Abstract
Background and Purpose
Mutations in the FIG4 gene have been linked to amyotrophic lateral sclerosis (ALS) type 11 in Caucasian populations. The purpose of this study was to identify FIG4 variants in a cohort of 15 familial ALS (FALS) indexes and 275 sporadic ALS (SALS) patients of Han Chinese origin.
Methods
All 23 exons of FIG4 were sequenced using targeted next-generation sequencing. An extensive literature review was performed to detect genotype-phenotype associations of FIG4 mutations.
Results
No FIG4 variants were identified in the FALS patients. One novel heterozygous missense variant (c.352G>T [p.D118Y]) and one novel heterozygous nonsense variant (c.2158G>T [p.E720X]) in FIG4 were identified in two SALS patients. The p.E720X variant is interpreted as likely pathogenic while the p.D118Y variant is a variant of uncertain significance. The patient carrying the p.E720X mutation developed lower-limb-onset slowly progressive ALS, and survived for 11.5 years. The patient harboring the FIG4 p.D118Y variant also presented with progressive ALS, with the score on the ALS Functional Rating Scale–Revised (ALSFRS-R) decreasing by 0.4 per month. The rate of decrease in the ALSFRS-R scores from symptom onset to diagnosis seemed to be lower in the patients carrying FIG4 variants than the no-FIG4-mutation ALS patients in this study.
Conclusions
Our findings suggest that ALS patients carrying FIG4 mutations are not common in the Chinese population and are more likely to exhibit slow progression.
Keywords: amyotrophic lateral sclerosis, FIG4, genetics
INTRODUCTION
Amyotrophic lateral sclerosis (ALS) is a fatal neurodegenerative disease characterized by the loss of upper motor neurons in the motor cortex and lower motor neurons in the brain stem and spinal cord, which leads to progressive muscular weakness, atrophy, and bulbar palsy. Affected patients typically die due to respiratory failure in 3–5 years after onset. Mutations in more than 20 genes have been linked to ALS.1 Mutations in the FIG4 gene were first linked to ALS type 11 (ALS11) in Caucasian populations in 2009.2 A subsequent study confirmed the association of FIG4 mutations with ALS in Caucasian populations.3 Both studies found that the mean disease duration was longer in patients with FIG4 mutations than in no-FIG4-mutation ALS patients.2,3 Two FIG4 missense variants of uncertain significance in sporadic ALS (SALS) patients of Chinese origin have been reported, but the phenotype was not described.4 Here we report novel variants identified in FIG4 in Chinese SALS patients with slow progression.
METHODS
Study population
The study included 15 familial ALS (FALS) probands and 275 SALS patients who were referred to Fujian Medical University Union Hospital and Henan Provincial People’s Hospital between January 2017 and December 2018. All patients were of Han Chinese origin. A diagnosis of definite, probable, or laboratory-supported probable ALS was established according to the revised El Escorial criteria. The research was performed following the Declaration of Helsinki and with the approval of the ethics boards of Fujian Medical University Union Hospital and Henan Provincial People’s Hospital (2017KY085). All patients had signed an informed-consent document.
Genetic studies
Targeted next-generation sequencing was performed on an Illumina HiSeq sequencer (Illumina, San Diego, CA, USA). The targeted regions were designed to include all 23 exons with intronic 50-bp flanking sites and 3′ and 5′ untranslated regions of FIG4 (NM_014845.6). As a result of sequencing, the mean on-target coverage was 600× with an average percentage of targets covered ≥100× of 99.8%. The identified variants were Sanger sequenced for confirmation. Patients carrying FIG4 variants were also screened for mutations in other genes that are known to be related to ALS (SOD1, FUS, TARDBP, OPTN, VAPB, SPG11, VCP, PFN1, ANG, ALS2, DAO, UBQLN2, SIGMAR1, SETX, DCTN1, SQSTM1, CHCHD10, MATR3, TUBA4A, TBK1, hnRNPA1, hnRNPA2B1, KIF5A, ANXA11, TIA1, CCNF, and NEK1), as well as the presence of the GGGGCC repeat expansions in C9orf72.
The methods have been described in detail previously.5 Variant frequencies were determined in three public sequencing databases: gnomAD (http://gnomad.broadinstitute.org/),6 Exome Aggregation Consortium (ExAC), and the online Chinese Millionome Database (CMDB, https://db.cngb.org/cmdb/). The CMDB contains whole-genome sequencing data from 141,431 unrelated healthy Chinese individuals.7 Variant frequencies were also determined in the Running Gene (China) in-house database that includes whole-exome sequencing data of 4,000 ethnicity-matched healthy controls.
RESULTS
Clinical features of the included ALS patients
The age at disease onset in the included ALS patients was 55.3±11.6 years (mean±SD). The male-to-female ratio was 1.5:1 (173 males, 117 females). The sites of symptom onset were the bulbar region (19.3%), limbs (79.0%), and respiratory muscles (1.7%).
Genetic analysis
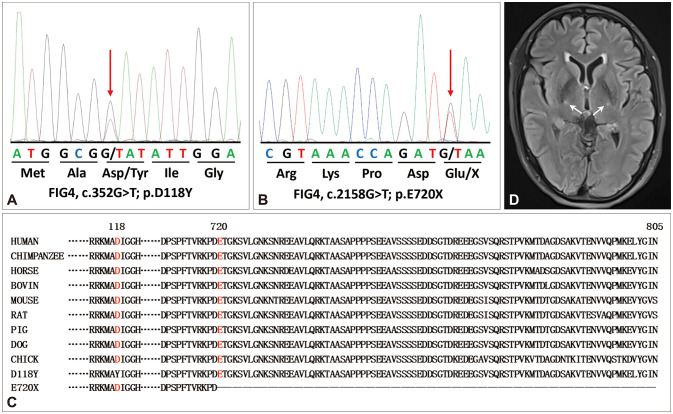
No variants in FIG4 were identified in the FALS probands. One novel missense variant (c.352G>T [p.D118Y]) (Fig. 1A), one novel nonsense variant (c.2158G>T [p.E720X]) (Fig. 1B), and one known frameshift variant (c.2661dupG [p.Q888Afs*34]) (Supplementary Fig. 1 in the online-only Data Supplement) in FIG4 were identified in three SALS patients. All of these variants were heterozygous. No further analysis could be performed because DNA was not available for the parents of the patients with FIG4 variants. None of the three patients harboring FIG4 variants carried a second mutation of other ALS-related genes. The p.D118Y and p.E720X variants are not present in the public gnomAD, ExAC, or CMDB database, or in the 4,000 ethnicity-matched controls. In contrast, the p.Q888Afs*34 variant is present in public databases (genome East Asian=0.0025, ExAC=0.000025). Evolutionary conservation of the FIG4 protein showed that the D118 and E720 residues are highly conserved across species (Fig. 1C). The PolyPhen-2 and Mutation Taster tools predicted that the p.D118Y variant had a deleterious effect, whereas the SIFT tool predicted that it was tolerable.
Fig. 1. Sequencing chromatograms and pathogenicity analysis of the FIG4 variants identified in this study. A and B: Sequencing chromatograms of the FIG4 p.D118Y (A) and p.E720X (B) variants. C: Evolutionary conservation of the FIG4 protein show that the D118 residue and C-terminal are highly conserved across species. D: Brain MRI showed a symmetric hyperintensity on fluid-attenuated inversion recovery sequences in the bilateral posterior limb of the internal capsule (white arrows).
Phenotype of the patients with FIG4 variants
The patient carrying the FIG4 p.E720X variant presented with weakness of the left foot at the age of 62 years. The muscle weakness progressed gradually to involve the left leg and left hand 1 year later. He developed weakness of the right lower limb and right hand at 3 years after the onset. He felt unsteady when walking and had difficulty climbing stairs and handwriting. Fasciculations of the limbs were noticed. He developed dysarthria and dysphagia at 9 years after onset, which had resulted in obvious weight loss. One year later he began to experience shortness of breath at night and so was referred to our hospital. A neurological examination revealed decreased muscle strength in both upper limbs and in the right lower limb (Medical Research Council Scale for Muscle Strength [MRC] 0/5 in the muscles of both hands, MRC 3/5 in the muscles of both arms, MRC 2/5 in the left foot, and MRC 3/5 in the left leg and right lower limb). Obvious muscle atrophy was observed in all extremities, which was more severe distally. Deep tendon reflexes were decreased in all four extremities. The Babinski sign was absent on both sides, while the palm-chin reflex was present on the right side. Neither sensory disturbance nor cognitive impairment was observed. Motor nerve conduction studies revealed decreased amplitudes of compound muscle action potentials in the nerves of all limbs. The findings of sensory nerve conduction studies were normal. Electromyography demonstrated both acute and chronic neurogenic changes in proximal and distal muscles of the four limbs, and in the sternocleidomastoid and thoracic paraspinal muscles. Brain MRI revealed a symmetric hyperintensity on fluid-attenuated inversion recovery sequences in the bilateral posterior limb of the internal capsule (Fig. 1D). The ALS Functional Rating Scale–Revised (ALSFRS-R) score was 21/48, with an estimated decrease in the score of 0.2 per month since symptom onset. The Mini-Mental State Examination (MMSE) score was 29/30. The patient refused to receive noninvasive ventilation and died 11.5 years after symptom onset due to respiratory failure. The patient’s parents had died in their 80s and had not developed muscle weakness or dementia.
The patient carrying the FIG4 p.D118Y variant presented with weakness of the right thumb and complained of difficulty in turning a key and clicking the fingers at the age of 58 years. He developed weakness and atrophy in his right hand 6 months later. The symptoms progressed gradually to involve the right arm and left hand at 1 year after onset. He presented with weakness of the right lower limb at 18 months after onset. He felt tired after walking 500 meters, and could only climb stairs slowly. A neurological examination showed intact cranial nerves. Obvious atrophy of upper limbs was observed. The muscle strength was MRC 3/5 in both hands, MRC 4/5 in both arms, MRC 4/5 in the right lower limb, and MRC 5/5 in the left lower limb. Deep tendon reflexes were brisk in the upper limbs and normal in the lower limbs. No sensory disturbance or cognitive impairment was detected. Needle electromyography showed both active and chronic denervation in the cervical, thoracic, and lumbosacral regions. Brain MRI was unremarkable. The ALSFRS-R score was 40/48, with an estimated rate of decrease in the score of 0.4 per month since symptom onset. At the last follow-up at 30 months after disease onset, the patient could take care of himself in daily life and walk independently, but had difficulty climbing stairs, with no bulbar symptoms. The ALSFRS-R score was 37/48, with an estimated rate of decrease in the score of 0.4 per month since symptom onset and 0.25 per month since the first visit. The patient’s mother was healthy and alive, and his father died at the age of 75 years due to cerebral hemorrhage.
The female harboring the FIG4 p.Q888Afs*34 variant demonstrated isolated bulbar ALS when she was 42 years old. Her detailed clinical features are presented in the Supplementary Material (in the online-only Data Supplement).
DISCUSSION
This study screened for FIG4 variants using targeted next-generation sequencing in a cohort comprising 15 unrelated FALS probands and 275 SALS patients of Chinese origin. One novel missense variant (p.D118Y), one novel nonsense variant (p.E720X), and one known variant (p.Q888Afs*34) were identified in three SALS patients.
As per the guidelines of the American College of Medical Genetics and Genomics for interpreting sequence variants,8 the p.E720X variant is interpreted as likely pathogenic since it leads to a null allele and possibly a deleterious effect (PVS1); it is not present in the control databases (gnomAD, ExAC, or CMBD) (PM2) or in the in-house 4,000 ethnicity-matched healthy controls. The p.D118Y variant is one of uncertain significance since it is not present in the control databases (PM2). However, further functional experiments are warranted to determine the pathogenicity of both FIG4 variants. The p.Q888Afs*34 variant is present in the gnomAD and ExAC population databases, and so it is likely to be a benign variant.
Nine missense and null mutations in FIG4 were first detected by Chow et al.2: 3 out of 109 FALS patients and 6 out of 364 SALS cases in a Caucasian population. The frequency of FIG4 mutations was 2.8% in FALS and 1.6% in SALS in the present study. Osmanovic et al.3 found 6 missense and frameshift mutations in 6 out of 200 SALS patients in a European population, corresponding to a mutation frequency of 3.0%. The frequency of FIG4 mutations in our study was 0.4% (1/275) in Chinese SALS patients, which is lower than the mutation frequencies found in those previous studies involving Caucasian populations.2,3
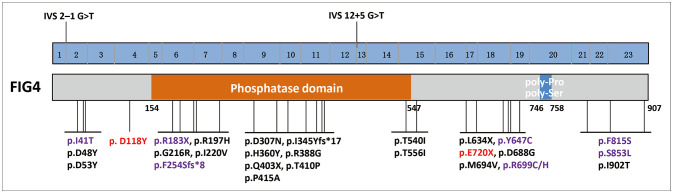
FIG4 encodes a phosphoinositide 5-phosphatase regulating PI(3,5)P2, which is an intracellular signaling lipid that plays a key role in endosomal vesicle trafficking. FIG4 is a multidomain protein with a protein interaction domain at the N-terminal, a SAC phosphatase domain in the central region, and poly-Pro and poly-Ser domains at the C-terminal.9 To date, 28 FIG4 mutations have been identified in ALS patients,2,3,10,11,12,13,14,15,16,17,18 all of which are heterozygous: 21 missense mutations, 3 nonsense mutations, 2 frameshift mutations, and 2 splicing mutations. Only five patients with FIG4 mutations had a definite family history of ALS and demonstrated an autosomal dominant hereditary pattern. Nearly one-half of ALS-associated FIG4 mutations affect the SAC phosphatase domain (Fig. 2). The p.E720X mutation is located at the C-terminal of FIG4, while the p.D118Y variant lies at the N-terminal. Further functional experiments are needed to determine the pathogenic mechanisms of mutations in different domains of FIG4.
Fig. 2. Schematic graph of the FIG4 protein and overview of the FIG4 mutations linked to amyotrophic lateral sclerosis (ALS). Variants identified in our study are marked in red. Mutations identified in both ALS and Charcot-Marie-Tooth disease patients are marked in purple.
Twenty-three FIG4-mutated ALS patients with detailed clinical features were identified. The clinical characteristics of the ALS cases with FIG4 mutations are summarized in Table 1. The mean age at onset was 55.2 years (range 25–81 years), and the sex ratio was approximately 1.0. Bulbar onset was reported in 39% (n=9) of the 23 patients carrying FIG4 mutations. Patients with FIG4 mutations presented with a wide range of phenotypes, including classical ALS, upper-motor-neuron-predominant ALS, primary lateral sclerosis, flail arm syndrome, and combined ALS and behavioral variant frontotemporal dementia.
Table 1. Summary of clinical features of ALS patients with FIG4 mutations.
| Exon | Nucleoid change | Amino acid change | Population | Sex | AAO (yr) | SOO | Phenotype | Disease duration (yr) | Familial history | Reference |
|---|---|---|---|---|---|---|---|---|---|---|
| 2 | c.67-1G>T | p.R23fs*30 | Caucasian | Male | 77 | UL | ALS | 1.3 | Yes | Chow et al.2 |
| 2 | c.122T>C | p.I41T | Italian | Female | 43 | Spinal | PLS | >12.25 (alive) | No | Osmanovic et al.3 |
| 2 | c.143A>G | p.D48G | Caucasian | Female | 29 | LL | PLS | NA | No | Chow et al.2 |
| 2 | c.157G>T | p.D53Y | Caucasian | Female | 56 | Bulbar | ALS | 2.6 | Yes | Chow et al.2 |
| 6 | c.547C>T | p.R183X | Caucasian | Male | 62 | Bulbar | ALS | 8.9 | No | Chow et al.2 |
| 6 | c.590G>A | p.R197H | Caucasian | Male | 60 | Cognition | ALS, bvFTD | >1 (alive) | No | Dols-Icardo et al.14 |
| 7 | c.646G>A | p.G216R | Caucasian | NA | NA | Spinal | PMA, sensory neuropathy | NA | No | Pensato et al.18 |
| 7 | c.759delG | p.F254Sfs*8 | German | Male | 40 | Spinal | ALS | >2.67 (alive) | Yes | Osmanovic et al.3 |
| 9 | c.919G>A | p.D307N | German | Male | 78 | Bulbar | UMN-ALS, MSAN | >5.25 (alive) | No | Osmanovic et al.3 |
| 9 | NA | p.I345Yfs*17‡ | Italian | Female | 65 | Bulbar | ALS | 3 | Yes | Lamp et al.15 |
| 10 | NA | p.H360Y† | Italian | Male | 81 | Bulbar | ALS, dementia | 3 | No | Lamp et al.15 |
| 11 | c.1162A>G | p.R388G | Caucasian | Male | 42 | LL | PLS | >29 (alive) | No | Chow et al.2 |
| 11 | c.1207C>T | p.Q403X | Caucasian | Female | 60 | Bulbar | ALS | 25 | No | Chow et al.2 |
| 11 | c.1243C>G | p.P415A | Caucasian | Female | 29 | Spinal | UMN-ALS | >10 (alive) | No | Pensato et al.18 |
| 12 | c.1386+5G>T | p.S424_K462 del insR | Caucasian | Female | 57 | UL | ALS | >2 (alive) | No | Chow et al.2 |
| 15 | c.1619C>T | p.T540I | German | Female | 72 | Bulbar | PLS | >3.25 (alive) | No | Osmanovic et al.3 |
| 15 2 |
c.1667C>T c.122T>C |
p.T556I p.I41T |
Caucasian | Female | 25 | LL | ALS, mental retardation | 1.5 | NA | Bertolin et al.13 |
| 17 | c.1940A>G | p.Y647C | Caucasian | Female | 65 | Bulbar | ALS | >2 (alive) | No | Chow et al.2 |
| 17 | c.1940A>G | p.Y647C | German | Male | 66 | Spinal | FAS, MSAN | 5.25§ | No | Osmanovic et al.3 |
| 19 | c.2158G>T | p.E720X | Chinese | Male | 62 | LL | LMN-ALS | 11.5 | No | This study |
| 23 | c.2558C>T | p.S853L | German | Female | 48 | Spinal | ALS, mild MSAN | 0.92 | No | Osmanovic et al.3 |
| 23 | c.2705T>C | p.I902T | Caucasian | Male | 55 | Bulbar | ALS | 1.7 | Yes | Chow et al.2 |
†Co-occurrence of SETX p.L158V variant; ‡Co-occurrence of C9orf72 repeat expansions, §Died of traumatic subdural hemorrhage.
AAO, age at onset; ALS, amyotrophic lateral sclerosis; bvFTD, behavioral variant frontotemporal dementia; FAS, flail arm syndrome; LL, lower limb; LMN-ALS, lower-motor-neuron-predominant ALS; MSAN, motor-sensory axonal neuropathy; NA, not available; PLS, primary lateral sclerosis; PMA, primary muscular atrophy; SOO, site of onset; UL, upper limb; UMN-ALS, upper-motor-neuron-predominant ALS.
FIG4-mutation ALS patients exhibit heterogeneity in disease progression. Our patient carrying the FIG4 p.E720X mutation had a phenotype of slowly progressive lower-motor-neuron-predominant ALS (rate of decrease in the ALSFRS-R score of 0.2 per month), and survived for 11.5 years. The patient with the FIG4 p.D118Y variant also presented with slowly progressive ALS (rate of decrease in the ALSFRS-R score of 0.4 per month). The rate of decrease in the ALSFRS-R scores from symptom onset to diagnosis seemed to be lower in the patients carrying FIG4 variants (mean of 0.30 per month) than in the no-FIG4-mutation ALS patients (0.82±1.02 per month) in this study. These findings are consistent with Chow et al.2 finding a long mean disease duration of 9.1 years in eight ALS patients with FIG4 mutations. The reported FIG4-mutation ALS patients included five cases with slow progression and a long duration (8.9-29 years), which included three cases with a phenotype of primary lateral sclerosis or upper-motor-neuron-dominant ALS.2,3,18 Seven ALS patients with FIG4 mutations demonstrated rapid progression and a survival duration of shorter than 3 years (Table 1). Among these patients, one harbored biallelic mutations of FIG4 (p.T556I and p.I41T),13 another had co-occurrence of the SETX p.L158V variant, and the other third had co-occurrence of C9orf72 repeat expansions.15 Additional extramotor symptoms were reported in six ALS patients carrying FIG4 mutations: one presented with mental retardation,13 one had dementia,15 one developed behavioral variant frontotemporal dementia,14 and the other three cases demonstrated motor-sensory axonal neuropathy or sensory axonal neuropathy.3,18 The patient with ALS plus dementia carried the FIG4 p.H360Y and SETX p.L158V variants,15 while the one with ALS and mental retardation harbored the FIG4 p.T556I and p.I41T mutations.13 These findings suggest that the disease phenotype and severity of FIG4-mutation ALS patients can be influenced by functional levels of the FIG4 protein or other genetic factors. Extensive genetic studies involving different populations to identify more ALS patients with FIG4 mutations may elucidate the genotype-phenotype relationships.
Biallelic mutations of FIG4 have been reported to cause Charcot-Marie-Tooth disease (CMT) type 4J (CMT4J), which is an autosomal recessive hereditary motor and sensory neuropathy where early onset and aggressive disease progression are not uncommon. The combination of a heterozygous missense p.I41T mutation with a protein truncation mutation is characteristic of patients with recessive CMT4J.19,20 Several frameshift and nonsense mutations (p.T556Nfs*20,19 p.H599Ifs*24,21 p.S730Kfs*3,22 p.E767Gfs*17, and p.Q796X23) at the C-terminal of FIG4 have been reported in patients with CMT4J. CMT4J patients with frameshift and nonsense mutations at the C-terminal of FIG4 tend to have a later onset and less-severe disability.19,21,22 A key factor linked to the slow progression of the present patient with the FIG4 p.E720X mutation may be the p.E720X protein truncation mutation not involving the phosphatase domain and maintaining some degree of function of the FIG4 protein. One particularly interesting observation was that one patient presenting with flail arm syndrome and motor-sensory axonal neuropathy harbored the FIG4 p.Y647C mutation,3 which was also identified in a patient with CMT4J.23 An additional seven FIG4 mutations identified in ALS patients have also been reported in patients with CMT (Fig. 2).19,20,23 These findings confirm that ALS11 and CMT4J belong to the FIG4-related disease spectrum and share some common symptoms.
The present findings suggest that ALS patients with FIG4 mutations are not common in the Chinese population and are more likely to exhibit slow progression. More independent large-sample studies involving different populations are warranted to confirm the associations between FIG4 variants and ALS.
Acknowledgements
The authors thank the patients and their families for their cooperation in this study. We are also grateful to Dr. Zhenghua Cao from Running Gene Inc. for the help in the interpretation of the variants.
Footnotes
- Conceptualization: Zhang-Yu Zou, Chang-Yun Liu.
- Data curation: Zhang-Yu Zou, Chang-Yun Liu.
- Formal analysis: Ji-Lan Lin, Shu-Yan Feng.
- Funding acquisition: Zhang-Yu Zou.
- Investigation: Zhang-Yu Zou, Chang-Yun Liu.
- Methodology: Zhang-Yu Zou, Chang-Yun Liu.
- Project administration: Zhang-Yu Zou.
- Resources: Chang-Yun Liu.
- Software: Ji-Lan Lin.
- Supervision: Zhang-Yu Zou.
- Validation: Chang-Yun Liu.
- Visualization: Chun-Hui Che, Hua-Pin Huang.
- Writing—original draft: Zhang-Yu Zou, Chang-Yun Liu.
- Writing—review & editing: all authors.
Conflicts of Interest: The authors have no potential conflicts of interest to disclose.
Funding Statement: This study was supported by grants from the National Natural Science Foundation of China (grant number 81671271, 81974199), Joint Funds for the innovation of science and Technology, Fujian province (grant number 2017Y91010015).
Availability of Data and Material
The datasets generated or analyzed during the study are available from the corresponding author on reasonable request.
Supplementary Materials
The online-only Data Supplement is available with this article at https://doi.org/10.3988/jcn.2022.18.1.41.
Sequencing chromatograms of the FIG4 p.Q888Afs*34 variant.
References
- 1.Brown RH, Al-Chalabi A. Amyotrophic lateral sclerosis. N Engl J Med. 2017;377:162–172. doi: 10.1056/NEJMra1603471. [DOI] [PubMed] [Google Scholar]
- 2.Chow CY, Landers JE, Bergren SK, Sapp PC, Grant AE, Jones JM, et al. Deleterious variants of FIG4, a phosphoinositide phosphatase, in patients with ALS. Am J Hum Genet. 2009;84:85–88. doi: 10.1016/j.ajhg.2008.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Osmanovic A, Rangnau I, Kosfeld A, Abdulla S, Janssen C, Auber B, et al. FIG4 variants in central European patients with amyotrophic lateral sclerosis: a whole-exome and targeted sequencing study. Eur J Hum Genet. 2017;25:324–331. doi: 10.1038/ejhg.2016.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang H, Cai W, Chen S, Liang J, Wang Z, Ren Y, et al. Screening for possible oligogenic pathogenesis in Chinese sporadic ALS patients. Amyotroph Lateral Scler Frontotemporal Degener. 2018;19:419–425. doi: 10.1080/21678421.2018.1432659. [DOI] [PubMed] [Google Scholar]
- 5.Feng SM, Che CH, Feng SY, Liu CY, Li LY, Li YX, et al. Novel mutation in optineurin causing aggressive ALS+/-frontotemporal dementia. Ann Clin Transl Neurol. 2019;6:2377–2383. doi: 10.1002/acn3.50928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu S, Huang S, Chen F, Zhao L, Yuan Y, Francis SS, et al. Genomic analyses from non-invasive prenatal testing reveal genetic associations, patterns of viral infections, and Chinese population history. Cell. 2018;175:347–359.e14. doi: 10.1016/j.cell.2018.08.016. [DOI] [PubMed] [Google Scholar]
- 8.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baulac S, Lenk GM, Dufresnois B, Ouled Amar Bencheikh B, Couarch P, Renard J, et al. Role of the phosphoinositide phosphatase FIG4 gene in familial epilepsy with polymicrogyria. Neurology. 2014;82:1068–1075. doi: 10.1212/WNL.0000000000000241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cady J, Allred P, Bali T, Pestronk A, Goate A, Miller TM, et al. Amyotrophic lateral sclerosis onset is influenced by the burden of rare variants in known amyotrophic lateral sclerosis genes. Ann Neurol. 2015;77:100–113. doi: 10.1002/ana.24306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gibson SB, Downie JM, Tsetsou S, Feusier JE, Figueroa KP, Bromberg MB, et al. The evolving genetic risk for sporadic ALS. Neurology. 2017;89:226–233. doi: 10.1212/WNL.0000000000004109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krüger S, Battke F, Sprecher A, Munz M, Synofzik M, Schöls L, et al. Rare variants in neurodegeneration associated genes revealed by targeted panel sequencing in a German ALS cohort. Front Mol Neurosci. 2016;9:92. doi: 10.3389/fnmol.2016.00092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bertolin C, Querin G, Bozzoni V, Martinelli I, De Bortoli M, Rampazzo A, et al. New FIG4 gene mutations causing aggressive ALS. Eur J Neurol. 2018;25:e41–e42. doi: 10.1111/ene.13559. [DOI] [PubMed] [Google Scholar]
- 14.Dols-Icardo O, García-Redondo A, Rojas-García R, Borrego-Hernández D, Illán-Gala I, Muñoz-Blanco JL, et al. Analysis of known amyotrophic lateral sclerosis and frontotemporal dementia genes reveals a substantial genetic burden in patients manifesting both diseases not carrying the C9orf72 expansion mutation. J Neurol Neurosurg Psychiatry. 2018;89:162–168. doi: 10.1136/jnnp-2017-316820. [DOI] [PubMed] [Google Scholar]
- 15.Lamp M, Origone P, Geroldi A, Verdiani S, Gotta F, Caponnetto C, et al. Twenty years of molecular analyses in amyotrophic lateral sclerosis: genetic landscape of Italian patients. Neurobiol Aging. 2018;66:179.e5–179.e16. doi: 10.1016/j.neurobiolaging.2018.01.013. [DOI] [PubMed] [Google Scholar]
- 16.Morgan S, Shatunov A, Sproviero W, Jones AR, Shoai M, Hughes D, et al. A comprehensive analysis of rare genetic variation in amyotrophic lateral sclerosis in the UK. Brain. 2017;140:1611–1618. doi: 10.1093/brain/awx082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Müller K, Brenner D, Weydt P, Meyer T, Grehl T, Petri S, et al. Comprehensive analysis of the mutation spectrum in 301 German ALS families. J Neurol Neurosurg Psychiatry. 2018;89:817–827. doi: 10.1136/jnnp-2017-317611. [DOI] [PubMed] [Google Scholar]
- 18.Pensato V, Magri S, Bella ED, Tannorella P, Bersano E, Sorarù G, et al. Sorting rare ALS genetic variants by targeted re-sequencing panel in Italian patients: OPTN, VCP, and SQSTM1 variants account for 3% of rare genetic forms. J Clin Med. 2020;9:412. doi: 10.3390/jcm9020412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nicholson G, Lenk GM, Reddel SW, Grant AE, Towne CF, Ferguson CJ, et al. Distinctive genetic and clinical features of CMT4J: a severe neuropathy caused by mutations in the PI(3,5)P2 phosphatase FIG4. Brain. 2011;134:1959–1971. doi: 10.1093/brain/awr148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chow CY, Zhang Y, Dowling JJ, Jin N, Adamska M, Shiga K, et al. Mutation of FIG4 causes neurodegeneration in the pale tremor mouse and patients with CMT4J. Nature. 2007;448:68–72. doi: 10.1038/nature05876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Michaelidou K, Tsiverdis I, Erimaki S, Papadimitriou D, Amoiridis G, Papadimitriou A, et al. Whole exome sequencing establishes diagnosis of Charcot-Marie-Tooth 4J, 1C, and X1 subtypes. Mol Genet Genomic Med. 2020;8:e1141. doi: 10.1002/mgg3.1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zimmermann M, Schuster S, Boesch S, Korenke GC, Mohr J, Reichbauer J, et al. FIG4 mutations leading to parkinsonism and a phenotypical continuum between CMT4J and Yunis Varón syndrome. Parkinsonism Relat Disord. 2020;74:6–11. doi: 10.1016/j.parkreldis.2020.03.021. [DOI] [PubMed] [Google Scholar]
- 23.DiVincenzo C, Elzinga CD, Medeiros AC, Karbassi I, Jones JR, Evans MC, et al. The allelic spectrum of Charcot-Marie-Tooth disease in over 17,000 individuals with neuropathy. Mol Genet Genomic Med. 2014;2:522–529. doi: 10.1002/mgg3.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sequencing chromatograms of the FIG4 p.Q888Afs*34 variant.
Data Availability Statement
The datasets generated or analyzed during the study are available from the corresponding author on reasonable request.