Abstract
Nipah virus (NiV) is an emerging and deadly zoonotic paramyxovirus that is responsible for periodic epidemics of acute respiratory illness and encephalitis in humans. Previous studies have shown that the NiV V protein antagonizes host antiviral immunity, but the molecular mechanism is incompletely understood. To address this gap, we biochemically characterized NiV V binding to the host pattern recognition receptor MDA5. We find that the C-terminal domain of NiV V (VCTD) is sufficient to bind the MDA5SF2 domain when recombinantly co-expressed in bacteria. Analysis by hydrogen–deuterium exchange mass spectrometry (HDX-MS) studies revealed that NiV VCTD is conformationally dynamic, and binding to MDA5 reduces the dynamics of VCTD. Our results also suggest that the β-sheet region in between the MDA5 Hel1, Hel2, and Hel2i domains exhibits rapid HDX. Upon VCTD binding, these β-sheet and adjacent residues show significant protection. Collectively, our findings suggest that NiV V binding disrupts the helicase fold and dynamics of MDA5 to antagonize host antiviral immunity.
Keywords: Nipah virus, V protein, MDA5, IFN antagonist, hydrogen−deuterium exchange mass spectrometry
Nipah virus (NiV) is an emerging zoonotic pathogen that causes severe respiratory disease and encephalitis associated with human fatalities.1−5 NiV was first identified in Malaysia and Singapore as the source of an outbreak of disease among pigs and subsequently in humans.6,7 Since then, recurring outbreaks have been documented in other parts of Asia, primarily in Bangladesh and India, with case fatality rates as high as 75%. Isolates from Malaysia and Bangladesh are genetically distinct. While there is high nucleotide homology (>91%) between the Malaysia and Bangladesh strains, the sequence heterogeneity in NiV genes or proteins may account for the differences in the transmission mode and rates.8,9 Currently, there are no approved, effective prophylactics or therapeutics available for NiV infection.
NiV, along with Hendra virus (HeV), are henipaviruses within the paramyxovirus family of the nonsegmented, negative-strand RNA viruses. These also include parainfluenza virus 5 (PIV5), measles virus (MV), mumps virus (MuV), and Sendai virus (SeV).10−13 The NiV genome encodes for four nonstructural proteins (P, V, W, and C) that are generated from the P gene and that function to inhibit host innate immune responses. V and W proteins are produced through RNA editing during transcription, and the C protein is expressed through leaky scanning and an alternative initiation codon site during ribosomal translation.2,10,14−17 Thus, P, V, and W proteins share a common N-terminal domain (NTD), but each has distinct C-terminal domains (CTDs). NiV VCTD also contains a conserved histidine and several cysteine residues, which likely form two zinc finger (ZnF) motifs based upon sequence homology to PIV5 and MV VCTDs.18−22
Like other paramyxoviruses, NiV V proteins function as interferon (IFN) antagonists to inhibit host antiviral signaling. Earlier studies showed that the NiV V protein is a major determinant of pathogenesis.23−25 Recombinant Nipah virus Malaysia with V protein knockout (rNiVM-VKO) is severely attenuated compared to WT virus or virus with W protein knockout due to reduced inhibition of the innate immune response; this allows for a potent neutralizing antibody response and viral clearance.23 The lack of serious disease in animals infected with rNiVM-VKO points to the V protein having a prominent and early role in modulating immune responses and controlling disease progression. NiV V targets the host cytoplasmic pattern-recognition receptor (PRR) melanoma differentiation-associated protein 5 (MDA5) that detects viral RNAs and triggers IFN production and antiviral gene expression to control viral infections.26−34 MDA5 contains a superfamily 2 (SF2) helicase domain preceded by two tandem N-terminal caspase activation and recruitment domains (CARDs) that are involved in autoregulatory activities and a CTD that binds double-stranded RNA (dsRNA). Previous studies showed that V proteins of several paramyxoviruses, including NiV, PIV5, HeV, SeV, and MuV, all bind MDA5 and not RIG-I.33−35 In addition, NiV V targets the STAT1 transcription factor. NiV P/VNTD binds to STAT1 and prevents its tyrosine phosphorylation and activation,14,36−38 whereas NiV WCTD contains a nuclear localization sequence that allows it to also sequester STAT1 in the nucleus and prevent transcriptional activation of IFN-stimulated genes.38−41 Although these studies show that NiV can target MDA5 and STAT1, how NiV proteins mediate these functions remains incompletely defined. Furthermore, the amino acid sequence encoded by NiV P/V/WNTD is different from other paramyxovirus P proteins, suggesting that NiV may use a distinct mechanism to antagonize IFN responses.
Here, we employed a combined approach to validate and to characterize biochemically the interaction between NiV V and MDA5. Co-immunoprecipitation (co-IP) and in vitro pulldown assays demonstrated direct binding between NiV V and MDA5 proteins. Further analysis defined the regions NiV VCTD and the MDA5 SF2 (MDA5SF2) domain as sufficient for interactions. Hydrogen–deuterium exchange mass spectrometry (HDX-MS) on NiV VCTD revealed a dynamic protein that becomes protected from HDX upon binding to the MDA5SF2 domain. HDX-MS studies on MDA5SF2 identified a highly dynamic β-sheet region that exposes a potential site for VCTD binding. Additional HDX-MS studies on the NiV VCTD/MDA5SF2 complex suggest that NiV VCTD binds to surfaces adjacent to this dynamic region within the SF2 domain. Altogether, our results support that NiV V binding disrupts the MDA5 fold and dynamics to antagonize host antiviral immunity.
Results
NiV VCTD Binds to MDA5SF2
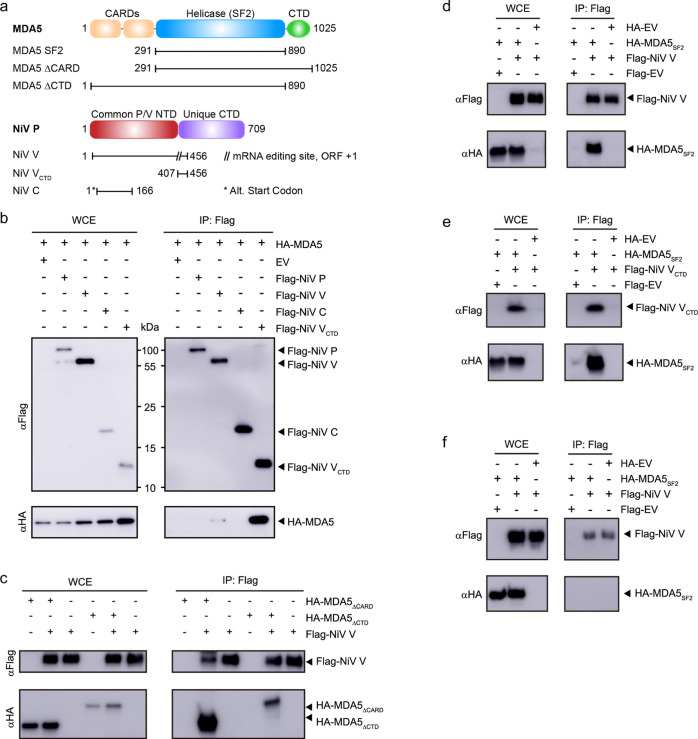
To further characterize the interaction between NiV V and MDA5, we first co-transfected plasmids encoding HA-tagged MDA5 and Flag-tagged P, V, or C proteins from the Malaysia strain (accession AAK50551) into HEK293T cells. We used the Malaysia strain sequence, since this was the first Nipah virus identified, even though the Bangladesh strain has been shown to be more pathogenic. Flag co-IP revealed that the NiV V protein immunoprecipitates with full-length MDA5 (Figures 1a,b and S1a), as shown previously.18,33−35,42−47 Transfections with the NiV V CTD (VCTD; encoding residues G407–G456) displayed stronger co-IP of full-length MDA5 relative to full-length V. Previous studies on full-length V revealed that the V protein alone is largely intrinsically disordered with some residual secondary structure and that regions in V can undergo a structural transition upon interactions with a binding partner.48 Our results here raise the possibility that other regions in the full-length V protein may prevent the VCTD from binding MDA5 through interactions with other host factors during co-IP that can restrict the conformation of V or the accessibility of the VCTD. While this needs further testing, the isolated VCTD is sufficient and strongly binds toMDA5, consistent with earlier observations.33−35,43
Figure 1.
NiV VCTD protein binds the MDA5SF2 domain. (a) Domain architecture and constructs of NiV P/V/C and MDA5 proteins. (b) co-IP of NiV P gene-encoded proteins with MDA5. NiV P, V, C, VCTD, or empty vector (EV) plasmids encoding a fusion N-terminal FLAG epitope tag were co-transfected with the human MDA5 or EV plasmid encoding a fusion N-terminal HA epitope tag. HEK293T cells were harvested at 48 h after co-transfection. Cells were lysed, and the resultant lysate was clarified as a whole cell extract (WCE). Anti-FLAG M2 beads were incubated with each WCE and washed before elution with the FLAG peptide for Western blot. (c) NiV V co-immunoprecipitates with MDA5 truncations. (d) NiV V co-immunoprecipitates with the MDA5SF2 domain. (e) NiV VCTD co-immunoprecipitates with the MDA5SF2 domain. (f) NiV V cannot pull-down MDA5SF2 when expressed separately, and lysates were mixed together prior to co-IP. Images show representative experimental data of three biological repeats.
To define the minimal region in MDA5 that interacts with the NiV VCTD, we generated a series of MDA5 truncations that were co-transfected with NiV VCTD and subjected to co-IP studies. Our results indicate that both MDA5ΔCARD (encoding residues 291–1025) and MDA5ΔCTD (encoding residues 1–890) co-immunoprecipitates with NiV V, suggesting that the CARDs and the C-terminal RNA binding domain are not required for the NiV V interaction (Figures 1c and S1b). We next tested binding to the MDA5SF2 domain (encoding residues 291–890) that contains the DECH helicase and pincer region. We found that MDA5SF2 co-immunoprecipitates with both NiV V full length (Figures 1d and S1c) and to its isolated VCTD (Figures 1e and S1c).
To further validate the interaction between NiV VCTD and the MDA5SF2 domain and to show direct binding, we next transfected the MDA5SF2 and NiV V constructs into separate pools of HEK293T cells, mixed the WCEs together, and performed the co-IP. However, we found that NiV V does not co-IP with MDA5SF2 under these conditions (Figures 1f and S1d). We next tested if recombinant maltose binding protein (MBP)-tagged NiV VCTD and the MDA5SF2 domain expressed separately in Escherichia coli bind in in vitro pulldown assays. When we mixed lysates containing each protein together prior to pulldown, we observed no binding (Figure 2a), consistent with our observations in HEK293T cells. When we performed pulldown assays using individually purified proteins, we also observed no detectable binding (Figure 2b). Together, these results suggest that the interaction between NiV VCTD and the MDA5SF2 domain requires additional host factors that may be present during co-IP or requires co-translational assembly. We next tested co-expression of MBP–NiV VCTD with the MDA5SF2 domain in bacteria and found that MBP-tagged NiV VCTD binds directly to the MDA5SF2 domain by pulldown assay (Figure 2c, left panel), eliminating the possibility that other factors are involved. Because the NiV VCTD contains highly conserved cysteine-rich domains that are predicted to form two Zn finger motifs,20,21,49 analogous to that observed in the PIV5 crystal structures18,50 (Figure S1e), we also performed in vitro pulldown assays in the presence of ethylenediaminetetraacetic acid (EDTA). We find that addition of EDTA had no impact on binding (Figure 2c, right panel), suggesting the formation of a stable complex. Together, our results indicate that simultaneous expression of the MDA5SF2 domain with NiV VCTD may be necessary for proper folding and stability.
Figure 2.
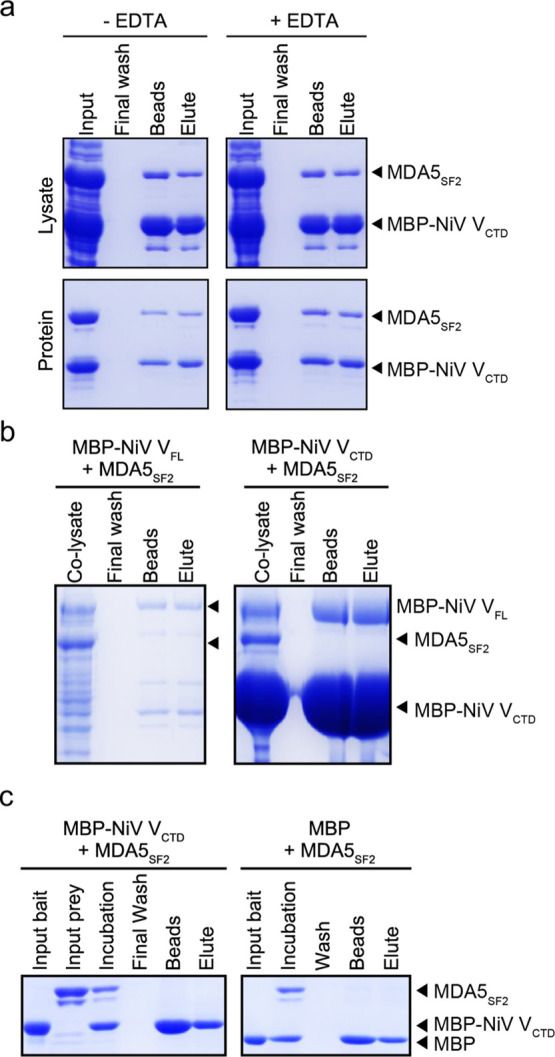
Co-expression of NiV VCTD with MDA5SF2 produces a soluble protein complex. (a) NiV VCTD co-expressed with MDA5SF2 showed direct interactions by in vitro pulldown assay. E. coli cells co-expressing both MBP–NiV VCTD and MDA5SF2 were lysed and clarified prior to a MBP pulldown assay on amylose resin in the absence (left) and presence (right) of 5 mM EDTA. (b) Co-lysis of bacterial cells containing NiV VCTD and MDA5SF2 that do not pull-down each other. E. coli cells expressing NiV VCTD were mixed with cells expressing MDA5SF2 prior to homogenization. The resultant supernatant after clarification of the co-lysates was applied to amylose resin and washed as in (a). (c) Individually purified NiV VCTD in E. coli cells does not pull-down MDA5SF2. Amylose beads were incubated with either the MBP tag alone or with the MBP–NiV VCTD protein as bait mixed with a separately purified MDA5SF2 domain as prey. After stringent washing, proteins bound on the resin were eluted with buffer containing 15 mM maltose.
MDA5SF2 Can Modulate the Dynamics of the NiV VCTD
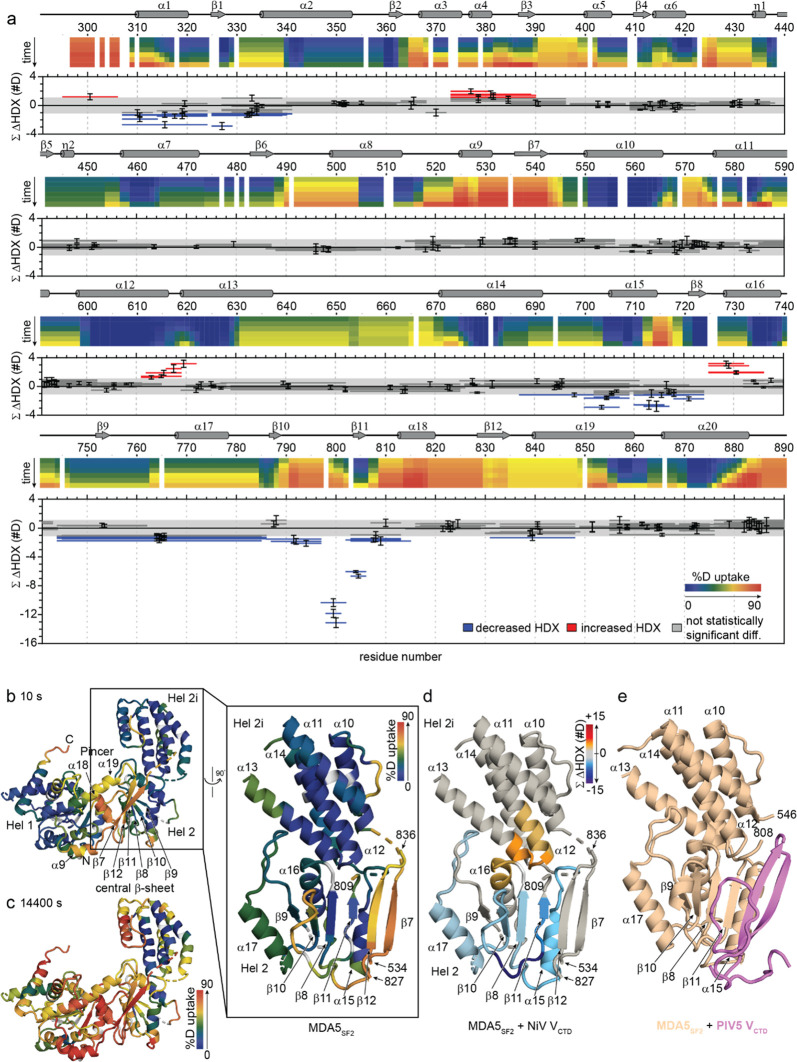
Because the structures for NiV V or VCTD are not currently known, we used HDX-MS to structurally characterize the NiV VCTD. HDX-MS reports on changes in solvent accessibility or hydrogen bonding of backbone amide groups; thus, HDX is sensitive to protein dynamics, conformational changes, and binding sites.51,52 Peptide digestion and bottom-up analysis of the isolated MBP-tagged NiV VCTD produced 24 unique peptides, reporting on 84% of the construct (Figure S2a). The most N-terminal residue of every peptide is prone to fast back-exchange and thus is not considered in the interpretation.53 More N-terminal residues show very low HDX at short incubation times (residues 405–434) (Figure 3a). The C-terminal end of VCTD (residues 433–454) shows a high degree of HDX, even after short HDX incubation times, indicating a more disordered structure.
Figure 3.
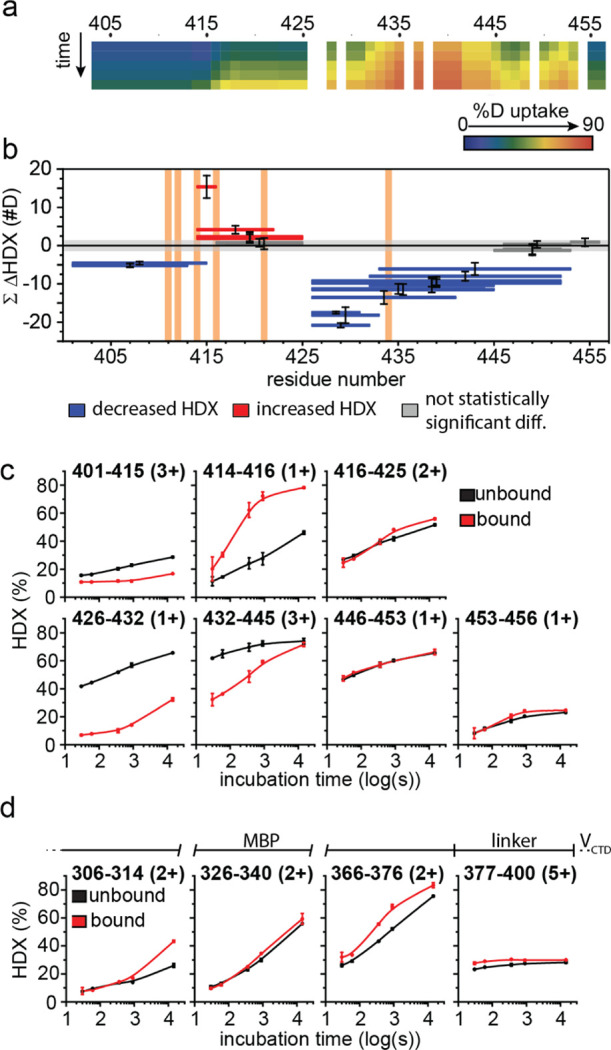
HDX-MS highlights the disordered nature of NiV VCTD and confirms strong binding with MDA5SF2. (a) Heat map showing the deuterium uptake extent of free MBP–NiV VCTD. HDX-MS of unbound MBP–NiV VCTD after 10 s through 14,400 s, where the extent of deuterium incorporation is indicated by using the color gradient. (b) Woods’ plot showing peptide-level HDX differences between unbound MBP–NiV VCTD and MDA5SF2-bound MBP–NiV VCTD. The horizontal bars depict each peptide length spanning a region of the sequence and showing cumulative differences in absolute deuterium uptake between the bound vs unbound state of NiV VCTD. Decreases in deuterium uptake are shown as negative values (blue), and increases are shown as positive values (red). Gray shading indicates the global significance range (p < 0.01) for which differences are considered statistically significant. Those peptides not reporting a significant difference are shown in dark gray. Error bars depict the propagated error in technical duplicate measurements across all time points. Vertical orange shading shows previously reported binding residues from the PIV5 binding study and sequence alignment. (c) Representative HDX kinetic plots showing peptides covering NiV VCTD. Unbound NiV VCTD (black) vs NiV VCTD bound with the MDA5SF2 domain (red). (d) Representative kinetic plots showing HDX differences observed of the N-terminal MBP tag. Kinetic plots are shown for the MBP tag region of MHT-NiV VCTD for free MBP–NiV VCTD (black) vs MBP–NiV V-CTD co-expressed with the MDA5SF2 domain (red). The bar given above shows the identity of the peptide with reference to the composition of the protein construct.
Analysis of the MBP–NiV VCTD/MDA5SF2 complex reveals that the N-terminus of NiV VCTD is protected from HDX and throughout much of the VCTD (Figure 3b). One region of the protein (represented by two peptides: 402–415 aa and 426–432 aa) indicate a static binding site (with slow on/off rates), where the HDX kinetic curves of the unbound (free state) and bound (in complex with MDA5SF2) protein run in parallel (Figure 3b,c). The intervening region (415–416 aa and 417–425 aa) shows increased HDX for the bound state, suggesting that these peptides become less hydrogen-bonded or more exposed for the MBP–NiV VCTD/MDA5SF2 complex than in the unbound state; in fact, part of this region of VCTD (residues 415–416) in the complex shows very high HDX after longer incubation times, consistent with little hydrogen bonding. In the region containing residues 433–445, the kinetic plots for unbound and bound converge with time, suggesting that the region becomes more dynamic and less bonded (faster off rate) than the more N-terminal binding regions. The C-terminal end of VCTD (residues 445–456) shows little to no difference in HDX between bound and unbound. The VCTD contains a histidine (H408) and seven cysteine (C415, C427, C431, C443, C445, C448, and C452) residues that are conserved in all paramyxovirus VCTDs.22,45 Parallels to PIV5 V proteins suggest that these residues are important for zinc coordination and formation of a Zn finger motif.18,19 The data shown here reveal that H408, C415, C448, and C452, which form the larger first Zn finger, undergo less changes in HDX upon MDA5SF2 binding compared to the four central cysteine residues C427, C431, C443, and C445, which form the second Zn finger, that are more protected from HDX upon MDA5SF2 binding. Our results here confirm that regions encompassing the central cysteine residues are important for binding to MDA5.
Given that our NiV VCTD is fused to an MBP-tag, we wanted to rule out the possibility that MBP was binding to MDA5. Most regions of the MBP tag show little to no statistically significant differences between MBP–NiV VCTD unbound or bound with the MDA5SF2 domain (Figure 3d). However, some regions of the MBP-tag show increased HDX, including the region encompassing the His6-tag and linker residues, which may be a result of steric effects between the larger MBP molecule and NiV VCTD; these interactions are lost when MBP–NiV VCTD is bound to the MDA5SF2 domain. This would manifest as an increase in HDX of some regions on NiV VCTD when comparing to the bound complex. Thus, the importance of the N-terminus of NiV VCTD, where the low HDX in the free state (residues 405–425) may be caused by the presence of MBP, is underestimated, and the observed large increase in the exposure of NiV VCTD in the free state (residues 415–422) is likely to be a consequence of the MBP-tag. While previous studies demonstrated the importance of residue I414 in interference with MDA5 activity,45 we cannot draw conclusions confirming or refuting the importance of I414; however, it is clear that the intrinsically disordered VCTD adopts a more ordered structure, especially with respect to the second Zn finger, in a relatively static binding interaction with MDA5SF2.
NiV VCTD Binds and Modulates MDA5SF2 Dynamics
Peptic digestion and bottom-up analysis of the MDA5SF2 domain, containing Hel1, Hel2, Hel2i, and pincer domains, produced 237 unique peptides (Figure S2b). Most of the MDA5SF2 showed low HDX rates, consistent with the high α helical content reported in the crystal structure (PDB 4GL2) (Figure 4a,b). Several regions, however, showed high deuterium exchange rates in parts of Hel2 (β7 and β12 of the central β-sheet region and α18), Hel1 (α9), and pincer (α19) domains, even early in the HDX experiment (Figure 4a,b), suggesting that these regions are more dynamic and conformationally flexible than those reported in the dsRNA-bound MDA5SF2 crystal structure. This is consistent with the role of SF2 domains in binding RNA and adenosine 5′-triphosphate that were described previously.54−56 Interestingly, the central β sheet shows mixed dynamic motion; we observed low HDX for β9, β10, and β8 throughout the experiment, low HDX for β11 at short times that increases with longer times, and high HDX for β12 and β7 even after short times [Figure 4b (inset)]. After 4 h of HDX, the Hel1, Pincer, and part of the Hel2 domains eventually accumulate a high degree of deuterium, but the Hel2i domain remains largely unexchanged, suggesting little conformational diversity and dynamic motion of this α-helix bundle (Figure 4c).
Figure 4.
HDX-MS reveals changes in solvent accessibility in the MDA5SF2 structure when compared to the MDA5SF2/NiV VCTD complex. (a) Heat map showing the deuterium uptake extent of free MDA5SF2 relative to secondary structural elements (top) combined with segmented Woods’ plot showing peptide-level HDX differences between unbound MDA5SF2 and MDA5SF2 bound with MBP–NiV VCTD (bottom). HDX of the unbound MDA5SF2 construct after 10 s through 14,400 s deuterium exchange times, where deuterium incorporation is indicated using a color scale. Secondary structures extracted from the dsRNA-bound MDA5SF2 crystal structure (PDB 4GL2) are shown above for reference. In the Woods’ plot, the horizontal bars depict each peptide spanning a region of the sequence and showing cumulative differences in absolute deuterium uptake between the bound and unbound states of the MDA5SF2 domain. Decreases in differential deuterium uptake are shown as negative values (blue), and increases are shown as positive values (red). Gray shading indicates the global significance range (p < 0.01) for which differences are considered statistically significant. Those peptides not reporting a significant difference are shown in gray. Error bars depict the propagated error in technical duplicate measurements across all time points. (b) HDX of free MDA5SF2 after 10 s of HDX mapped onto the MDA5SF2 structure. The % HDX was mapped onto the MDA5SF2 in the crystal structure bound to dsRNA (PDB 4GL2, ΔCTD, left) and onto MDA5SF2 (displaying residues 534–809 and827–836) rotated 90° (right). The HDX extent is indicated by the scale bar to the right: n.d., not determined deuterium uptake due to limited peptide coverage (white). (c) Deuterium uptake of free MDA5SF2 after 14,400 s of HDX mapped onto the MDA5SF2 structure. The % HDX was mapped onto the MDA5SF2 in the crystal structure bound to dsRNA (PDB 4GL2, ΔCTD, left). (d) Peptide-level differential HDX mapped to the dsRNA-bound MDA5 crystal structure. The statistically significant differential HDX for representative peptides shown in a (bottom) were mapped onto the dsRNA-bound MDA5SF2 (displaying residues 534–809 and827–836) structure as shown in (b) (right). Differential HDX is indicated by the color bar. (e) Comparison with the PIV5 VCTD-bound MDA5SF2 Hel2 and Hel2i crystal structure (PDB 4I1S).
We next evaluated the MDA5SF2 domain in the context of the MDA5SF2/MBP–NiV VCTD complex. Binding to MBP–NiV VCTD resulted in both increases and decreases in HDX of several regions of the MDA5SF2 domain, as shown in the Woods’ differential HDX plot (Figure 4a). Within the Hel1 domain, decreased HDX is observed in the regions of α1 through β1 (residues 306–328), whereas increased HDX is observed around α3 through α4 (residues 373–382). The most pronounced difference in HDX is the protection observed in the Hel2 domain (Figure 4a,d) on the periphery of the slow exchanging core noted in the free state (Figure 4b); the largest decreases in HDX are clustered around the region surrounding β11, including α17 through β10 and β11 (residues 763–806), and the flexible region between α14 and α15 through β8 (residues 700–724) (Figure 4a,d). Note that the long peptides encompassing residues 744–784 and covering α17 show decreased HDX, but the shorter peptides encompassing residues 744–762 and before α17 do not. Interestingly, the more dynamic β-strands, β7 and β12, show no evidence of differential HDX within the MDA5SF2 domain/MBP–NiV VCTD complex. Overall, all these observations are consistent with the crystal structure of MDA5SF2 Hel2 and Hel2i domains in complex with the V protein of another paramyxovirus, PIV5 (Figure 4e).18 PIV5 V protein displaces the MDA5SF2 β7 and β12 in the Hel2 domain and forms a continuous β-sheet with β11. Comparison of our cumulative differential HDX results for the MDA5SF2/MBP–NiV VCTD complex with the MDA5SF2/PIV5 V structure (PDB 4I1S) reveals that the same decreased HDX around β11 observed in the presence of NiV VCTD would be expected for the PIV5 V protein (Figure 4d,e), suggesting that like the PIV5 V protein, NiV VCTD likely binds to these regions adjacent to Hel2 and pincer domains, causing HDX protection.
MDA5 regions with increased HDX are also observed in the MDA5SF2/MBP–NiV VCTD complex, in particular at the ends of α-helices at the interface between Hel2 and Hel2i (Figure 4a,d). Increased HDX, suggesting increased dynamics or solvent exposure, occurs upon binding between α12 and α13 (residues 610–622) and between β8 and α16 (residues 725–732). In other words, these regions show relatively less HDX in the unbound state with the exception of the linker regions already reported in the crystal structure (Figure 4a,b), indicating that they undergo conformation changes distal to the binding sites, and these changes open the structure.57 Collectively, our data suggest that NiV VCTD not only binds MDA5SF2 similar to the PIV5 V protein but also induces changes in the dynamics in other regions within MDA5; these latter changes may impact its antiviral activity.
Discussion
The NiV V protein is a critical virulence factor that potently inhibits host innate immune responses.22−25 The importance of the NiV V protein in this process was established by earlier studies where rNiVM strains deficient in the V protein failed to suppress immune responses and caused disease in an animal model.23 The NiV V protein directly binds to the PRR MDA5 to prevent IFN signaling.34,35,43,46,47 To develop a better understanding of the role of NiV V in this process, we performed a biochemical and structural characterization of the interaction between NiV V and MDA5. Co-IP assays demonstrated that the NiV VCTD binds to the MDA5SF2 domain when co-transfected and not to other domains. In vitro pulldown assays using purified recombinant proteins and co-purification of cell lysates containing individually expressed proteins do not show direct interactions. However, co-expression and co-purification of the recombinant MBP–NiV VCTD/MDA5SF2 domain complex provide evidence of a soluble, intact complex. This is not surprising as heterologous protein overexpression in E. coli can result in misfolded or insoluble proteins. Several factors contribute to the production of stable proteins, including cofactors and post-translational modifications, which may be absent in E. coli. Co-expression with a binding partner can help overcome some of these barriers and facilitate production of functional proteins by reducing conformational flexibility.58 Moreover, the NiV V protein has been recently described as an intrinsically disordered protein (IDP) that lacks a significantly folded structure and conformational stability.48 IDPs like the NiV V protein can undergo binding-induced folding during complex formation that is dependent on the nature of the binding partner.59−61 Thus, co-expression and co-translation in a recombinant system can generate complexes that may not form by mixing individual components, especially when involving IDPs.
Structural analysis of the co-expressed and co-purified NiV VCTD/MDA5SF2 domain complex revealed extensive HDX protection throughout NiV VCTD. Differential HDX-MS analysis for residues covering residues 408–415 and 426–432 indicates static binding with MDA5SF2 (i.e., low on–off rates). A region encompassing residues 433–445 suggests weaker bonding or a more dynamic interaction with nearby MDA5SF2 residues (i.e., higher off rates). The importance of conserved residues R409, E411, I414, and C431 that attenuate NiV V binding to MDA545 is consistent with the static localized binding shown by the HDX-MS analysis; C443 and C445 are within the more dynamic binding site, but there is no evidence of C448 participation. Notably, these regions also cover the four central cysteine residues that form the second zinc finger (i.e., C427, C431, C443, and C445). While these results cannot confirm the importance of the cysteine side chains, the HDX results are evidence of direct binding of these regions with MDA5SF2 or binding-induced stabilization of the intrinsically disordered unbound VCTD backbone. Despite this observation, addition of EDTA did not disrupt the interaction between NiV VCTD and MDA5SF2, suggesting that the Zn motif is either protected within the complex or that higher concentrations are needed to extract the zinc ions.
HDX-MS analysis of free MDA5SF2 is largely consistent with the reported crystal structure of MDA5SF2 bound to dsRNA;54,62 however, the HDX data also identified a highly exchanging region comprising β7, β12, and α18 of the Hel2 domain, α9 of the Hel1 domain, and much of α19 of the pincer domain. The extensive HDX observed for β7 and β12 at short times and that observed for β11 at longer times indicates that this β-sheet structure is not static in the free state but rather very dynamic.
Differential HDX-MS analysis of NiV VCTD binding to the MDA5SF2 domain indicates a relatively static binding interaction covering the vicinity of β11 (Figure 4) but no effect on the dynamic strands of the β-sheet region, β7 and β12. The location of this binding site is interesting because if the β-sheet structure is maintained in the unbound state, binding-induced protection should not be observed for β11. The lack of statistically significant differences in HDX between bound and unbound within this dynamic region indicates that this region is locked in an exposed or open conformation.
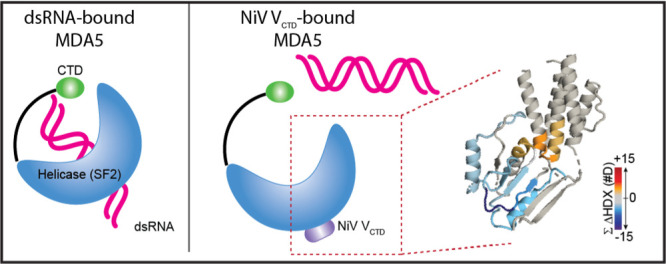
There are significant parallels to what we observe by HDX-MS of the NiV VCTD/MDA5SF2 domain complex and the reported crystal structure of PIV5 VCTD/MDA5SF2 (PDB 4I1S). In the PIV5 VCTD/MDA5SF2 structure, the PIV5 VCTD replaces the MDA5SF2 β7 and β12 strands in the central β-sheet. PIV5 V binding with MDA5SF2 is associated with decreased formation of RNA-bound signaling MDA5 oligomers, possibly disrupting SF2–SF2 or SF2–RNA-binding domain interactions.18 The results reported here are consistent with NiV VCTD, similarly displacing the MDA5SF2 β7 and β12 strands. Furthermore, the HDX results suggest that this displacement does not change the conformational preference of the MDA5SF2 β-strands from the RNA-free unbound state but would prevent formation of the central β-sheet observed in the RNA-bound MDA5SF2 crystal structure (PDB 4GL2).54 This suggests that NiV V may sterically hinder “closing” of the RNA-free dynamic conformation of MDA5SF2.
Our HDX-MS results revealed binding-induced conformational changes in regions distal to the binding interface of NiV VCTD/MDA5SF2 that may have additional implications for MDA5SF2 binding with dsRNA. Regions near α13 and α14 of the Hel2i subdomain and α6 of the Hel2 subdomain show increased HDX when MDA5SF2 is bound to NiV VCTD, indicating either a binding-induced decrease in the α-helical character and/or an increase in dynamic motion. We observe decreased HDX of a remote region near α17 of the Hel2 subdomain and α1 and β1 of the Hel1 domain and some exposure near α3 and α4. These remote conformational changes present important potential implications for NiV VCTD binding with the MDA5SF2 domain, wherein NiV VCTD may impede dsRNA binding to the MDA5SF2 domain despite its remote binding site. Thus, our HDX results indicate that the NiV V protein binding site on the MDA5SF2 domain is similar but not entirely the same as that of PIV5 V.18,46
We have shown here that the β-sheet region within the MDA5SF2 domain is dynamic, and the regions at the interface become protected upon binding with NiV VCTD. Formation of the NiV VCTD/MDA5SF2 domain complex stabilizes the VCTD, including in the region encompassing the conserved central cysteine residues, and leads to allosteric changes in MDA5 that likely disrupts its activity. Thus, modulation of the conformational dynamics within MDA5 may present a potential mechanism for immune evasion by a highly pathogenic RNA virus.
Methods
Recombinant Protein Production
The gene encoding the NiV V protein (accession AAK50551) was codon-optimized for expression in E. coli (GeneScript, NJ) prior to subcloning into a modified pET15b plasmid containing a maltose-binding protein (MBP) tag followed by a His6 tag. MDA5 and MDA5SF2 were amplified from human complementary DNA by polymerase chain reaction. All sequences were confirmed with Sanger sequencing. Recombinant proteins were expressed in E. coli BL21(DE3) cells at 18 °C for 16 h with 0.25 mM IPTG induction. Cells were pelleted down by centrifugation and re-suspended in lysis buffer containing 50 mM Tris-HCl (pH 8.0), 300 mM NaCl, and protease inhibitors prior to lysis with an EmulsiFlex-C5 homogenizer (Avestin, Canada). Lysate was clarified by centrifugation at 60,000g and then used for pulldown assays or further protein purification using a series of affinity, ion exchange, and gel filtration columns (GE Healthcare, IL). The MBP-tag on MDA5SF2 was cleaved using TEV protease and removed by affinity purification. Final protein samples were assessed by SDS-PAGE.
Co-immunoprecipitation
HEK293T cells were grown in 6-well plates in Dulbecco’s modified Eagle medium containing 10% fetal bovine serum. Cells were transfected at approximately 80% confluency with a mixture of one plasmid encoding NiV proteins and the other encoding MDA5 protein for co-IP experiments. Cells were harvested by centrifugation at 48 h after transfection. After washing once with phosphate-buffered saline, cells were either stored in −80 °C for use or suspended in lysis buffer containing 50 mM Tris-HCl, pH 8.0, 280 mM NaCl, 2 mM EGTA, 0.4 mM EDTA, 10% glycerol, 0.5% IGEPAL CA630 (Sigma-Aldrich), and a protease inhibitor cocktail. The lysate was clarified with centrifugation at 14,000 rpm for 15 min, and the clarification was repeated three times. The resultant supernatant, WCE hereafter, was used to incubate with anti-FLAG M2 magnetic beads (Sigma-Aldrich, M8823) at 4 °C. The beads were pelleted down using a magnetic rack after 1 h of incubation and washed five times with the lysis buffer. Proteins bound on the beads were eluted with 150 ng/mL FLAG peptide (Sigma-Aldrich, F3290). Samples from the WCE and the elution were transferred to the poly(vinylidene difluoride) membrane for Western blot after resolving by SDS-PAGE. Mouse anti-FLAG (F3165) and anti-HA (Sigma-Aldrich, H3663) monoclonal antibodies were used as primary antibodies to detect viral proteins and human proteins with the corresponding epitope, respectively. Goat anti-mouse IgG–HRP conjugated antibody (Santa Cruz, SC-2005) was used as a secondary antibody for the western blot. Blot images were taken from Amersham Imager 600 (GE Healthcare).
MBP Pulldown Assay
MBP pulldown assay was similar to that previously reported.63 Briefly, E. coli lysate clarified by centrifugation or purified proteins was incubated with amylose resin at high flow (New England Biolabs, E8022L) in binding buffer for 30 min with moderate agitation in a cold room. The resin was washed six times with washing buffer and eluted with binding buffer containing 15 mM maltose. Samples were resolved by SDS-PAGE and stained with Coomassie blue for visualization. For co-lysate preparation, cells expressing either the viral protein or human MDA5SF2 were re-suspended in lysis buffer containing 150 mM NaCl and mixed before homogenization.
Hydrogen–Deuterium Exchange Mass Spectrometry (HDX-MS)
All chemical reagents used in HDX-MS experiments were purchased from Sigma-Aldrich (St. Louis, MO) unless otherwise noted. HDX-MS analyses were carried out, as previously described,64,65 on purified MBP–NiV VCTD, MDA5SF2, and the co-expressed MBP–NiV V-CTD/MDA5SF2 protein complex (stock solutions 40 μM per protein in PBS, pH 7.4). Briefly, HDX was initiated by diluting 4 μL of a protein stock solution with a ninefold excess of PBS in D2O (Cambridge Isotope Laboratories, Inc., Tewksbury, MA) (final composition 90/10% v/v D2O/H2O). The mixture was exchanged on ice for 0, 10, 30, 60, 360, 900, or 144,000 s, and the HDX was quenched by adding 30 μL of a quench buffer (3 M urea in PBS, pH 2.4). In-solution digestion was performed using protease type XIII from Aspergillus saitoi (fungal XIII, PBS pH 2.4) immediately after quenching and incubated for 3 min at 22 °C.
All liquid chromatography/mass spectrometry (LC/MS) analyses were performed using a custom-built liquid chromatography (LC) assembly coupled to an LTQ-FTICR mass spectrometer (Thermo Fisher, Waltham, MA). The LC system incorporated online pepsin protease digestion using a custom-prepared immobilized pepsin column (2 × 20 mm), followed by peptide trapping and desalting on a ZORBAX Eclipse XDB C8 column (2.1 × 15 mm, Agilent, Santa Clara, CA) and reversed-phase peptide separation with an XSelect CSH C18 column (2.1 × 50 mm, Waters, Manchester, UK). All HDX-MS time points were analyzed in duplicate.
Using the same setup and procedure, proteolytic mapping analyses were performed prior to HDX on nondeuterated aliquots of the protein stock solutions, except that tandem MS in the data-dependent mode was used. The top six most abundant fragment ions from each scan were selected for MS/MS analysis. The MS/MS files were analyzed using Byonic and Byologic (Protein Metrics, San Carlos, CA) to generate a list of identified peptides to be followed in the HDX-MS analyses. The search was also carried out against reverse sequences of the proteins to discard ambiguous assignments.
All HDX-MS analyses were processed using HDExaminer (version 2.5.1, Sierra Analytics, Modesto, CA); adjustment of LC boundaries and validation of MS interferences were performed manually on all peptides. Woods’ plot peptide cumulative HDX differences were calculated as the sum of all differences at all time points in daltons [∑(Dcoexpressed) – ∑(Dfree)]. The standard errors of the mean were propagated through the calculation and shown as error bars. The global significance limit was calculated as previously described.66,67 Briefly, the pooled standard deviation was calculated for all time points and all peptides within each state (i.e., bound and unbound). The pooled standard deviations were used to calculate the population standard error of the mean, and this standard error was used with a two-tailed t-test value (p < 0.01) to calculate the confidence interval distribution for differences in HDX across the dataset. Statistically significant differences for representative peptides were mapped onto structures using PyMOL. For a summary of all HDX data, see the Supporting Information.
Acknowledgments
This work was supported by grants from the NIH (R01AI140758 to D.W.L.; P41 GM103422 and R24GM136766 to M.L.G.; and P01AI120943 and R01AI109945 to G.K.A.). We thank members of the Amarasinghe, Gross, and Leung laboratories for support and comments.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsinfecdis.1c00403.
Western blot images, coverage maps for HDX-MS, additional views of the HDX-MS results mapped onto the structures, and compilation of raw HDX values (PDF)
Author Present Address
∥ Department of Integrative Structural and Computational Biology, The Scripps Research Institute, La Jolla, CA 92037, USA
Author Contributions
△ N.D.W. and H.L. contributed equally. D.W.L. and G.K.A. conceived the project. D.W.L., G.K.A., and M.L.G. designed the experimental plans. H.L. generated samples. H.L., N.D.W., and H.W.R. performed experiments and analyzed results. H.L. wrote an initial draft manuscript with N.D.W. N.D.W. analyzed data and completed manuscript. N.D.W., M.L.G., G.K.A., and D.W.L. participated in editing the final version of the manuscript.
The authors declare no competing financial interest.
Supplementary Material
References
- Farrar J. J. Nipah-virus encephalitis-investigation of a new infection. Lancet 1999, 354, 1222–1223. 10.1016/s0140-6736(99)90124-1. [DOI] [PubMed] [Google Scholar]
- Chua K. B.; Bellini W. J.; Rota P. A.; Harcourt B. H.; Tamin A.; Lam S. K.; Ksiazek T. G.; Rollin P. E.; Zaki S. R.; Shieh W.-J.; Goldsmith C. S.; Gubler D. J.; Roehrig J. T.; Eaton B.; Gould A. R.; Olson J.; Field H.; Daniels P.; Ling A. E.; Peters C. J.; Anderson L. J.; Mahy B. W. J. Nipah virus: a recently emergent deadly paramyxovirus. Science 2000, 288, 1432–1435. 10.1126/science.288.5470.1432. [DOI] [PubMed] [Google Scholar]
- Sarji S. A.; Abdullah B. J. J.; Goh K. J.; Tan C. T.; Wong K. T. MR imaging features of Nipah encephalitis. AJR, Am. J. Roentgenol. 2000, 175, 437–442. 10.2214/ajr.175.2.1750437. [DOI] [PubMed] [Google Scholar]
- Pelissier R.; Iampietro M.; Horvat B. Recent advances in the understanding of Nipah virus immunopathogenesis and anti-viral approaches. F1000Research 2019, 8, F1000. 10.12688/f1000research.19975.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anam A. M.; Ahmad J.; Huq S. M. R.; Rabbani R. Nipah virus encephalitis: MRI findings. J. R. Coll. Physicians Edinb. 2019, 49, 227–228. 10.4997/jrcpe.2019.312. [DOI] [PubMed] [Google Scholar]
- Clayton B. A. Nipah virus: transmission of a zoonotic paramyxovirus. Curr. Opin. Virol. 2017, 22, 97–104. 10.1016/j.coviro.2016.12.003. [DOI] [PubMed] [Google Scholar]
- Epstein J. H.; Field H. E.; Luby S.; Pulliam J. R. C.; Daszak P. Nipah virus: impact, origins, and causes of emergence. Curr. Infect. Dis. Rep. 2006, 8, 59–65. 10.1007/s11908-006-0036-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harcourt B. H.; Lowe L.; Tamin A.; Liu X.; Bankamp B.; Bowden N.; Rollin P. E.; Comer J. A.; Ksiazek T. G.; Hossain M. J.; Gurley E. S.; Breiman R. F.; Bellini W. J.; Rota P. A. Genetic characterization of Nipah virus, Bangladesh, 2004. Emerging Infect. Dis. 2005, 11, 1594–1597. 10.3201/eid1110.050513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mire C. E.; Satterfield B. A.; Geisbert J. B.; Agans K. N.; Borisevich V.; Yan L.; Chan Y.-P.; Cross R. W.; Fenton K. A.; Broder C. C.; Geisbert T. W. Pathogenic Differences between Nipah Virus Bangladesh and Malaysia Strains in Primates: Implications for Antibody Therapy. Sci. Rep. 2016, 6, 30916. 10.1038/srep30916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L.-F.; Harcourt B. H.; Yu M.; Tamin A.; Rota P. A.; Bellini W. J.; Eaton B. T. Molecular biology of Hendra and Nipah viruses. Microbes Infect. 2001, 3, 279–287. 10.1016/s1286-4579(01)01381-8. [DOI] [PubMed] [Google Scholar]
- Marsh G. A.; de Jong C.; Barr J. A.; Tachedjian M.; Smith C.; Middleton D.; Yu M.; Todd S.; Foord A. J.; Haring V.; Payne J.; Robinson R.; Broz I.; Crameri G.; Field H. E.; Wang L.-F. Cedar virus: a novel Henipavirus isolated from Australian bats. PLoS Pathog. 2012, 8, e1002836 10.1371/journal.ppat.1002836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z.; Yang L.; Yang F.; Ren X.; Jiang J.; Dong J.; Sun L.; Zhu Y.; Zhou H.; Jin Q. Novel Henipa-like virus, Mojiang Paramyxovirus, in rats, China, 2012. Emerging Infect. Dis. 2014, 20, 1064–1066. 10.3201/eid2006.131022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amarasinghe G. K.; Ayllón M. A.; Bào Y.; Basler C. F.; Bavari S.; Blasdell K. R.; Briese T.; Brown P. A.; Bukreyev A.; Balkema-Buschmann A.; Buchholz U. J.; Chabi-Jesus C.; Chandran K.; Chiapponi C.; Crozier I.; de Swart R. L.; Dietzgen R. G.; Dolnik O.; Drexler J. F.; Dürrwald R.; Dundon W. G.; Duprex W. P.; Dye J. M.; Easton A. J.; Fooks A. R.; Formenty P. B. H.; Fouchier R. A. M.; Freitas-Astúa J.; Griffiths A.; Hewson R.; Horie M.; Hyndman T. H.; Jia̅ng D.; Kitajima E. W.; Kobinger G. P.; Kondo̅ H.; Kurath G.; Kuzmin I. V.; Lamb R. A.; Lavazza A.; Lee B.; Lelli D.; Leroy E. M.; Lǐ J.; Maes P.; Marzano S.-Y. L.; Moreno A.; Mühlberger E.; Netesov S. V.; Nowotny N.; Nylund A.; Økland A. L.; Palacios G.; Pályi B.; Pawęska J. T.; Payne S. L.; Prosperi A.; Ramos-González P. L.; Rima B. K.; Rota P.; Rubbenstroth D.; Shi̅ M.; Simmonds P.; Smither S. J.; Sozzi E.; Spann K.; Stenglein M. D.; Stone D. M.; Takada A.; Tesh R. B.; Tomonaga K.; Tordo N.; Towner J. S.; van den Hoogen B.; Vasilakis N.; Wahl V.; Walker P. J.; Wang L.-F.; Whitfield A. E.; Williams J. V.; Zerbini F. M.; Zha̅ng T.; Zhang Y.-Z.; Kuhn J. H. Taxonomy of the order Mononegavirales: update 2019. Arch. Virol. 2019, 164, 1967–1980. 10.1007/s00705-019-04247-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park M.-S.; Shaw M. L.; Munoz-Jordan J.; Cros J. F.; Nakaya T.; Bouvier N.; Palese P.; Garcia-Sastre A.; Basler C. F. Newcastle disease virus (NDV)-based assay demonstrates interferon-antagonist activity for the NDV V protein and the Nipah virus V, W, and C proteins. J. Virol. 2003, 77, 1501–1511. 10.1128/jvi.77.2.1501-1511.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni S.; Volchkova V.; Basler C. F.; Palese P.; Volchkov V. E.; Shaw M. L. Nipah virus edits its P gene at high frequency to express the V and W proteins. J. Virol. 2009, 83, 3982–3987. 10.1128/jvi.02599-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo M. K.; Harcourt B. H.; Mungall B. A.; Tamin A.; Peeples M. E.; Bellini W. J.; Rota P. A. Determination of the henipavirus phosphoprotein gene mRNA editing frequencies and detection of the C, V and W proteins of Nipah virus in virus-infected cells. J. Gen. Virol. 2009, 90, 398–404. 10.1099/vir.0.007294-0. [DOI] [PubMed] [Google Scholar]
- Shaw M. Henipaviruses employ a multifaceted approach to evade the antiviral interferon response. Viruses 2009, 1, 1190–1203. 10.3390/v1031190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motz C.; Schuhmann K. M.; Kirchhofer A.; Moldt M.; Witte G.; Conzelmann K.-K.; Hopfner K.-P.; Paramyxovirus V. Paramyxovirus V Proteins Disrupt the Fold of the RNA Sensor MDA5 to Inhibit Antiviral Signaling. Science 2013, 339, 690–693. 10.1126/science.1230949. [DOI] [PubMed] [Google Scholar]
- Li T.; Chen X.; Garbutt K. C.; Zhou P.; Zheng N. Structure of DDB1 in complex with a paramyxovirus V protein: Viral hijack of a propeller cluster in ubiquitin ligase. Cell 2006, 124, 105–117. 10.1016/j.cell.2005.10.033. [DOI] [PubMed] [Google Scholar]
- Liston P.; Briedis D. J. Measles virus V protein binds zinc. Virology 1994, 198, 399–404. 10.1006/viro.1994.1050. [DOI] [PubMed] [Google Scholar]
- Paterson R. G.; Leser G. P.; Shaughnessy M. A.; Lamb R. A. The paramyxovirus SV5 V protein binds two atoms of zinc and is a structural component of virions. Virology 1995, 208, 121–131. 10.1006/viro.1995.1135. [DOI] [PubMed] [Google Scholar]
- Audsley M. D.; Moseley G. W. Paramyxovirus evasion of innate immunity: Diverse strategies for common targets. World J. Virol. 2013, 2, 57–70. 10.5501/wjv.v2.i2.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satterfield B. A.; Cross R. W.; Fenton K. A.; Agans K. N.; Basler C. F.; Geisbert T. W.; Mire C. E. The immunomodulating V and W proteins of Nipah virus determine disease course. Nat. Commun. 2015, 6, 7483. 10.1038/ncomms8483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneda M.; Guillaume V.; Sato H.; Fujita K.; Georges-Courbot M.-C.; Ikeda F.; Omi M.; Muto-Terao Y.; Wild T. F.; Kai C. The nonstructural proteins of Nipah virus play a key role in pathogenicity in experimentally infected animals. PLoS One 2010, 5, e12709 10.1371/journal.pone.0012709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satterfield B. A.; Cross R. W.; Fenton K. A.; Borisevich V.; Agans K. N.; Deer D. J.; Graber J.; Basler C. F.; Geisbert T. W.; Mire C. E. Nipah Virus C and W Proteins Contribute to Respiratory Disease in Ferrets. J. Virol. 2016, 90, 6326–6343. 10.1128/jvi.00215-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos H. J.; Gale M. Jr. RIG-I like receptors and their signaling crosstalk in the regulation of antiviral immunity. Curr. Opin. Virol. 2011, 1, 167–176. 10.1016/j.coviro.2011.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkins C.; Gale M. Jr. Recognition of viruses by cytoplasmic sensors. Curr. Opin. Immunol. 2010, 22, 41–47. 10.1016/j.coi.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneyama M.; Kikuchi M.; Natsukawa T.; Shinobu N.; Imaizumi T.; Miyagishi M.; Taira K.; Akira S.; Fujita T. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat. Immunol. 2004, 5, 730–737. 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- Kang D.-c.; Gopalkrishnan R. V.; Wu Q.; Jankowsky E.; Pyle A. M.; Fisher P. B. mda-5: An interferon-inducible putative RNA helicase with double-stranded RNA-dependent ATPase activity and melanoma growth-suppressive properties. Proc. Natl. Acad. Sci. U.S.A. 2002, 99, 637–642. 10.1073/pnas.022637199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneyama M.; Kikuchi M.; Matsumoto K.; Imaizumi T.; Miyagishi M.; Taira K.; Foy E.; Loo Y.-M.; Gale M. Jr.; Akira S.; Yonehara S.; Kato A.; Fujita T. Shared and unique functions of the DExD/H-box helicases RIG-I, MDA5, and LGP2 in antiviral innate immunity. J. Immunol. 2005, 175, 2851–2858. 10.4049/jimmunol.175.5.2851. [DOI] [PubMed] [Google Scholar]
- Miyoshi K.; Cui Y.; Riedlinger G.; Robinson P.; Lehoczky J.; Zon L.; Oka T.; Dewar K.; Hennighausen L. Structure of the mouse Stat 3/5 locus: evolution from Drosophila to zebrafish to mouse. Genomics 2001, 71, 150–155. 10.1006/geno.2000.6433. [DOI] [PubMed] [Google Scholar]
- Hur S. Double-Stranded RNA Sensors and Modulators in Innate Immunity. Annu. Rev. Immunol. 2019, 37, 349–375. 10.1146/annurev-immunol-042718-041356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrejeva J.; Childs K. S.; Young D. F.; Carlos T. S.; Stock N.; Goodbourn S.; Randall R. E. The V proteins of paramyxoviruses bind the IFN-inducible RNA helicase, mda-5, and inhibit its activation of the IFN-beta promoter. Proc. Natl. Acad. Sci. U.S.A. 2004, 101, 17264–17269. 10.1073/pnas.0407639101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Childs K.; Stock N.; Ross C.; Andrejeva J.; Hilton L.; Skinner M.; Randall R.; Goodbourn S. mda-5, but not RIG-I, is a common target for paramyxovirus V proteins. Virology 2007, 359, 190–200. 10.1016/j.virol.2006.09.023. [DOI] [PubMed] [Google Scholar]
- Childs K. S.; Andrejeva J.; Randall R. E.; Goodbourn S. Mechanism of mda-5 Inhibition by paramyxovirus V proteins. J. Virol. 2009, 83, 1465–1473. 10.1128/jvi.01768-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez J. J.; Parisien J.-P.; Horvath C. M. Nipah virus V protein evades alpha and gamma interferons by preventing STAT1 and STAT2 activation and nuclear accumulation. J. Virol. 2002, 76, 11476–11483. 10.1128/jvi.76.22.11476-11483.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez J. J.; Cruz C. D.; Horvath C. M. Identification of the nuclear export signal and STAT-binding domains of the Nipah virus V protein reveals mechanisms underlying interferon evasion. J. Virol. 2004, 78, 5358–5367. 10.1128/jvi.78.10.5358-5367.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciancanelli M. J.; Volchkova V. A.; Shaw M. L.; Volchkov V. E.; Basler C. F. Nipah virus sequesters inactive STAT1 in the nucleus via a P gene-encoded mechanism. J. Virol. 2009, 83, 7828–7841. 10.1128/jvi.02610-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw M. L.; Garcia-Sastre A.; Palese P.; Basler C. F. Nipah virus V and W proteins have a common STAT1-binding domain yet inhibit STAT1 activation from the cytoplasmic and nuclear compartments, respectively. J. Virol. 2004, 78, 5633–5641. 10.1128/jvi.78.11.5633-5641.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith K. M.; Tsimbalyuk S.; Edwards M. R.; Cross E. M.; Batra J.; Soares da Costa T. P.; Aragão D.; Basler C. F.; Forwood J. K. Structural basis for importin alpha 3 specificity of W proteins in Hendra and Nipah viruses. Nat. Commun. 2018, 9, 3703. 10.1038/s41467-018-05928-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw M. L.; Cardenas W. B.; Zamarin D.; Palese P.; Basler C. F. Nuclear localization of the Nipah virus W protein allows for inhibition of both virus- and toll-like receptor 3-triggered signaling pathways. J. Virol. 2005, 79, 6078–6088. 10.1128/jvi.79.10.6078-6088.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Aparicio M. T.; Feinman L. J.; Garcia-Sastre A.; Shaw M. L. Paramyxovirus V proteins interact with the RIG-I/TRIM25 regulatory complex and inhibit RIG-I signaling. J. Virol. 2018, 92, e01960–17. 10.1128/jvi.01960-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisien J.-P.; Bamming D.; Komuro A.; Ramachandran A.; Rodriguez J. J.; Barber G.; Wojahn R. D.; Horvath C. M. A shared interface mediates paramyxovirus interference with antiviral RNA helicases MDA5 and LGP2. J. Virol. 2009, 83, 7252–7260. 10.1128/jvi.00153-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Childs K.; Randall R.; Goodbourn S. Paramyxovirus V proteins interact with the RNA Helicase LGP2 to inhibit RIG-I-dependent interferon induction. J. Virol. 2012, 86, 3411–3421. 10.1128/jvi.06405-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran A.; Horvath C. M. Dissociation of paramyxovirus interferon evasion activities: universal and virus-specific requirements for conserved V protein amino acids in MDA5 interference. J. Virol. 2010, 84, 11152–11163. 10.1128/jvi.01375-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez K. R.; Horvath C. M. Amino acid requirements for MDA5 and LGP2 recognition by paramyxovirus V proteins: a single arginine distinguishes MDA5 from RIG-I. J. Virol. 2013, 87, 2974–2978. 10.1128/jvi.02843-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez K. R.; Horvath C. M. Paramyxovirus V protein interaction with the antiviral sensor LGP2 disrupts MDA5 signaling enhancement but is not relevant to LGP2-mediated RLR signaling inhibition. J. Virol. 2014, 88, 8180–8188. 10.1128/jvi.00737-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiavina M.; Salladini E.; Murrali M. G.; Tria G.; Felli I. C.; Pierattelli R.; Longhi S. Ensemble description of the intrinsically disordered N-terminal domain of the Nipah virus P/V protein from combined NMR and SAXS. Sci. Rep. 2020, 10, 19574. 10.1038/s41598-020-76522-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath C. M. Weapons of STAT destruction. Interferon evasion by paramyxovirus V protein. Eur. J. Biochem. 2004, 271, 4621–4628. 10.1111/j.1432-1033.2004.04425.x. [DOI] [PubMed] [Google Scholar]
- Angers S.; Li T.; Yi X.; MacCoss M. J.; Moon R. T.; Zheng N. Molecular architecture and assembly of the DDB1-CUL4A ubiquitin ligase machinery. Nature 2006, 443, 590–593. 10.1038/nature05175. [DOI] [PubMed] [Google Scholar]
- Chalmers M. J.; Busby S. A.; Pascal B. D.; West G. M.; Griffin P. R. Differential hydrogen/deuterium exchange mass spectrometry analysis of protein-ligand interactions. Expert Rev. Proteomics 2011, 8, 43–59. 10.1586/epr.10.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weis D. D.Hydrogen Exchange Mass Spectrometry of Proteins: Fundamentals, Methods, and Applications; Wiley: Chichester, West Sussex, 2016; p 1 online resource. [Google Scholar]
- Masson G. R.; Burke J. E.; Ahn N. G.; Anand G. S.; Borchers C.; Brier S.; Bou-Assaf G. M.; Engen J. R.; Englander S. W.; Faber J.; Garlish R.; Griffin P. R.; Gross M. L.; Guttman M.; Hamuro Y.; Heck A. J. R.; Houde D.; Iacob R. E.; Jørgensen T. J. D.; Kaltashov I. A.; Klinman J. P.; Konermann L.; Man P.; Mayne L.; Pascal B. D.; Reichmann D.; Skehel M.; Snijder J.; Strutzenberg T. S.; Underbakke E. S.; Wagner C.; Wales T. E.; Walters B. T.; Weis D. D.; Wilson D. J.; Wintrode P. L.; Zhang Z.; Zheng J.; Schriemer D. C.; Rand K. D. Recommendations for performing, interpreting and reporting hydrogen deuterium exchange mass spectrometry (HDX-MS) experiments. Nat. Methods 2019, 16, 595–602. 10.1038/s41592-019-0459-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu B.; Peisley A.; Richards C.; Yao H.; Zeng X.; Lin C.; Chu F.; Walz T.; Hur S. Structural basis for dsRNA recognition, filament formation, and antiviral signal activation by MDA5. Cell 2013, 152, 276–289. 10.1016/j.cell.2012.11.048. [DOI] [PubMed] [Google Scholar]
- Rawling D. C.; Pyle A. M. Parts, assembly and operation of the RIG-I family of motors. Curr. Opin. Struct. Biol. 2014, 25, 25–33. 10.1016/j.sbi.2013.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalinski E.; Lunardi T.; McCarthy A. A.; Louber J.; Brunel J.; Grigorov B.; Gerlier D.; Cusack S. Structural basis for the activation of innate immune pattern-recognition receptor RIG-I by viral RNA. Cell 2011, 147, 423–435. 10.1016/j.cell.2011.09.039. [DOI] [PubMed] [Google Scholar]
- Englander S. W. Hydrogen exchange and mass spectrometry: A historical perspective. J. Am. Soc. Mass Spectrom. 2006, 17, 1481–1489. 10.1016/j.jasms.2006.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolia N. H.; Joshua-Tor L. Strategies for protein coexpression in Escherichia coli. Nat. Methods 2006, 3, 55–64. 10.1038/nmeth0106-55. [DOI] [PubMed] [Google Scholar]
- Arai M.; Sugase K.; Dyson H. J.; Wright P. E. Conformational propensities of intrinsically disordered proteins influence the mechanism of binding and folding. Proc. Natl. Acad. Sci. U.S.A. 2015, 112, 9614–9619. 10.1073/pnas.1512799112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toto A.; Malagrinò F.; Visconti L.; Troilo F.; Pagano L.; Brunori M.; Jemth P.; Gianni S. Templated folding of intrinsically disordered proteins. J. Biol. Chem. 2020, 295, 6586–6593. 10.1074/jbc.rev120.012413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uversky V. N. Paradoxes and wonders of intrinsic disorder: Stability of instability. Intrinsically Disord. Proteins 2017, 5, e1327757 10.1080/21690707.2017.1327757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchikawa E.; Lethier M.; Malet H.; Brunel J.; Gerlier D.; Cusack S. Structural Analysis of dsRNA Binding to Anti-viral Pattern Recognition Receptors LGP2 and MDA5. Mol. Cell 2016, 62, 586–602. 10.1016/j.molcel.2016.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H.; Zhang M.; He W.; Zhu Z.; Teng M.; Gao Y.; Niu L. Structural insights into yeast histone chaperone Hif1: a scaffold protein recruiting protein complexes to core histones. Biochem. J. 2014, 462, 465–473. 10.1042/bj20131640. [DOI] [PubMed] [Google Scholar]
- Ramakrishnan D.; Xing W.; Beran R. K.; Chemuru S.; Rohrs H.; Niedziela-Majka A.; Marchand B.; Mehra U.; Zabransky A.; Dolezal M.; Hubalek M.; Pichova I.; Gross M. L.; Kwon H. J.; Fletcher S. P. Hepatitis B Virus X Protein Function Requires Zinc Binding. J. Virol. 2019, 93, e00250–19. 10.1128/jvi.00250-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amatya P.; Wagner N.; Chen G.; Luthra P.; Shi L.; Borek D.; Pavlenco A.; Rohrs H.; Basler C. F.; Sidhu S. S.; Gross M. L.; Leung D. W. Inhibition of Marburg Virus RNA Synthesis by a Synthetic Anti-VP35 Antibody. ACS Infect. Dis. 2019, 5, 1385–1396. 10.1021/acsinfecdis.9b00091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houde D.; Berkowitz S. A.; Engen J. R. The Utility of Hydrogen/Deuterium Exchange Mass Spectrometry in Biopharmaceutical Comparability Studies. J. Pharm. Sci. 2011, 100, 2071–2086. 10.1002/jps.22432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weis D. D. Comment on Houde, D.; Berkowitz, S. A.; Engen, J. R., The Utility of Hydrogen/Deuterium Exchange Mass Spectrometry in Biopharmaceutical Comparability Studies. J. Pharm. Sci. 2011, 100, 2071-2086. J. Pharm. Sci. 2019, 108, 807–810. 10.1016/j.xphs.2018.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.