Abstract
Plasmodium falciparum causes the most lethal form of malaria. Peroxide antimalarials based on artemisinin underpin the frontline treatments for malaria, but artemisinin resistance is rapidly spreading. Synthetic peroxide antimalarials, known as ozonides, are in clinical development and offer a potential alternative. Here, we used chemoproteomics to investigate the protein alkylation targets of artemisinin and ozonide probes, including an analogue of the ozonide clinical candidate, artefenomel. We greatly expanded the list of proteins alkylated by peroxide antimalarials and identified significant enrichment of redox-related proteins for both artemisinins and ozonides. Disrupted redox homeostasis was confirmed by dynamic live imaging of the glutathione redox potential using a genetically encoded redox-sensitive fluorescence-based biosensor. Targeted liquid chromatography-mass spectrometry (LC-MS)-based thiol metabolomics also confirmed changes in cellular thiol levels. This work shows that peroxide antimalarials disproportionately alkylate proteins involved in redox homeostasis and that disrupted redox processes are involved in the mechanism of action of these important antimalarials.
Keywords: Plasmodium falciparum, peroxide antimalarials, ozonides, artemisinins, redox homeostasis, glutathione
Malaria is a major global health challenge, and an estimated 409 000 deaths and 229 million new malaria cases occurred worldwide in 2019.1 The majority of malaria-related mortality is due to infection with the deadliest parasite species, Plasmodium falciparum. Currently, antimalarial drugs are one of the most powerful tools for combating malaria. The first-line treatment options for uncomplicated malaria are artemisinin-based combination therapies (ACTs). However, the emergence and spread of parasite strains that are resistant to multiple antimalarial drugs are major challenges to malarial treatment.
The clinically used artemisinins, such as artemether, artesunate, and dihydroartemisinin (DHA), are semisynthetic derivatives of the natural product artemisinin and possess a 1,2,4-trioxane core incorporating a peroxide bond that is essential for activity.2 Artemisinins clear P. falciparum infections rapidly and provide prompt resolution of malarial symptoms in patients with both uncomplicated and severe infections.3 However, artemisinins are limited by poor biopharmaceutical properties and short in vivo half-lives (typically <1 h in humans).4 This necessitates a 3 day treatment regimen and coadministration with a long-acting partner drug.
To overcome some of these limitations, the peroxide bond of artemisinin was used as inspiration for the design of synthetic peroxide antimalarials, collectively known as ozonides. The first-generation ozonide, OZ277 (arterolane), was found to be rapidly acting and exhibits similar blood-stage activity to clinically used artemisinin derivatives, both in vitro and in vivo.5 In 2012, OZ277 was the first synthetic peroxide antimalarial to be approved for clinical use. Continued development of the ozonides led to a second-generation series of compounds that had a significantly improved in vivo exposure profile and maintained the potent in vitro and in vivo activities of the first-generation compounds.6,7 The second-generation ozonide clinical candidate, OZ439 (artefenomel), is the first long half-life peroxide antimalarial to be tested clinically8,9 and has reached advanced stages of development in combination with ferroquine.
Artemisinins and ozonides are thought to act through similar mechanisms as both classes require the peroxide pharmacophore for activity.2,10,11 The proposed mechanism of action involves initial bioactivation by heme released through the parasite digestion of hemoglobin.12−15 This process results in cleavage of the peroxide bond and generation of carbon-centered radicals11,16 that alkylate heme17,18 and proteins19−26 and induce widespread oxidative stress.27−31 Death of the parasite likely results from disruption to multiple vital processes, including hemoglobin degradation in the food vacuole.32
Concerningly, parasites with decreased sensitivity to artemisinins have emerged in the Greater Mekong Subregion33 and more recently in eastern India,34 Africa,35 and Papua New Guinea.36 Clinically, this manifests as delayed parasite clearance following treatment with an ACT37,38 and can lead to approximately 50% treatment failure in areas with concomitant partner drug resistance.39,40 Delayed parasite clearance is associated with point mutations in the propeller domain of the P. falciparum Kelch13 protein41 and decreased parasite susceptibility to short exposures of artemisinin in vitro.42,43 Although the delayed parasite clearance phenotype does not represent complete resistance,44 the emergence of these parasites poses a major threat to global malarial control efforts. For this reason, it is critically important to understand the mechanism of action of antimalarial peroxides, with a view to overcoming resistance. The potential for cross-resistance between artemisinins and ozonides has also been the subject of considerable debate,45 and some reports suggest that the second-generation series of ozonides, e.g., OZ439, may be less susceptible to the mechanisms underpinning decreased artemisinin sensitivity.46−48 This raises the prospect that first-generation ozonides, second-generation ozonides, and artemisinins could alkylate unique targets within the parasite. Previous click chemistry-based proteomics studies aimed at identifying the proteins alkylated by peroxides have shown that artemisinin and simple ozonide probe compounds (containing the 1,2,4-trioxolane core flanked by a cyclohexane moiety on one side and an adamantane group on the other) share a common alkylation signature,22 whereas others have proposed that protein alkylation is indiscriminate and stochastic.20,23
Here, we used chemical proteomics to directly compare the temporal protein alkylation profile of clickable peroxide probes based on artemisinin, the simple ozonide core structure, and the ozonide clinical candidate, OZ439. We employed a unique data-independent acquisition mass spectrometry (DIA-MS) approach and extensive controls to generate an extended and robust list of protein alkylation targets for peroxide antimalarials, identifying redox homeostasis proteins as key targets. The impact on redox homeostasis was confirmed by a ratiometric redox sensor consisting of human glutaredoxin 1 fused to a reduction–oxidation-sensitive green fluorescent protein (GFP) (hGrx1-roGFP2)49 and targeted LC-MS-based thiol metabolomics. Taken together, we demonstrate that the mechanism of action of peroxide antimalarials involves disproportionate alkylation of proteins involved with redox processes and significant disruption to P. falciparum redox homeostasis.
Results
Design and Synthesis of Alkyne Peroxide Probes for the Identification of Alkylated Proteins
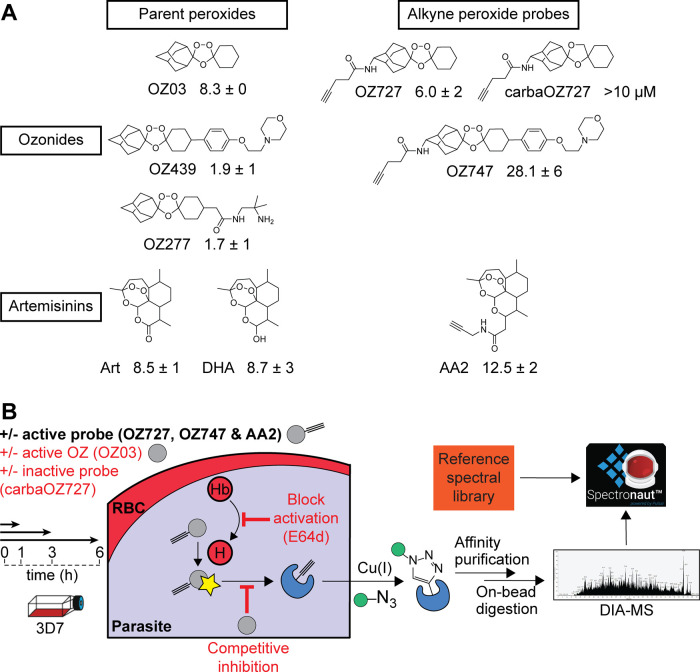
To profile and identify alkylated proteins of peroxide antimalarials, we designed and synthesized a series of artemisinin and ozonide probe compounds with a clickable alkyne tag, including an alkyne probe based on the ozonide clinical candidate, OZ439 (Figure 1A). The alkyne tag can be further appended with a fluorescent dye or azide agarose beads through click chemistry, allowing proteins alkylated by the probes to be visualized on sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) or affinity-purified for mass spectrometric analysis (Figure 1B). The active peroxide probes (AA2, OZ727, and OZ747) containing alkyne functionality retained potency against P. falciparum 3D7 parasites as determined by their 50% inhibition concentration (IC50) in vitro, whereas the alkyne-modified nonperoxide control probe, carbaOZ727, was inactive (Figure 1A). These findings confirmed that the addition of the clickable alkyne tag does not interfere with probe efficacy and that antimalarial activity is peroxide-bond-dependent.
Figure 1.
Identification of proteins alkylated by peroxide antimalarials using click chemistry-based chemical proteomics. (A) Molecular structures of clickable alkyne probes OZ727, OZ747, AA2, and carbaOZ727 and their associated nonclickable controls OZ03, OZ277, OZ439, artemisinin (Art), and dihydroartemisinin (DHA). Activity of test compounds against P. falciparum 3D7 (nM) is shown as the mean ± standard error of the mean (SEM) of data from three independent replicates with at least two technical repeats. (B) General workflow of the copper-catalyzed click chemistry approach used to identify proteins alkylated by peroxide antimalarials. P. falciparum parasites are incubated with the alkyne probes, which get activated (star) by free heme (H) derived from hemoglobin (Hb) digestion to form free radicals that can alkylate protein targets in the cell. The alkyne-modified proteins are affinity-purified by covalent attachment to azide agarose beads using copper-catalyzed click chemistry and identified using data-independent acquisition (DIA) LC-MS/MS. The complex spectra obtained from the MS are then processed using a reference in-house P. falciparum spectral library in Spectronaut Software. As an additional strategy to identify the biologically relevant alkylated proteins, peroxide activation was blocked in situ with the falcipain hemoglobinase inhibitor, E64d, which antagonizes the antimalarial effects of peroxides, and the nonclickable parent ozonide, OZ03, served as a competitive inhibitor of probe binding in situ to distinguish specific versus nonspecific protein hits. Control conditions are shown in the red text.
Fluorescence Labeling of Peroxide-Alkylated Proteins
As an initial step, in vitro cultures of trophozoite-stage P. falciparum 3D7 were treated for 3 h with 300 nM of the active (OZ727 and OZ747) or inactive (carbaOZ727) alkyne ozonide probes, a time and concentration previously shown to be pharmacologically relevant under the incubation conditions used in this study (10% parasitemia and 2% hematocrit (Hct)).12,32 Cultures incubated with an equivalent volume of dimethyl sulfoxide (DMSO) acted as an additional negative control. Protein extracts were prepared from the treated live parasites, clicked with a fluorescent dye, and resolved by SDS-PAGE. Only alkylated proteins with the alkyne handle can be labeled and visualized with fluorescence scanning. Many proteins were fluorescently labeled in the samples containing the active ozonide probes (Figure S1). OZ727 showed the strongest signal after 3 h, agreeing with the known faster iron activation kinetics of simple ozonides, when compared to slower-acting second-generation compounds (e.g., OZ439), which possess a more sterically restricted peroxide bond.6,66−68 Importantly, fluorescent labeling was not observed for the inactive negative control probe, carbaOZ727, or untreated (DMSO) samples (Figure S1), confirming the effectiveness and specificity of our chemoproteomic strategy.
Identification of Proteins Alkylated by Peroxide Antimalarials
For the identification of peroxide covalent binding proteins by mass spectrometry, we employed a time course treatment and comprehensive control strategy to ensure specific and high confidence identification of biologically relevant alkylated proteins (Figure 1B). We have previously shown that artemisinins have a faster onset of activity compared to ozonides and that this correlates with their relative rates of activation by iron.13,69 To reflect the difference in peroxide activity kinetics, we exposed synchronized trophozoite cultures to only a short 1 h treatment with the alkyne artemisinin probe, AA2 (300 nM). The exposure time was extended to up to 6 h for the simple alkyne ozonide probe, OZ727 (similar to the exposures used in other chemoproteomic studies with similar ozonide probes),20,22 and the OZ439-like probe, OZ747 (both 300 nM). Cultures incubated for 1 h with the inactive (clickable) nonperoxide probe, carbaOZ727, and untreated cultures (DMSO) were included to control for protein identifications that are unrelated to peroxide antimalarial activity and proteins that bind nonspecifically to the agarose beads used for affinity purification. We have previously shown that peroxide treatment causes significant upregulation of parasite proteins.32 Therefore, the treatment of P. falciparum cultures with the active, nonclickable parent ozonide, OZ03, that are time-matched to each alkyne probe incubation (1–6 h) was also included to further control for false-positive identifications resulting from the nonspecific binding of proteins to the agarose beads. OZ03 also served as a competitive inhibitor of probe binding in situ to distinguish specific versus nonspecific protein hits (Figure 1B). As parasite degradation of hemoglobin provides the iron source for peroxide activation, we blocked peroxide activation in situ with E64d (Figure 1B), a falcipain hemoglobinase inhibitor known to antagonize the antimalarial effects of peroxides,12,13,27 to further eliminate proteins unlikely to be involved with the action of peroxides in parasite killing.
Following probe treatment of synchronized trophozoite cultures, parasite isolation, and protein extraction, alkyne-labeled proteins were affinity-purified by copper-catalyzed click chemistry onto azide agarose beads and probe-alkylated proteins were identified with DIA-MS (Figure 1B). Protein intensities were used to provide semiquantitative analysis of protein abundances for a total of 928 parasite proteins identified across all sample groups (Supporting Data 1). Proteins in the OZ727, OZ747, and AA2 groups with a fold-change ≥2 compared to the DMSO, carbaOZ727, and time-matched OZ03 controls, and with a p-value <0.05 (Mann–Whitney U test) compared to those in carbaOZ727 or DMSO, were considered as alkylated proteins. After the removal of false-positive protein identifications and nonspecific protein hits, a total of 182, 94, and 261 parasite proteins were, respectively, enriched for OZ727, OZ747, and AA2 after 1 h of treatment (at least four independent experiments) (Figure 2A). OZ727 labeling was markedly decreased by coincubation with increasing concentrations of OZ03 (active nonclickable parent ozonide), indicating that the engineered alkyne probes alkylate the same proteins as their nonclickable parents (Figure 2A and Supporting Data 1). Furthermore, preincubation of parasite cultures with E64d drastically decreased the labeling of proteins with OZ727 (Figure 2A and Supporting Data 1), consistent with the reports that inhibiting hemoglobin digestion abrogates peroxide activity.12,13,27,70 Taken together, these results indicate that the alkyne probes are pharmacologically similar to the parent compounds and the alkylated proteins identified in this study are associated with their antimalarial peroxide activity.71
Figure 2.
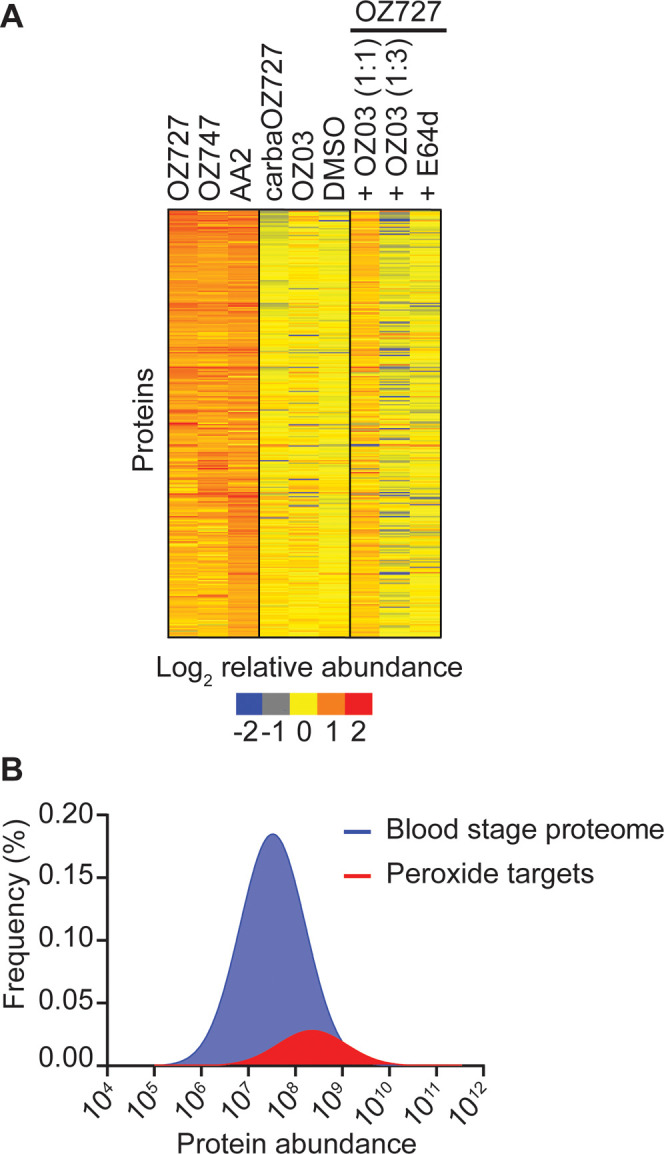
Proteins enriched by peroxide antimalarials in P. falciparum. (A) Heatmap representation of all proteins enriched by OZ727, OZ747, and AA2 following the treatment of P. falciparum infected RBCs for 1 h (all 300 nM). Heatmap analysis was performed by plotting the average log-transformed protein abundance for each protein and normalizing by the mean for that protein across all samples. Proteins in the OZ727 (n = 6), OZ747 (n = 5), and AA2 (n = 4) groups with a fold-change ≥2 compared to those in the DMSO (n = 9), carbaOZ727 (n = 7), and OZ03 (n = 3) controls, and with a p-value <0.05 (Mann–Whitney U test) compared to that of carbaOZ727 or DMSO, were considered as alkylated proteins. Protein labeling by OZ727 was decreased in the presence of increasing concentrations of the active nonclickable parent ozonide, OZ03 (n = 3–4), and by preincubation of parasite cultures with the cysteine (Cys) hemoglobinase inhibitor, E64d (n = 4). For additional protein information, see Supporting Data 1. (B) Histogram showing that the 436 proteins enriched by the peroxide probes across all study time points (red) are distributed across the P. falciparum blood-stage protein abundance range and not limited to the most abundant proteins within the parasite. The frequency distribution of proteins in the P. falciparum blood-stage proteome (Siddiqui et al., unpublished data) is shown in blue.
Importantly, the proteins labeled by OZ727, OZ747, and AA2 were not simply the proteins that are reproducibly detected as the most abundant proteins in untargeted proteomic studies of P. falciparum parasites (Figure 2B). Approximately 45% of the proteins identified as being alkylated by peroxides were among the top 500 most abundant proteins in the P. falciparum blood-stage proteome. This contrasts with previous peroxide chemical proteomics studies, where between 60 and 80% of proteins identified were among the most abundant proteins.21,23 A significant proportion of the proteins identified as being alkylated by peroxides were also detected by Wang et al.23 (55%) and Ismail et al.2121 (36%) (Figure S2A). However, only 13 proteins were common between all three studies and these were all highly expressed P. falciparum blood-stage proteins, with the exception of the putative Fe–S cluster assembly protein DRE2 (PF3D7_0824600) (Figure S2B). In another peroxide chemical proteomic study, where the NF54 parasite line was used, 25 alkylated proteins were identified with 100 ng/mL of the chemical probes (similar to the probe concentration used in this study). Of the 25 P. falciparum alkylated proteins, 8 were also identified in our list of proteins; however, these 8 proteins were not the same as the common 13 proteins identified across the other studies.20 In addition to these commonly identified proteins, our unique data acquisition approach with extensive controls added a further 352 biologically relevant proteins to the list of proteins alkylated by peroxides in P. falciparum (Figure S2B).
In our data set, approximately 30% of the combined 435 proteins alkylated by OZ727, OZ747, and AA2 were enriched by all three compounds. A range of proteins were also identified as unique alkylation targets for OZ727 (79 proteins), OZ747 (35 proteins), or AA2 (59 proteins). For example, AA2 alkylated several proteins involved in vesicle-mediated transport (PF3D7_0303000, PF3D7_1231100, and PF3D7_1429800) that were not binding targets for OZ727 or OZ747. Similarly, several proteins involved in nucleotide biosynthesis (PF3D7_0206700, PF3D7_0629100, PF3D7_1012600, and PF3D7_1437200), an essential pathway for parasite growth and survival, were only alkylated by OZ727, while OZ747 uniquely alkylated proteins involved in ion transport (PF3D7_0715900 and PF3D7_1362300). Some of the major proteins previously identified as targets of artemisinin and simple ozonide probe compounds, such as translationally controlled tumor protein (PF3D7_0511000),72 actin 1 (PF3D7_1246200), ornithine aminotransferase (PF3D7_0608800), and the glycolytic enzymes, lactate dehydrogenase (PF3D7_1324900), enolase (PF3D7_1015900), and glyceraldehyde-3-phosphate dehydrogenase (PF3D7_1462800),21−23 were confirmed to be alkylation targets for AA2 and OZ727 but were not alkylated by the OZ439 analogue, OZ747.
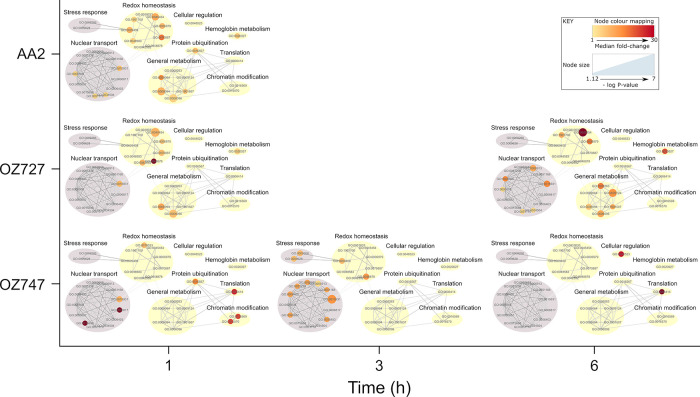
We performed gene ontology (GO) enrichment analysis to identify the major cellular processes and compartments targeted by the peroxide antimalarials. These data showed that the alkylation targets of OZ727, OZ747, and AA2 were significantly enriched in several cellular components, including the cytosol, ribosome, nucleus, and food vacuole (Supporting Data 2). Notably, the food vacuole (GO:0020020) was the most significantly enriched compartment for both OZ727 (p-value = 9.90 × 10–12) and AA2 (p-value = 1.30 × 10–15) at the 1 h time point and was the second most significantly enriched compartment for OZ747 (p-value = 0.00019) (Supporting Data 2). Pretreatment with E64d and coincubation with increasing concentrations of OZ03 inhibited OZ727 binding to food vacuole proteins (Figure S3), confirming that these targets are important for peroxide activity. These findings are consistent with peroxides initially acting by disrupting parasite hemoglobin digestion,32 an essential process that occurs within the parasite food vacuole. GO analysis further revealed that the proteins alkylated by the peroxide probes were involved in numerous essential biological processes of the parasite (Figure 3 and Supporting Data 3). Several significantly enriched biological processes were identified in redox homeostasis, metabolism, nuclear transport, and stress response (Figure 3). To control for nonspecific protein binding, the enrichment analysis of proteins identified in the control samples found GO terms associated with nuclear transport and stress response, suggesting that those functions may not be specifically targeted by these drugs. However, the enrichment of GO terms associated with redox homeostasis and metabolism was specific to the peroxide-treated parasites and not the controls (Figure 3 and Supporting Data 3). Of these, cell redox homeostasis (GO:0045454) was one of the most significantly enriched biological process GO terms for both OZ727 (p = 0.00015) and AA2 (p = 0.0016) after 1 h and was not enriched in the control data set (Figure S4 and Supporting Data 3).
Figure 3.
Gene ontology (GO) biological process enrichment of P. falciparum proteins covalently interacting with peroxide antimalarials. Proteins alkylated by the peroxide probes are involved in multiple vital biological processes. GO analysis was conducted using topGO. PlasmoDB GO terms were used for GO term mapping, and the elim algorithm was applied to navigate the topology of the GO graph. The Fisher exact test was used for testing the enrichment of GO terms. P. falciparum proteins from the in-house spectral library acted as the genomic background for statistical testing, and GO terms represented by fewer than five proteins in this library were excluded from the analysis. Significantly enriched GO terms (p < 0.05) were filtered to exclude those with <1.5-fold enrichment of protein identifications relative to the background library. These were then visualized with Cytoscape 3.6 and manually clustered based on the alkylated protein targets shared between GO terms and the semantic similarity measure of Schlicker et al.59 Node size represents p-value. Node color represents the median fold-change (versus DMSO) for proteins within each GO term. GO term clusters that were also represented in an identical analysis of proteins identified in control samples are shaded gray.
A targeted analysis of the 28 proteins within the cell redox homeostasis GO term identified nine alkylated proteins that were enriched by both OZ727 and AA2 within 1 h of treatment (Figure S5A), and alkylation was decreased upon coincubation of OZ727 with OZ03 or preincubation with E64d (Figure S5B). The enriched proteins included several thioredoxin proteins, protein disulfide isomerases, and peroxiredoxin, which are important for maintaining parasite antioxidant defenses and regulating redox activity of proteins.73 Notably, similar sets of proteins were recently identified as being alkylated by related ozonide clickable probes in mammalian cells.74 Similar trends were seen for some of these proteins in the OZ747 group (Figure S5A). Differences in the extent of redox homeostasis protein labeling by the probes likely reflect the difference in peroxide activity kinetics between faster-acting artemisinin (AA2) and simple ozonide (OZ727) probes, compared to that of the slower-acting compounds based on second-generation ozonides (e.g., OZ439-like compounds such as OZ747).12,13,67,68 Oxidation–reduction (GO:0055114) and response to oxidative stress (GO:0006979) processes were also significantly enriched biological process GO terms (Supporting Data 3). Combined, these data suggest that the altered redox regulation is an important mechanism for the activity of antimalarial peroxides.
Validation of Recombinant hGrx1-roGFP2 Probe with Peroxide Antimalarial Drugs In Vitro
To further investigate the effects of peroxide antimalarials on the redox status of P. falciparum parasites, we used the genetically integrated ratiometric redox sensor hGrx1-roGFP2 (human glutaredoxin 1 fused to reduction–oxidation-sensitive green fluorescent protein), expressed in the cytosol of the NF54attBP. falciparum parasite line (NF54attB[hGrx1-roGFP2]).49 To exclude a direct influence of the drugs on the probe, we initially characterized the interaction of the peroxides with recombinant hGrx1-roGFP2 in vitro. DHA and the ozonides, OZ277 and OZ439, were tested at concentrations ranging from 0.1 μM to 1 mM for up to 10 h in a standard reaction buffer containing ferrous iron (to activate the endoperoxide bond) in a plate reader. When compared to DMSO control, the drugs did not significantly affect the fluorescence ratio of recombinant hGrx1-roGFP2 over time (Figure S6), which is consistent with previously published data for peroxides and recombinant hGrx1-roGFP2.61,62 The recombinant protein was fully oxidized with 1 mM diamide (DIAM) and fully reduced with 10 mM dithiothreitol (DTT) (Figure S6).
Time-Dependent Effects of Peroxides on the Glutathione Redox Potential of P. falciparum
The redox effects of clinically relevant concentrations and exposures of DHA, OZ277, and OZ439 were assessed using NF54attB[hGrx1-roGFP2] transgenic parasites in both confocal laser scanning microscopy (CLSM) and plate reader assays (Figures 4 and S7A,B). To measure the completely oxidized and reduced state of the probe, 30–34 h post invasion (h.p.i.) trophozoites (6–8% parasitemia and 2% Hct) were incubated for 5 min with 1 mM DIAM or 10 mM DTT, respectively, blocked with 2 mM N-ethylmaleimide (NEM), and then magnetically enriched. Peroxide-induced effects on parasite oxidative status were monitored at concentrations ranging from 0.1 to 1 μM and at intervals between 10 min and 9 h, after which free thiol groups were blocked with 2 mM NEM and trophozoite-infected RBCs were magnetically enriched (Figures 4 and S7A,B).
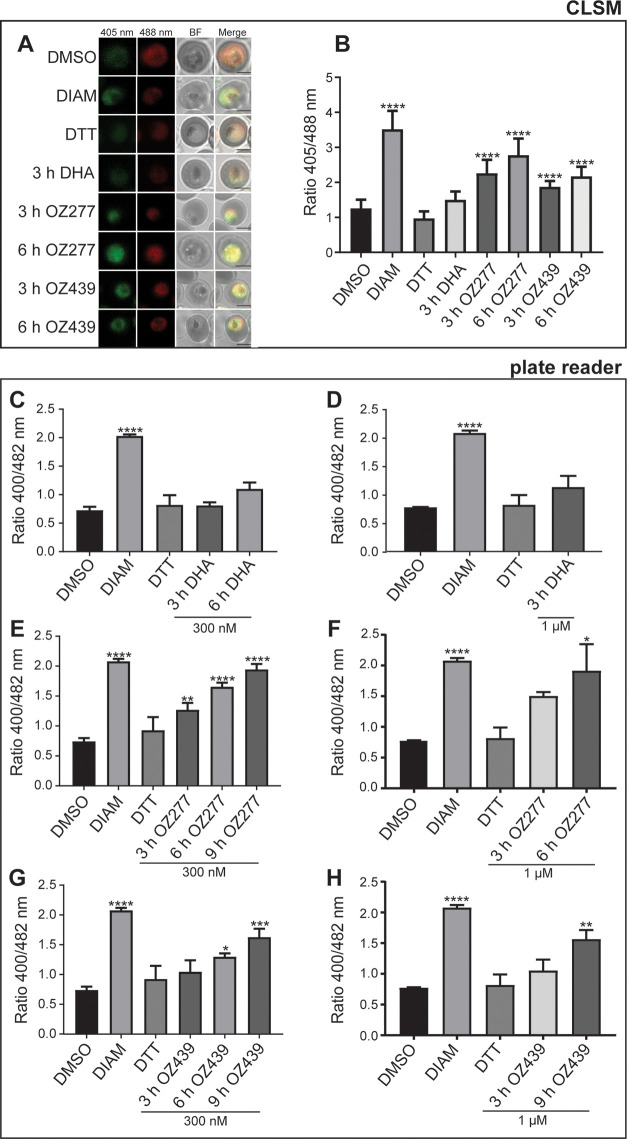
Figure 4.
Effects of peroxide antimalarials on the redox ratio of P. falciparum NF54attB[hGrx1-roGFP2] parasites. NF54attB[hGrx1-roGFP2] transgenic parasites were treated with DHA, OZ277, and OZ439 at 300 nM, and samples were taken at 3 h post treatment for DHA, OZ277, and OZ439 and at 6 h for OZ277 and OZ439 for the measurement of the fluorescence ratio of the redox sensor. Fluorescence ratio of the redox sensor increased for parasites treated with OZ277 and OZ439 for 3 and 6 h treatments, while DHA treatment did not change the redox ratio as measured with confocal laser scanning microscopy (CLSM) (A, B). Oxidized (405 nm; green), reduced (488 nm; red), BF: bright field, merge: is an overlay of all of the panels. The black magnification bar represents 2 μm. Plate reader measurement of the redox ratio following the treatment of transgenic parasites with DHA (300 nM (C), 1 μM (D)), OZ277 (300 nM (E), 1 μM (F)), and OZ439 (300 nM (G), 1 μM (H)) showed the same results as CLSM, where only the ozonides affected the redox ratio and this ratio increased over the duration of drug exposure. DMSO-treated parasites at the longest drug incubation duration acted as control, DTT (10 mM, 5 min treatment) was the fully reduced control, while DIAM (1 mM, 5 min treatment) was the fully oxidized control. Columns represent the mean of at least two independent experiments with the error bars expressed as SEM. CLSM data were composed of 15–35 trophozoites analyzed per experiment for each condition. p-Value was calculated using one-way analysis of variance (ANOVA) (*, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001).
Both OZ277 and OZ439 caused time- and concentration-dependent oxidation of the NF54attB[hGrx1-roGFP2] parasite cytosol in the CLSM and plate reader when tested at 300 nM for 1–9 h (Figures 4 and S7B). OZ277 at 300 nM increased the fluorescence ratio of NF54attB[hGrx1-roGFP2] parasites by 1.27-fold via plate reader (p-value <0.01) (Figure 4E) and 2.25-fold via CLSM detection (Figures 4A,B and S7B) (p-value <0.0001) within 3 h of drug treatment and oxidation peaked after 9 h (2.0-fold increase, p-value <0.0001), when compared to the DMSO control. In contrast, OZ439 at 300 nM increased cytosolic oxidation after 6 h of exposure (1.3-fold increase via plate reader detection (p-value <0.05) (Figure 4G) and 1.8-fold increase via CLSM detection (p-value <0.0001)) (Figures 4A,B and S7B) and had a less pronounced impact compared to OZ277 (2.0-fold) on the fluorescence ratio over a 9 h period (1.63-fold increase, p-value <0.001), consistent with OZ439 having slower activity kinetics within the parasite.32 A similar temporal increase in cytosolic oxidation was observed in parasites treated with 1 μM of OZ277 or OZ439 (Figure 4F,H).
Surprisingly, DHA showed no significant effect on the fluorescence ratio of NF54attB[hGrx1-roGFP2] parasites in either the CLSM or plate reader when tested at 300 nM for 1, 3, or 6 h (Figures 4A–D and S7B). To rule out a concentration-dependent effect on the fluorescence ratio and the possibility that DHA induced a rapid oxidative change in the cytosol that quickly returned to basal levels, we also tested DHA concentrations as high as 1 μM (Figure 4D) and exposures as short as 10 min (100 nM) (Figure S7A) in the plate reader assay. No significant change in the fluorescence ratio was detected with short DHA exposures or at a concentration up to 1 μM (Figure 4C,D), suggesting that DHA had no effect on the cytosolic glutathione-dependent redox ratio of NF54attB[hGrx1-roGFP2] parasites under these conditions.
As expected, treatment with 1 mM DIAM led to a significant increase of the fluorescence ratio of NF54attB[hGrx1-roGFP2] parasites with both the CLSM and plate reader, whereas there was no change in the fluorescence ratio after treatment with 10 mM DTT when compared to that of the untreated control (Figures 4 and S7A,B). This is consistent with the basal cytosolic environment of P. falciparum being strongly reducing.61
Free Thiol and Glutathione Levels in Peroxide-Treated P. falciparum Infected RBCs
To further interrogate peroxide-mediated changes to the intracellular redox milieu, we employed a thiol derivatization-targeted mass spectrometry-based metabolomics approach to accurately measure the abundance of small-molecule thiols and related metabolites within peroxide-treated infected RBCs (P. falciparum 3D7 strain (Figures 5 and S8A,B)) (NF54attB[hGrx1-roGFP2] strain (Figure S8C)). Trophozoite-infected RBCs (30–34 h.p.i.) at 6–8% parasitemia and 2% Hct were treated with DHA (100 nM for 3 h), OZ277 (300 nM for 3 h), or OZ439 (300 nM for 6 h). Equivalent cultures treated with DIAM (1 mM for 5 min), DTT (10 mM for 5 min), or an equal volume of DMSO (6 h) acted as the oxidation, fully reduced, and untreated control, respectively. Following the drug incubation, the cells were magnetically enriched and extracted with 50 mM NEM to rapidly and irreversibly derivatize thiol metabolites and protect them from oxidation during subsequent sample preparation, thus preserving their redox state (method 2) (Figure S8B). As the magnetic enrichment step could alter the cellular redox state prior to NEM stabilization, we also performed the same drug treatments on magnetically purified cultures that were extracted and derivatized with NEM immediately after the drug incubation (method 3). NEM stabilization was also performed after drug treatment and prior to the magnet enrichment step (method 1). In all cases, results were compared to those of DMSO-treated infected RBCs. For peroxide treatments performed on magnetically purified cultures, the activity of the peroxide antimalarials was confirmed to be in the expected nanomolar range for a 3 or 6 h pulse and the IC50 values were equivalent in both 3D7 and NF54attB[hGrx1-roGFP2] parasites, confirming that the peroxides act similarly in both lines (Figure S8D).
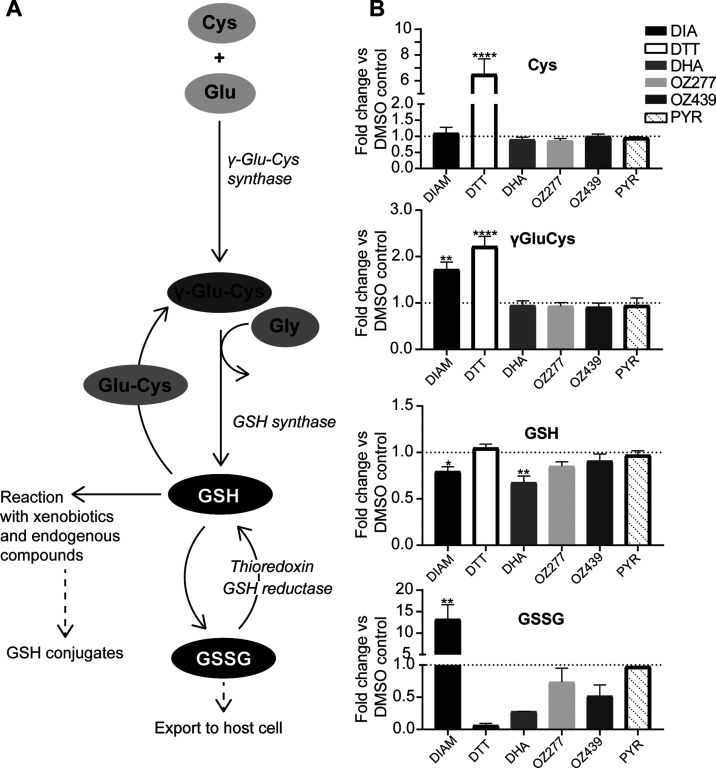
Figure 5.
Effect of peroxide antimalarials on thiol levels in P. falciparum 3D7 infected RBCs. (A) Schematic of glutathione biosynthesis pathway. (B) Determination of relative levels of NEM-derivatized cysteine (Cys), γ-glutamyl cysteine (γ-GluCys), reduced glutathione (GSH), and oxidized glutathione (GSSG) compared to that of DMSO control in P. falciparum 3D7 parasites following treatment with DHA (100 nM, 3 h treatment), OZ277 (300 nM, 3 h treatment), OZ439 (300 nM, 6 h treatment), and pyrimethamine (PYR) (1 μM, 5 h treatment) using method 3. DTT (10 mM, 5 min treatment) was the fully reduced control, while DIAM (1 mM, 5 min treatment) was the fully oxidized control. Bars represent the fold-change (mean ± SEM) compared to DMSO control. Thiol measurement is from two to five biological replicates (with four technical within each biological). p-Value was calculated using one-way ANOVA (*, p < 0.05; **, p < 0.01; ****, p < 0.0001).
In cultures immediately stabilized with NEM (method 3), the oxidant DIAM significantly depleted GSH (p-value <0.05) and there was a corresponding increase in the levels of oxidized glutathione (GSSG) (p-value <0.01), while the reductant DDT depleted GSSG only (Figure 5). This is consistent with DIAM causing an oxidative shift and DTT causing a reducing shift in the free cellular glutathione redox balance and agrees with the fluorescence ratio observed in DIAM- and DTT-treated NF54attB[hGrx1-roGFP2] parasites (Figure 4). The glutathione precursor γ-glutamyl cysteine (γ-GluCys) was increased after DIAM (p-value <0.01) and DTT (p-value <0.0001) exposure, likely as a response to restore cellular glutathione levels via increased synthesis (Figure 5). However, cysteine (Cys) levels were not affected by DIAM treatment, but DTT treatment caused a significant increase in Cys levels (p-value <0.0001) (Figure 5). This significant increase in Cys levels is likely a result of all cystine (the oxidized dimer of Cys) in the infected RBCs being converted to the reduced form (Cys).
Both OZ277 and OZ439 showed a trend toward depleted GSH levels (not significant), with GSSG levels being unaffected (Figure 5). The ozonides caused no detectable changes in Cys or γ-GluCys levels. DHA treatment depleted the total pool of free glutathione, and this included a significant decrease in reduced GSH (p-value <0.01) and a trend toward decreased oxidized GSSG (1.5- and 3.7-fold decreases, respectively) levels (Figure 5). Cys and γ-GluCys levels were unaffected by DHA treatment (Figure 5). Pyrimethamine, an antimalarial that targets the parasite’s dihydrofolate reductase, did not affect the relative abundance of thiols in our assay (Figure 5). For most of the compounds tested, similar results were also observed with 3D7 cultures magnetically enriched and stabilized after drug treatment (method 1 and method 2; Figure S8A,B) and in the NF54attB[hGrx1-roGFP2] parasite strain (method 3; Figure S8C).
Discussion
Here, we have used click chemistry-based chemical proteomics with an extensive control strategy to show that peroxide antimalarials based on artemisinin and ozonides alkylate similar sets of proteins within P. falciparum and that these proteins are involved in numerous essential biological processes. These findings agree with previous studies demonstrating that ozonides and artemisinins have a similar multifaceted mode of action and that they disrupt multiple processes important for parasite survival.20−23,32 We also identified a set of proteins that were alkylated exclusively by specific peroxides. This suggests that the structures around the peroxide bond may lead to subtle differences in the underlying mechanisms by which artemisinins, first-generation ozonides, and second-generation ozonides kill the malaria parasite and could have important implications for ozonide cross-resistance with artemisinins.
Functional enrichment analysis of the peroxide-alkylated protein targets identified food vacuole proteins to be significantly enriched, consistent with this compartment being an important initial site of damage for peroxides,32 as well as proteins involved in redox processes. Peroxide-induced perturbation of the parasite antioxidant defense system was then confirmed by (1) a parasite cytosolic-specific redox probe, which determined glutathione-dependent redox state in real time, and (2) targeted liquid chromatography-mass spectrometry (LC-MS)-based thiol metabolomics, involving derivatization of thiol metabolites with NEM, to accurately measure total parasite glutathione in both its reduced (GSH) and oxidized (GSSG) forms and its precursors.
Previous studies using clickable artemisinin and ozonide probes to identify peroxide alkylation targets have demonstrated a minimal overlap in proteins (25 in total).21,23 When including the alkylation targets identified in our study, only 13 proteins were common between all three data sets (Figure S2). Further analysis of these 13 proteins identified them to all be highly abundant in the P. falciparum blood-stage proteome (Figure S2), with the exception of PF3D7_0824600 (Fe–S cluster assembly protein DRE2). This minimal overlap could be due to random alkylation of proteins by the peroxides, as proposed by Jourdan et al.20 However, we identify a significant proportion of proteins from each of these studies individually (36 and 55%), which argues against this hypothesis. An alternative explanation is likely due to the type of mass spectrometry analysis, which in all previous studies utilized the data-dependent acquisition approach (DDA). DDA selects precursor ions, corresponding to peptides, for fragmentation in the mass spectrometer based on their abundances, so only the most abundant peptides will produce fragment spectra, which are necessary for protein identification. In a complex sample, this data-dependent approach can lead to inconsistent peptide identification, which limits the number of proteins that can be reproducibly identified and quantified, often favoring highly abundant proteins.75 In contrast, the mass spectrometry approach used in our study (DIA-MS) selects all precursor ions (peptides) within a mass window and fragments them regardless of their intensity.76 This approach results in an extremely high run-to-run reproducibility and a more comprehensive data set that is not biased toward highly abundant proteins. This is evident from our data, where we have significantly expanded the list of peroxide alkylation targets by ∼64% and the identified proteins ranged in abundance across the P. falciparum blood-stage proteome (Figure 2). The potential limitation of our DIA approach is the increased identification of low-abundance non-drug-specific proteins that bind to the agarose resin. However, this potential issue was overcome by the inclusion of an extensive set of six control conditions that accounted for any background binding. Overall, the analysis of a total of 36 control samples, alongside the 23 samples from active alkyne-tagged peroxides, provides extremely high confidence in the alkylated proteins identified in this study.
GO analysis of the proteins alkylated by the peroxide probes identified the food vacuole and redox homeostasis as the most significant cellular compartment and biological process to be enriched, respectively (Figure 3 and Supporting Data 2 and 3). Confirming that these proteins are important for the activity of peroxides, alkylation of these proteins with OZ727 was decreased in the presence of the nonclickable parent ozonide, OZ03, and following pretreatment with the falcipain hemoglobinase inhibitor, E64d, which blocks peroxide activation (Figures S3 and S5). This also confirms that heme derived from hemoglobin digestion is the major iron source responsible for peroxide activation. The food vacuole is a site considered important for the heme-dependent activation of peroxides; heme itself is a known small molecule alkylated by peroxides,14,17,18 and alkylated proteins functioning in the food vacuole were also shown to be enriched in previous peroxide mode of action studies.21,23 Furthermore, the hemoglobin digestion pathway within the food vacuole is known to be significantly perturbed following treatment with the peroxides.32
GO terms associated with general metabolic processes were also specifically enriched in peroxide-treated parasites and not the controls (Figure 3 and Supporting Data 3). These enriched terms included glycolysis (GO:0006096), which is consistent with previous studies identifying glycolytic proteins as alkylation targets of peroxides.21−23 However, our previous metabolomic analyses revealed that the abundance of glycolysis metabolites32 and flux through the glycolysis pathway77 are unaffected by peroxide exposure. It is possible that a corresponding increase in the abundance of glycolytic proteins32 compensates for the loss of enzyme function, raising questions as to the functional importance of these proteins to the mechanism of action of peroxides.
The antioxidant system is essential within the parasite not only for the maintenance of redox homeostasis but also for the parasite’s ability to respond to drug-induced stress.78 During blood-stage development, reactive oxygen species produced as a byproduct of hemoglobin digestion induces intraparasitic oxidative stress, which the parasite manages by engaging a highly efficient antioxidant defense system,78 which includes glutathione- and thioredoxin-related proteins.78 Parasite glutathione is crucial for maintaining redox balance and has been linked to various drug resistance mechanisms, including resistance against the antimalarial drug chloroquine78,79 and more recently artemisinins.80,81 As glutathione is the major intraparasitic antioxidant, we used a parasite cytosolic-expressed fluorescence-based glutathione biosensor, hGrx1-roGFP2, to monitor glutathione redox potential in infected RBCs treated with various peroxide antimalarials. Interestingly, we found that the ozonides perturbed the parasite cytosolic redox ratio in a concentration- and time-dependent manner, while DHA had no measurable impact on the redox ratio. This was shown using both plate reader and microscopy (Figure 4) and was somewhat surprising considering the involvement of free radicals in the mode of action of artemisinins.82−84 Interestingly, DHA was shown to increase the redox ratio in a time-dependent manner when the same probe was localized to the parasite mitochondria,62,80,85,86 suggesting that artemisinins differentially affect the redox status of parasite organelles. We therefore measured the total glutathione and its associated metabolites using a targeted LC-MS-based metabolomics method, where we derivatized parasite thiols using NEM to capture the parasite redox state. Our redox analytical method demonstrated that peroxides generally had a trend toward the depletion of total glutathione (GSH and GSSG), with DHA having the most profound effect, compared to control (Figure 5). DHA significantly depleted both the reduced (GSH) and oxidized (GSSG) forms of glutathione, which likely explains the lack of impact on the glutathione redox balance (based on the ratiometric fluorescent biosensor), despite clear depletion of reduced glutathione as measured by LC-MS. In addition to its direct role in redox homeostasis, glutathione plays an important role in repair mechanisms for proteins that have undergone oxidative damage, in addition to potential roles in heme degradation and conjugation of small molecules. While it is possible that glutathione is consumed by direct quenching of the peroxide-derived radicals,84 we propose that a significant amount of glutathione is consumed by the parasite’s stress response32 to this extensive protein alkylation (Figure 2).
DHA demonstrated significantly more depletion of total glutathione levels than the ozonides, which is consistent with the faster onset of activity of artemisinins compared to ozonides13 and their temporal effects on parasite biochemical pathways.32 More extensive direct alkylation of free thiols by artemisinins may also contribute to the greater depletion of parasite glutathione levels by DHA compared to that of the ozonides.25 While the ozonides had a greater impact than DHA on the glutathione redox ratio (Figure 5), all tested peroxides alkylated several proteins involved in redox homeostasis (Figure 3), suggesting that redox imbalance plays a critical role in the mode of action of these two classes of peroxide antimalarials, albeit the specific role differs between classes.31,87,88 In addition to the protein alkylation events, drug conjugates with the redox active small-molecule heme,14,17,18 are also expected to contribute to redox imbalance in peroxide-treated parasites.89 Supporting this redox stress hypothesis, pro-oxidant molecules are known to enhance the activity of artemisinins28 and parasites with a mutant Kelch13 propeller allele, linked to artemisinin resistance both in vitro and in vivo, have elevated glutathione levels.81 Glutathione has also been associated with artemisinin sensitivity in other parasite lines,90−92 and we propose that glutathione metabolism may provide opportunities for intervention with pro-oxidant combinations to enhance peroxide sensitivity in the context of artemisinin resistance.93
Conclusions
We have demonstrated with chemical proteomics that peroxide antimalarials alkylate a range of Plasmodium proteins that are vital for parasite growth and survival. The parasite antioxidant system was one of the crucial pathways targeted. Using genetically encoded redox sensors and LC-MS-based targeted thiol measurements, we showed that peroxide antimalarials cause significant disruption to P. falciparum glutathione metabolism. This detailed description of the alkylated protein targets of artemisinins and ozonides, and the important role of redox metabolism in their modes of action, will underpin future research to optimize their efficacy against increasingly common artemisinin-resistant parasites.
Methods
P. falciparum Cell Culture
3D7 and the NF54attB[hGrx1-roGFP2]P. falciparum transgenic line49 were cultured as previously described.50 Parasites were tightly synchronized by double sorbitol lysis 14 h apart.
Peroxide Antimalarials and Alkyne-Tagged Peroxide Probes
DHA and artemisinin were purchased from Jomar Bioscience and Sigma-Aldrich, respectively. OZ277 and OZ439 were provided by the Medicines for Malaria Venture (Geneva). OZ03, the artemisinin alkyne, AA2, and ozonide alkynes, OZ727 and carbaOZ727, were obtained by previously published procedures.20,21,51 Synthesis of the alkyne ozonide OZ747, the click chemistry probe analogous to the advanced-stage antimalarial clinical candidate, OZ439 (artefenomel), is described below.
Synthesis of OZ747
cis-6-(Pent-4-ynamido)adamantane-2-spiro-3′-8′-[4-(2-morpholinoethoxy)phenyl]-1′,2′,4′-trioxaspiro[4.5]decane Mesylate (OZ747)
Step 1: to a solution of cis-6-oxoadamantane-2-spiro-3′-8′-(4-hydroxyphenyl)-1′,2′,4′-trioxaspiro[4.5]decane52 (500 mg, 1.35 mmol) in dry DME (25 mL) were added powdered NaOH (324 mg, 8.01 mmol) and Bu4NHSO4 (93 mg, 0.50 mmol). The resulting mixture was stirred at room temperature (rt) for 30 min before the addition of N-(2-chloroethyl)morpholine hydrochloride (415 mg, 2.22 mmol). The resulting reaction mixture was stirred at 60 °C for 12 h. After filtration of the solid material, the filtrate was concentrated in vacuo to afford a residue, which was dissolved in ethyl acetate (EA) (100 mL), washed with H2O (50 mL) and brine (50 mL), dried over MgSO4, and filtered and concentrated in vacuo to afford cis-6-oxoadamantane-2-spiro-3′-8′-[4-(2-morpholinoethoxy)phenyl]-1′,2′,4′-trioxaspiro[4.5]decane (560 mg, 86%) as a white solid. mp 145–146 °C. 1H NMR (500 MHz, CDCl3) δ 1.67–1.75 (m, 2H), 1.84–1.92 (m, 4H), 1.94 (d, J = 13.0 Hz, 2H), 1.99 (d, J = 13.0 Hz, 2H), 2.06 (d, J = 13.0 Hz, 2H), 2.15 (s, 2H), 2.26 (d, J = 13.0 Hz, 2H), 2.35 (d, J = 13.0 Hz, 2H), 2.50 (m, 3H), 2.57 (s, 4H), 2.79 (t, J = 5.5 Hz, 2H), 3.73 (t, J = 4.5 Hz, 4H), 4.09 (t, J = 5.5 Hz, 2H), 6.84 (d, J = 8.5 Hz, 2H), 7.12 (d, J = 8.5 Hz, 2H); 13C NMR (125 MHz, CDCl3) δ 31.57, 34.62, 35.68, 35.75, 35.90, 41.97, 44.72, 45.15, 54.09, 57.70, 65.82, 66.95, 109.04, 108.28, 114.53, 127.62, 138.30, 157.16, 215.88. Anal. calcd for C28H37NO6: C, 69.54; H, 7.71; N, 2.90. Found: C, 70.00; H, 7.47; N, 2.77. Step 2: following the method of Wu et al.,52 a one-pot reductive amination/acylation of cis-6-oxoadamantane-2-spiro-3′-8′-[4-(2-morpholinoethoxy)phenyl]-1′,2′,4′-trioxaspiro[4.5]decane (430 mg, 0.89 mmol) afforded the free base of OZ747 (170 mg, 34%). Step 3: to a solution of the free base of OZ747 (170 mg, 0.30 mmol) in EA (5 mL) was added dropwise a solution of methanesulfonic acid (58 mg, 0.60 mmol) in diethyl ether (1 mL). The mixture was stirred at rt for 30 min, and the resulting solid was filtered, washed with cold EA, and dried in vacuo at 50 °C to afford OZ747 as a white solid (157 mg, 79%). mp 151–152 °C. 1H NMR (500 MHz, DMSO-d6) δ 9.91 (s, 1H), 7.84 (d, J = 7.3 Hz, 1H), 7.17 (d, J = 8.4 Hz, 2H), 6.93 (d, J = 8.3 Hz, 2H), 4.32 (t, J = 4.7 Hz, 2H), 3.98 (dd, J = 12.9, 3.4 Hz, 2H), 3.77 (d, J = 7.0 Hz, 1H), 3.71 (t, J = 12.5 Hz, 2H), 3.61–3.53 (m, 2H), 3.50 (d, J = 12.5 Hz, 2H), 3.20 (q, J = 11.5 Hz, 2H), 2.75 (s, 1H), 2.57 (t, J = 11.9 Hz, 1H), 2.40–2.24 (m, 7H), 2.09–1.62 (m, 17H), 1.55 (m, 3H); 13C NMR (125 MHz, DMSO-d6) δ 170.39, 156.30, 139.39, 128.04, 115.16, 110.74, 108.70, 84.24, 71.74, 63.70, 62.43, 55.57, 52.48, 52.18, 41.17, 40.24, 35.50, 35.35, 34.54, 34.51, 34.20, 31.75, 30.62, 30.22, 28.83, 28.78, 14.93. Anal. calcd for C34H48N2O9S: C, 61.80; H, 7.32; N, 4.24. Found: C, 61.56; H, 7.10; N 4.00.
Drug Sensitivity Assays
Parasite growth inhibition assays with the peroxide antimalarials and the clickable alkyne-tagged peroxide probes were performed as previously described.12,53 Briefly, for clickable alkyne-tagged peroxides (OZ727, OZ747, AA2, and carbaOZ727) and their associated nonclickable controls (OZ03, OZ277, OZ439, DHA, and artemisinin), IC50 assays53 were performed on P. falciparum (3D7) infected cultures at 1% parasitemia and 2% hematocrit (Hct) in a 96-well plate for 48 h at 37 °C to confirm that alkyne modification does not impact antimalarial activity. Pulse activity assays12 were also performed on OZ277, OZ439, and DHA in both 3D7 and NF54attB[hGrx1-roGFP2]P. falciparum lines that were magnet-harvested (90% parasitemia) and adjusted to a final Hct of 0.12%. This was performed to confirm that the peroxides act similarly in both 3D7 and the P. falciparum line used to measure glutathione redox potential (NF54attB[hGrx1-roGFP2]). Briefly, following a drug pulse of 3 h for DHA and OZ277 and 6 h for OZ439, peroxides were washed off as previously described12 with minor modification. Following four washes with 200 μL of complete Roswell Park Memorial Institute (RPMI) culture media containing 5% Albumax II, an extra wash with complete RPMI media was also included. After the washes, parasitemia was adjusted to 0.6% and the Hct was returned to 2% using uninfected RBCs. Cultures were then transferred to a flat-bottom 96-well microplate and incubated at 37 °C for 48 h. Following the 48 h incubation, parasite drug susceptibility was assessed by the measurement of SYBR green I fluorescence, as previously described.12,53 The data were analyzed using GraphPad Prism software version 8.0.2, as previously described.12
Parasite Treatment for Chemical Proteomics
P. falciparum parasites (3D7) synchronized to the trophozoite stage (28–32 h post invasion (h.p.i.)) were diluted to 10% parasitemia and adjusted to 2% Hct. For each treatment condition, 30 mL of infected RBC culture was used. Cultures were treated with 300 nM of the active clickable peroxides AA2, OZ727, or OZ747 for 1–6 h. Control cultures were incubated with 300 nM of the inactive (clickable) nonperoxide probe, carbaOZ727 (1 h), 300 nM of the nonclickable parent ozonide, OZ03 (1, 3, and 6 h), or an equivalent volume of DMSO (1 h, <0.01% final concentration). For competition experiments, the cultures were treated together with 300 nM of OZ727 and either an equivalent (300 nM) or excessive (900 nM) concentration of OZ03 for 1 h. In experiments using the cysteine protease inhibitor E64d to antagonize peroxide activity, cultures were pretreated for 30 min with 10 μM of E64d, followed by incubation with OZ727 for an additional 1 h. For all experiments, cultures were maintained at 37 °C under a gas atmosphere of 94% N2, 5% CO2, and 1% O2 for the duration of drug exposure.
After incubation with the alkyne probes, all subsequent steps were performed on ice or at 4 °C. The cultures were centrifuged at 700g for 3 min to remove the media, and parasites were isolated from the RBC by incubating with 0.1% saponin in phosphate-buffered saline (PBS) containing protease (Roche) and phosphatase inhibitors (Sigma-Aldrich, 20 mM sodium fluoride, 0.1 mM sodium orthovanadate, and 10 mM β glycerophosphate) for 10 min. The cell lysates were then centrifuged at 2576g to remove RBC proteins, and the resulting parasite pellets were washed a further three times in PBS containing protease and phosphatase inhibitors to ensure the removal of RBC membrane debris. Parasite pellets were stored at −80 °C until protein extraction and click chemistry enrichment of probe-labeled proteins.
In-Gel Fluorescence Analysis of Peroxide-Alkylated Proteins
Proteins were extracted by solubilizing parasite proteins using 1% sodium deoxycholate (SDC) in 100 mM N-(2-hydroxyethyl)piperazine-N′-ethanesulfonic acid (HEPES) buffer and heated at 90 °C for 5 min. This was followed by 3 × 30 s cycles of probe sonication on ice. The soluble fraction was separated by centrifugation at 14 800g for 5 min, and the SDC concentration was diluted to 0.5% with 100 mM HEPES buffer. The protein concentration was determined using the Pierce BCA assay kit (Thermo Scientific Pierce), and an equal amount of protein was used for each reaction condition. Click reactions were performed with 44.2 μL of sample. Alkyne-labeled proteins were labeled by the addition of 0.5 μL of Alexa Fluor 647 azide reporter (1 mM stock), followed by 1 μL of freshly prepared Tris(2-carboxyethyl)phosphine (TCEP) (50 mM in Milli-Q water), 3.3 μL of Tris-hydroxypropyltriazolylmethylamine (THPTA) ligand (1.7 mM in DMSO/t-butanol, 1:4 ratio), and 1 μL of CuSO4 (50 mM in Milli-Q water). Each reaction was briefly vortexed and allowed to proceed for 1 h in the dark at room temperature with regular mixing. The reaction was quenched by the addition of 2× SDS-PAGE reducing buffer followed by heating at 90 °C for 8 min. An equal volume of each sample was then loaded onto 12% polyacrylamide gels and resolved by SDS-PAGE. Fluorescence detection was performed by scanning with the Cy5 filter (excitation/emission: 649/670 nm) with an Amersham Typhoon 5 Biomolecular Imager (GE Healthcare Life Sciences). Coomassie staining was used to confirm equal protein loading.
Copper-Catalyzed Click Chemistry Enrichment of Alkylated Proteins for Identification by Mass Spectrometry
Proteins were extracted by solubilizing parasite proteins using 1% SDC in 100 mM HEPES buffer and heating each sample at 90 °C for 5 min to detect chemically modified proteins (alkylated) as per the previous publication.54 This was followed by 3 × 30 s cycles of probe sonication on ice. The soluble fraction was separated by centrifugation at 14 800g for 5 min, and the protein concentration was determined using the Pierce BCA assay kit (Thermo Scientific Pierce). The protein concentration of each sample was adjusted to 693 μg/mL, and the SDC concentration was diluted to 0.5% with 100 mM HEPES buffer. Alkyne-labeled proteins were enriched and affinity-purified by direct attachment to azide agarose beads (Jena Bioscience) using copper-catalyzed click chemistry. For each reaction, 50 μL of azide agarose beads (prewashed with 1400 μL of Milli-Q water), 22.6 μL of TCEP (50 mM in Milli-Q water), 68 μL of THPTA ligand (1.7 mM in DMSO/t-butanol, 1:4 ratio), and 22.6 μL of CuSO4 (50 mM in Milli-Q water) were sequentially added to 837 μL of the cell lysate. The lysate was incubated at room temperature for 1 h while rotating end-over-end. Following the click reaction, the agarose resins containing the clicked proteins were washed with 1 mL of Milli-Q water.
Washing and On-Bead Digestion of Clicked Proteins
The washed resins with the clicked proteins attached were resuspended in 1 mL of agarose wash buffer (100 mM HEPES, 1% SDS, 250 mM NaCl, 5 mM ethylenediaminetetraacetic acid (EDTA), pH 8). Samples were reduced by the addition of 10 μL of 1 M DTT and heated at 70 °C for 15 min, before being allowed to cool for a further 15 min. The supernatant was then removed by centrifugation at 4000g for 5 min, and the samples were alkylated in the dark with 40 mM iodoacetamide in 1 mL of agarose wash buffer for 30 min at room temperature. The beads were then extensively washed five times with agarose wash buffer, five times with 8 M urea, and a further five times with 20% acetonitrile (ACN). Between each wash, samples were centrifuged at 4000g for 3 min. Following the final wash, the resins were resuspended in 500 μL of digestion buffer (100 mM HEPES, 2 mM CaCl2, 10% ACN) and the contents were transferred to a fresh tube. The tube was rinsed with a further 500 μL of digestion buffer, and this was transferred to the same fresh tube. The resins were then pelleted by centrifugation at 4000g for 5 min and ∼800 μL of supernatant was removed. On-bead protein digestion was performed by adding 1 μg of sequencing-grade trypsin (Promega) to the samples, which were incubated overnight at 37 °C while spinning end-over-end. On the following day, the beads were separated from the digested peptides by centrifuging the samples at 4000g for 5 min and the supernatant was transferred to a clean tube. The resins were briefly vortexed and washed with a further 500 μL of Milli-Q water, which was then transferred to the same clean tube containing the digested peptides. Each sample was made up to 1 mL volume with Milli-Q water and acidified by the addition of 2 μL of formic acid. The samples were then desalted using in-house-generated StageTips,55 dried, and resuspended in 10 μL of 2% ACN and 0.1% formic acid containing indexed retention time (RT) peptides for LC-MS/MS analysis.
Liquid Chromatography-Mass Spectrometry (LC-MS/MS) Analysis
Proteomics LC-MS/MS data were acquired using a Dionex Ultimate 3000 RSLCnano system coupled to a Q-Exactive HF mass spectrometer (Thermo Scientific) carried out as described previously,56 with minor modifications. MS2 data were collected in data-independent mode (DIA) with a 33-fixed window setup of an 18 m/z effective precursor isolation over the m/z range of 375–975 Da.
Chemical Proteomics Data Analysis
Raw files were processed using Spectronaut 13.0 against an in-house-generated asexual P. falciparum spectral library. The library contained 44 449 peptides corresponding to 4730 proteins, of which 3113 were P. falciparum proteins.56 For processing, raw files were loaded and Spectronaut calculated the ideal mass tolerances for data extraction and scoring based on extensive mass calibration using a correction factor of 1. At both the precursor and fragment levels, the highest data point within the selected m/z tolerance was chosen for targeted data extraction. Identification of peptides against the library was based on the default Spectronaut settings (Manual for Spectronaut 13.0, available on the Biognosis website). Briefly, the Q-value cutoff at the precursor and protein levels was set at 1%; therefore, only those that passed this threshold were considered as identified and used for other subsequent processes. RT prediction type was set to dynamic-indexed RT. Interference correction was on the MS2 level. For quantification, the interference correction was activated and a significance filter of 0.01 was used for Q-value filtering, with the Q-value sparse setting applied. The imputing strategy was set to no imputing.
Identified proteins in the OZ727 and OZ747 groups with a fold-change ≥2 compared to those of the DMSO, carbaOZ727, and time-matched OZ03 controls, and with a p-value <0.05 (Mann–Whitney U test) compared to that of carbaOZ727, were considered as alkylated proteins. The same thresholds were applied to proteins in the AA2 group, except that p-value filtering was based on comparison with the DMSO control.
The identified alkylated proteins for OZ727, OZ747, and AA2 were subjected to gene ontology (GO) enrichment analysis with topGO.57 PlasmoDB GO terms were used for GO term mapping, and the elim algorithm was applied to navigate the topology of the GO graph.58 The Fisher exact test was used for testing the enrichment of GO terms. P. falciparum proteins from the in-house spectral library were used as the genomic background for statistical testing, and GO terms represented by fewer than five proteins in this library were excluded from the analysis. Significantly enriched GO terms (p < 0.05 and enrichment >1.5-fold) were visualized in Cytoscape 3.6 and manually clustered based on the alkylated proteins shared between GO terms and the semantic similarity measure of Schlicker et al.59
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE60 partner repository with the data set identifier PXD027334.
In Vitro Interaction of Peroxide Antimalarials with the hGrx1-roGFP2 Recombinant Protein
In vitro measurements with the recombinant hGrx1-roGFP2 redox sensor were carried out, as previously described.49,61,62 Peroxides, diamide (DIAM), and dithiothreitol (DTT) were diluted with a degassed reaction buffer (100 mM potassium phosphate, 1 mM ethylenediaminetetraacetic acid, EDTA, pH 7.0) and used immediately. The purified recombinant protein was prepared as previously described49,62 and diluted in reaction buffer to a concentration of 1.25 μM, so that in the plate assay, the final concentration is 1 μM. Peroxides at different concentrations combined with DIAM and DTT in the presence of iron(II) chloride (Sigma-Aldrich) (8.7 mM final concentration) were mixed with hGrx1-roGFP2 in a 96-well plate (black, μClear TC Greiner Bio-One). The emission at 510 nm after excitation at 405 and 480 nm was measured after 5 min, 30 min, 1 h, 3 h, 5 h, and 10 h in a plate reader with optimal reading settings. Data from two independent experiments with two technical replicates were analyzed for each concentration.
Effects of the Peroxides and Other Antimalarial Drugs on Redox Homeostasis
The effect of peroxides on P. falciparum redox homeostasis was investigated on late trophozoites (30–34 h.p.i.) using the NF54attB[hGrx1-roGFP2] parasite line and was carried out as previously described.49 Briefly, parasites (8% parasitemia, 2% Hct) were incubated with 0.1–1 μM of each peroxide antimalarial for 10 min to 9 h, depending on the compound. Following the appropriate incubation period, free thiol groups were blocked with 2 mM N-ethylmaleimide (NEM) for 15 min at 37 °C. After incubation with NEM, the infected RBC cultures were washed and resuspended in Ringer’s solution. Resuspended cells were then magnetically enriched (80% parasitemia) (Miltenyi Biotech, Bergisch Gladbach, Germany), and the parasite redox ratio was measured using either the confocal live-cell imaging or plate reader analyses. All peroxide incubation experiments included an untreated control group (“mock-treated” with DMSO for 9 h), as well as controls for complete oxidation (1 mM DIAM for 5 min) and complete reduction (10 mM DTT for 5 min). Peroxide incubation times were based on the kinetics of activity as per previous publications.12,13 Times for the fully oxidized and reduced controls were also based on a previous publication.49
Confocal Live-Cell Imaging and Image Processing
Live-cell imaging was performed as previously described.49 Briefly, drug-treated cells that had been magnet-harvested and resuspended in prewarmed Ringer’s solution were seeded onto poly-l-lysine-coated flat μ-slides VI (Ibidi, Martinsried, Germany). The probes were excited at 405 and 488 nm, and emissions were detected at 500–550 nm. Images were analyzed with a custom-written macro in the Fiji distribution of ImageJ.63 The macro was as follows: regions of interest (ROIs) of the cells were generated by applying a Gaussian blur (Sigma = 4), generating a binary image and applying a binary watershed. A 32-bit ratio image was generated by dividing the two fluorescent channels. The ROIs were used to measure the fluorescent intensity ratio in each cell. Images of the analyzed cells were saved out and manually curated to select only the infected RBCs that showed fluorescent signals at both 405 and 488 nm excitation and had an intact host cell. The 405/488 nm ratios were calculated, and graphs were plotted using GraphPad Prism 8.0.2 software. One-way ANOVA was applied for statistical analysis of significance (****, p < 0.0001).
Plate Reader Analyses of Redox Homeostasis
Plate reader analyses were performed as previously described.49 Briefly, magnet-harvested drug-treated NF54attB[hGrx1-roGFP2] parasites (80% parasitemia) with their free thiols blocked by NEM were counted using the improved Neubauer hemocytometer (Brand GmbH, Wertheim, Germany) and adjusted to 2.0 × 105 iRBCs/μL. Parasites (20 μL) were transferred to a 384-well plate, and emission (530 nm) was measured with excitation wavelengths of 400 and 482 nm using a Clariostar plate reader (multichromatic) (BMG Labtech, Ortenberg, Germany). The calculated ratio of 400/482 nm was then plotted using GraphPad Prism 8.0.2 software. One-way ANOVA was applied for statistical analysis of significance (*, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001).
LC-MS Determination of Free Thiol and Glutathione Levels in Peroxide-Treated P. falciparum Infected RBCs
Levels of reduced glutathione (GSH) and other thiol metabolites were determined by LC-MS for 3D7 and NF54attB[hGrx1-roGFP2]P. falciparum parasites following treatment with peroxides. Samples were prepared via several different methodologies to obtain the most reproducible output and ensure that no major experimental artifacts were introduced during the sample processing. For simplicity, these are referred to as methods 1–3. The results from method 3 are reported in the main manuscript (Figure 5), while the results from methods 1 and 2 are shown in the Supporting Information (Figure S8).
Method 1 sample preparation was the same as experiments used to measure the parasite redox ratio. Briefly, infected RBC cultures were incubated with the test compounds for the desired period (100 nM DHA for 3 h, 300 nM OZ277 for 3 h, 300 nM OZ439 for 6 h, 1 mM DIAM for 5 min, and DMSO for 6 h) and thiols were quenched by adding freshly prepared NEM solution to the final concentration of 2 mM in culture media. The cultures were then magnet-harvested, and the parasitemia for each condition was adjusted to be 80%. The cells (5 × 107) were pelleted by centrifugation for 5 min at 1000g, and the supernatant was removed. The cells were then washed with PBS, prior to the addition of 100 μL ice-cold methanol for the extraction of thiols.
For method 2 sample preparation, the initial sample preparation was the same as described for method 1, except that NEM was added to the extraction solvent (50 μL of 50 mM NEM in 80% methanol, 20% 10 mM ammonium formate, pH 7.0, and 50 μL of 100% ACN). Therefore, free thiols were derivatized at the extraction step.
For method 3, magnetically enriched cultures at 90% parasitemia and 0.12% Hct were incubated with the test compounds for the desired times (drug treatments, 100 nM DHA for 3 h, 300 nM OZ277 for 3 h, 300 nM OZ439 for 6 h, 1 μM pyrimethamine for 5 h, and three control conditions, 1 mM DIAM for 5 min, 10 mM DTT for 5 min, and DMSO for 6 h). Following the drug incubation, cells were washed with PBS and 5 × 107 cells were used for thiol metabolite extraction. Thiols were extracted using NEM-containing extraction solvent (50 μL of 50 mM NEM in 80% methanol, 20% of 10 mM ammonium formate, pH 7.0, and 50 μL of 100% ACN).
For all methods, following thiol extraction with the appropriate solvent, cells were incubated on a shaker for 1 h at 4 °C and then centrifuged at 20 000g to remove the insoluble material. Supernatants were transferred to glass high-performance liquid chromatography (HPLC) vials and stored at −80 °C until analysis.
LC-MS Thiol Analysis and Data Processing
LC-MS data were acquired on a Q-Exactive Orbitrap mass spectrometer (Thermo Scientific, Waltham, Massachusetts) coupled with a Dionex Ultimate 3000 RS (Thermo Scientific) HPLC system. The method was adapted from the previously described procedure64,65 with some changes. Chromatographic separation was performed on a Poroshel Infinity HILIC-Z column (2.7 μm, 2.1 × 100 mm2, Agilent Technologies) kept at 25 °C. The total run time was 20 min at 0.3 mL/min flowrate with an injection volume of 10 μL. Buffer A was 20 mM ammonium carbonate, buffer B was acetonitrile, and syringe wash solvent was 50% isopropanol. The gradient started 90% B, decreased to 65% B at 10 min, further decreased to 20% B at 11.5 min, kept at 20% B until 13 min, returned to 90% B at 14 min, and kept at 90% B until 20 min. HESI ion source parameters were as follows: sheath gas 50, aux gas 20 and sweep gas 2 arbitrary units, spray voltage was set to 4.0 kV, capillary temperature to 300 °C, S-Lens RF level to 50, and auxiliary gas heater temperature to 120 °C. The mass spectrometer was operated in a full scan mode in a positive detection mode at 35 000 resolution at 200 m/z with a detection range of 85–1275 m/z. The samples were analyzed as a single batch to reduce batch-to-batch variation and randomized to account for the LC-MS system drift over time.
Data were processed using peak areas, which were obtained with QuanBrowser (Thermo Scientific) software, version 4.2 by integrating MS1 peaks. Peak identities were then confirmed by NEM-derivatized authentic standards. The calculated peak area was plotted using GraphPad Prism 8.0.2 software, and one-way ANOVA was applied for statistical analysis (*, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001).
Acknowledgments
Funding support was provided by the NHMRC (APP#1160705, APP1128003, APP1185354, and APP1148700) and the NIH (AI116723-01). The authors acknowledge the Monash Proteomics and Metabolomics Facility (Parkville Node) for providing LC-MS technical assistance. The Australian Red Cross Blood Service in Melbourne donated human red blood cells for in vitro parasite cultivation.
Glossary
Abbreviations
- ACN
acetonitrile
- ACTs
artemisinin-based combination therapies
- Cys
cysteine
- DHA
dihydroartemisinin
- DIAM
diamide
- DIA-MS
data-independent acquisition mass spectrometry
- DDA
data-dependent analysis
- DMSO
dimethyl sulfoxide
- DTT
dithiothreitol
- GO
gene ontology
- GSH
reduced glutathione
- GSSG
oxidized glutathione
- h
hour
- Hct
hematocrit
- hGrx1-roGFP2
glutaredoxin 1 fused to a reduction–oxidation-sensitive GFP
- IC50
50% inhibition concentration
- LC-MS
liquid chromatography-mass spectrometry
- m/z
mass-to-charge ratio
- mM
millimolar
- NEM
N-ethylmaleimide
- PBS
phosphate-buffered saline
- P. falciparum
Plasmodium falciparum
- RBCs
red blood cells
- SDC
sodium deoxycholate
- γ-GluCys
γ-glutamyl cysteine
- μL
microliter
- μM
micromolar
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsinfecdis.1c00550.
In-gel fluorescence analysis of ozonide alkylated proteins in P. falciparum; comparison of pulldown results with previously published data sets; heatmap representation of the effect of OZ03 coincubation or E64d pretreatment on the OZ727 labeling of food vacuole proteins (GO:0020020); GO biological process enrichment analysis of control data set; alkylation of redox homeostasis proteins (GO:0045454) by peroxide probes and effect of OZ03 coincubation or E64d pretreatment on the OZ727 labeling of redox homeostasis proteins; effects of peroxide antimalarial on reduced hGrx1-roGFP2; effect of peroxide antimalarials on the redox ratio of P. falciparum NF54attB[hGrx1-roGFP2] parasites; effect on thiols (using different sample preparation methods) and activity of peroxide antimalarials in P. falciparum 3D7 and NF54attB[hGrx1-roGFP2] parasites (PDF)
Complete proteomics data set for the identification of alkylated proteins of peroxide antimalarials and heatmap representation of all enriched proteins for OZ727, OZ747, and AA2 following the treatment of P. falciparum infected RBCs for 1 h (all 300 nM) with protein IDs and significant proteins (+) (Supporting Data 1) (XLSX)
GO enrichment analysis results for cellular component (Supporting Data 2) (XLSX)
GO enrichment analysis results for biological process (Supporting Data 3) (XLSX)
Author Contributions
⊥ G.S. and C.G. contributed equally to this work. D.J.C., K.B., G.S., and C.G. directed the overall research program; G.S., C.G., A.D.P., A.K.S., K.C.H., and D.A. performed the experiments; G.S., C.G., A.D.P., T.G.B., and C.A.M. analyzed the data and prepared figures; J.W., X.W., Y.D., and J.L.V. made the click chemistry probes; all authors contributed to experimental design and wrote and edited the manuscript.
The authors declare no competing financial interest.
Supplementary Material
References
- WHO . World Malaria Report 2020; World Health Organisation: Geneva, 2020.
- O’Neill P. M.; Barton V. E.; Ward S. A. The molecular mechanism of action of artemisinin--the debate continues. Molecules 2010, 15, 1705–1721. 10.3390/molecules15031705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodrow C. J.; Haynes R. K.; Krishna S. Artemisinins. Postgrad. Med. J. 2005, 81, 71–78. 10.1136/pgmj.2004.028399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris C. A.; Duparc S.; Borghini-Fuhrer I.; Jung D.; Shin C.-S.; Fleckenstein L. Review of the clinical pharmacokinetics of artesunate and its active metabolite dihydroartemisinin following intravenous, intramuscular, oral or rectal administration. Malar. J. 2011, 10, 263 10.1186/1475-2875-10-263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vennerstrom J. L.; Arbe-Barnes S.; Brun R.; Charman S. A.; Chiu F. C.; Chollet J.; Dong Y.; Dorn A.; Hunziker D.; Matile H.; McIntosh K.; Padmanilayam M.; Santo Tomas J.; Scheurer C.; Scorneaux B.; Tang Y.; Urwyler H.; Wittlin S.; Charman W. N. Identification of an antimalarial synthetic trioxolane drug development candidate. Nature 2004, 430, 900–904. 10.1038/nature02779. [DOI] [PubMed] [Google Scholar]
- Charman S. A.; Arbe-Barnes S.; Bathurst I. C.; Brun R.; Campbell M.; Charman W. N.; Chiu F. C.; Chollet J.; Craft J. C.; Creek D. J.; Dong Y.; Matile H.; Maurer M.; Morizzi J.; Nguyen T.; Papastogiannidis P.; Scheurer C.; Shackleford D. M.; Sriraghavan K.; Stingelin L.; Tang Y.; Urwyler H.; Wang X.; White K. L.; Wittlin S.; Zhou L.; Vennerstrom J. L. Synthetic ozonide drug candidate OZ439 offers new hope for a single-dose cure of uncomplicated malaria. Proc. Natl. Acad. Sci. U.S.A. 2011, 108, 4400–4405. 10.1073/pnas.1015762108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y.; Wang X.; Kamaraj S.; Bulbule V. J.; Chiu F. C. K.; Chollet J.; Dhanasekaran M.; Hein C. D.; Papastogiannidis P.; Morizzi J.; Shackleford D. M.; Barker H.; Ryan E.; Scheurer C.; Tang Y.; Zhao Q.; Zhou L.; White K. L.; Urwyler H.; Charman W. N.; Matile H.; Wittlin S.; Charman S. A.; Vennerstrom J. L. Structure–activity relationship of the antimalarial ozonide artefenomel (OZ439). J. Med. Chem. 2017, 60, 2654–2668. 10.1021/acs.jmedchem.6b01586. [DOI] [PubMed] [Google Scholar]
- Phyo A. P.; Jittamala P.; Nosten F. H.; Pukrittayakamee S.; Imwong M.; White N. J.; Duparc S.; Macintyre F.; Baker M.; Möhrle J. J. Antimalarial activity of artefenomel (OZ439), a novel synthetic antimalarial endoperoxide, in patients with Plasmodium falciparum and Plasmodium vivax malaria: an open-label phase 2 trial. Lancet Infect. Dis. 2016, 16, 61–69. 10.1016/S1473-3099(15)00320-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moehrle J. J.; Duparc S.; Siethoff C.; van Giersbergen P. L.; Craft J. C.; Arbe-Barnes S.; Charman S. A.; Gutierrez M.; Wittlin S.; Vennerstrom J. L. First-in-man safety and pharmacokinetics of synthetic ozonide OZ439 demonstrates an improved exposure profile relative to other peroxide antimalarials. Br. J. Clin. Pharmacol. 2013, 75, 535–548. 10.1111/j.1365-2125.2012.04368.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser M.; Wittlin S.; Nehrbass-Stuedli A.; Dong Y.; Wang X.; Hemphill A.; Matile H.; Brun R.; Vennerstrom J. L. Peroxide bond-dependent antiplasmodial specificity of artemisinin and OZ277 (RBx11160). Antimicrob. Agents Chemother. 2007, 51, 2991–2993. 10.1128/AAC.00225-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fügi M. A.; Wittlin S.; Dong Y.; Vennerstrom J. L. Probing the antimalarial mechanism of artemisinin and OZ277 (arterolane) with nonperoxidic isosteres and nitroxyl radicals. Antimicrob. Agents Chemother. 2010, 54, 1042–1046. 10.1128/AAC.01305-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannangelo C.; Stingelin L.; Yang T.; Tilley L.; Charman S. A.; Creek D. J. Parasite-mediated degradation of synthetic ozonide antimalarials impacts in vitro antimalarial activity. Antimicrob. Agents Chemother. 2018, 62, e01566-17 10.1128/aac.01566-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T.; Xie S. C.; Cao P.; Giannangelo C.; McCaw J.; Creek D. J.; Charman S. A.; Klonis N.; Tilley L. Comparison of the exposure time-dependence of the activities of synthetic ozonide antimalarials and dihydroartemisinin against K13 wild-type and mutant Plasmodium falciparum strains. Antimicrob. Agents Chemother. 2016, 60, 4501–4510. 10.1128/AAC.00574-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meshnick S. R.; Thomas A.; Ranz A.; Xu C. M.; Pan H. Z. Artemisinin (qinghaosu): the role of intracellular hemin in its mechanism of antimalarial action. Mol. Biochem. Parasitol. 1991, 49, 181–189. 10.1016/0166-6851(91)90062-B. [DOI] [PubMed] [Google Scholar]
- Meshnick S. R.; Yang Y. Z.; Lima V.; Kuypers F.; Kamchonwongpaisan S.; Yuthavong Y. Iron-dependent free radical generation from the antimalarial agent artemisinin (qinghaosu). Antimicrob. Agents Chemother. 1993, 37, 1108–1114. 10.1128/aac.37.5.1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y.; Dong Y.; Wang X.; Sriraghavan K.; Wood J. K.; Vennerstrom J. L. Dispiro-1,2,4-trioxane analogues of a prototype dispiro-1,2,4-trioxolane: mechanistic comparators for artemisinin in the context of reaction pathways with iron(II). J. Org. Chem. 2005, 70, 5103–5110. 10.1021/jo050385+. [DOI] [PubMed] [Google Scholar]
- Giannangelo C.; Anderson D.; Wang X.; Vennerstrom J. L.; Charman S. A.; Creek D. J. Ozonide antimalarials alkylate heme in the malaria parasite Plasmodium falciparum. ACS Infect. Dis. 2019, 5, 2076–2086. 10.1021/acsinfecdis.9b00257. [DOI] [PubMed] [Google Scholar]
- Robert A.; Benoit-Vical F.; Claparols C.; Meunier B. The antimalarial drug artemisinin alkylates heme in infected mice. Proc. Natl. Acad. Sci. U.S.A. 2005, 102, 13676–13680. 10.1073/pnas.0500972102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jourdan J.; Matile H.; Reift E.; Biehlmaier O.; Dong Y.; Wang X.; Mäser P.; Vennerstrom J. L.; Wittlin S. Monoclonal antibodies that recognize the alkylation signature of antimalarial ozonides OZ277 (arterolane) and OZ439 (artefenomel). ACS Infect. Dis. 2016, 2, 54–61. 10.1021/acsinfecdis.5b00090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jourdan J.; Walz A.; Matile H.; Schmidt A.; Wu J.; Wang X.; Dong Y.; Vennerstrom J. L.; Schmidt R. S.; Wittlin S.; Mäser P. Stochastic protein alkylation by antimalarial peroxides. ACS Infect. Dis. 2019, 2067. 10.1021/acsinfecdis.9b00264. [DOI] [PubMed] [Google Scholar]
- Ismail H. M.; Barton V.; Phanchana M.; Charoensutthivarakul S.; Wong M. H. L.; Hemingway J.; Biagini G. A.; O’Neill P. M.; Ward S. A. Artemisinin activity-based probes identify multiple molecular targets within the asexual stage of the malaria parasites Plasmodium falciparum 3D7. Proc. Natl. Acad. Sci. U.S.A. 2016, 113, 2080–2085. 10.1073/pnas.1600459113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ismail H. M.; Barton V. E.; Panchana M.; Charoensutthivarakul S.; Biagini G. A.; Ward S. A.; O’Neill P. M. A click chemistry-based proteomic approach reveals that 1, 2, 4-trioxolane and artemisinin antimalarials share a common protein alkylation profile. Angew. Chem., Int. Ed. 2016, 55, 6401–6405. 10.1002/anie.201512062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J.; Zhang C.-J.; Chia W. N.; Loh C. C.; Li Z.; Lee Y. M.; He Y.; Yuan L.-X.; Lim T. K.; Liu M.; Liew C. X.; Lee Y. Q.; Zhang J.; Lu N.; Lim C. T.; Hua Z.-C.; Liu B.; Shen H.-M.; Tan K. S. W.; Lin Q. Haem-activated promiscuous targeting of artemisinin in Plasmodium falciparum. Nat. Commun. 2015, 6, 10111 10.1038/ncomms10111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridgford J. L.; Xie S. C.; Cobbold S. A.; Pasaje C. F. A.; Herrmann S.; Yang T.; Gillett D. L.; Dick L. R.; Ralph S. A.; Dogovski C.; Spillman N. J.; Tilley L. Artemisinin kills malaria parasites by damaging proteins and inhibiting the proteasome. Nat. Commun. 2018, 9, 3801 10.1038/s41467-018-06221-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisewski A. M.; Quiros J. P.; Ng C. L.; Adikesavan A. K.; Miura K.; Putluri N.; Eastman R. T.; Scanfeld D.; Regenbogen S. J.; Altenhofen L.; Llinás M.; Sreekumar A.; Long C.; Fidock D. A.; Lichtarge O. Supergenomic network compression and the discovery of EXP1 as a glutathione transferase inhibited by artesunate. Cell 2014, 158, 916–928. 10.1016/j.cell.2014.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mbengue A.; Bhattacharjee S.; Pandharkar T.; Liu H. N.; Estiu G.; Stahelin R. V.; Rizk S. S.; Njimoh D. L.; Ryan Y.; Chotivanich K.; Nguon C.; Ghorbal M.; Lopez-Rubio J. J.; Pfrender M.; Emrich S.; Mohandas N.; Dondorp A. M.; Wiest O.; Haldar K. A molecular mechanism of artemisinin resistance in Plasmodium falciparum malaria. Nature 2015, 520, 683–687. 10.1038/nature14412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klonis N.; Crespo-Ortiz M. P.; Bottova I.; Abu-Bakar N.; Kenny S.; Rosenthal P. J.; Tilley L. Artemisinin activity against Plasmodium falciparum requires hemoglobin uptake and digestion. Proc. Natl. Acad. Sci. U.S.A. 2011, 108, 11405–11410. 10.1073/pnas.1104063108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krungkrai S. R.; Yuthavong Y. The antimalarial action on Plasmodium falciparum of qinghaosu and artesunate in combination with agents which modulate oxidant stress. Trans. R. Soc. Trop. Med. Hyg. 1987, 81, 710–714. 10.1016/0035-9203(87)90003-4. [DOI] [PubMed] [Google Scholar]
- Hartwig C. L.; Lauterwasser E. M. W.; Mahajan S. S.; Hoke J. M.; Cooper R. A.; Renslo A. R. Investigating the antimalarial action of 1,2,4-trioxolanes with fluorescent chemical probes. J. Med. Chem. 2011, 54, 8207–8213. 10.1021/jm2012003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwig C. L.; Rosenthal A. S.; D’Angelo J.; Griffin C. E.; Posner G. H.; Cooper R. A. Accumulation of artemisinin trioxane derivatives within neutral lipids of Plasmodium falciparum malaria parasites is endoperoxide-dependent. Biochem. Pharmacol. 2009, 77, 322–336. 10.1016/j.bcp.2008.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meshnick S. R. Artemisinin: mechanisms of action, resistance and toxicity. Int. J. Parasitol. 2002, 32, 1655–1660. 10.1016/S0020-7519(02)00194-7. [DOI] [PubMed] [Google Scholar]
- Giannangelo C.; Siddiqui G.; De Paoli A.; Anderson B. M.; Edgington-Mitchell L. E.; Charman S. A.; Creek D. J. System-wide biochemical analysis reveals ozonide antimalarials initially act by disrupting Plasmodium falciparum haemoglobin digestion. PLoS Pathog. 2020, 16, e1008485 10.1371/journal.ppat.1008485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dondorp A. M.; Nosten F.; Yi P.; Das D.; Phyo A. P.; Tarning J.; Lwin K. M.; Ariey F.; Hanpithakpong W.; Lee S. J.; Ringwald P.; Silamut K.; Imwong M.; Chotivanich K.; Lim P.; Herdman T.; An S. S.; Yeung S.; Singhasivanon P.; Day N. P.; Lindegardh N.; Socheat D.; White N. J. Artemisinin resistance in Plasmodium falciparum malaria. N. Engl. J. Med. 2009, 361, 455–467. 10.1056/NEJMoa0808859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S.; Saha B.; Hati A. K.; Roy S. Evidence of artemisinin-resistant Plasmodium falciparum malaria in eastern India. N. Engl. J. Med. 2018, 379, 1962–1964. 10.1056/NEJMc1713777. [DOI] [PubMed] [Google Scholar]
- Uwimana A.; Legrand E.; Stokes B. H.; Ndikumana J.-L. M.; Warsame M.; Umulisa N.; Ngamije D.; Munyaneza T.; Mazarati J.-B.; Munguti K.; Campagne P.; Criscuolo A.; Ariey F.; Murindahabi M.; Ringwald P.; Fidock D. A.; Mbituyumuremyi A.; Menard D. Emergence and clonal expansion of in vitro artemisinin-resistant Plasmodium falciparum kelch13 R561H mutant parasites in Rwanda. Nat. Med. 2020, 1602. 10.1038/s41591-020-1005-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miotto O.; Sekihara M.; Tachibana S.-I.; Yamauchi M.; Pearson R. D.; Amato R.; Gonçalves S.; Mehra S.; Noviyanti R.; Marfurt J.; Auburn S.; Price R. N.; Mueller I.; Ikeda M.; Mori T.; Hirai M.; Tavul L.; Hetzel M. W.; Laman M.; Barry A. E.; Ringwald P.; Ohashi J.; Hombhanje F.; Kwiatkowski D. P.; Mita T. Emergence of artemisinin-resistant Plasmodium falciparum with kelch13 C580Y mutations on the island of New Guinea. PLoS Pathog. 2020, 16, e1009133 10.1371/journal.ppat.1009133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashley E. A.; Dhorda M.; Fairhurst R. M.; Amaratunga C.; Lim P.; Suon S.; Sreng S.; Anderson J. M.; Mao S.; Sam B.; Sopha C.; Chuor C. M.; Nguon C.; Sovannaroth S.; Pukrittayakamee S.; Jittamala P.; Chotivanich K.; Chutasmit K.; Suchatsoonthorn C.; Runcharoen R.; Hien T. T.; Thuy-Nhien N. T.; Thanh N. V.; Phu N. H.; Htut Y.; Han K. T.; Aye K. H.; Mokuolu O. A.; Olaosebikan R. R.; Folaranmi O. O.; Mayxay M.; Khanthavong M.; Hongvanthong B.; Newton P. N.; Onyamboko M. A.; Fanello C. I.; Tshefu A. K.; Mishra N.; Valecha N.; Phyo A. P.; Nosten F.; Yi P.; Tripura R.; Borrmann S.; Bashraheil M.; Peshu J.; Faiz M. A.; Ghose A.; Hossain M. A.; Samad R.; Rahman M. R.; Hasan M. M.; Islam A.; Miotto O.; Amato R.; MacInnis B.; Stalker J.; Kwiatkowski D. P.; Bozdech Z.; Jeeyapant A.; Cheah P. Y.; Sakulthaew T.; Chalk J.; Intharabut B.; Silamut K.; Lee S. J.; Vihokhern B.; Kunasol C.; Imwong M.; Tarning J.; Taylor W. J.; Yeung S.; Woodrow C. J.; Flegg J. A.; Das D.; Smith J.; Venkatesan M.; Plowe C. V.; Stepniewska K.; Guerin P. J.; Dondorp A. M.; Day N. P.; White N. J. Spread of artemisinin resistance in Plasmodium falciparum malaria. N. Engl. J. Med. 2014, 371, 411–423. 10.1056/NEJMoa1314981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thriemer K.; Van Hong N.; Rosanas-Urgell A.; Phuc B. Q.; Ha D. M.; Pockele E.; Guetens P.; Van Van N.; Duong T. T.; Amambua-Ngwa A.; D’Alessandro U.; Erhart A. Delayed parasite clearance after treatment with dihydroartemisinin-piperaquine in Plasmodium falciparum malaria patients in central Vietnam. Antimicrob. Agents Chemother. 2014, 58, 7049–7055. 10.1128/AAC.02746-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaratunga C.; Lim P.; Suon S.; Sreng S.; Mao S.; Sopha C.; Sam B.; Dek D.; Try V.; Amato R.; Blessborn D.; Song L.; Tullo G. S.; Fay M. P.; Anderson J. M.; Tarning J.; Fairhurst R. M. Dihydroartemisinin-piperaquine resistance in Plasmodium falciparum malaria in Cambodia: a multisite prospective cohort study. Lancet Infect. Dis. 2016, 16, 357–365. 10.1016/S1473-3099(15)00487-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spring M. D.; Lin J. T.; Manning J. E.; Vanachayangkul P.; Somethy S.; Bun R.; Se Y.; Chann S.; Ittiverakul M.; Sia-ngam P.; Kuntawunginn W.; Arsanok M.; Buathong N.; Chaorattanakawee S.; Gosi P.; Ta-aksorn W.; Chanarat N.; Sundrakes S.; Kong N.; Heng T. K.; Nou S.; Teja-isavadharm P.; Pichyangkul S.; Phann S. T.; Balasubramanian S.; Juliano J. J.; Meshnick S. R.; Chour C. M.; Prom S.; Lanteri C. A.; Lon C.; Saunders D. L. Dihydroartemisinin-piperaquine failure associated with a triple mutant including kelch13 C580Y in Cambodia: an observational cohort study. Lancet Infect. Dis. 2015, 15, 683–691. 10.1016/S1473-3099(15)70049-6. [DOI] [PubMed] [Google Scholar]
- Ariey F.; Witkowski B.; Amaratunga C.; Beghain J.; Langlois A.-C.; Khim N.; Kim S.; Duru V.; Bouchier C.; Ma L.; Lim P.; Leang R.; Duong S.; Sreng S.; Suon S.; Chuor C. M.; Bout D. M.; Menard S.; Rogers W. O.; Genton B.; Fandeur T.; Miotto O.; Ringwald P.; Le Bras J.; Berry A.; Barale J.-C.; Fairhurst R. M.; Benoit-Vical F.; Mercereau-Puijalon O.; Menard D. A molecular marker of artemisinin-resistant Plasmodium falciparum malaria. Nature 2014, 505, 50–55. 10.1038/nature12876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witkowski B.; Amaratunga C.; Khim N.; Sreng S.; Chim P.; Kim S.; Lim P.; Mao S.; Sopha C.; Sam B.; Anderson J. M.; Duong S.; Chuor C. M.; Taylor W. R.; Suon S.; Mercereau-Puijalon O.; Fairhurst R. M.; Menard D. Novel phenotypic assays for the detection of artemisinin-resistant Plasmodium falciparum malaria in Cambodia: in-vitro and ex-vivo drug-response studies. Lancet Infect. Dis. 2013, 13, 1043–1049. 10.1016/s1473-3099(13)70252-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaratunga C.; Witkowski B.; Dek D.; Try V.; Khim N.; Miotto O.; Menard D.; Fairhurst R. M. Plasmodium falciparum founder populations in western Cambodia have reduced artemisinin sensitivity in vitro. Antimicrob. Agents Chemother. 2014, 58, 4935–4937. 10.1128/aac.03055-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellems T. E.; Sá J. M.; Su X.-Z.; Connelly S. V.; Ellis A. C. “Artemisinin Resistance”: Something New or Old? Something of a Misnomer?. Trends Parasitol. 2020, 735. 10.1016/j.pt.2020.05.013. [DOI] [PubMed] [Google Scholar]
- Giannangelo C.; Fowkes F. J. I.; Simpson J. A.; Charman S. A.; Creek D. J. Ozonide antimalarial activity in the context of artemisinin-resistant malaria. Trends Parasitol. 2019, 35, 529–543. 10.1016/j.pt.2019.05.002. [DOI] [PubMed] [Google Scholar]
- Straimer J.; Gnädig N. F.; Stokes B. H.; Ehrenberger M.; Crane A. A.; Fidock D. A. Plasmodium falciparum K13 mutations differentially impact ozonide susceptibility and parasite fitness in vitro. mBio 2017, 8, e00172-17 10.1128/mBio.00172-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgärtner F.; Jourdan J.; Scheurer C.; Blasco B.; Campo B.; Mäser P.; Wittlin S. In vitro activity of anti-malarial ozonides against an artemisinin-resistant isolate. Malar. J. 2017, 16, 45 10.1186/s12936-017-1696-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siriwardana A.; Iyengar K.; Roepe P. D. Endoperoxide drug cross-resistance patterns for Plasmodium falciparum exhibiting an artemisinin delayed-clearance phenotype. Antimicrob. Agents Chemother. 2016, 60, 6952–6956. 10.1128/aac.00857-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuh A. K.; Rahbari M.; Heimsch K. C.; Mohring F.; Gabryszewski S. J.; Weder S.; Buchholz K.; Rahlfs S.; Fidock D. A.; Becker K. Stable Integration and Comparison of hGrx1-roGFP2 and sfroGFP2 Redox Probes in the Malaria Parasite Plasmodium falciparum. ACS Infect. Dis. 2018, 4, 1601–1612. 10.1021/acsinfecdis.8b00140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trager W.; Jensen J. B. Human malaria parasites in continuous culture. Science 1976, 193, 673–675. 10.1126/science.781840. [DOI] [PubMed] [Google Scholar]
- Dong Y.; Chollet J.; Matile H.; Charman S. A.; Chiu F. C.; Charman W. N.; Scorneaux B.; Urwyler H.; Santo Tomas J.; Scheurer C.; et al. Spiro and dispiro-1, 2, 4-trioxolanes as antimalarial peroxides: charting a workable structure-activity relationship using simple prototypes. J. Med. Chem. 2005, 48, 4953–4961. 10.1021/jm049040u. [DOI] [PubMed] [Google Scholar]
- Wu J.; Wang X.; Chiu F. C.; Haberli C.; Shackleford D. M.; Ryan E.; Kamaraj S.; Bulbule V. J.; Wallick A. I.; Dong Y.; et al. Structure–activity relationship of antischistosomal ozonide carboxylic acids. J. Med. Chem. 2020, 63, 3723–3736. 10.1021/acs.jmedchem.0c00069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smilkstein M.; Sriwilaijaroen N.; Kelly J. X.; Wilairat P.; Riscoe M. Simple and inexpensive fluorescence-based technique for high-throughput antimalarial drug screening. Antimicrob. Agents Chemother. 2004, 48, 1803–1806. 10.1128/AAC.48.5.1803-1806.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clements J. L.; Pohl F.; Muthupandi P.; Rogers S. C.; Mao J.; Doctor A.; Birman V. B.; Held J. M. A clickable probe for versatile characterization of S-nitrosothiols. Redox Biol. 2020, 37, 101707 10.1016/j.redox.2020.101707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappsilber J.; Ishihama Y.; Mann M. Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal. Chem. 2003, 75, 663–670. 10.1021/ac026117i. [DOI] [PubMed] [Google Scholar]
- Birrell G. W.; Challis M. P.; De Paoli A.; Anderson D.; Devine S. M.; Heffernan G. D.; Jacobus D. P.; Edstein M. D.; Siddiqui G.; Creek D. J. Multi-omic characterisation of the mode of action of a potent new antimalarial compound, JPC-3210, against Plasmodium falciparum. Mol. Cell. Proteomics 2019, 308. 10.1074/mcp.RA119.001797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexa A.; Rahnenführer J.. topGO: Enrichment Analysis for Gene Ontology, R package version 2.42.0, 2020.
- Alexa A.; Rahnenführer J.; Lengauer T. Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics 2006, 22, 1600–1607. 10.1093/bioinformatics/btl140. [DOI] [PubMed] [Google Scholar]
- Schlicker A.; Domingues F. S.; Rahnenführer J.; Lengauer T. A new measure for functional similarity of gene products based on Gene Ontology. BMC Bioinf. 2006, 7, 302 10.1186/1471-2105-7-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Riverol Y.; Csordas A.; Bai J.; Bernal-Llinares M.; Hewapathirana S.; Kundu D. J.; Inuganti A.; Griss J.; Mayer G.; Eisenacher M.; Perez E.; Uszkoreit J.; Pfeuffer J.; Sachsenberg T.; Yilmaz S.; Tiwary S.; Cox J.; Audain E.; Walzer M.; Jarnuczak A. F.; Ternent T.; Brazma A.; Vizcaino J. A. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 2019, 47, D442–D450. 10.1093/nar/gky1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasozi D.; Mohring F.; Rahlfs S.; Meyer A. J.; Becker K. Real-time imaging of the intracellular glutathione redox potential in the malaria parasite Plasmodium falciparum. PLoS Pathog. 2013, 9, e1003782 10.1371/journal.ppat.1003782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohring F.; Rahbari M.; Zechmann B.; Rahlfs S.; Przyborski J. M.; Meyer A. J.; Becker K. Determination of glutathione redox potential and pH value in subcellular compartments of malaria parasites. Free Radical Biol. Med. 2017, 104. 10.1016/j.freeradbiomed.2017.01.001. [DOI] [PubMed] [Google Scholar]
- Schindelin J.; Arganda-Carreras I.; Frise E.; Kaynig V.; Longair M.; Pietzsch T.; Preibisch S.; Rueden C.; Saalfeld S.; Schmid B.; et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behringer S.; Wingert V.; Oria V.; Schumann A.; Grunert S.; Cieslar-Pobuda A.; Kolker S.; Lederer A. K.; Jacobsen D. W.; Staerk J.; Schilling O.; Spiekerkoetter U.; Hannibal L. Targeted Metabolic Profiling of Methionine Cycle Metabolites and Redox Thiol Pools in Mammalian Plasma, Cells and Urine. Metabolites 2019, 9, 235 10.3390/metabo9100235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortmayr K.; Schwaiger M.; Hann S.; Koellensperger G. An integrated metabolomics workflow for the quantification of sulfur pathway intermediates employing thiol protection with N-ethyl maleimide and hydrophilic interaction liquid chromatography tandem mass spectrometry. Analyst 2015, 140, 7687–7695. 10.1039/C5AN01629K. [DOI] [PubMed] [Google Scholar]
- Creek D. J.; Chalmers D. K.; Charman W. N.; Duke B. J. Quantum chemical study of the intermediate complex required for iron-mediated reactivity and antimalarial activity of dispiro-1, 2, 4-trioxolanes. J. Mol. Graphics Modell. 2008, 27, 394–400. 10.1016/j.jmgm.2008.06.008. [DOI] [PubMed] [Google Scholar]
- Creek D. J.; Charman W. N.; Chiu F. C.; Prankerd R. J.; Dong Y.; Vennerstrom J. L.; Charman S. A. Relationship between antimalarial activity and heme alkylation for spiro- and dispiro-1,2,4-trioxolane antimalarials. Antimicrob. Agents Chemother. 2008, 52, 1291–1296. 10.1128/aac.01033-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creek D. J.; Charman W. N.; Chiu F. C.; Prankerd R. J.; McCullough K. J.; Dong Y.; Vennerstrom J. L.; Charman S. A. Iron-mediated degradation kinetics of substituted dispiro-1,2,4-trioxolane antimalarials. J. Pharm. Sci. 2007, 96, 2945–2956. 10.1002/jps.20958. [DOI] [PubMed] [Google Scholar]
- Wang X.; Creek D. J.; Schiaffo C. E.; Dong Y.; Chollet J.; Scheurer C.; Wittlin S.; Charman S. A.; Dussault P. H.; Wood J. K.; Vennerstrom J. L. Spiroadamantyl 1,2,4-trioxolane, 1,2,4-trioxane, and 1,2,4-trioxepane pairs: relationship between peroxide bond iron(II) reactivity, heme alkylation efficiency, and antimalarial activity. Bioorg. Med. Chem. Lett. 2009, 19, 4542–4545. 10.1016/j.bmcl.2009.07.013. [DOI] [PubMed] [Google Scholar]
- Xie S. C.; Dogovski C.; Hanssen E.; Chiu F.; Yang T.; Crespo M. P.; Stafford C.; Batinovic S.; Teguh S.; Charman S.; Klonis N.; Tilley L. Haemoglobin degradation underpins the sensitivity of early ring stage Plasmodium falciparum to artemisinins. J. Cell Sci. 2016, 129, 406–416. 10.1242/jcs.178830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui L.; Wang Z.; Jiang H.; Parker D.; Wang H.; Su X.-Z.; Cui L. Lack of association of the S769N mutation in Plasmodium falciparum SERCA (PfATP6) with resistance to artemisinins. Antimicrob. Agents Chemother. 2012, 56, 2546–2552. 10.1128/AAC.05943-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhisutthibhan J.; Meshnick S. R. Immunoprecipitation of [(3)H]dihydroartemisinin translationally controlled tumor protein (TCTP) adducts from Plasmodium falciparum-infected erythrocytes by using anti-TCTP antibodies. Antimicrob. Agents Chemother. 2001, 45, 2397–2399. 10.1128/aac.45.8.2397-2399.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller S. Role and Regulation of Glutathione Metabolism in Plasmodium falciparum. Molecules 2015, 20, 10511–10534. 10.3390/molecules200610511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.-C.; Oses-Prieto J. A.; Pope L. E.; Burlingame A. L.; Dixon S. J.; Renslo A. R. Reactivity-Based Probe of the Iron(II)-Dependent Interactome Identifies New Cellular Modulators of Ferroptosis. J. Am. Chem. Soc. 2020, 142, 19085–19093. 10.1021/jacs.0c06709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalski A.; Cox J.; Mann M. More than 100,000 detectable peptide species elute in single shotgun proteomics runs but the majority is inaccessible to data-dependent LC-MS/MS. J. Proteome Res. 2011, 10, 1785–1793. 10.1021/pr101060v. [DOI] [PubMed] [Google Scholar]
- Gillet L. C.; Navarro P.; Tate S.; Rost H.; Selevsek N.; Reiter L.; Bonner R.; Aebersold R. Targeted data extraction of the MS/MS spectra generated by data-independent acquisition: a new concept for consistent and accurate proteome analysis. Mol. Cell. Proteomics 2012, 11, O111.016717 10.1074/mcp.O111.016717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobbold S. A.; Chua H. H.; Nijagal B.; Creek D. J.; Ralph S. A.; McConville M. J. Metabolic dysregulation induced in Plasmodium falciparum by dihydroartemisinin and other front line antimalarial drugs. J. Infect. Dis. 2016, 213, 276–286. 10.1093/infdis/jiv372. [DOI] [PubMed] [Google Scholar]
- Becker K.; Tilley L.; Vennerstrom J. L.; Roberts D.; Rogerson S.; Ginsburg H. Oxidative stress in malaria parasite-infected erythrocytes: host-parasite interactions. Int. J. Parasitol. 2004, 34, 163–189. 10.1016/j.ijpara.2003.09.011. [DOI] [PubMed] [Google Scholar]
- Patzewitz E. M.; Salcedo-Sora J. E.; Wong E. H.; Sethia S.; Stocks P. A.; Maughan S. C.; Murray J. A.; Krishna S.; Bray P. G.; Ward S. A.; Muller S. Glutathione transport: a new role for PfCRT in chloroquine resistance. Antioxid. Redox Signaling 2013, 19, 683–695. 10.1089/ars.2012.4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnädig N. F.; Stokes B. H.; Edwards R. L.; Kalantarov G. F.; Heimsch K. C.; Kuderjavy M.; Crane A.; Lee M. C. S.; Straimer J.; Becker K.; Trakht I. N.; Odom John A. R.; Mok S.; Fidock D. A. Insights into the intracellular localization, protein associations and artemisinin resistance properties of Plasmodium falciparum K13. PLoS Pathog. 2020, 16, e1008482 10.1371/journal.ppat.1008482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqui G.; Srivastava A.; Russell A. S.; Creek D. J. Multi-omics Based Identification of Specific Biochemical Changes Associated With PfKelch13-Mutant Artemisinin-Resistant Plasmodium falciparum. J. Infect. Dis. 2017, 215, 1435–1444. 10.1093/infdis/jix156. [DOI] [PubMed] [Google Scholar]
- Wu W.-M.; Wu Y.; Wu Y.-L.; Yao Z.-J.; Zhou C.-M.; Li Y.; Shan F. Unified mechanistic framework for the Fe (II)-induced cleavage of qinghaosu and derivatives/analogues. The first spin-trapping evidence for the previously postulated secondary C-4 radical. J. Am. Chem. Soc. 1998, 120, 3316–3325. 10.1021/ja973080o. [DOI] [Google Scholar]
- Robert A.; Coppel Y.; Meunier B. Alkylation of heme by the antimalarial drug artemisinin. Chem. Commun. 2002, 5, 414–415. 10.1039/b110817b. [DOI] [PubMed] [Google Scholar]
- Wang D.-Y.; Wu Y.-L. A possible antimalarial action mode of qinghaosu (artemisinin) series compounds. Alkylation of reduced glutathione by C-centered primary radicals produced from antimalarial compound qinghaosu and 12-(2,4-dimethoxyphenyl)-12-deoxoqinghaosu. Chem. Commun. 2000, 22, 2193–2194. 10.1039/b006906j. [DOI] [Google Scholar]
- Mohring F.; Jortzik E.; Becker K. Comparison of methods probing the intracellular redox milieu in Plasmodium falciparum. Mol. Biochem. Parasitol. 2016, 206, 75–83. 10.1016/j.molbiopara.2015.11.002. [DOI] [PubMed] [Google Scholar]
- Rahbari M.; Rahlfs S.; Jortzik E.; Bogeski I.; Becker K. H2O2 dynamics in the malaria parasite Plasmodium falciparum. PLoS One 2017, 12, e0174837 10.1371/journal.pone.0174837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoine T.; Fisher N.; Amewu R.; O’Neill P. M.; Ward S. A.; Biagini G. A. Rapid kill of malaria parasites by artemisinin and semi-synthetic endoperoxides involves ROS-dependent depolarization of the membrane potential. J. Antimicrob. Chemother. 2014, 69, 1005–1016. 10.1093/jac/dkt486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coertzen D.; Reader J.; van der Watt M.; Nondaba S. H.; Gibhard L.; Wiesner L.; Smith P.; D’Alessandro S.; Taramelli D.; Wong H. N.; du Preez J. L.; Wu R. W. K.; Birkholtz L. M.; Haynes R. K. Artemisone and Artemiside Are Potent Panreactive Antimalarial Agents That Also Synergize Redox Imbalance in Plasmodium falciparum Transmissible Gametocyte Stages. Antimicrob. Agents Chemother. 2018, 62, e02214-17 10.1128/AAC.02214-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman P. A.; Adams P. A. Artemisinin enhances heme-catalysed oxidation of lipid membranes. Free Radical Biol. Med. 1997, 22, 1283–1288. 10.1016/S0891-5849(96)00508-4. [DOI] [PubMed] [Google Scholar]
- Witkowski B.; Lelievre J.; Nicolau-Travers M. L.; Iriart X.; Njomnang Soh P.; Bousejra-Elgarah F.; Meunier B.; Berry A.; Benoit-Vical F. Evidence for the contribution of the hemozoin synthesis pathway of the murine Plasmodium yoelii to the resistance to artemisinin-related drugs. PLoS One 2012, 7, e32620 10.1371/journal.pone.0032620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phompradit P.; Chaijaroenkul W.; Na-Bangchang K. Cellular mechanisms of action and resistance of Plasmodium falciparum to artemisinin. Parasitol. Res. 2017, 116, 3331–3339. 10.1007/s00436-017-5647-z. [DOI] [PubMed] [Google Scholar]
- Chandra R.; Tripathi L. M.; Saxena J. K.; Puri S. K. Implication of intracellular glutathione and its related enzymes on resistance of malaria parasites to the antimalarial drug arteether. Parasitol. Int. 2011, 60, 97–100. 10.1016/j.parint.2010.09.009. [DOI] [PubMed] [Google Scholar]
- Jortzik E.; Becker K. Thioredoxin and glutathione systems in Plasmodium falciparum. Int. J. Med. Microbiol. 2012, 302, 187–194. 10.1016/j.ijmm.2012.07.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.