S&C is not consistent with the highly reproducible pace of mitotic progression [18]. These inconsistencies indicate that S&C is facilitated by additional mechanisms and several of such mechanisms have been characterized in recent years. Among them are the regulated changes in the shape of the cell [19], pre-positioning of spindle components [20, 21], nucleation of non-centrosomal microtubules near kinetochore as well as guidance of astral microtubule growth towards kinetochores [22–29] and adaptive changes of the kinetochore architecture [30–33]. While none of these mechanisms is essential, their combined contributions ensure that stochastic encounters between microtubules and kinetochores occur with high efficiency (rapidly) and fidelity (low rate of erroneous attachments). In this chapter we review roles of various facilitating mechanisms in S&C-driven mitotic spindle assembly.
1.1. Mechanisms that maintain chromosomes within the reach of microtubules
Rapid oscillation between short periods of growth and shrinkage at the plus end is the foundation of microtubule’s ability to explore space [16]. A limitation of this behaviour is that the short period of growth limits microtubules’ outreach to < 15 μm [34,35], which is not sufficient to cover even a moderately large cell. This hindrance would be particularly pronounced if the complex shapes displayed by interphase cells were maintained during cell division. Not surprisingly, cells round during division (Fig. 1C), which helps to lower the surface to volume ratio. This change is driven by rapid remodelling of actin cytoskeleton and reorganization of the microtubule network initiated during prophase [19,36,37]. Perturbation of the simplistic shape of mitotic cells causes defects in spindle assembly as microtubules struggle to reach all chromosomes [38]. Similarly, spindle assembly tends to take longer in cells that naturally remain flat during cell division. For example, in the extremely flat newt lung cells, incorporation of peripherally located chromosomes may take hours which is several folds longer than the entire duration of mitosis in a rounded cell [8].
Although mitotic rounding limits excessive chromosome scattering, this morphological change is not always sufficient. In larger cells such as oocytes, chromosomes are actively gathered within a limited space by contractility of actin filaments distributed around the nucleus at the onset of cell division [34,39]. Further, actin filaments contribute to the organization of space within the forming spindle which promotes formation of stable microtubule attachments [40,41]. Indeed, cooperative action of actin and microtubules appears to be important for speedy spindle assembly in various cell types and species [42,43].
While the direct involvement of the contractile actin network in gathering chromosomes has been demonstrated only in the extremely large oocytes, similar structural barriers exist in somatic cells. In epithelia, where cells tend to remain flat during mitosis, a rigid perinuclear cage of keratin intermediate filaments helps to avert excessive dispersion of chromosomes [44]. In cells that lack the keratin cage, more ubiquitous albeit less rigid barriers derived from the nuclear envelope have been observed [45,46].
Although the nuclear envelope breaks down at the onset of spindle assembly (nuclear envelope breakdown, NEB), constituents of this structure continue to surround the spindle as a structurally inconspicuous yet functionally important barrier between the ‘clear zone’ where the spindle assembles and the rest of the cytoplasm. Formation of this barrier is regulated by a mechanism that involves the small GTPase Ran and the microtubule motor dynein [45,46]. Depletion of Lamin B, the principal structural component of nuclear envelope, causes defects in mitotic spindle assembly and leads to chromosome mis-segregation [45]. In addition to restricting scattering of chromosomes and preventing invasion of large cytoplasmic organelle into the clear zone, remnants of the nuclear envelope operate as a matrix responsible for localized accumulation of soluble protein complexes required for spindle assembly, for example, αβ-tubulin dimers [47]. Germane here is that cells employ multiple mechanisms to ensure that stochastic interactions between kinetochores and microtubules occur within a relatively small subcellular domain defined by structural and biochemical barriers.
1.2. Mechanisms that present kinetochores to microtubules
The necessity to maintain chromosomes within a small volume searchable by the dynamic microtubules leads to a steric constraint that, left unchecked, may significantly impede S&C-driven spindle assembly. In most animal cells, kinetochores are several folds smaller than the chromosome arms. Therefore, corralling numerous kinetochores within the reach of microtubules comes at the expense of crowding this space with chromatin that is impenetrable to growing microtubules. As a result, kinetochores of chromosomes positioned deeper inside the searchable volume are shielded from microtubules by the arms of chromosomes residing closer to the centrosomes. Computational analysis suggests that only ~3% of kinetochores would have a direct line of sight to the centrosomes when 46 human chromosomes are packed into a spherical volume with the diameter of a nucleus [17]. In reality, the situation is more complex as in cells adhered to rigid substrates, the nucleus is shaped as a disc rather than a sphere (Fig. 2A,B). Due to this asymmetry, the number of kinetochores presented to microtubules at the onset of spindle assembly depends on the position of centrosomes during NEB (Fig. 2A’B’). The highest fraction of kinetochores is exposed when the centrosomes reside on the opposite sides of the larger nuclear surfaces (Fig. 2A,A’). In this configuration, chromosomes arrange in a thin layer orthogonal to the spindle axis which offers kinetic advantages for S&C (Fig. 2C,C’). In contrast, the multilayer distribution of chromosomes when spindle axis sets parallel to the longer axis of the nucleus impedes direct access of microtubules to the kinetochores residing in the middle layers (Fig. 2D,D’) and delays congression of chromosomes onto a metaphase plate (cf., Fig. 2E,F). Not surprisingly, orientation of the spindle axis along the shorter axis of the nucleus at NEB appears to be a common feature in various types of mammalian cells adhered to rigid substrates [21,48]. Preferred spatial pattern is achieved by active movements of the centrosomes along the nuclear envelope as well as by changes in the position of the nucleus during prophase, driven by microtubule motors recruited to the nuclear surface and coordinated with changes in the dynamics of microtubules and actin filaments [48].
Fig. 2.
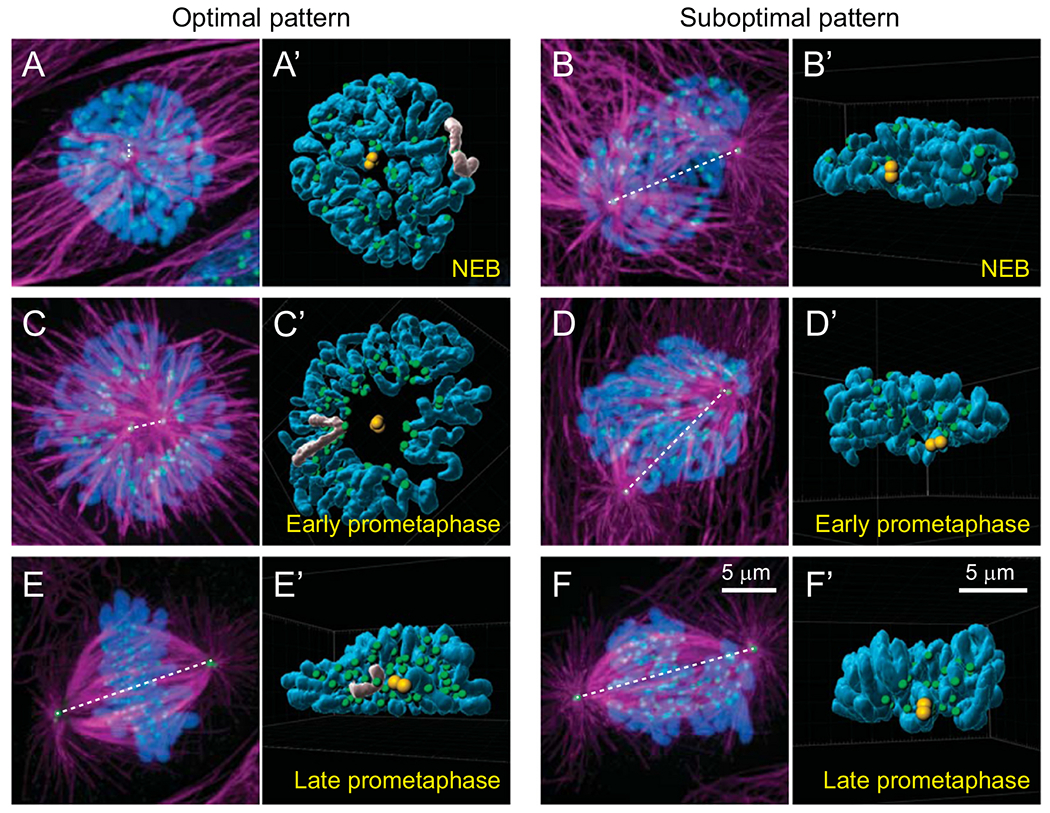
Steric constraints arising from different relative positions of centrosomes and chromosomes at the onset of spindle assembly. (A,B) The number of kinetochores with a direct line of sight to the spindle poles depends on whether the centrosomes separate along the shorter vs. longer axis of the nucleus. In the ‘Optimal pattern’ (A), chromosomes form a thin layer orthogonal to the spindle axis while in the ‘Suboptimal pattern’ (B) many kinetochores are shielded from microtubules by chromosome arms (compare the number of green spots visible in A’ vs. B’). (C,D) Optimal positioning of centrosomes at NEB promotes toroidal distribution of chromosomes during early prometaphase (C). Within the toroid, kinetochores (green) reside near the surface of a microtubule-dense ‘nascent spindle’ (magenta) while chromosome arms are ejected outwards (compare shape of chromosomes greyed in A’ vs. C’). Suboptimal pattern of centrosome orientation leads to multiple layers of chromosomes. (E,F) In the Optimal pattern, chromosomes rapidly repopulate the inner parts of the spindle (E,E’). In contrast, wider distribution of chromosomes in the Suboptimal pattern delays chromosome alignment at the spindle equator (F,F’). (A-F) Maximal intensity projections of the entire cell volumes with microtubules shown in magenta (α-Tubulin), kinetochores and centrosomes in green (CenpA-GFP and Centrin1-GFP), and chromosomes in blue (Hoechst 33342). Orientation of the spindle axis is denoted by a dashed white line. (A’-F’) Surface-rendered models of presenting positions of chromosome arms (blue), kinetochores (green), and centrosomes (yellow) in 3D space. Each model is oriented to present the view from one centrosome towards the other.
While a functional spindle eventually assembles even when centrosomes remain within a single complex until NEB, this configuration leads to a higher frequency of merotelic attachments [49,50]. Similarly, the number of errors is elevated when supernumerary centrosomes assume positions on several sides of the nucleus. Although most cells ultimately coalesce supernumerary centrosomes into precisely two clusters and restore the bipolar architecture of the spindle, chromosome mis-segregation is rampant under these conditions [50–53]. Thus, even transient deviations from the proper bipolar geometry during early prometaphase are detrimental for the fidelity of spindle assembly.
A second set of mechanisms that facilitates exposure of kinetochores to the searching microtubules arises from the interplay of microtubule-mediated forces acting along the chromosome arms vs. the kinetochores. Because of their large size, chromosome arms are likely to encounter microtubules earlier than the kinetochores and these early interactions play an important role in defining position and orientation of the chromosome within the forming spindle. Due to ubiquitous presence of plus-end directed kinesins on chromatin (‘chromokinesins’, kinesins 4 and 10) [54], direct contacts with microtubules produce a force that pushes the chromatin towards the plus ends, i.e., away from the spindle poles [55–59]. The large number of momentous encounters with microtubules on chromosome arms rapidly clear the central part of the forming spindle from chromatin making kinetochores accessible to the microtubules (Fig. 2C,C’). As the ejection force along the arms is counterbalanced by the inward-directed forces produced by the molecular motors at the kinetochores [30,60], chromosomes arrange in a toroid around the nascent spindle packed with microtubules during early prometaphase (Fig 2C,C’). Within the toroid, kinetochores are adjacent to the spindle surface while the arms, straightened by the ejection force, point away towards the cell periphery [21,61] (Fig. 2C’). Transient arrangement of chromosomes into a ring around the spindle during early prometaphase occurs consistently but this pattern is quite transient as the chromosomes rapidly repopulate the hollow centre of the spindle by moving inward towards the spindle axis (Fig. 2E,E’). Various molecular pathways, such as inactivation of chromokinesins [21] or deviation from the optimal centrosome separation pattern prior to NEB (Fig. 2F,F’), prevent formation of the toroid and lead to prolonged spindle assembly and increased frequency of chromosome mis-segregation [21,30,59,60]. This common outcome of perturbations in biochemically unrelated molecular cascades, supports the notion that the efficiency and fidelity of S&C depend on achieving an optimal pattern in spatial distribution of centrosomes and kinetochores during the critical period of spindle assembly when amphitelic attachments are formed.
Electron microscopy (EM) analyses of early prometaphase reveal that kinetochores residing on the surface of the nascent spindle are in direct contact with walls of numerous microtubules; however, end-on attachments to microtubule bundles extending towards the spindle poles (K-fibers) are rarely observed at this stage [25,30]. Further, the toroidal pattern forms and persists for a much longer time in cells with kinetochores incapable of attaching to microtubules in the end-on fashion [21,61,62]. These observations suggest that prior to the capture of the plus ends, kinetochores interact with microtubules ‘laterally’ at various points along the entire length of a microtubule [11,21,63–65]. Whether these lateral interactions are subsequently converted into end-on attachments or whether they indirectly facilitate direct capture of microtubule plus ends remains unknown. However, transition from lateral interactions to end-on attachments appear to involve coordinated activities of various molecular motors, microtubule depolymerases residing at the kinetochore as well as chromokinesins acting upon chromosome arms [66–70].
An important role of lateral interactions in the context of S&C is that kinetochore gliding alongside of microtubules may deliver centromeres to the areas of the spindle favourable for formation of proper end-on attachments. For example, gliding alongside of astral microtubules, towards their minus ends facilitates incorporation of peripheral chromosomes into the microtubule-dense spindle [63]. Gliding towards plus end of bundled microtubules brings monotelic chromosomes to the spindle equator, where attachments to microtubules produced by the distal pole can rapidly form [63,71] These poleward and antipoleward movements driven by lateral interactions are mediated by the two molecular motors residing at the kinetochores, namely the plus-end-directed kinesin CenpE (kinesin 7) [71,72] and the minus-end-directed dynein [12,66,73]. Both motors are present in high concentrations at unattached kinetochores, which could lead to tag-of-war situation where poleward and antipoleward forces would be simultaneously exerted. This potentially dangerous situation is averted by the different preferences of CenpE vs. dynein to various post-translational modifications (PTMs) that exist on different classes of spindle microtubules [74]. This mechanism ensures that lateral interactions with detyrosinated α-tubulin within relatively stable microtubule bundles, such as K-fibers, engages CenpE activity that transports kinetochores toward the plus ends, i.e., to the spindle equator [75]. In contrast, dynein/dynactin’s preference for tyrosinated microtubules [76,77] seems to be important for the initiation of minus-end directed gliding towards the end of astral microtubules [77].
However, large displacements of kinetochores are rarely observed during early prometaphase in human cells [21]. Instead, kinetochores tend to exhibit directionally unstable brief movements that do not significantly change position of the chromosome [25] yet rotate the centromere which orients sister kinetochores roughly parallel to the spindle axis [30]. These changes in the centromere orientation appear to accelerate formation of amphitelic attachments and suppress the number of errors during early stages of spindle assembly [20,30].
1.3. Regulation of kinetochore architecture at various stages of spindle assembly
It is self-evident that larger objects have greater chances of being discovered in a random exploration of space than smaller ones. In the context of S&C, this implies that larger kinetochores would encounter microtubules with higher frequency and thus accelerate spindle assembly. However, this acceleration comes at a cost as a large kinetochore may simultaneously capture microtubules from both spindle poles, forming a merotelic attachment (Fig. 3A). Further, large sister kinetochores would encircle the centromere and be prone to capturing microtubules produced by the same spindle pole, which leads to syntelic attachments (Fig. 3A). Intuitively, efficiency and fidelity of S&C-driven spindle assembly appear to be in a reciprocal relation: a higher rate of microtubule capture inevitably increases frequency of errors. A ramification of this relationship is that sufficiently fast and error-free S&C is possible only when all kinetochores adhere to an ‘optimal’ architecture with minimal variability in their size and shape.
Fig. 3.
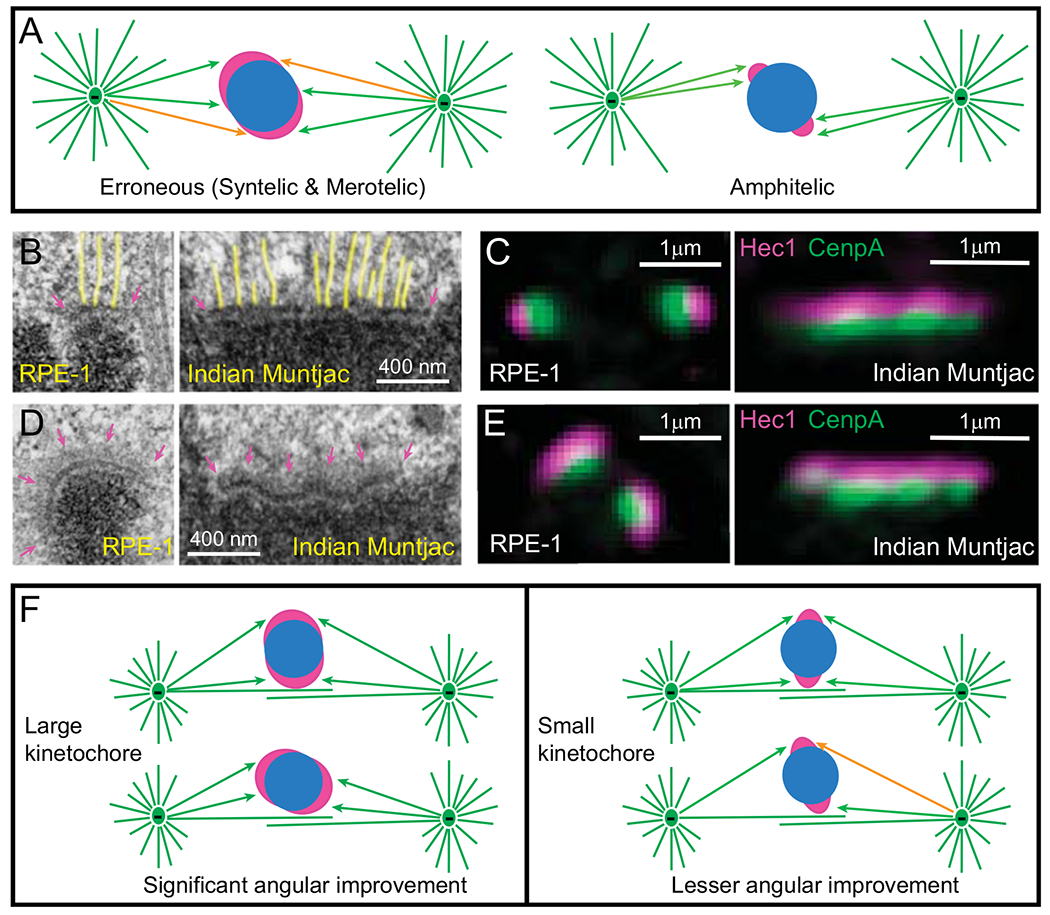
Effects of kinetochore architecture on the efficiency and fidelity of Search and Capture. (A) Reciprocity in the efficiency vs. fidelity of microtubule capture in the original S&C hypothesis. (Left cartoon) Larger kinetochores are likely to encounter microtubules faster; however, they are likely to be exposed to microtubules emanating from both spindle poles. This results in a mixture of proper (green arrows) and erroneous (orange arrows) interactions (syntelic and merotelic). (Right cartoon) Smaller kinetochores are protected from the exposure to ‘wrong’ spindle poles; however, microtubule search for small kinetochores is longer. (B–C) Architecture of attached kinetochores. (B) EM of metaphase kinetochores in human (RPE-1) and Indian muntjac cells. Arrows denote the trilaminar morphology of the plate comprising two electron-dense layers separated by a translucent middle layer. The length of the plate varies greatly. Notice end-on attached microtubules (highlighted yellow). (C) In fluorescence LM, mammalian kinetochores appear near diffraction-limited spots, while IM kinetochores form thin lines. Layered distribution of proteins within the kinetochore is apparent with DNA-binding proteins (CenpA-GFP, green) residing in the inner layer and microtubule-binding proteins (Hec1, magenta) in the outer layers. (D-E) Architecture of unattached kinetochores. (D) EM reveals fibrous corona (arrows in magenta) adjacent to the outer layer in the absence of microtubule attachments. In human cells (RPE-1), the plate elongates and encircles a large part of the centromere. In contrast, the length and shape of the plate does not change significantly in Indian muntjac. (E) Fluorescence LM is consistent with the elongation and shape changes in human (RPE-1) but not in Indian muntjac kinetochores. (F) Effects of kinetochore architecture on rotational alignment of the centromere in early prometaphase. Larger kinetochores support a higher degree of rotation on the surface of nascent spindle as they maintain constant contact with microtubules (left). In contrast, rotation angles of smaller kinetochores are sterically limited (right). As a result of extensive rotation, larger sister kinetochores have higher probability of forming proper attachments (green arrows) while less-aligned smaller kinetochores remain exposed to microtubules from ‘wrong’ spindle poles (orange arrow).
In conventional EM, kinetochores appear as ~75 nm thin discoid plates situated on the surface of the centromere [65,78,79]. The plate comprises electron-opaque inner and outer layers separated by a translucent middle layer. Each layer is ~25-nm thin (Fig. 3B). The ‘trilaminar’ morphology is well conserved; however, the length of the plate varies significantly among the chromosomes within a cell as well as among different organisms. EM reconstructions of human cells suggest ~4-fold variation in the plate length during metaphase (~150–600 nm) with the average value of ~300 nm [80]. This range is consistent with ~8 fold variation observed in volumetric analysis [81]. Similar ~3-fold variability in linear dimension of the plate exists in the cell of Indian muntjac [82], although kinetochores in this species are much larger with the longest plate exceeding 1.5 μm (Fig. 3B). The budding yeast S. cerevisiae presents the opposite extreme as its kinetochores are not much larger than a single microtubule [83,84]. Such a wide range of sizes implies that mechanisms responsible for the formation of proper microtubule attachments are sufficiently robust to cope with morphological irregularity of the kinetochore.
The inconspicuous morphology of the plate in EM does not do justice to the staggering biochemical complexity of this organelle that incorporates numerous proteins organized in an intricate network [5,85]. Attachment to microtubules is enacted by a multimeric assembly containing dozens of proteins [86]. The load-bearing connection at the microtubule tip is maintained primarily by Hec1, a member of the Ndc80 complex [87,88]; however, several additional protein complexes contribute as well [89–91]. Significant progress has been achieved towards solving the molecular structure of the microtubule attachment site in vitro [86,92].
In the context of spindle assembly, the ability to capture microtubules that randomly approach the kinetochore from various directions depends on the 3-D architecture of the kinetochore plate as well on the distribution and orientation of molecular ensembles within the plate. Consistent with the trilaminar morphology observed in EM, light microscopy (LM) suggests a layered distribution of proteins within the kinetochore with microtubule-binding components concentrating near the outer surface (Fig. 3C). Two major models of the outer layer organization have been proposed. In one view, the outer layer comprises tandem repeats, each containing the entire molecular machinery required for a single microtubule attachment. The number of these repeats ranges from one in the budding yeast whose kinetochores capture a single microtubule [83,84] to dozens required for the formation of K-fibers in higher eukaryotes [93,94]. A corollary of this model is that the outer layer is a highly ordered structure where molecular interactions with each attached microtubule are constrained within a binding site [95,96]. In an alternative view, no predefined microtubule-binding site exists prior to microtubule attachment. Instead, the outer layer comprises an unstructured assortment of complexes that act as a “molecular lawn” with random sets of molecules attaching to microtubules that accidentally come within their reach [97–99]. The latter model appears to gain popularity. Most recently, FRET analyses of protein distribution within the outer layer demonstrate that Ndc80 molecules are uniformly distributed and variably oriented in the absence of microtubules but they cluster after formation of end-on attachments [100]. This architecture is advantageous for S&C as it permits capture of microtubules approaching the plate from various directions.
The classic early EM images obtained primarily from metaphase cells [78,79] have formed a perception of the kinetochore plate as a rigid flat disk; however, more recent whole-cell reconstructions suggest that the plate is pliable. On average, length and curvature of the plate increase when microtubule dynamics are suppressed by Taxol in human cells [80]. Similar yet larger scale changes occur when metaphase kinetochores lose their attachment to microtubules [30,101–104].
In the absence of end-on microtubule attachment, kinetochores gradually recruit additional proteins, primarily to its outer layers [31–33]. Morphologically, this process is manifested as progressive elongation of the plate and formation of ‘fibrous corona’ [65,78], a mat of fibrous material adjacent to the electron dense outer layer of the plate (Fig. 3D,E). Interestingly, outer kinetochore enlargement appears to be more pronounced on smaller kinetochores. After full enlargement induced by the microtubule-depolymerizing drug nocodazole, kinetochores of the largest human chromosome do not differ in size or the amount of outer kinetochore components from kinetochores of other chromosomes [105]. In contrast, end-on attached kinetochores tend to be larger on chromosome 1 than on other chromosomes in human cells [106]. 3-D EM reconstructions demonstrate that the plate of unattached human kinetochores form crescents (Fig. 3D) with sister kinetochores encircle the centromere, which creates a large surface capable of capturing microtubule on all chromosomes [30].
Molecular mechanism behind the elongation of plate involves the outer-kinetochore complex comprising the trio of ROD, Zwilch, and Zw10 proteins, acting as unified ‘RZZ’ complex [107] as well as the RZZ-binding protein Spindly [108]. Oligomerization of the RZZ and Spindly appears to trigger formation of a meshwork that gradually extends from the core of the kinetochore outwards, increasing the length of the plate [31–33]. The process is controlled by various post-translational modifications, such as farnesylation of Spindly and phosphorylation of RZZ components that are in turn governed by the mitosis-specific kinases [109]. Importantly, growth of the plate is reversed upon formation of end-on attachments likely by the removal of the RZZ complex as well as other outer layer components via dynein mediated transport along the attached microtubules [110,111]. As a result, end-on attached kinetochores rapidly compact down to the size and morphology typical for metaphase cells [30,104]. The cycle ofkinetochore enlargement at the onset of spindle assembly followed by rapid compaction upon formation of end-on microtubule attachment bears pronounced functional significance in the context of S&C.
As discussed earlier in this chapter, capture of microtubules by large kinetochores is expected to be efficient yet error prone (Fig. 3A). Consistent with this expectation, perpetual enlargement of kinetochores in human cells with the mutant SpindlyΔN protein increases the frequency of merotelic attachments and chromosome mis-segregation [31]. Similarly, in Indian muntjac cells, where, in sharp contrast to human cells, the length of kinetochore plate does not change in response to microtubule attachment (compare Fig. 3B with 3D and 3C with 3E) larger kinetochores have higher propensity of forming merotelic connections [82]. However, no linear relationship between the size of the centromere and the probability of attachment errors has been found in cells where transient enlargement of the plate at the onset of mitosis is followed by a rapid compaction upon formation of end-on microtubule attachments. Indeed, while mis-segregation rates are not equal among human chromosomes, similar frequencies have been reported for the largest and smallest chromosomes [105,112–115]. This suggests that regulated architecture of the kinetochore somehow facilitates formation of proper attachments.
Computational modelling predicts that forces acting upon the enlarged kinetochores during early stages of spindle assembly that are dominated by lateral interactions, rotationally align centromeres roughly parallel to the spindle axis. During this process, larger crescent-shaped kinetochores support a greater angular improvement as they remain in contact with microtubules at a wider range of angles (Fig. 3F). As lateral interactions are established rapidly, by the time a kinetochore has a reasonable chance of capturing a microtubule plus end, the centromere is already ‘pre-aligned’ on the surface of the nascent spindle. Rapid compaction of the kinetochore, triggered by the end-on attachment, further decreases probability of errors in the formation of amphitelic attachment [30]. Rotation of centromeres leading to roughly parallel orientation of the centromere prior to the formation of end-on attachments has been observed in live cells, lending experimental support to the computational predictions [30,31].
It is important to emphasize that transient enlargement of the kinetochore is advantageous only when S&C occurs in the context of spatial cues and constraints provided by the facilitating mechanisms described in Sections 1.1–1.2. As the rotation of enlarged kinetochores is driven by lateral interactions with microtubules on the surface of the nascent spindle, conditions that perturb spindle architecture or impede lateral interactions, inevitably increase the number of erroneous attachments [21,31]. Conversely, conditions that affect kinetochores’ ability to enlarge or compact at the appropriate stages of spindle assembly become disadvantageous when other facilitating mechanism are functional [31].
1.4. Role of non-centrosomal microtubules
In the classic formulation of S&C hypothesis, every microtubule captured by a kinetochore was expected to form a direct connection with a spindle pole, as the duplicated centrosomes were assumed to be the only source of microtubules [6]. Although well justified at the time, this assumption proved to be incorrect. We now know that a significant fraction of microtubules within the spindle are nucleated at locales other than the centrosomes. Of particular importance in the context of S&C are non-centrosomal microtubules that assemble near unattached kinetochores. Due to their spatial proximity kinetochores are more likely to capture these microtubules rather than astral microtubules radially spreading from a distantly located centrosome. However, this capture does not connect the kinetochores with the poles. Instead, it leads to the formation of a ‘nascent K-fiber’, a set of microtubules with minus ends protruding from the kinetochore. Due to continuous incorporation of tubulin subunits into the plus ends of these microtubules at the kinetochore, nascent K-fibers gradually elongate [23,116].
Nascent K-fibers allow kinetochores to establish indirect connections with the spindle poles via poleward ‘microtubule on microtubule’ gliding that is a major transport property of the spindle [24]. This transport is driven by the ubiquitous flow of cytoplasmic dynein, a large minus-end directed molecular motor [23,117]. As dynein uses microtubules as both the track and the cargo, it efficiently pulls microtubules with free minus ends towards the spindle poles. Nascent K-fibers experimentally created by severing a full-size K-fiber in mammalian cells connect to the neighbouring microtubules in less than a minute [118–120]. Several minutes later, continuous elongation of microtubules within a nascent K-fiber delivers its minus ends to the pole. Such a rapid incorporation of chromosomes into the spindle is clearly advantageous over direct capture of astral microtubules by ‘naked’ kinetochores. However, the advantage exists only when short microtubules attached to the kinetochore are organized in a single bundle oriented roughly parallel to the spindle axis. A bundle orthogonal to the spindle axis or a set of individual microtubules emanating from the kinetochore in various directions would be prone to forming simultaneous connections with both spindle poles (i.e., merotelic attachment). Indeed, erroneous reattachment of nascent K-fibers has been observed [118]. Thus, mechanisms must be in place to ensure that capture of non-centrosomal microtubules does not lead to intolerable levels of erroneous attachments.
Assembly and organization of non-centrosomal microtubules near chromosomes are regulated by two major molecular pathways. One is governed by a small GTPase Ran that acts upon a range of ‘Spindle Assembly Factors’ (SAF), molecular complexes responsible for localized microtubule assembly. Due to the high affinity of RCC1, Ran’s guanine exchange factor, to the DNA, concentration of GTP-bound Ran (RanGTP) is maximal in the proximity of chromatin. As a result, SAFs, sequestered in a complex with importin-β throughout the cytoplasm, are released near chromosomes and their activity triggers microtubule growth [121–125]. In the context of S&C, microtubule arrays produced by the RanGTP gradient are spatially organized rather than random. This organization arises from ‘branching microtubule nucleation’ promoted by the interplay between the downstream target of RanGTP, TPX2 [126, 127], microtubule branching complex Augmin [128–131], and the γ-Tubulin Ring Complex (γ-TuRC) [132,133] that nucleates microtubules. TPX2-mediated recruitment of Augmin and γ-TuRCs to the walls of existing microtubules leads to the formation of branched microtubule networks in which new microtubules grow in the same general direction as the older ‘mother’ microtubules [129,134–136]. ‘Branching microtubule nucleation’ has a potential to rapidly convert a single microtubule end-on attached to a kinetochore into a K-fiber, accelerating spindle assembly [137]. There is direct evidence that TPX2 accumulates within older microtubule lattices near chromosomes, such as K-fiber, which is consistent with the potential role of this process in facilitation of spindle assembly [136].
A second chromatin-mediated pathway that appears to function independently of RanGTP is driven by the chromosome passenger complex (CPC) [138] which resides within the centromere and acts via local regulation of microtubule dynamics [139–142]. CPC complex consists of inner centrosome protein (INCENP), borealin, survivin and Aurora B kinase and is recruited directly to the centromeres. The kinase subunit of CPC has been proposed to phosphorylate and inactivates locally microtubule-depolymerizing proteins, such as MCAK [139–141] and Op18/Stathmin [142], thereby facilitating the stabilization of microtubules and initiating spindle formation from chromosomes [141]. Although the CPC concentrates at each centromere, an Aurora B diffusion gradient extends over the length of the spindle, which prevents microtubule depolymerization within the spindle [143,144].
Importantly, in sharp contrast to in vitro systems such as Xenopus egg extracts, in mammalian cells, non-centrosomal microtubules appear to form exclusively near the centromeres and not along the chromosome arms [22,145,146] although the mechanism(s) that spatially restrict microtubule nucleation are not fully understood. Rapid appearance of short randomly oriented microtubules in the immediate proximity of kinetochores was originally observed in EM reconstructions of cells recovering from microtubule-depolymerizing drugs [145,146]. More recent serial-section EM analyses demonstrate that ~75% of kinetochores are in contact with non-centrosomal microtubules during early prometaphase in human cells. Interestingly, these microtubules tend to orient either parallel or orthogonal to the kinetochore plate, the angles typical for lateral interactions and end-on attachments [25]. However, orthogonal orientation is suppressed upon inactivation of the kinetochore-associated kinesin CenpE (kinesin-7). These observations suggest that CenpE transports the plus ends of laterally attached microtubules towards the kinetochore promoting formation of end-on attachments [25]. Consistent with this proposed role, CenpE has been shown to attach firmly to microtubule plus ends for an extended time in vitro [147]. Importantly, EM analyses suggest that sorting of non-centrosomal microtubules into nascent K-fibers occurs after centromeres rotationally align on the spindle surface [25] so that the K-fibers growing from the sister kinetochores extend roughly towards the opposite spindle poles.
2. Conclusions
For over 30 years, S&C hypothesis has stood the test of time as discoveries of numerous facilitating mechanisms provided a clue on how cells manage to overcome the impediments arising from the stochastic nature of interactions between kinetochores and microtubules. These mechanisms ensure that spindle assembly takes place in well-defined and structured space, where components are optimally distributed, and steric hindrances are minimized. Proper patterns of centrosome separation during prophase and ejection of chromosome arms during the earliest stages of spindle assembly enable rapid rotational alignment of centromeres to spindle axis via lateral interactions between the enlarged kinetochores and microtubules on the surface of nascent spindle (Fig. 4). A large number of microtubules nucleated in the immediate proximity of the centromere promote end-on attachments. Together, concerted contributions from these mechanisms allow the spindle to assemble rapidly yet with the minimal number of errors. Importantly, while molecular machinery employed by various facilitating mechanisms may appear dissimilar, contributions of these mechanisms into spindle assembly are interrelated. Indeed, deficiency in one of the facilitators may render other mechanisms counterproductive. For example, while kinetochore enlargement increases fidelity of spindle assembly when capture is preceded by rotational alignment on the surface of nascent spindle; this adaptive change in kinetochore architecture would increase the number of errors if the architecture of nascent spindle is perturbed. Indeed, erroneous attachments become numerous when chromosomes and centrosomes are intermixed at the onset of spindle assembly in cells recovering from microtubule-depolymerizing drugs [30,148]. This interdependency highlights wholistic nature of spindle assembly and the necessity of strict coordination among the contributing mechanisms.
Fig. 4.
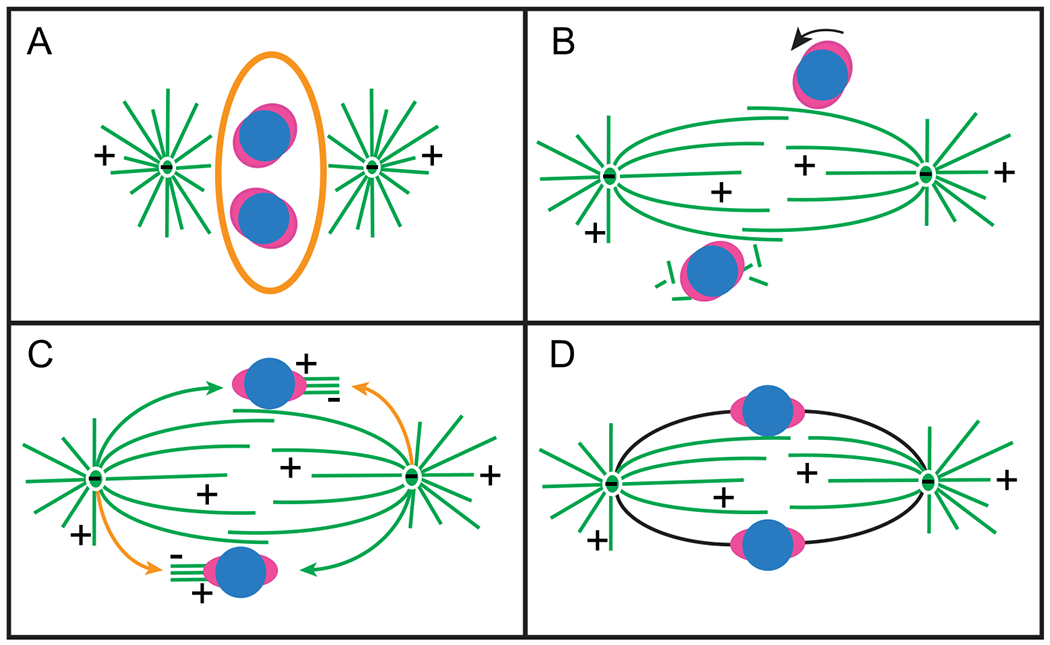
Spindle assembly via facilitated Search and Capture. (A) Positioning of the duplicated centrosomes on the shorter axis prior to NEB facilitates rapid formation of nascent spindle and maximal exposure of kinetochores to microtubules at the onset of spindle assembly. (B) Lateral interactions between spindle microtubules and enlarged kinetochores orient centromeres roughly parallel to the spindle axis while nucleation of non-centrosomal microtubules near centromeres promotes capture of microtubule plus ends. (C) Capture of astral microtubules by enlarged kinetochores aligned on the spindle surface ensures rapid formation of direct connections between kinetochores and spindle poles (green arrows). Nascent K-fibers produced by the kinetochores are transported poleward by dynein-mediated gliding astral microtubules (orange arrows). (D) Concerted action of multiple mechanisms results in rapid formation of amphitelic connections (black lines).
Acknowledgements
We thank Ms. Irina Tikhonenko and Rebecca Fisher for their expert help with images used in the illustrations Electron microscopy was performed at the Wadsworth Center’s 3D Electron Microscopy Core Facility. Our work is supported by the National Institutes of Health, US, National Institute of General Medical Sciences, R35 grant GM130298 to Alexey Khodjakov.
Footnotes
Conflict of interest
The authors declare no conflict of interest.
References
- [1].Potapova T, Gorbsky GJ, The consequences of chromosome segregation errors in mitosis and meiosis, Biology (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Bakhoum SF, Compton DA, Kinetochores and disease: keeping microtubule dynamics in check!, Curr. Opin. Cell Biol 24 (1) (2012) 64–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Gordon DJ, Resio B, Pellman D, Causes and consequences of aneuploidy in cancer, Nat. Rev. Genet 13 (3) (2012) 189–203. [DOI] [PubMed] [Google Scholar]
- [4].O’Toole E, Morphew M, McIntosh JR, Electron tomography reveals aspects of spindle structure important for mechanical stability at metaphase, Mol. Biol. Cell 31 (3) (2020) 184–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Musacchio A, Desai A, A Molecular View of Kinetochore Assembly and Function, Biology (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Kirschner M, Mitchison T, Beyond self-assembly: from microtubules to morphogenesis, Cell 45 (3) (1986) 329–342. [DOI] [PubMed] [Google Scholar]
- [7].Mitchison T, Kirschner M, Dynamic instability of microtubule growth, Nature 312 (5991) (1984) 237–242. [DOI] [PubMed] [Google Scholar]
- [8].Hayden JH, Bowser SS, Rieder CL, Kinetochores capture astral microtubules during chromosome attachment to the mitotic spindle: direct visualization in live newt lung cells, J. Cell Biol 111 (3) (1990) 1039–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Alexander SP, Rieder CL, Chromosome motion during attachment to the vertebrate spindle: initial saltatory-like behavior of chromosomes and quantitative analysis of force production by nascent kinetochore fibers, J. Cell Biol 113 (4) (1991) 805–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Merdes A, De May J, The mechanism of kinetochore-spindle attachment and polewards movement analyzed in PtK2 cells at the prophase-prometaphase transition, Eur. J. Cell Biol 53 (2) (1990) 313–325. [PubMed] [Google Scholar]
- [11].Tanaka K, Mukae N, Dewar H, van Breugel M, James EK, Prescott AR, Antony C, Tanaka TU, Molecular mechanisms of kinetochore capture by spindle microtubules, Nature 434 (2005) 987–994. [DOI] [PubMed] [Google Scholar]
- [12].Yang Z, Tulu US, Wadsworth P, Rieder CL, Kinetochore dynein is required for chromosome motion and congression independent of the spindle checkpoint, Curr. Biol 17 (11) (2007) 973–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Hendricks AG, Lazarus JE, Perlson E, Gardner MK, Odde DJ, Goldman YE, Holzbaur EL, Dynein tethers and stabilizes dynamic microtubule plus ends, Curr. Biol 22 (7) (2012) 632–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Bomont P, Maddox P, Shah JV, Desai AB, Cleveland DW, Unstable microtubule capture at kinetochores depleted of the centromere-associated protein CENP-F, EMBO J. 24 (22) (2005) 3927–3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Feng J, Huang H, Yen TJ, CENP-F is a novel microtubule-binding protein that is essential for kinetochore attachments and affects the duration of the mitotic checkpoint delay, Chromosoma 115 (4) (2006) 320–329. [DOI] [PubMed] [Google Scholar]
- [16].Holy TE, Leibler S, Dynamic instability of microtubules as an efficient way to search in space, Proc. Natl. Acad. Sci. U.S.A 91 (12) (1994) 5682–5685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Wollman R, Cytrynbaum EN, Jones JT, Meyer T, Scholey JM, Mogilner A, Efficient chromosome capture requires a bias in the ‘search-and-capture’ process during mitotic-spindle assembly, Curr. Biol 15 (9) (2005) 828–832. [DOI] [PubMed] [Google Scholar]
- [18].Jones JT, Myers JW, Ferrell JE, Meyer T, Probing the precision of the mitotic clock with a live-cell fluorescent biosensor, Nat. Biotechnol 22 (3) (2004) 306–312. [DOI] [PubMed] [Google Scholar]
- [19].Cadart C, Zlotek-Zlotkiewicz E, Le Berre M, Piel M, Matthews HK, Exploring the function of cell shape and size during mitosis, Dev. Cell 29 (2) (2014) 159–169. [DOI] [PubMed] [Google Scholar]
- [20].Paul R, Wollman R, Silkworth WT, Nardi IK, Cimini D, Mogilner D, Computer simulations predict that chromosome movements and rotations accelerate mitotic spindle assembly without compromising accuracy, Proc. Natl. Acad. Sci. U.S.A 106 (37) (2009) 15708–15713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Magidson V, O’Connell CB, Loncarek J, Paul R, Mogilner A, Khodjakov A, The spatial arrangement of chromosomes during prometaphase facilitates spindle assembly, Cell 146 (4) (2011) 555–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].O’Connell CB, Loncarek J, Kalab P, Khodjakov A, Relative contributions of chromatin and kinetochores to mitotic spindle assembly, J. Cell Biol 187 (1) (2009) 43–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Maiato H, Rieder CL, Khodjakov A, Kinetochore-driven formation of kinetochore fibers contributes to spindle assembly during animal mitosis, J. Cell Biol 167 (5) (2004) 831–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Tulu US, Fagerstrom C, Ferenz NP, Wadsworth P, Molecular requirements for kinetochore-associated microtubule formation in mammalian cells, Curr. Biol 16 (5) (2006) 536–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Sikirzhytski V, Renda F, Tikhonenko I, Magidson V, McEwen BF, Khodjakov A, Microtubules assemble near most kinetochores during early prometaphase in human cells, J. Cell Biol 217 (8) (2018) 2647–2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Petry S, Vale RD, Microtubule nucleation at the centrosome and beyond, Nat. Cell Biol 17 (9) (2015) 1089–1093. [DOI] [PubMed] [Google Scholar]
- [27].Scrofani J, Sardon T, Meunier S, Vernos I, Microtubule nucleation in mitosis by a RanGTP-dependent protein complex, Curr. Biol 25 (2) (2015) 131–140. [DOI] [PubMed] [Google Scholar]
- [28].Meunier S, Vernos I, Acentrosomal microtubule assembly in mitosis: the where, when, and how, Trends Cell Biol. 26 (2) (2016) 80–87. [DOI] [PubMed] [Google Scholar]
- [29].Khodjakov A, Copenagle L, Gordon MB, Compton DA, Kapoor TM, Minus-end capture of preformed kinetochore fibers contributes to spindle morphogenesis, J. Cell Biol 160 (5) (2003) 671–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Magidson V, Paul R, Yang N, Ault JG, O’Connell CB, Tikhonenko I, McEwen BE, Mogilner A, Khodjakov A, Adaptive changes in the kinetochore architecture facilitate proper spindle assembly, Nat. Cell Biol 17 (9) (2015) 1134–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Sacristan C, Ahmad MUD, Keller J, Fermie J, Groenewold V, Tromer E, Fish A, Melero R, Carazo JM, Klumperman J, Musacchio A, Perrakis A, Kops GJ, Dynamic kinetochore size regulation promotes microtubule capture and chromosome biorientation in mitosis, Nat. Cell Biol 20 (2018) 800–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Rodriguez-Rodriguez JA, Lewis C, McKinley KL, Sikirzhytski V, Corona J, Maciejowski J, Khodjakov A, Cheeseman IM, Jallepalli PV, Distinct roles of RZZ and Bub1-KNL1 in mitotic checkpoint signaling and kinetochore expansion, Curr. Biol 28 (21) (2018) 3422–3429, 3422,–3429 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Pereira C, Reis RM, Gama JB, Celestino R, Cheerambathur DK, Carvalho AX, Gassmann R, Self-assembly of the RZZ complex into filaments drives kinetochore expansion in the absence of microtubule attachment, Curr. Biol 28 (21) (2018) 3408–3421, 3408,–3421 e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Lenart P, Daigle N, Hand AR, Eils R, Terasaki M, Ellenberg J, A contractile nuclear actin network drives chromosome congression in oocytes, Nature 436 (7052) (2005) 812–818. [DOI] [PubMed] [Google Scholar]
- [35].Piehl M, Cassimeris L, Organization and dynamics of growing microtubule plus ends during early mitosis, Mol. Biol. Cell 14 (3) (2003) 916–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Bakhshandeh S, Taieb HM, Schlussler R, Kim K, Beck T, Taubenberger A, Guck J, Cipitria A, Optical quantification of intracellular mass density and cell mechanics in 3D mechanical confinement, Soft Matter (2020). [DOI] [PubMed] [Google Scholar]
- [37].Hosseini K, Taubenberger A, Werner C, Fischer-Friedrich E, EMT-induced cell-mechanical changes enhance mitotic rounding strength, Adv. Sci 7 (19) (2020), 2001276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Lancaster OM, Le Berre M, Dimitracopoulos A, Bonazzi D, Zlotek-Zlotkiewicz E, Picone R, Duke T, Piel M, Baum B, Mitotic rounding alters cell geometry to ensure efficient bipolar spindle formation, Dev. Cell 25 (3) (2013) 270–283. [DOI] [PubMed] [Google Scholar]
- [39].Field CM, Lenart P, Bulk cytoplasmic actin and its functions in meiosis and mitosis, Curr. Biol 21 (19) (2011) R825–R830. [DOI] [PubMed] [Google Scholar]
- [40].Mogessie B, Schuh M, Actin protects mammalian eggs against chromosome segregation errors, Science 357 (6353) (2017), eaal1647. [DOI] [PubMed] [Google Scholar]
- [41].Mogessie B, Scheffler K, Schuh M, Assembly and positioning of the oocyte meiotic spindle, Annu Rev. Cell Dev. Biol 34 (2018) 381–403. [DOI] [PubMed] [Google Scholar]
- [42].Sandquist JC, Kita AM, Bement WM, And the dead shall rise: actin and myosin return to the spindle, Dev. Cell 21 (3) (2011) 410–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Burdyniuk M, Callegari A, Mori M, Nedelec F, Lenart P, F-Actin nucleated on chromosomes coordinates their capture by microtubules in oocyte meiosis, J. Cell Biol 217 (8) (2018) 2661–2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Mandeville EC, Rieder CL, Keratin filaments restrict organelle migration into the forming spindle of newt pneumocytes, Cell Motil. Cytoskelet 15 (2) (1990) 111–120. [DOI] [PubMed] [Google Scholar]
- [45].Tsai MY, Wang S, Heidinger JM, Shumaker DK, Adam SA, Goldman RD, Zheng Y, A mitotic lamin B matrix induced by RanGTP required for spindle assembly, Science 311 (5769) (2006) 1887–1893. [DOI] [PubMed] [Google Scholar]
- [46].Ma L, Tsai MY, Wang S, Lu B, Chen R, III JR, Zhu X, Zheng Y, Requirement for Nudel and dynein for assembly of the lamin B spindle matrix, Nat. Cell Biol 11 (3) (2009) 247–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Schweizer N, Pawar N, Weiss M, Maiato H, An organelle-exclusion envelope assists mitosis and underlies distinct molecular crowding in the spindle region, J. Cell Biol 210 (5) (2015) 695–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Nunes V, Dantas M, Castro D, Vitiello E, Wang I, Carpi N, Balland M, Piel M, Aguiar P, Maiato H, Ferreira JG, Centrosome-nuclear axis repositioning drives the assembly of a bipolar spindle scaffold to ensure mitotic fidelity, Mol. Biol. Cell 31 (16) (2020) 1675–1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Kaseda K, McAinsh AD, Cross RA, Dual pathway spindle assembly increases both the speed and the fidelity of mitosis, Biol. Open 1 (1) (2012) 12–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Silkworth WT, Nardi IK, Paul R, Mogilner A, Cimini D, Timing of centrosome separation is important for accurate chromosome segregation, Mol. Biol. Cell 23 (3) (2012) 401–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Silkworth WT, Cimini D, Transient defects of mitotic spindle geometry and chromosome segregation errors, Cell Div. 7 (1) (2012) 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Silkworth WT, Nardi IK, Scholl LM, Cimini D, Multipolar spindle pole coalescence is a major source of kinetochore mis-attachment and chromosome mis-segregation in cancer cells, PLoS One 4 (8) (2009), e6564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Ganem NJ, Godinho SA, Pellman D, A mechanism linking extra centrosomes to chromosomal instability, Nature 460 (7252) (2009) 278–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Almeida AC, Maiato H, Chromokinesins, Curr. Biol 28 (19) (2018) R1131–R1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Rieder CL, Salmon ED, Motile kinetochores and polar ejection forces dictate chromosome position on the vertebrate mitotic spindle, J. Cell Biol 124 (3) (1994) 223–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Vanneste D, Ferreira V, Vernos I, Chromokinesins: localization-dependent functions and regulation during cell division, Biochem. Soc. Trans 39 (5) (2011) 1154–1160. [DOI] [PubMed] [Google Scholar]
- [57].Levesque AA, Compton DA, The chromokinesin Kid is necessary for chromosome arm orientation and oscillation, but not congression, on mitotic spindles, J. Cell Biol 154 (6) (2001) 1135–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Wandke C, Barisic M, Sigl R, Rauch V, Wolf F, Amaro AC, Tan CH, Pereira AJ, Kutay U, Maiato H, Meraldi P, Geley S, Human chromokinesins promote chromosome congression and spindle microtubule dynamics during mitosis, J. Cell Biol 198 (5) (2012) 847–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Cane S, Ye AA, Luks-Morgan SJ, Maresca TJ, Elevated polar ejection forces stabilize kinetochore-microtubule attachments, J. Cell Biol 200 (2) (2013) 203–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Barisic M, Aguiar P, Geley S, Maiato H, Kinetochore motors drive congression of peripheral polar chromosomes by overcoming random arm-ejection forces, Nat. Cell Biol 16 (12) (2014) 1249–1256. [DOI] [PubMed] [Google Scholar]
- [61].Itoh G, Ikeda M, Iemura K, Amin MA, Kuriyama S, Tanaka M, Mizuno N, Osakada H, Haraguchi T, Tanaka K, Lateral attachment of kinetochores to microtubules is enriched in prometaphase rosette and facilitates chromosome alignment and bi-orientation establishment, Sci. Rep 8 (1) (2018) 3888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].DeLuca JG, Dong Y, Hergert P, Strauss J, Hickey JM, Salmon ED, McEwen BF, Hec1 and Nuf2 are core components of the kinetochore outer plate essential for organizing microtubule attachment sites, Mol. Biol. Cell 16 (2) (2005) 519–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Rieder CL, Alexander SP, Kinetochores are transported poleward along a single astral microtubule during chromosome attachment to the spindle in newt lung cells, J. Cell Biol 110 (1) (1990) 81–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Kalinina I, Nandi A, Delivani P, Chacon MR, Klemm AH, Ramunno-Johnson D, Krull A, Lindner B, Pavin N, Tolic-Norrelykke IM, Pivoting of microtubules around the spindle pole accelerates kinetochore capture, Nat. Cell Biol 15 (1) (2013) 82–87. [DOI] [PubMed] [Google Scholar]
- [65].Roos UP, Light and electron microscopy of rat kangaroo cells in mitosis. II. Kinetochore structure and function, Chromosoma 41 (2) (1973) 195–220. [DOI] [PubMed] [Google Scholar]
- [66].Vorozhko VV, Emanuele MJ, Kallio MJ, Stukenberg PT, Gorbsky GJ, Multiple mechanisms of chromosome movement in vertebrate cells mediated through the Ndc80 complex and dynein/dynactin, Chromosoma 117 (2) (2008) 169–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Radford SJ, Hoang TL, Gluszek AA, Ohkura H, McKim KS, Lateral and End-On kinetochore attachments are coordinated to achieve bi-orientation in drosophila oocytes, PLoS Genet. 11 (10) (2015), e1005605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Shrestha RL, Draviam VM, Lateral to end-on conversion of chromosome-microtubule attachment requires kinesins CENP-E and MCAK, Curr. Biol 23 (16) (2013) 1514–1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Shrestha RL, Conti D, Tamura N, Braun D, Ramalingam RA, Cieslinski K, Ries J, Draviam VM, Aurora-B kinase pathway controls the lateral to end-on conversion of kinetochore-microtubule attachments in human cells, Nat. Commun 8 (1) (2017) 150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Drpic D, Pereira AJ, Barisic M, Maresca TJ, Maiato H, Polar ejection forces promote the conversion from lateral to end-on kinetochore-microtubule attachments on mono-oriented chromosomes, Cell Rep. 13 (3) (2015) 460–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Kapoor TM, Lampson MA, Hergert P, Cameron L, Cimini D, Salmon ED, McEwen BF, Khodjakov A, Chromosomes can congress to the metaphase plate before biorientation, Science 311 (5759) (2006) 388–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Cai S, O’Connell CB, Khodjakov A, Walczak CE, Chromosome congression in the absence of kinetochore fibers, Nat. Cell Biol 11 (7) (2009) 832–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Li Y, Yu W, Liang Y, Zhu X, Kinetochore dynein generates a poleward pulling force to facilitate congression and full chromosome alignment, Cell Res. 17 (8) (2007) 701–712. [DOI] [PubMed] [Google Scholar]
- [74].Barisic M, Maiato H, The Tubulin Code: A Navigation System for Chromosomes during Mitosis, Trends Cell Biol. 26 (10) (2016) 766–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Barisic M, Silva e Sousa R, Tripathy SK, Magiera MM, Zaytsev AV, Pereira AL, Janke C, Grishchuk EL, Maiato H, Mitosis. Microtubule detyrosination guides chromosomes during mitosis, Science 348 (6236) (2015) 799–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Peris L, Thery M, Faure J, Saoudi Y, Lafanechere L, Chilton JK, Gordon-Weeks P, Galjart N, Bornens M, Wordeman L, Wehland J, Andrieux A, Job D, Tubulin tyrosination is a major factor affecting the recruitment of CAP-Gly proteins at microtubule plus ends, J. Cell Biol 174 (6) (2006) 839–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].McKenney RJ, Huynh W, Vale RD, Sirajuddin M, Tyrosination of α-tubulin controls the initiation of processive dynein–dynactin motility, EMBO J. 35 (2016) 1175–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Rieder CL, The formation, structure, and composition of the mammalian kinetochore and kinetochore fiber, Int. Rev. Cytol 79 (1982) 1–58. [DOI] [PubMed] [Google Scholar]
- [79].Brinkley BR, Stubblefield E, The fine structure of the kinetochore of a mammalian cell in vitro, Chromosoma 19 (1966) 28–43. [DOI] [PubMed] [Google Scholar]
- [80].Renda F, Magidson V, Tikhonenko I, Fisher R, Miles C, Mogilner A, Khodjakov A, Effects of malleable kinetochore morphology on measurements of intrakinetochore tension, Open Biol. 10 (7) (2020), 200101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Nixon FM, Honnor TR, Clarke NI, Starling GP, Beckett AJ, Johansen AM, Brettschneider JA, Prior IA, Royle SJ, Microtubule organization within mitotic spindles revealed by serial block face scanning electron microscopy and image analysis, J. Cell Sci 130 (10) (2017) 1845–1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Drpic D, Almeida AC, Aguiar P, Renda F, Damas J, Lewin HA, Larkin DM, Khodjakov A, Maiato H, Chromosome segregation is biased by kinetochore size, Curr. Biol 28 (9) (2018) 1344–1356, 1344,–1356 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Gonen S, Akiyoshi B, Iadanza MG, Shi D, Duggan N, Biggins S, Gonen T, The structure of purified kinetochores reveals multiple microtubule-attachment sites, Nat. Struct. Mol. Biol 19 (9) (2012) 925–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Joglekar AP, Bloom K, Salmon ED, In vivo protein architecture of the eukaryotic kinetochore with nanometer scale accuracy, Curr. Biol 19 (8) (2009) 694–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Cheeseman IM, The kinetochore, Cold Spring Harb. Perspect. Biol 6 (7) (2014), a015826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Pesenti ME, Prumbaum D, Auckland P, Smith CM, Faesen AC, Petrovic A, Erent M, Maffini S, Pentakota S, Weir JR, Lin YC, Raunser S, McAinsh AD, Musacchio A, Reconstitution of a 26-subunit human kinetochore reveals cooperative microtubule binding by CENP-OPQUR and NDC80, Mol. Cell 71 (6) (2018) 923–939, 923,–939 e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Cheeseman IM, Chappie JS, Wilson-Kubalek EM, Desai A, The conserved KMN network constitutes the core microtubule-binding site of the kinetochore, Cell 127 (5) (2006) 983–997. [DOI] [PubMed] [Google Scholar]
- [88].DeLuca JG, Gall WE, Ciferri C, Cimini D, Musacchio A, Salmon ED, Kinetochore microtubule dynamics and attachment stability are regulated by Hec1, Cell 127 (5) (2006) 969–982. [DOI] [PubMed] [Google Scholar]
- [89].Helgeson LA, Zelter A, Riffle M, MacCoss MJ, Asbury CL, Davis TN, Human ska complex and Ndc80 complex interact to form a load-bearing assembly that strengthens kinetochore-microtubule attachments, Proc. Natl. Acad. Sci. U.S.A 115 (11) (2018) 2740–2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Auckland P, Roscioli E, Coker HLE, McAinsh AD, CENP-F stabilizes kinetochore-microtubule attachments and limits dynein stripping of corona cargoes, J. Cell Biol 219 (5) (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Vitre B, Gudimchuk N, Borda R, Kim Y, Heuser JE, Cleveland DW, Grishchuk EL, Kinetochore-microtubule attachment throughout mitosis potentiated by the elongated stalk of the kinetochore kinesin CENP-E, Mol. Biol. Cell 25 (15) (2014) 2272–2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Pesenti ME, Weir JR, Musacchio A, Progress in the structural and functional characterization of kinetochores, Curr. Opin. Struct. Biol 37 (2016) 152–163. [DOI] [PubMed] [Google Scholar]
- [93].McDonald KL, O’Toole ET, Mastronarde DN, McIntosh JR, Kinetochore microtubules in PTK cells, J. Cell Biol 118 (2) (1992) 369–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].McEwen BF, Heagle AB, Cassels GO, Buttle KF, Rieder CL, Kinetochore fiber maturation in PtK1 cells and its implications for the mechanisms of chromosome congression and anaphase onset, J. Cell Biol 137 (7) (1997) 1567–1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Roscioli E, Germanova TE, Smith CA, Embacher PA, Erent M, Thompson AI, Burroughs NJ, McAinsh AD, Ensemble-level organization of human kinetochores and evidence for distinct tension and attachment sensors, Cell Rep. 31 (4) (2020), 107535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Suzuki A, Badger BL, Salmon ED, A quantitative description of Ndc80 complex linkage to human kinetochores, Nat. Commun 6 (2015) 8161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Zaytsev AV, Mick JE, Maslennikov E, Nikashin B, DeLuca JG, Grishchuk EL, Multisite phosphorylation of the NDC80 complex gradually tunes its microtubule-binding affinity, Mol. Biol. Cell 26 (10) (2015) 1829–1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Zaytsev AV, Sundin LJ, DeLuca KF, Grishchuk EL, DeLuca JG, Accurate phosphoregulation of kinetochore-microtubule affinity requires unconstrained molecular interactions, J. Cell Biol 206 (1) (2014) 45–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Dong Y, Vanden Beldt KJ, Meng X, Khodjakov A, McEwen BF, The outer plate in vertebrate kinetochores is a flexible network with multiple microtubule interactions, Nat. Cell Biol 9 (5) (2007) 516–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Kukreja AA, Kavuri S, Joglekar AP, Microtubule attachment and centromeric tension shape the protein architecture of the human kinetochore, Curr. Biol 30 (2020) 4869–4881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Thrower DA, Jordan MA, Wilson L, Modulation of CENP-E organization at kinetochores by spindle microtubule attachment, Cell Motil. Cytoskelet 35 (1996) 121–133. [DOI] [PubMed] [Google Scholar]
- [102].McEwen BF, Hsieh CE, Mattheyses AL, Rieder CL, A new look at kinetochore structure in vertebrate somatic cells using high-pressure freezing and freeze substitution, Chromosoma 107 (1998) 366–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Hoffman DB, Pearson CG, Yen TJ, Howell BJ, Salmon ED, Microtubule-dependent changes in assembly of microtubule motor proteins and mitotic spindle checkpoint proteins at PtK 1 kinetochores, Mol. Biol. Cell 12 (7) (2001) 1995–2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Wynne DJ, Funabiki H, Kinetochore function is controlled by a phospho-dependent coexpansion of inner and outer components, J. Cell Biol 210 (6) (2015) 899–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Worrall JT, Tamura N, Mazzagatti A, Shaikh N, van Lingen T, Bakker B, Spierings DCJ, Vladimirou E, Foijer F, McClelland SE, Non-random mis-segregation of human chromosomes, Cell Rep. 23 (11) (2018) 3366–3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Sanchez L, Martinez P, Goyanes V, Analysis of centromere size in human chromosomes 1, 9, 15, and 16 by electron microscopy, Genome 34 (5) (1991) 710–713. [DOI] [PubMed] [Google Scholar]
- [107].Karess R, Rod-Zw10-Zwilch: a key player in the spindle checkpoint, Trends Cell Biol. 15 (7) (2005) 386–392. [DOI] [PubMed] [Google Scholar]
- [108].Griffis ER, Stuurman N, Vale RD, Spindly, a novel protein essential for silencing the spindle assembly checkpoint, recruits dynein to the kinetochore, J. Cell Biol 177 (6) (2007) 1005–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Kops G, Gassmann R, Crowning the kinetochore: the fibrous corona in chromosome segregation, Trends Cell Biol. 30 (2020) 653–667. [DOI] [PubMed] [Google Scholar]
- [110].Howell BJ, McEwen BF, Canman JC, Hoffman DB, Farrar EM, Rieder CL, Salmon ED, Cytoplasmic dynein/dynactin drives kinetochore protein transport to the spindle poles and has a role in mitotic spindle checkpoint inactivation, J. Cell Biol 155 (7) (2001) 1159–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Wojcik E, Basto R, Serr M, Scaerou F, Karess R, Hays T, Kinetochore dynein: its dynamics and role in the transport of the Rough deal checkpoint protein, Nat. Cell Biol 3 (2001) 1001–1008. [DOI] [PubMed] [Google Scholar]
- [112].Cherry LM, Johnston DA, Size variation in kinetochores of human chromosomes, Hum. Genet 75 (2) (1987) 155–158. [DOI] [PubMed] [Google Scholar]
- [113].Tovini L, McClelland SE, Impaired CENP-E function renders large chromosomes more vulnerable to congression failure, Biomolecules 9 (2) (2019) 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Fachinetti D, Han JS, McMahon MA, Ly P, Abdullah A, Wong AJ, Cleveland DW, DNA sequence-specific binding of CENP-B enhances the fidelity of human centromere function, Dev. Cell 33 (3) (2015) 314–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Sullivan LL, Boivin CD, Mravinac B, Song IY, Sullivan BA, Genomic size of CENP-A domain is proportional to total alpha satellite array size at human centromeres and expands in cancer cells, Chromosom. Res 19 (4) (2011) 457–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Maiato H, Khodjakov A, Rieder CL, Drosophila CLASP is required for the incorporation of microtubule subunits into fluxing kinetochore fibres, Nat. Cell Biol 7 (1) (2005) 42–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Rusan NM, Tulu US, Fagerstrom CJ, Wadsworth P, Reorganization of the microtubule array in prophase/prometaphase requires cytoplasmic dyneindependent microtubule transport, J. Cell Biol 158 (6) (2002) 997–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Sikirzhytski V, Magidson V, Steinman JB, He J, Le Berre M, Tikhonenko I, Ault JG, McEwen BF, Chen JK, Sui H, Piel M, Kapoor TM, Khodjakov A, Direct kinetochore-spindle pole connections are not required for chromosome segregation, J. Cell Biol 206 (2) (2014) 231–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Elting MW, Hueschen CL, Udy DB, Dumont S, Force on spindle microtubule minus ends moves chromosomes, J. Cell Biol 206 (2) (2014) 245–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Elting MW, Prakash M, Udy DB, Dumont S, Mapping load-bearing in the mammalian spindle reveals local kinetochore fiber anchorage that provides mechanical isolation and redundancy, Curr. Biol 27 (14) (2017) 2112–2122, 2112,–2122 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Carazo-Salas RE, Gruss OJ, Mattaj IW, Karsenti E, Ran-GTP coordinates regulation of microtubule nucleation and dynamics during mitotic-spindle assembly, Nat. Cell Biol 3 (3) (2001) 228–234. [DOI] [PubMed] [Google Scholar]
- [122].Wilde A, Lizarraga SB, Zhang L, Wiese C, Gliksman NR, Walczak CE, Zheng Y, Ran stimulates spindle assembly by altering microtubule dynamics and the balance of motor activities, Nat. Cell Biol 3 (3) (2001) 221–227. [DOI] [PubMed] [Google Scholar]
- [123].Kalab P, Weis K, Heald R, Visualization of a Ran-GTP gradient in interphase and mitotic Xenopus egg extracts, Science 295 (5564) (2002) 2452–2456. [DOI] [PubMed] [Google Scholar]
- [124].Kalab P, Pralle A, Isacoff EY, Heald R, Weis K, Analysis of a RanGTP-regulated gradient in mitotic somatic cells, Nature 440 (7084) (2006) 697–701. [DOI] [PubMed] [Google Scholar]
- [125].Clarke PR, Zhang C, Spatial and temporal coordination of mitosis by Ran GTPase, Nat. Rev. Mol. Cell Biol 9 (6) (2008) 464–477. [DOI] [PubMed] [Google Scholar]
- [126].Gruss OJ, Carazo-Salas RE, Schatz CA, Guarguaglini G, Kast J, Wilm M, Le Bot N, Vernos I, Karsenti E, Mattaj IW, Ran induces spindle assembly by reversing the inhibitory effect of importin alpha on TPX2 activity, Cell 104 (1) (2001) 83–93. [DOI] [PubMed] [Google Scholar]
- [127].Wittmann T, Wilm M, Karsenti E, Vernos I, TPX2, a novel Xenopus MAP involved in spindle pole organization, J. Cell Biol 149 (7) (2000) 1405–1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Goshima G, Mayer M, Zhang N, Stuurman N, Vale RD, Augmin: a protein complex required for centrosome-independent microtubule generation within the spindle, J. Cell Biol 181 (3) (2008) 421–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Petry S, Groen AC, Ishihara K, Mitchison TJ, Vale RD, Branching microtubule nucleation in xenopus egg extracts mediated by augmin and TPX2, Cell 152 (4) (2013) 768–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Lawo S, Bashkurov M, Mullin M, Ferreria MG, Kittler R, Habermann B, Tagliaferro A, Poser I, Hutchins JR, Hegemann B, Pinchev D, Buchholz F, Peters JM, Hyman AA, Gingras AC, Pelletier L, HAUS, the 8-subunit human augmin complex, regulates centrosome and spindle integrity, Curr. Biol 19 (10) (2009) 816–826. [DOI] [PubMed] [Google Scholar]
- [131].Kamasaki T, O’Toole E, Kita S, Osumi M, Usukura J, McIntosh JR, Goshima G, Augmin-dependent microtubule nucleation at microtubule walls in the spindle, J. Cell Biol 202 (1) (2013) 25–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Oegema K, Wiese C, Martin OG, Iwamatsu A, Mitchison TJ, Zheng Y, Characterization of two related Drosophila g -tubulin complexes that differ in their ability to nucleate microtubules, J. Cell Biol 144 (4) (1999) 721–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Zheng Y, Wong ML, Alberts B, Mitchison T, Nucleation of microtubule assembly by a gamma-tubulin-containing ring complex, Nature 378 (1995) 578–583. [DOI] [PubMed] [Google Scholar]
- [134].Alfaro-Aco R, Thawani A, Petry S, Biochemical reconstitution of branching microtubule nucleation, eLife 9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Song JG, King MR, Zhang R, Kadzik RS, Thawani A, Petry S, Mechanism of how augmin directly targets the gamma-tubulin ring complex to microtubules, J. Cell Biol 217 (7) (2018) 2417–2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Thawani A, Stone HA, Shaevitz JW, Petry S, Spatiotemporal organization of branched microtubule networks, eLife 8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].David AF, Roudot P, Legant WR, Betzig E, Danuser G, Gerlich DW, Augmin accumulation on long-lived microtubules drives amplification and kinetochore-directed growth, J. Cell Biol 218 (7) (2019) 2150–2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Zierhut C, Funabiki H, Nucleosome functions in spindle assembly and nuclear envelope formation, Bioessays 37 (10) (2015) 1074–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Andrews PD, Ovechkina Y, Morrice N, Wagenbach M, Duncan K, Wordeman L, Swedlow J, Aurora B regulates MCAK at the mitotic centromere, Dev. Cell 6 (2) (2004) 253–268. [DOI] [PubMed] [Google Scholar]
- [140].Lan W, Zhang X, Kline-Smith SL, Rosasco SE, Barrett-Wilt GA, Shabanowitz J, Hunt DF, Walczak CE, Stukenberg PT, Aurora B phosphorylates centromeric MCAK and regulates its localization and microtubule depolymerization activity, Curr. Biol 14 (4) (2004) 273–286. [DOI] [PubMed] [Google Scholar]
- [141].Sampath SC, Ohi R, Leismann O, Salic A, Pozniakovski A, Funabiki H, The chromosomal passenger complex is required for chromatin-induced microtubule stabilization and spindle assembly, Cell 118 (2) (2004) 187–202. [DOI] [PubMed] [Google Scholar]
- [142].Kelly AE, Sampath SC, Maniar TA, Woo EM, Chait BT, Funabiki H, Chromosomal enrichment and activation of the aurora B pathway are coupled to spatially regulate spindle assembly, Dev. Cell 12 (1) (2007) 31–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143].Tan L, Kapoor TM, Examining the dynamics of chromosomal passenger complex (CPC)-dependent phosphorylation during cell division, Proc. Natl. Acad. Sci. U.S.A 108 (40) (2011) 16675–16680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].Wang E, Ballister ER, Lampson MA, Aurora B dynamics at centromeres create a diffusion-based phosphorylation gradient, J. Cell Biol 194 (4) (2011) 539–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [145].Snyder JA, McIntosh JR, Initiation and growth of microtubules from mitotic centers in lysed mammalian cells, J. Cell Biol 67 (3) (1975) 744–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [146].Witt PL, Ris H, Borisy GG, Origin of kinetochore microtubules in Chinese hamster ovary cells, Chromosoma 81 (3) (1980) 483–505. [DOI] [PubMed] [Google Scholar]
- [147].Gudimchuk N, Vitre B, Kim Y, Kiyatkin A, Cleveland DW, Ataullakhanov FI, Grishchuk EL, Kinetochore kinesin CENP-E is a processive bi-directional tracker of dynamic microtubule tips, Nat. Cell Biol 15 (9) (2013) 1079–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [148].Cimini D, Moree B, Canman JC, Salmon ED, Merotelic kinetochore orientation occurs frequently during early mitosis in mammalian tissue cells and error correction is achieved by two different mechanisms, J. Cell Sci 116 (20) (2003) 4213–4225. [DOI] [PubMed] [Google Scholar]


