Abstract
Mycobacterium tuberculosis isolates with identical IS6110 restriction fragment length polymorphism (RFLP) patterns are considered to originate from the same ancestral strain and thus to reflect ongoing transmission. In this study, we investigated 1,277 IS6110 RFLP patterns for the presence of multiple low-intensity bands (LIBs), which may indicate infections with multiple M. tuberculosis strains. We did not find any multiple LIBs, suggesting that multiple infections are rare in the Netherlands. However, we did observe a few LIBs in 94 patterns (7.4%) and examined the nature of this phenomenon. With single-colony cultures it was found that LIBs mostly represent mixed bacterial populations with slightly different RFLP patterns. Mixtures were expressed in RFLP patterns as LIBs when 10 to 30% of the DNA analyzed originated from a bacterial population with another RFLP pattern. Presumably, a part of the LIBs did not represent mixed bacterial populations, as in some clusters all strains exhibited LIBs in their RFLP patterns. The occurrence of LIBs was associated with increased age in patients. This may reflect either a gradual change of the bacterial population in the human body over time or IS6110-mediated genetic adaptation of M. tuberculosis to changes in the environmental conditions during the dormant state or reactivation thereafter.
The standardized IS6110 restriction fragment length polymorphism (RFLP) typing of Mycobacterium tuberculosis isolates (28) is based on the concept that RFLP patterns reflect the presence of the IS6110 element at different sites in the genome of M. tuberculosis complex strains (22). RFLP typing has permitted the differentiation of clinical M. tuberculosis isolates in many parts of the world (1, 8, 11, 17, 19, 20, 30, 31, 32). M. tuberculosis isolates with identical IS6110 RFLP patterns are considered to be clonally related and thus to represent ongoing transmission (14, 26, 33). In the early 1990s, comparing RFLP patterns of M. tuberculosis isolates proved useful in investigating outbreaks in closed communities, such as hospitals and prisons (6, 7, 12, 15, 21). Several population-based studies have also been carried out (2, 4, 25, 29), providing information on risk factors for transmission (2, 25, 29) and transmission dynamics (3).
As most clinical M. tuberculosis isolates have distinctive RFLP patterns with clearly defined bands, it is commonly assumed that a pattern reflects the presence of IS6110 elements in the genome of a single M. tuberculosis strain. However, multiple M. tuberculosis infections have been observed (34). RFLP typing of an isolate from multiple M. tuberculosis infections will result in a mixture of RFLP patterns. If one of the bacterial populations predominates, the other(s) will be reflected as a background pattern of usually multiple low-intensity bands (LIBs) in the mixed RFLP pattern.
Recently, a study of the instability of IS6110 RFLP patterns of M. tuberculosis showed that RFLP patterns of consecutive isolates can differ in one or two bands from the patterns of initial isolates (9). In this study, it was assumed that after a period of time the bacterial population in an M. tuberculosis isolate had or had not changed as revealed in the IS6110 RFLP patterns of follow-up isolates. However, it is likely that the change in an M. tuberculosis population is gradual, so that only part of a bacterial population shows a genetic change in the IS6110 RFLP. This would result in a mixture of two RFLP patterns with and without the changed IS6110 element and hence result in one or two LIBs.
Analysis of the IS6110 RFLP patterns of M. tuberculosis isolates has been standardized to a large extent: autoradiograms are scanned, converted to computerized images, and normalized by software such as the GelCompar program (Applied Maths, Kortrijk, Belgium). However, LIBs hamper the interpretation of RFLP typing results for epidemiological investigations, as the interpretation of LIBs is subject to variation. Some LIBs will be detected automatically; others will not. It is common practice to manually correct the bands detected by the computer. This correction procedure is not standardized, and therefore a potential inaccuracy is introduced in studying the transmission of tuberculosis when using IS6110 RFLP typing.
In this study, the prevalence and nature of LIBs in the IS6110 RFLP patterns of M. tuberculosis are investigated. The inaccuracy in matching M. tuberculosis isolates to the Dutch database of RFLP patterns, related to the interpretation of LIBs, is determined. Finally, possible associations between LIBs in the RFLP patterns and patient and strain characteristics are studied.
MATERIALS AND METHODS
General.
Since January 1993, all M. tuberculosis complex isolates in the Netherlands have been subjected to IS6110 RFLP typing (28) and have been analyzed by computer (18) using GelCompar software (version 4.1). In the GelCompar program, 700 positions per lane are scanned. IS6110 RFLP patterns of M. tuberculosis isolates are considered clustered if their patterns are identical (within a position tolerance of 1%). When an epidemiological link between patients is confirmed, isolates are also considered clustered if they differ by at most one band.
LIBs: prevalence and between-reader agreement.
Two fingerprint readers independently studied all 1,277 M. tuberculosis complex isolates fingerprinted in the Netherlands between June 1997 and May 1998 by eye to detect multiply banded background patterns and to score IS6110 RFLP patterns with LIBs. None of these were “repeat” isolates from the same patient. The results from each reader, blinded to the findings of the other, were compared. LIBs identified by either reader were assessed by an experienced third reader. The level of agreement between readers was studied further by comparing the number of LIBs detected by four experienced readers in 64 RFLP patterns. Of these patterns, 24 had been selected by an experienced reader to represent RFLP patterns with bands that were difficult to interpret and another 40 had been selected randomly from the 1,277 RFLP patterns. Objective criteria for the assessment of LIBs were constructed using the computerized RFLP patterns: percentages of the average intensity of bands in a pattern were compared to the experts' assignment of LIBs.
Consequences of interpretation of LIBs in RFLP patterns for clustering.
Whether or not a band is assigned to the computerized RFLP pattern at the position of the LIB will influence the clustering of RFLP patterns of M. tuberculosis isolates. This was studied for 94 RFLP patterns by matching the patterns with all bands assigned and again without the LIBs. The resulting percentages of clustered strains were compared with the actual percentage of clustered patterns.
Nature of LIBs in RFLP patterns.
To investigate whether the occurrence of LIBs in RFLP patterns could be due to a heterogeneous bacterial population in M. tuberculosis isolates, single-colony cultures (SCCs) of isolates with LIBs in their RFLP patterns were prepared. Eight isolates with different RFLP patterns exhibiting LIBs were selected from the isolates collected since 1993, and 3 to 10 SCCs of each isolate were produced by single-colony strikes on 7H10 plates and reculturing of individual colonies. The SCCs were subjected to standard RFLP typing (28). To study whether M. tuberculosis isolates without LIBs in the RFLP patterns would reveal heterogeneity in RFLP patterns of SCCs, 3 to 10 SCCs per isolate were produced from six isolates with different RFLP patterns without LIBs and subjected to standard RFLP analysis.
To study in more detail whether LIBs in RFLP patterns could represent mixed bacterial populations, two experiments were undertaken. First, we assessed whether LIBs could be produced by mixing DNA from two different M. tuberculosis isolates with completely different RFLP patterns. Second, we studied at what ratios DNA mixtures would show LIBs. This was done by mixing in different ratios the DNAs of two M. tuberculosis isolates with RFLP patterns differing by only one band. All DNA mixtures were typed using standard IS6110 RFLP.
To investigate whether LIBs could be a fixed phenomenon, present in identical RFLP patterns of M. tuberculosis isolates from epidemiologically related patients, the RFLP patterns of a total of 726 clusters were reviewed by an experienced reader to score LIBs. The clusters consisted of two or more strains with identical IS6110 RFLP patterns, comprising five or more bands, found in The Netherlands since January 1993. If more than 10 RFLP patterns per cluster were available, the 10 most recently found were reviewed.
To further study the possibility that LIBs represent a fixed phenomenon, three patients with identical RFLP patterns containing LIBs were selected from each of two clusters. SCCs of their M. tuberculosis isolates were prepared and typed using IS6110 RFLP.
Preferential band positions of LIBs.
To determine whether LIBs were more prevalent at certain restriction fragment positions, the relative frequencies of band positions of LIBs in IS6110 RFLP patterns were investigated. In this investigation, RFLP patterns of unique isolates and, in the case of clustered isolates, patterns of the first isolates of clusters were included. This amounted to a total of 838 RFLP patterns, containing 6,189 normal-intensity bands and 69 LIBs.
Characteristics of patients and mycobacteria associated with LIBs.
A comparison was made between patients infected by M. tuberculosis with and without LIBs in the RFLP pattern. Patient variables were age, sex, infection at extrapulmonary site, and being in a cluster of patients with identical RFLP patterns. Bacterial variables were the resistance profile and the number of IS6110 copies in the RFLP pattern. Fifteen to 50 RFLP patterns of isolates with a certain resistance profile analyzed in the period from January 1993 to July 1998 were reviewed at random to score LIBs.
Associations were tested by the χ2 test or the median two-sample test (normal approximation) as appropriate. Statistical significance was accepted if P was <0.05.
RESULTS
LIBs: prevalence and between-reader agreement.
In 1,277 IS6110 RFLP patterns of M. tuberculosis complex isolates fingerprinted in the Netherlands between June 1997 and May 1998, no multiply banded background patterns, indicative of multiple infections, were found. This suggests that the M. tuberculosis complex strains from this period were all clonal.
LIBs were detected in 94 of the 1,277 IS6110 RFLP patterns of M. tuberculosis complex isolates (7.4%) when the laboratory technician most experienced in RFLP typing reviewed 97 RFLP patterns with possible LIBs. These 97 RFLP patterns were found by adding the RFLP patterns with LIBs found by reader A to those found by reader B. Reader A detected LIBs in 73 out of 1,277 RFLP patterns (5.7%), whereas reader B detected such bands in 55 (4.3%) patterns. Both readers agreed upon LIBs in 31 RFLP patterns, whereas their findings were not in agreement for 66 patterns. A further study of interreader agreement on 64 RFLP patterns revealed that four readers found LIBs in 9, 14, 15, and 19 patterns. The levels of agreement among these four readers on LIBs in 64 RFLP patterns ranged from a kappa of 0.42 to 0.64.
In GelCompar, the intensity per band is recorded in binary form as the height (indicative of the brightness of the band) and the sigma (indicative of the size of the band). To decide whether height and/or sigma was indicative of a band having a low intensity, the average height and sigma of LIBs (identified by experts) were compared to the average height and sigma of normal-intensity bands. Only the average height of LIBs differed from that of normal intensity bands, so the height was considered to be a reliable reflection of the intensity. Percentages of the average intensity per RFLP pattern were compared to LIBs assigned by expert readers. The level of agreement, corrected for the agreement attributable to chance, between the expert assignment of LIBs and percentages of the average intensity per RFLP pattern was highest (kappa, >0.4) for 10 to 15% of the average intensity of the bands in the RFLP patterns of 10 to 15%.
Consequences of interpretation of LIBs in RFLP patterns for clustering.
From 94 M. tuberculosis isolates with computerized RFLP patterns that were identified as having one or more LIBs, on the basis of routine reading of RFLP patterns 42 were clustered with other RFLP patterns in the entire Dutch database. If none of the LIBs were assigned, 35 patterns were clustered, of which 3 were not considered clustered before. If all LIBs were assigned, 15 patterns were clustered. Five RFLP patterns would be considered clustered either with or without the LIB. However, assigning all or no LIBs, in both cases leading to a decreased number of clustered RFLP patterns, did not significantly reduce the clustering percentage in the entire Dutch database in the study period. In the period from June 1997 to May 1998, 49.3% of all RFLP patterns were clustered on the basis of routine reading of RFLP patterns. When none of the LIBs or all LIBs were assigned, this percentage decreased to 48.8 and 47.1%, respectively.
Nature of LIBs in RFLP patterns.
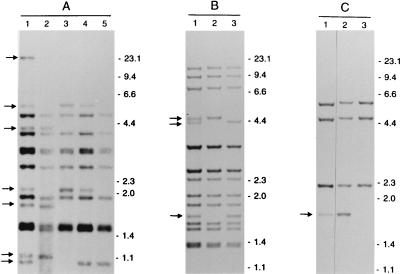
In order to investigate whether genetic heterogeneity could be the reason for the occurrence of LIBs, SCCs were prepared from clinical isolates exhibiting LIBs. All RFLP patterns of the SCCs produced from the eight isolates with LIBs did not show the LIBs of the parental cultures but either showed no band at all or a band with a normal intensity at the position of the LIB of the parental culture. Eight out of eight isolates with LIBs consisted of mixed bacterial populations (100%; 95% confidence interval, 63 to 100%). Three of the analyzed cultures are shown in Fig. 1. Remarkably, one of the clinical isolates with LIBs (isolate A in Fig. 1) revealed four different RFLP patterns in the SCCs. This isolate was taken from a patient from whom two other M. tuberculosis isolates, AI and AII (data not shown), were obtained on the same day. The RFLP pattern of isolate AI was identical to the pattern of one of the SCCs of isolate A. All other patterns of either isolates or SCCs from this patient were different. The RFLP pattern of isolate A had 16 bands (of which seven were LIBs), the pattern of isolate AI had 12 bands (one LIB), and the pattern of isolate AII had 14 bands (four LIBs). All four RFLP patterns had 11 bands in common. The SCCs of the six parental cultures without LIBs had RFLP patterns identical to those of the parental cultures.
FIG. 1.
IS6110 RFLP patterns of isolates, showing LIBs, and SCCs, from three different patients (A to C). Lanes 1 show the banding patterns of the isolates, with LIBs indicated by arrows. Lanes 2 to 5 show the banding patterns of SCCs of these isolates. The numbers on the right indicate the sizes of standard DNA fragments in kilobase pairs.
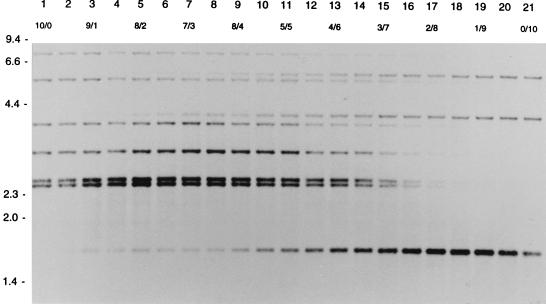
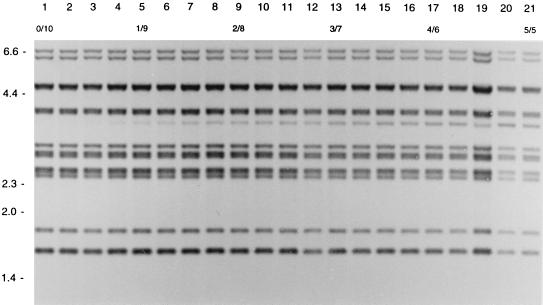
In order to measure the detection level of DNA of one strain mixed with DNA of another strain, different ratios of DNA from two strains with different RFLP patterns were tested in RFLP typing. If DNA for RFLP typing consisted of DNA of strain A and DNA of strain B in the ratios 1/9 to 3/7, the IS6110 RFLP pattern of strain A occurred as LIBs (Fig. 2). If the DNA mixture consisted of DNA of a strain with an additional band and DNA of a strain without that band, an LIB was visible in the ratios 1/9 to 2/8 (Fig. 3).
FIG. 2.
IS6110 RFLP patterns of different mixtures of the DNAs of two M. tuberculosis strains. Lanes 1 and 21 show the RFLP patterns of the pure DNAs of the two strains. Lanes 2 to 20 show the patterns of mixtures of the DNAs of these strains. The numbers in the second horizontal row indicate the ratios of the DNA mixtures. The numbers on the left indicate the sizes of standard DNA fragments in kilobase pairs.
FIG. 3.
IS6110 RFLP patterns of different mixtures of the DNAs of two SCCs of an M. tuberculosis strain differing in a single IS6110 element. Lane 1 shows the pattern of pure DNA of one SCC. Lanes 2 to 21 depict patterns of mixtures of this DNA with an increasing amount of DNA of another SCC containing an additional IS6110 copy at a PvuII restriction fragment of approximately 3.5 kb. The numbers in the second horizontal row indicate the ratios of the DNA mixtures. The numbers on the left indicate the sizes of standard DNA fragments in kilobase pairs.
To investigate whether LIBs could represent a phenomenon not associated with mixed bacterial populations, LIBs were scored in RFLP patterns of 726 clusters. At least two isolates with LIBs in the RFLP patterns were found in 18 of the 726 clusters. In 12 (1.7%) of these clusters, all the isolates showed an LIB at the same band position. For two clusters of strains with LIB-containing RFLP patterns, three SCCs of isolates were made, and these showed the same pattern as that of the parental cultures. Figure 4 shows the LIB-containing RFLP patterns of three individual patient isolates of a cluster.
FIG. 4.
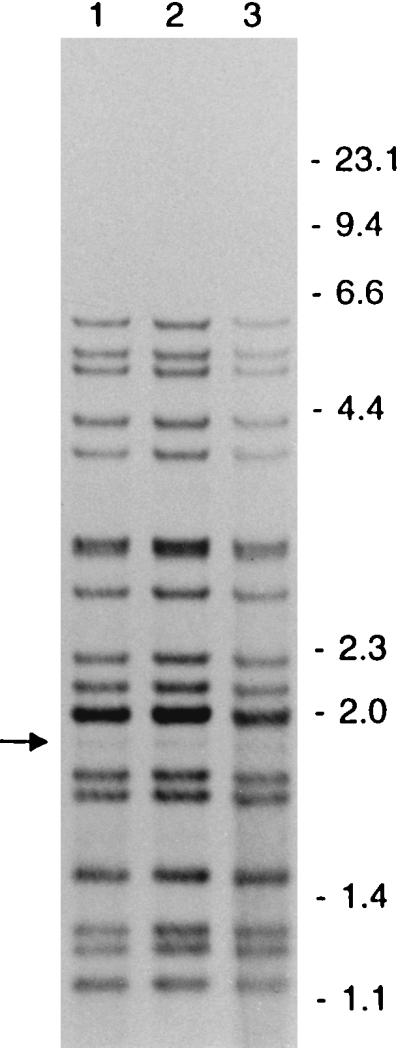
IS6110 RFLP patterns of three patient isolates of a cluster showing an LIB (indicated by the arrow) at the same PvuII restriction fragment. The numbers on the right indicate the sizes of standard DNA fragments in kilobase pairs.
Preferential band positions of LIBs.
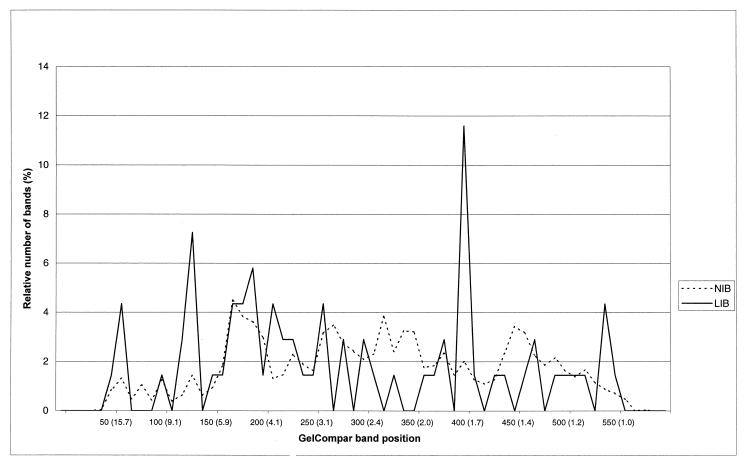
When the band positions of 6,189 normal-intensity bands and 69 LIBs were studied, LIBs were found more often at particular band positions, namely, band positions 125 (7.2 kb) and 400 (1.7 kb) (Fig. 5).
FIG. 5.
Frequency distribution of 6,189 normal-intensity bands (NIB) and 69 LIBs per band position category (GelCompar band position and sizes of standard DNA fragments in kilobase pairs).
Characteristics of patients and mycobacteria associated with LIBs.
Of the 1,277 RFLP patterns of M. tuberculosis complex isolates, 1,207 were included in the analysis of characteristics of patients and strains. A total of 70 isolates were excluded (44 Mycobacterium bovis BCG isolates, 1 Mycobacterium microti isolate, 11 isolates of unknown species, and 14 laboratory cross contamination isolates).
LIBs in the RFLP patterns of M. tuberculosis isolates were more often observed in older than in younger patients (Table 1; the trend was statistically significant [P < 0.05]). LIBs also occurred slightly more often in isolates from resistant strains (not significant). The occurrence of LIBs in the RFLP patterns of M. tuberculosis isolates was not associated with the patient's sex, the extrapulmonary sampling site, clustering of tuberculosis patients based on identical RFLP patterns, or the number of IS6110 copies in the RFLP pattern (Table 1).
TABLE 1.
Patient characteristics and strain characteristics for isolates with one or more LIBs in the IS6110 RFLP pattern of M. tuberculosis and for isolates with normal-intensity bands in the RFLP pattern
| Characteristic | Isolates with LIBs in RFLP patterns
|
Isolates without LIBs in RFLP patterns
|
||
|---|---|---|---|---|
| n | No. positivea | n | No. positivea | |
| Age (yr) | 77 | 1,087 | ||
| <25 | 9 (11.7) | 205 (18.9) | ||
| 25–44 | 33 (42.9) | 523 (48.1) | ||
| 45–64 | 17 (22.1) | 163 (15.0) | ||
| ≥65 | 18 (23.4) | 196 (18.0)b | ||
| Median (P5; P95)c | 39 (19;80) | 35 (17;81) | ||
| Male | 72 | 46 (63.9) | 1,029 | 617 (60.0) |
| Extrapulmonary isolate | 72 | 9 (12.5) | 911 | 120 (13.2) |
| Clustered | 83 | 46 (55.4) | 1,119 | 573 (51.2) |
| Drug-resistant isolate | 82 | 17 (20.7) | 1,116 | 156 (14.0) |
| Median no. of IS6110 bands (P5;P95) | 82 | 11 (1;17) | 1,113 | 10 (1;17) |
Values are number (percentage) unless otherwise indicated.
χ2trend = 4.7; P < 0.05.
(P5;P95), 5th percentile; 95th percentile.
Because LIBs seemed to be associated with resistance of M. tuberculosis to tubercular drugs, the prevalence of LIBs in the RFLP patterns of isolates with different resistance profiles was investigated. Table 2 presents the percentages of RFLP patterns with LIBs for different resistance profiles for which at least 15 RFLP patterns were investigated. The highest percentage of patterns with LIBs, 17%, was found among isolates resistant to isoniazid, rifampin, ethambutol, and streptomycin. However, this percentage was not significantly different from that of isolates with other resistance profiles. In addition, the percentage of patterns with LIBs for isolates with other resistance profiles ranged between 0 and 8%, indicating that LIBs were not associated with any particular resistance profile.
TABLE 2.
No. of RFLP patterns with LIBs for different resistance profiles
| Resistancea
|
No. of patterns studied | No. (%) of patterns with LIBs | |||
|---|---|---|---|---|---|
| Isoniazid | Rifampin | Ethambutol | Streptomycin | ||
| 50 | 4 (8) | ||||
| Resistant | 50 | 1 (2) | |||
| Resistant | 17 | 0 (0) | |||
| Resistant | 50 | 4 (8) | |||
| Resistant | Resistant | 50 | 3 (6) | ||
| Resistant | Resistant | 23 | 1 (4) | ||
| Resistant | Resistant | Resistant | Resistant | 18 | 3 (17) |
Resistant is defined as follows: isoniazid, >0.2 μg/ml; rifampin, >1 μg/ml; ethambutol, >5 μg/ml; streptomycin, >5 μg/ml (MICs).
DISCUSSION
This study showed that the prevalence of multiple M. tuberculosis complex infections in The Netherlands is very low. M. tuberculosis complex isolates analyzed from May 1997 to June 1998 in The Netherlands were screened for indications of multiple infections expressed by the presence of low-intensity background patterns. In 1,277 IS6110 RFLP patterns no multiply banded background patterns, indicative of tuberculosis infections with more than one strain, were observed. This conclusion seems to be valid, as a second strain could be detected if 10 to 30 or 70 to 90% of the DNA necessary for RFLP typing originated from a second strain. However, if mixtures were present in another proportion, it is possible that multiple infections were not detected. Still, as we did not find multiply banded background patterns and as they have only been documented anecdotally, for instance, during an episode of laboratory cross contamination (27), we conclude that multiple infections are rarely encountered. This is in agreement with our previous study of the stability of IS6110 RFLP, in which almost all initial and follow-up M. tuberculosis isolates of 546 patients had identical or nearly identical RFLP patterns (9). Further research is needed to study whether multiple infections, reflected in multiply banded background patterns, occur more often in areas with a high prevalence of tuberculosis infection.
Although we did not find any indication of multiple infections, a relatively high number of RFLP patterns (6.7%) contained one or two LIBs. We showed that these LIBs can be explained by genetic heterogeneity. In the SCCs produced from eight LIB-exhibiting isolates, we found either a normal-intensity band or no hybridization at all at the position of the LIB in the parental strain. It should be noted that we may have underestimated the percentage of RFLP patterns containing LIBs in this study, as 3 out of the 43 laboratories in The Netherlands submitted M. tuberculosis SCCs instead of full specimen cultures. As would be expected, the isolates from these three laboratories did not show any LIBs in the RFLP patterns.
We proved with RFLP typing of SCCs that LIBs in RFLP patterns can be explained by the presence of two bacterial populations, with and without a transposed IS6110 element. Assuming that a single infectious unit can be sufficient to transmit tuberculosis (10), LIBs resulting from mixed bacterial populations will not be observed in all isolates of clusters. However, we found RFLP patterns with LIBs to be reproduced in 1.7% of all clusters in The Netherlands. This finding was supported by analysis of two clusters of isolates with LIB-containing RFLP patterns; all RFLP patterns of the SCCs of three isolates of these clusters contained the same LIB as the parental cultures. This indicates that not all LIBs reflect mixed bacterial populations. The nature of these LIBs is not yet clear, but they could be due to truncation, as was observed in IS1081 (5), or to mutations in the DNA sequences of the respective IS elements, resulting in a lower hybridization signal (13). Another less likely explanation for LIBs occurring in clustered strains could be the transmission of a mixture of bacterial populations in fixed ratios.
The fact that M. tuberculosis isolates with LIBs in their RFLP patterns can be separated into bacterial populations with and without an additional band(s) suggests that changes in IS6110 elements occur as gradual shifts in the bacterial populations in a patient and not as favorable selective events. However, our findings suggest that selective pressure may play a role in these changes. The significant association between the occurrence of LIBs in RFLP patterns and advanced age of the patients could be due to long-term suppressed in vivo multiplication or the endogenous reactivation of M. tuberculosis thereafter. The specific growing conditions in microaerobic lesions may lead to an increased genetic diversification of offspring, as indicated by the study of Ghanekar et al. (16). This would also correspond to our previous study, in which we found that variant RFLP patterns of M. tuberculosis were more often observed in extrapulmonary isolates (9). In this respect, IS6110 transpositions may be a driving force of the genetic rearrangement in the adaptive process of M. tuberculosis during dormancy or the revival thereafter.
The LIBs were not equally distributed over the 700 band positions that were defined in the computer-assisted analysis of RFLP patterns but were clearly related to preferential band positions, especially those corresponding to 7.1 and 1.7 kb. These differed from the preferential band positions of normal-intensity bands. The examination of these preferential genomic sites, compared to the preferential sites for IS6110 in general (24), may shed light on the mechanisms that play a role in the evolution of M. tuberculosis. One of these mechanisms may be IS6110-related mutagenesis (13, 23). Further research is needed to reveal how genetic heterogeneity is related to the time between infection and isolation of M. tuberculosis and/or bacterial growth under specific conditions.
RFLP typing of M. tuberculosis isolates is highly standardized (28), but the fact that LIBs occur and how to interpret such bands have not been described. The recognition of LIBs depends on laboratory procedures, such as the exposure time of the autoradiogram and hybridization intensity. In general, we observed that overexposure of autoradiograms resulted in the detection of more LIBs. However, in our study the exposure time was adjusted to obtain clear RFLP patterns for computer-assisted analysis. Furthermore, hybridization procedures in our laboratory were highly standardized and were always carried out by experienced laboratory technicians. As laboratory procedures may play a role, we think the influence of laboratory artifacts on the occurrence or detection of LIBs needs further study.
We showed that the level of agreement between readers for the detection of LIBs was reasonable, but not very good. In addition, it was shown that the interpretation of patterns with such bands in routine laboratory settings could be biased towards clustering. Although the effect of this bias on the percentage of clustered RFLP patterns in the entire database was not very strong, we saw that this percentage was lower when none or all of the LIBs were assigned to the computerized RFLP patterns. Therefore, based on our study, we propose the standardization of the procedure for handling LIBs. Bands recorded in GelCompar with a height of >15% of the average height of all bands in the RFLP pattern should be considered normal-intensity bands. Bands recorded in GelCompar with a height of 10 to 15% of the average height of all bands in that RFLP pattern should be labeled as low-intensity bands. If RFLP patterns with LIBs cluster with other RFLP patterns, this clustering result, as well as its possible epidemiological confirmation, needs to be interpreted carefully.
ACKNOWLEDGMENTS
This study builds on many years of laboratory practice at the National Institute of Public Health and the Environment, Department of Mycobacteriology. We therefore thank all laboratory technicians who contributed to any part of the work resulting in the establishment of the IS6110 RFLP database. Marja Pospiech-Greijn is acknowledged for producing SCCs and, together with Joan Kwakkel and Mirjam Dessens-Kroon, for identifying LIBs in RFLP patterns. We also thank Jan van Embden and Nico Nagelkerke for critical review of the manuscript.
Financial support was obtained from the Ministry of Health, Welfare and Sports of the Netherlands and the European Union (Project Molecular Epidemiology and Control of Tuberculosis; BMH4-CT97-2102).
REFERENCES
- 1.Barnes P F, el-Hajj H, Preston-Martin S, Cave M D, Jones B E, Otaya M, Pogoda J, Eisenach K D. Transmission of tuberculosis among the urban homeless. JAMA. 1996;275:305–307. [PubMed] [Google Scholar]
- 2.Bauer J, Yang Z, Poulsen S, Andersen A B. Results from 5 years of nationwide DNA fingerprinting of Mycobacterium tuberculosis complex isolates in a country with a low incidence of M. tuberculosis infection. J Clin Microbiol. 1998;36:305–308. doi: 10.1128/jcm.36.1.305-308.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Borgdorff M W, Nagelkerke N, van Soolingen D, de Haas P E W, Veen J, van Embden J D A. Analysis of tuberculosis transmission between nationalities in the Netherlands in the period 1993–1995 using DNA fingerprinting. Am J Epidemiol. 1998;147:187–195. doi: 10.1093/oxfordjournals.aje.a009433. [DOI] [PubMed] [Google Scholar]
- 4.Butcher P D, Hutchinson N A, Doran T J, Dale J W. The application of molecular techniques to the diagnosis and epidemiology of mycobacterial diseases. Soc Appl Bacteriol Symp Ser. 1996;25:53S–71S. [PubMed] [Google Scholar]
- 5.Cole S T, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon S V, Eiglmeier K, Gas S, Barry III C E, Tekaia F, Badcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyd S, Hornsby T, Jagels K, Krogh A, McLean J, Moule S, Murphy L, Oliver K, Osborne J, Quail M A, Rajandream M A, Rogers J, Rutter S, Seeger K, Skelton J, Squares R, Squares S, Sulston J E, Taylor K, Whitehead S, Barrell B G. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 6.Coronado V G, Beck-Sague C M, Hutton M D, et al. Transmission of multidrug-resistant Mycobacterium tuberculosis among persons with human immunodeficiency virus infection in an urban hospital: epidemiologic and restriction fragment length polymorphism analysis. J Infect Dis. 1993;168:1052–1055. doi: 10.1093/infdis/168.4.1052. [DOI] [PubMed] [Google Scholar]
- 7.Daley C L, Small P M, Schechter G F, Schoolnik G K, McAdam R A, Jacobs W R, Jr, Hopewell P C. An outbreak of tuberculosis with accelerated progression among persons infected with the human immunodeficiency virus. N Engl J Med. 1992;326:231–235. doi: 10.1056/NEJM199201233260404. [DOI] [PubMed] [Google Scholar]
- 8.Das S, Paramasivan C N, Lowrie D B, Prabhakar R, Narayanan P R. IS6110 restriction fragment length polymorphism typing of clinical isolates of Mycobacterium tuberculosis from patients with pulmonary tuberculosis in Madras, South India. Tuber Lung Dis. 1995;76:550–554. doi: 10.1016/0962-8479(95)90533-2. [DOI] [PubMed] [Google Scholar]
- 9.De Boer A S, Borgdorff M W, de Haas P E W, Nagelkerke N, van Embden J D A, van Soolingen D. Rate of change of IS6110 genotypes of Mycobacterium tuberculosis based on serial patient isolates. J Infect Dis. 1999;180:1238–1244. doi: 10.1086/314979. [DOI] [PubMed] [Google Scholar]
- 10.Des Prez R M, Heim C R. Mycobacterium tuberculosis. In: Mandell G L, Douglas R G, Bennett J E, editors. Principles and practice of infectious diseases. 3rd ed. New York, N.Y: Churchill Livingstone Inc.; 1990. p. 1880. [Google Scholar]
- 11.Diaz R, Kremer K, de Haas P E W, Gomez R I, Marrero A, Valdivia J A, van Embden J D A, van Soolingen D. Molecular epidemiology of tuberculosis in Cuba outside of Havana, July 1994–June 1995: utility of spoligotyping versus IS6110 restriction fragment length polymorphism. Int J Tuber Lung Dis. 1998;2:743–750. [PubMed] [Google Scholar]
- 12.Edlin B R, Tokars J I, Grieco M H, et al. An outbreak of multidrug-resistant tuberculosis among hospitalized patients with the acquired immunodeficiency syndrome. N Engl J Med. 1992;326:1514–1521. doi: 10.1056/NEJM199206043262302. [DOI] [PubMed] [Google Scholar]
- 13.Fang Z, Doig C, Kenna D T, Smittipat N, Palittapongarnpim P, Watt B, Forbes K J. IS6110-mediated deletions of wild-type chromosomes of Mycobacterium tuberculosis. J Bacteriol. 1999;181:1014–1020. doi: 10.1128/jb.181.3.1014-1020.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frieden T R, Sherman L F, Maw K L, Fujiwara P I, Crawford J T, Nivin B, Sharp V, Hewlett D, Jr, Brudney K, Alland D, Kreisworth B N. A multi-institutional outbreak of highly drug-resistant tuberculosis. Epidemiology and clinical outcomes. JAMA. 1996;276:1229–1235. [PubMed] [Google Scholar]
- 15.Frieden T R, Woodley C L, Crawford J T, Lew D, Dooley S M. The molecular epidemiology of tuberculosis in New York City: the importance of nosocomial transmission and laboratory error. Tuber Lung Dis. 1996;77:407–413. doi: 10.1016/s0962-8479(96)90112-4. [DOI] [PubMed] [Google Scholar]
- 16.Ghanekar K, McBride A, Dellagostin O, Thorne S, Mooney R, McFadden J. Stimulation of transposition of the Mycobacterium tuberculosis insertion sequence IS6110 by exposure to a microaerobic environment. Mol Microbiol. 1999;33:982–993. doi: 10.1046/j.1365-2958.1999.01539.x. [DOI] [PubMed] [Google Scholar]
- 17.Gillespie S H, Kennedy N, Ngowi F I, Fomukong N G, al-Maamary S, Dale J W. Restriction fragment length polymorphism analysis of Mycobacterium tuberculosis isolates from patients with pulmonary tuberculosis in northern Tanzania. Trans R Soc Trop Med Hyg. 1995;89:335–338. doi: 10.1016/0035-9203(95)90571-5. [DOI] [PubMed] [Google Scholar]
- 18.Heersma H F, Kremer K, van Embden J D A. Computer analysis of IS6110 RFLP patterns of Mycobacterium tuberculosis. Methods Mol Biol. 1998;101:395–422. doi: 10.1385/0-89603-471-2:395. [DOI] [PubMed] [Google Scholar]
- 19.Hermans P W, Messadi F, Guebrexabher H, et al. Analysis of the population structure of Mycobacterium tuberculosis in Ethiopia, Tunisia, and the Netherlands: usefulness of DNA typing for global tuberculosis epidemiology. J Infect Dis. 1995;171:1504–1513. doi: 10.1093/infdis/171.6.1504. [DOI] [PubMed] [Google Scholar]
- 20.Horgen L, Sola C, Devallois A, Goh K S, Rastogi N. Follow up of Mycobacterium tuberculosis transmission in the French West Indies by IS6110-DNA fingerprinting and DR-based spoligotyping. FEMS Immunol Med Microbiol. 1998;21:203–212. doi: 10.1111/j.1574-695X.1998.tb01167.x. [DOI] [PubMed] [Google Scholar]
- 21.Jereb J A, Burwen D R, Dooley S W, et al. Nosocomial outbreak of tuberculosis in a renal transplant unit: application of a new technique for restriction fragment length polymorphism analysis of Mycobacterium tuberculosis isolates. J Infect Dis. 1993;168:1219–1224. doi: 10.1093/infdis/168.5.1219. [DOI] [PubMed] [Google Scholar]
- 22.Kremer K, van Soolingen D, Frothingham R, Haas W H, Hermans P W M, Martin C, Palittapongarnpim P, Plikaytis B B, Riley L W, Yakrus M A, Musser J M, van Embden J D A. Comparison of methods based on different molecular epidemiological markers for typing of Mycobacterium tuberculosis complex strains: interlaboratory study of discriminatory power and reproducibility. J Clin Microbiol. 1999;8:2607–2618. doi: 10.1128/jcm.37.8.2607-2618.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lari N, Rindi L, Lami C, Garzelli C. IS6110-based restriction fragment length polymorphism (RFLP) analysis of Mycobacterium tuberculosis H37Rv and H37Ra. Microbiol Pathog. 1999;26:281–286. doi: 10.1006/mpat.1998.0270. [DOI] [PubMed] [Google Scholar]
- 24.McHugh T D, Gillespie S H. Nonrandom association of IS6110 and Mycobacterium tuberculosis: implications for molecular epidemiological studies. J Clin Microbiol. 1998;36:1410–1413. doi: 10.1128/jcm.36.5.1410-1413.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pfyffer G E, Strassle A, Rose N, Wirth R, Brandli O, Shang H. Transmission of tuberculosis in the metropolitan area of Zurich: a 3-year survey based on DNA fingerprinting. Eur Respir J. 1998;11:804–808. doi: 10.1183/09031936.98.11040804. [DOI] [PubMed] [Google Scholar]
- 26.Small P M, Hopewell P C, Singh S P, Paz A, Parsonnet J, Ruston D C, Schecter G F, Daley C L, Schoolnik G K. The epidemiology of tuberculosis in San Francisco. A population-based study using conventional and molecular methods. N Engl J Med. 1994;330:1703–1709. doi: 10.1056/NEJM199406163302402. [DOI] [PubMed] [Google Scholar]
- 27.Van Duin J M, Pijnenburg J E M, van Rijswoud C M, de Haas P E W, Hendriks W D H, van Soolingen D. Investigation of cross contamination in a Mycobacterium tuberculosis laboratory using IS6110 DNA fingerprinting. Int J Tuber Lung Dis. 1998;2:425–429. [PubMed] [Google Scholar]
- 28.Van Embden J D A, Cave M D, Crawford J T, et al. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standard methodology. J Clin Microbiol. 1993;31:406–409. doi: 10.1128/jcm.31.2.406-409.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Van Soolingen D, Borgdorff M W, de Haas P E W, Sebek M M G G, Veen J, Dessens M, Kremer K, Van Embden J D A. Molecular epidemiology in the Netherlands: a nation-wide study from 1993 through 1997. J Infect Dis. 1999;180:726–736. doi: 10.1086/314930. [DOI] [PubMed] [Google Scholar]
- 30.Warren R, Hauman J, Beyers N, Richardson M, Schaaf H S, Donald P, van Helden P. Unexpectedly high strain diversity of Mycobacterium tuberculosis in a high-incidence community. S Afr Med J. 1996;86:45–49. [PubMed] [Google Scholar]
- 31.Yang Z H, de Haas P E W, van Soolingen D, van Embden J D A, Andersen A B. Restriction fragment length polymorphism of Mycobacterium tuberculosis strains isolated from Greenland during 1992: evidence of tuberculosis transmission between Greenland and Denmark. J Clin Microbiol. 1994;32:3018–3025. doi: 10.1128/jcm.32.12.3018-3025.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang Z H, de Haas P E W, Wachmann C H, van Soolingen D, van Embden J D A, Andersen A B. Molecular epidemiology of tuberculosis in Denmark in 1992. J Clin Microbiol. 1995;33:2077–2081. doi: 10.1128/jcm.33.8.2077-2081.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang Z, Barnes P F, Chaves F, Eisenach K D, Weis S E, Bates J H, Cave M D. Diversity of DNA fingerprints of Mycobacterium tuberculosis isolates in the United States. J Clin Microbiol. 1998;36:1003–1007. doi: 10.1128/jcm.36.4.1003-1007.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yeh R W, Hopewell P C, Daley C L. Simultaneous infection with two strains of Mycobacterium tuberculosis identified by restriction fragment length polymorphism analysis. Int J Tuber Lung Dis. 1999;3:537–539. [PubMed] [Google Scholar]