Abstract
Single quadrupole mass spectrometry (MS) with enhanced in-source multiple fragment ion monitoring was designed to perform high sensitivity quantitative mass analyses. Enhanced in-source fragmentation amplifies fragmentation from traditional soft electrospray ionization producing fragment ions that have been found to be identical to those generated in tandem MS. We have combined enhanced in-source fragmentation data with criteria established by the European Union Commission Directive 2002/657/EC for electron ionization single quadrupole quantitative analysis to perform quantitative analyses. These experiments were performed on multiple types of complex samples that included a mixture of 50 standards, as well as cell and plasma extracts. The dynamic range for these quantitative analyses was comparable to triple quadrupole multiple reaction monitoring (MRM) analyses at up to 5 orders of magnitude with the cell and plasma extracts showing similar matrix effects across both platforms. Amino acid and fatty acid measurements performed from certified NIST 1950 plasma with isotopically labelled standards demonstrated accuracy in the range of 91–110% for the amino acids, 76–129% for the fatty acids, and good precision (coefficient of variation < 10%). To enhance specificity, a newly developed correlated ion monitoring algorithm was designed to facilitate these analyses. This algorithm autonomously processes, aligns, filters, and compiles multiple ions within one chromatogram enabling both precursor and in-source fragment ions to be correlated within a single chromatogram, also enabling the detection of co-eluting species based on precursor and fragment ion ratios. Single quadrupole instrumentation can provide MRM level quantitative performance by monitoring/correlating precursor and fragment ions facilitating high sensitivity analysis on existing single quadrupole instrumentation that are generally inexpensive, easy to operate, and technically less complex.
Graphical Abstract
INTRODUCTION
The single quadrupole mass analyzer, first conceived by Nobel Laureate Wolfgang Paul over sixty years ago,1 represents a milestone invention in mass spectrometry (MS) and laid the foundation for most current MS and tandem MS technologies. For example, triple quadrupole2 tandem MS and multiple reaction monitoring3 (MRM), with the advent of soft atmospheric pressure ionization4 in the late 1980s, has become the primary approach for targeted small molecule analysis. In the last three decades MRM has dominated the MS landscape with its ultrahigh selectivity, sensitivity, and broad dynamic range, especially in the quantification of small molecules and peptides.5–7 MRM exploits the unique capability of tandem mass spectrometers to act as a double mass filter, facilitating the analysis of analytes from complex matrices. In MRM with a triple quadrupole mass spectrometer, a predetermined precursor ion is selected/isolated with the first quadrupole, then fragmented in the collision cell (second quadrupole) with a neutral gas (e.g., nitrogen).8 Thus, only precursor derived fragment ions pass to the third quadrupole and reach the detector. The precursor-fragment ion pairs are referred to as 'transitions' and over a hundred transitions can be recorded in (scheduled) MRM analyses, enabling the simultaneous targeted analysis of multiple analytes using liquid chromatography tandem MS.9 MRM is considered the gold standard in the quantitative analysis of small molecules10 however, these instruments are typically expensive and suffer from inherent sensitivity losses in the collision chamber. Less expensive instrumentation, offering quantitative analysis for broader implementation, would provide a technically advantageous alternative option.
Enhanced in-source fragmentation is an approach that promotes the generation of both molecular ions and their respective fragments by increasing in-source fragmentation voltages. These fragment ions are generally the same as those generated in tandem MS MRM experiments.11–16 This similarity makes possible the simultaneous monitoring of both precursor ion and its fragments with a single quadrupole mass spectrometer, in experiments typically performed with tandem mass spectrometers. Although any fragment ions could provide similar results. To streamline the processing of the enhanced in-source fragmentation data, and flag potentially co-eluting species, a correlated ion monitoring algorithm was developed to facilitate these analyses. Correlated ion monitoring autonomously processes, aligns, filters, and compiles multiple ions within one chromatogram enabling both precursor and in-source fragment ions to be correlated within a single chromatogram, also enabling the detection of co-eluting species based on precursor and fragment ion ratios.
Single quadrupole multiple fragment ion monitoring is a variation on an approach that has been widely used for high sensitivity detection and quantitative analysis of small molecules especially with gas chromatography MS.17–19 However, the combination of enhanced in-source fragmentation and multiple fragment ion monitoring in a single quadrupole LC/MS instrument provides enhanced selectivity, specificity, and sensitivity for the quantitative analyses of a broader range of compounds (over traditional gas chromatography MS).12 In addition, as compared with MRM, single quadrupole multiple fragment ion monitoring analysis is available at ~30% the cost and have enhanced signal since the single quadrupole instruments do not have a collision cell and therefore do not experience the associated collision cell ion losses.12
In this work, these three separate technologies have been combined and developed to provide quantitative analysis with single quadrupole instrumentation, they include: 1) soft atmospheric pressure ionization with enhanced in-source fragmentation, 2) precursor and multiple fragment ion monitoring, and 3) a newly created correlated ion monitoring algorithm. We demonstrate that single quadrupole mass spectrometers can have comparable quantitative performance to triple quadrupole mass spectrometers. Using a mixture of endogenous molecules, we evaluated key analytical merits in quantitative analysis including selectivity, sensitivity/dynamic range, matrix effects, accuracy and precision. Subsequently, we used both enhanced in-source multiple fragment ion monitoring and MRM with the same triple quadrupole mass spectrometer to determine the concentrations of metabolites in a plasma extract, a bacterial cell extract, and a mammalian cell extract and the results acquired using both methods were compared. To confirm the applicability of single quadrupole quantitative analysis, we further performed the analysis of a National Institute of Standards and Technology (NIST) certified plasma sample using a single quadrupole mass spectrometer.
EXPERIMENTAL SECTION
Materials.
A total of 50 endogenous molecules were selected to represent a broad range of physicochemical properties and chemical structures, including amino acids, lipids, and fatty acids. For the investigation of quantitative performance of enhanced in-source multiple fragment ion monitoring including the limit of quantification (LOQ) and dynamic range, the fifty molecules were prepared at 9 concentrations spanning 7 orders of magnitude: 0.2 nM, 0.5 nM, 1 nM, 10 nM, 100 nM, 1 μM, 10 μM, 100 μM, and 1 mM. Additional intermediate calibration standards were used when necessary, resulting in at least five calibration points. For investigating matrix effects in enhanced in-source multiple fragment ion monitoring analysis, plasma (both human male AB plasma and NIST SRM 1950 plasma), mixtures of two mammalian cell lines (VERO C1008 and L6 myocyte), and bacteria cells (Pantoea strain sp. MT058) isolated from the groundwater in Oak Ridge Reservation were used. Standards and both plasma samples were purchased from Sigma-Aldrich (St. Louis, MO). VERO C1008 cell line and L6 myocyte cell line were obtained from James E. Voss laboratory in the Scripps Research Institute (La Jolla, CA) and Olivia Osborn laboratory in the University of California San Diego, respectively. Bacteria cell pellets were shipped from Michael W. Adams laboratory at University of Georgia and Nitin S. Baliga laboratory in the Institute for Systems Biology.
Sample preparation.
Plasma samples were prepared using protein precipitation for both matrix preparation and amino acids extraction.20 Briefly, 400 μL solvent (acetonitrile:methanol; 1:1) was added to 100 μL plasma sample. After storage at –20 °C for 1 hour, the sample was sonicated on ice for 10 min, followed by centrifugation at 13,000 ×g at 4 °C for 15 min. The supernatant was collected and dried in a vacuum concentrator (LABCONCO) at 10 °C and reconstituted with 100 μL acetonitrile:water (1:1). After sonication (10 min, on ice) and centrifugation (13, 000 ×g, 4 °C, 15 min), the supernatant was transferred to a LC-MS glass vial with inserts for instrumental injection and for preparing matrix matched calibration lines. For the extraction of total fatty acids from plasma sample, a mixture of hexane/isopropanol (3:2) was used and 0.3 M KOH in 80% methanol was used for alkaline hydrolysis, with details documented elsewhere.21–22
Bacterial and mammalian cells were extracted using a solvent mixture of acetonitrile:methanol:water (2:2:1).20 In brief, cell samples (~1 million cells in each tube) were sonicated on ice for 15 min after shock-freezing in liquid nitrogen and subsequent thawing at room temperature. The operation was repeated three times. The samples were then incubated at –20 °C for one hour allowing protein precipitation, followed by centrifugation at 13,000 ×g at 4 °C for 15 min. Using the same procedure as mentioned above, the supernatant was collected, dried and reconstituted with 50 μL acetonitrile:water (1:1). After sonication (10 min, on ice) and centrifugation (13,000 ×g, 4 °C, 15 min), the supernatant was transferred to a LC-MS glass vial with inserts for instrumental injection and for preparing matrix matched calibration lines.
Triple quadrupole mass spectrometry analysis.
Single and triple quadrupole experiments were carried out on a Waters Xevo TQ-XS triple quadrupole mass spectrometer (Milford, MA) to allow for consistent comparison without the variability associated with using different instruments. Collision gas flow on the triple quadrupole was turned off when performing the single quadrupole analyses. The analyses were performed in both positive and negative electrospray ionization modes, and the single and triple quadrupole transitions measured for each molecule are shown in Table S1. In the single quadrupole experiments, the relationship between cone voltage and peak intensity of each ion (precursor ion and fragments) was investigated individually, and the optimal cone voltage at which maximum peak intensity is achieved was selected for each ion. In the triple quadrupole experiments, the optimal cone voltage of the precursor ion was determined for every corresponding transition, and the collision energy was optimized for fragment ion generation. A dwell time of 15 ms was used for each channel in the single quadrupole and each transition in triple quadrupole. The ion source desolvation temperature was set at 500°C.
ACQUITY UPLC BEH C18 column (2.1 × 100 mm, 1.7 μm, Waters) and ACQUITY UPLC BEH Amide column (2.1 × 100 mm, 1.7 μm, Waters) were used in the separation of metabolites in reverse phase and HILIC analysis, respectively. For the reversed phase analyses, metabolites were separated by gradient elution at a flow rate of 200 μL/min starting at 25% (v/v) B, held for 1 min, increased to 99% B within 8 min, held for 3 min, and reverted to 25% B at 12.1 min, held for 2.9 min, with a total run time of 15 min. The mobile phases comprised acetonitrile:water (60:40) containing 0.1% formic acid and 1 mM ammonium formate (A) and isopropanol:acetonitrile (90:10) containing 0.1% formic acid and 1 mM ammonium formate (B). For the HILIC analysis, metabolites were separated by gradient elution at a flow rate of 250 μL/min starting at 5% (v/v) A, increased to 80% A within 8 min, held for 0.5 min, and reverted to 5% A at 8.6 min, held for 3.9 min, with a total run time of 12.5 min. The mobile phases were comprised of water containing 10 mM ammonium acetate and 0.1% formic acid (A) and water/acetonitrile (5:95) containing 0.1% formic acid (B).
Single quadrupole mass spectrometry analysis.
The analysis of 11 amino acids (positive mode) and 5 fatty acids (negative mode) as well as their respective isotope labelled standards was performed on an Agilent 6130 Quadrupole LC-MS (Santa Clara, CA). The ESI source parameters in both positive and negative modes were set as follows: drying gas flow 9 L/min, drying gas temperature 350 °C, and nebulizer pressure 30 psig. Capillary voltage was set at 4000 V in positive mode and 3500 V in negative mode, respectively. The fragmentor voltage was optimized for each monitored ion and dwell time was 290 ms at each channel.
ACQUITY UPLC BEH C18 column (2.1 × 100 mm, 1.7 μm, Waters) was used in the analysis of fatty acids, and ACQUITY UPLC BEH Amide column (2.1 × 100 mm, 1.7 μm, Waters) was used in the analysis of amino acids. The gradient in fatty acid analysis was as follows: starting at 20% (v/v) B, held for 1 min, increased to 95% B within 12 min, held for 1 min, and reverted to 20% B at 14.5 min, held for 3.5 min, with a total run time of 18 min. The gradient in amino acid analysis was as follows: starting at 10% (v/v) A, held for 2 min, increased to 70% A within 11 min, and reverted to 10% A at 11.5 min, held for 3.5 min, with a total run time of 15 min. The mobile phases used in fatty acid and amino acid analysis were the same as those used in reverse phase and HILIC analysis in the triple quadrupole MS analysis, respectively. The mobile phase flow rate was set as 100 μL/min in the analysis of both fatty acids and amino acids.
Data analysis.
Extracted ion chromatograms (EICs) were generated using both the single and triple quadrupoles and manually inspected with vendor specific software. Targeted ions were carefully selected using precursor/fragment ions and retention time. The peak intensity and area were recorded for either comparison or quantification purpose. Each molecule typically has four associated EICs (one precursor and three fragment ions) in single quadrupole, which corresponds to three EICs (three transitions) in triple quadrupole. The peak area from each EIC was used separately in both the single and triple quadrupole, and the selection of the quantifier ion was based on the linear dynamic range of each ion (or transition) and the concentration of target metabolites in the test sample.
For the single and triple quadrupole experiments, molecular standards of all 50 molecules (10 μM) were injected into the same triple quadrupole mass spectrometer to compare peak intensities of the same fragment ions. Following these analyses, limits of quantification (LOQs) and the linear dynamic ranges (LDRs) of the 50 molecules were investigated for each acquisition method using a total of 9 concentration levels. The LOQ was defined as the lowest analyte concentration in the calibration that can be quantitatively detected within acceptable accuracy (back calculated 70–130%) and precision (coefficient of variation (CV) < 30%). This approach for determining LOQs was used since the signal-to-noise levels were expected to be impacted by the background levels generally being higher for single quadrupole instrumentation. Using the solvent-based calibration (1/x2 weighted) acquired with both methods, the concentrations of the fifty metabolites were determined in a bacteria cell extract; for metabolites not endogenously present in the bacteria cell extract, standards were spiked into the sample at varying concentrations ranging from 10 nM to 100 μM for quantitative performance evaluation. These metabolites were also analyzed from a plasma extract and a mammalian cell extract to investigate the performance of the single quadrupole in different types of matrices. Every test sample was replicated five times and the averaged results were used. CV, calculated as the percent ratio (%) between standard deviation of observed metabolite concentration divided by the mean measured concentration, was used to assess the precision of replicate analysis. It is worth noting that since we used solvent based calibration standards in determining the LOQs, the values reported in this manuscript only reflect the instrumental quantification limits.
The biological matrices mentioned above, including plasma, bacterial cell extract, and mammalian cell extract, were selected as typical biological matrices to investigate the matrix effect on the single quadrupole data. Based on the results, the 50 metabolites were classified into two groups: those endogenously present and those absent. For endogenous metabolites, standard addition method was used to investigate the matrix effects at seven concentration levels, namely 0.1, 1, 5, 10, 50, 100, and 200 μM. The matrix effect was further assessed by comparing the peak area of the analyte standard in solvent and matrix at the same concentration, defined as [(peak area in solvent/in matrix) × 100]. For those metabolites not present or at low levels (< 1 nM) in the target matrix, matrix matched calibration lines were prepared at concentrations ranging from 10 nM to 200 μM by spiking standards into the post-extraction samples, referred to as analyte free matrix method. The slopes of the calibration lines for the same set of standards generated in both solvent and matrix were compared to assess the matrix effect. Each standard was replicated five times and the averaged peak area or slope was used in the analysis.
The accuracy and precision of single quadrupole was further evaluated by measuring amino acids and fatty acids in a certified NIST SRM 1950 plasma sample using isotope dilution-based matrix matched calibration. The intra- and inter-day precision of the method was determined by replicate analyses (n=5): intra-day precision was estimated using five replicate injections performed on a single day; inter-day precision estimation was performed based on five replicate injections conducted in five consecutive days. The linearity of the calibration was confirmed by plotting the peak area ratio of each metabolite (area metabolite/area internal standard) against its concentration. The sample concentrations were calculated from the equation y = mx+b, as determined by weighted (1/x2) linear regression of the calibration line. The accuracy of the method was expressed as [(mean observed concentration)/(certified/reference mean concentration)] × 100.
Correlated ion monitoring:
The correlated ion monitoring algorithm was developed to perform peak picking, alignment, and data analysis (https://github.com/ricoderks/eisaCIM). In a typical experiment, chromatograms for each individual molecule are created by (1) peak picking each SIM trace within a pre-set intensity threshold (using XCMS v3.12.0). Determining the threshold for each SIM trace is a crucial step which allows for filtering co-eluting peaks. In the next step (2) the SIM traces are aligned and grouped within a retention time window (typically 3–5 seconds) where trace groups are retained only if they contain a peak for all four SIM traces (precursor and three fragment ions). (3) the filtered SIM traces are aligned, and a correlated ion monitoring chromatogram is created.
RESULTS AND DISCUSSION
Single Quadrupole Enhanced In-Source Multiple Fragment Ion Monitoring
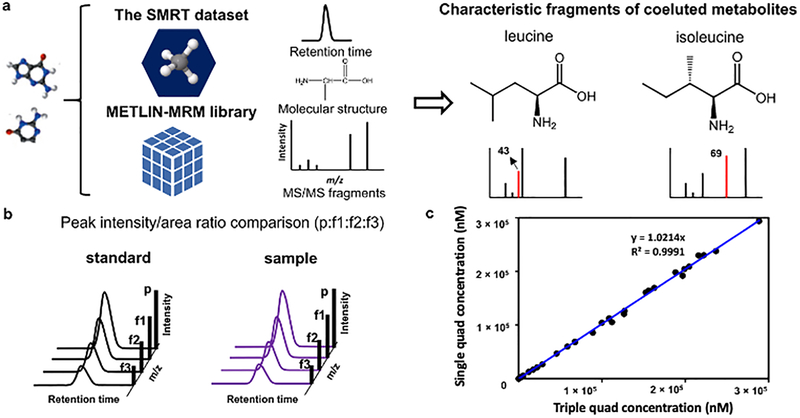
In earlier work,12 we demonstrated that enhanced in-source fragmentation allowed to produce both high abundance precursor ions and fragment ions that were characteristic of tandem MS data for the majority of molecules investigated, thus enabling untargeted metabolomic experiments with single mass analyzers. In this effort we extend enhanced in-source multiple fragment ion monitoring as a quantitative technology platform demonstrating that it can also facilitate the quantitative analysis of small molecules in a single quadrupole mass spectrometer by producing high abundant precursor ion and respective fragments simultaneously. This can significantly improve the applicability of a single quadrupole mass spectrometer (Figure 1a) in quantitative analysis, which typically only uses the precursor ion in selecting the target analyte.
Figure 1.
(a) Single quadrupole MS quantitive analysis general experimental design. (b) Fragment ion intensity ratio for enhanced in-source multiple fragment ion monitoring single quadrupole and MRM triple quadrupole comparison between electrospray ionization positive mode and negative mode. (c) Fragment ion intensity ratio comparison between lipids/fatty acids and other molecules. Log transformed intensity ratio was used in (b) and (c). (d) A typical extracted ion chromatogram of a single quadrupole channel (xanthine in solvent, fragment ion 108 at RT 2.80 min). (e) A typical extracted ion chromatogram of a triple quadrupole transition (xanthine in solvent, transition 151>108 at RT 2.84 min). The number next to the single quadrupole channel or triple quadrupole transition in (d) and (e) represents the maximum intensity of the EIC.
To investigate the in-source fragments generation performance with a single quadrupole, we prepared a group of fifty molecules containing a variety of chemical structures including amino acids, sugars, fatty acids, and lipids (Table S1). Except for the fatty acids analyzed in the negative mode, all other molecules produced at least 3 fragments (Table S1). In-source fragmentation was performed with a triple quadrupole to enable direct comparison of single and triple quadrupole performance on the same instrumentation. For example, the cone voltage in the Waters triple quadrupole mass spectrometer was optimized for each ion including both precursor and fragment ions to acquire their own maximum peak intensities. With the increase of cone voltage, peak intensity of ions either increase or a relationship of reversed U shape curve is observed (Figure S1). In enhanced in-source multiple fragment ion monitoring, the optimal cone voltage of each ion was used in preparing the quantitative analysis method to acquire the precursor ion and its fragments at their own peak intensities.
When performing the enhanced in-source multiple fragment ion monitoring experiments on the triple quadrupole mass spectrometer, the collision gas flow was turned off and the first quadrupole was used for these analyses. In addition, for direct comparison purposes, we used MRM to analyze the same collision-induced fragments generated in the collision cell for all the target molecules from the same instrument that the enhanced in-source multiple fragment ion monitoring experiments were performed. Maximum intensity was optimized for each fragment ion by adjusting the relevant parameters including collision energy. Comparison of enhanced in-source multiple fragment ion monitoring and MRM chromatograms of identical standards (10 μM each) revealed that 86% of the fragments acquired using single quadrupole enhanced in-source multiple fragment ion monitoring had higher peak intensity with a median increase of around 300% and 400% in electrospray ionization positive and negative mode, respectively (Figure 1b), with an overall median increase of 260% in peak intensity (Figure S2). We further investigated the impact of single quadrupole enhanced in-source multiple fragment ion monitoring on metabolites within different chemical classes and found that the median increase of fragment ion intensity for lipids and fatty acids were over 800%, while it was over 300% for other molecules, indicating that enhanced in-source multiple fragment ion monitoring provides different enhancements across different chemical classes (Figure 1c). For example, lipids and fatty acids appear to have enhanced fragmentation. This analyte dependent fragmentation in single quadrupole enhanced in-source multiple fragment ion monitoring can be further developed for optimizing quantitative analysis.
Selectivity/Specificity
Single quadrupole enhanced in-source multiple fragment ion monitoring would seemingly suffer from limited selectivity when compared to MRM analysis, lacking the ability to generate transitional mass spectrometric pairs (as in MRM) deriving from a selected precursor. However, single quadrupole enhanced in-source multiple fragment ion monitoring takes advantage of its ability to simultaneously produce both high abundant precursor and fragment ions originating from the same molecule in a single quadrupole mass spectrometer. Therefore, by applying criteria originally adopted for GC/MS based selected ion monitoring quantitation, as described in EU Commission Directive 2002/657/EC,18 we were able to generate quantitative data similar to what has been previously accomplished with GC/MS and MRM. While MRM exploits a tandem quadrupole mass analyzer’s ability to selectively and simultaneously analyze both precursor and fragment ions resulting in four identification points (according to 2002/657/EC Directive), single quadrupole enhanced in-source multiple fragment ion monitoring implements this concept by monitoring one precursor ion and three respective fragments (termed “channels”). The resultant enhanced in-source multiple fragment ion monitoring is composed of 4 channels, thereby equaling four identification points equivalent to two MRM transitions with one precursor and two fragment ions, facilitating effective quantitative analysis. Xanthine is provided as an example, with a typical EIC from an enhanced in-source multiple fragment ion monitoring channel and an MRM transition (using the same fragment ion) shown in Figure 1d and 1e, respectively.
The high specificity of MRM in quantitative analysis is primarily achieved using specific precursor/fragment ion pairs and retention time. Single quadrupole MS analysis utilizes one mass filter and thus it is not possible to directly link fragments with the associated precursor ion. However, we observed that single quadrupole enhanced in-source multiple fragment ion monitoring can significantly improve the selective capability of target analytes from the MS1 spectra by simultaneously monitoring the precursor ion and its high abundant characteristic fragments (n=3) that elute at the same retention time. The in-source fragments serve as an identity indicator and can also be used for quantification once specificity is confirmed.
The METLIN small molecule retention time (SMRT) dataset24 and METLIN-MRM10 library created from the METLIN tandem mass spectra database25 has also been implemented to facilitate single quadrupole enhanced in-source multiple fragment ion monitoring (Figure 2) and help deconvolve commonly co-eluting compounds (Figure 2a). The METLIN SMRT dataset is an experimentally acquired reverse-phase chromatography retention time data set generated from over 80,000 small molecules. Using METLIN SMRT in conjunction with METLIN MRM offers an opportunity to help predict and deconvolve coeluting species. Leucine and isoleucine were analyzed as examples (Figure 2a). In addition, interfering molecular ions can be detected by comparing the peak intensity/area ratios between the precursor ion and its fragments (p:f1:f2:f3) across standards and samples (Figure 2b). This takes advantage of the fact that a peak intensity/area ratio between precursor ion and fragment ions stemming from one compound stay constant between standard and sample.
Figure 2.
Single and triple quadrupole characteristic fragments of co-eluting metabolites can be selected using retention time data generated from the SMRT dataset and fragmentation data from the METLIN-MRM library (e.g., (a) leucine and isoleucine). This data can be used to monitor the specificity of quantifier based on (b) peak intensity ratios, for example. Concentrations of 50 metabolites calculated using single quadrupole monitoring aligned well with the results calculated using a triple quadrupole generated from identical cell sample (analytical standards were spiked into the cell sample at varying concentrations if the metabolites were not detected in the cell sample) (c). p: precursor ion; f1, f2 and f3: fragment ion 1, 2, and 3, respectively.
Another approach to enhance selectivity is through application of orthogonal separation technologies where the additional separate “channels” could be used for identification and/or quantification purposes. For example, ion mobility data could serve as an orthogonal set of information on the in-source fragment ions, as shown to be separated via drift tube-based ion mobility (Figure S3) and thus act as an alternative means for characterizing these ions in accordance with Directive 2002/657/EC,18 using CCS values. This is demonstrated in Figure S3 where tandem MS data is shown to coinciding with the in-source fragments where ion mobility could serve to provide additional discrimination between co-eluting molecules and therefore further enhance specificity. Nevertheless, great care must be taken in choosing selective fragments ions for both, enhanced in-source fragmentation with correlated ion monitoring as well as MRM analysis as some analyte classes (e.g., fatty acids) tend to produce ubiquitous fragment ions (e.g., carboxylic acids predominantly show a loss of water and CO2).
Sensitivity/Dynamic Range
A wide linear dynamic range (LDR) is important for small molecule quantitation in biological samples. According to the Human Metabolome Database, metabolite concentrations in human plasma and urine broadly vary from the pico- up to the millimolar range,26,27 Having a wide LDR is useful in helping to avoid re-analysis and additional dilutions. The LDR can be limited either by the LOQ or by saturation at the ion source and detector.28 Thus, we prepared solvent-based calibrations for all fifty molecules at nine concentration levels ranging from 0.2 nM to 1 mM to compare the LDRs for the same compound between enhanced in-source fragmentation multiple fragment ion monitoring and MRM.16 Weighted (1/x2) linear regression calibrations were established for each molecule by plotting the relationship between concentration level and the corresponding peak area. LOQs were determined as the lowest concentration level in the calibration with acceptable accuracy and precision (Table S2), which were also used as the lower limit of the LDR. The upper limit in the LDR was determined using the same criterion. Here, only those molecules containing 3 fragments were analyzed and several molecules which didn’t provide linear relationship between the concentration level and peak area were excluded. Finally, LDRs of a total of 43 molecules acquired using both single quadrupole multiple fragment ion monitoring and triple quadrupole MRM are reported (Table S3).
We first acquired the LDRs for each molecular ion (single quadrupole enhanced in-source multiple fragment ion monitoring) or triple quadrupole MRM transition, then an integrated LDR was reported for each molecule in each method, combining the lower limit of the LDR of a primary sensitive ion and the upper limit of the LDR of a less sensitive ion. It was expected that sensitivity in single quadrupole enhanced in-source multiple fragment ion monitoring was going to be lower compared to triple quadrupole MRM for the same molecule because of higher background noise levels observed in the EIC when only one mass filter is used. However, our results showed that the single quadrupole enhanced in-source multiple fragment ion monitoring can achieve the same sensitivity for most analytes in quantitative analysis. Of the 43 analytes, only 30% of the compounds tested had one order of magnitude lower LOQs in single quadrupole enhanced in-source multiple fragment ion monitoring than MRM. In addition, we observed that five molecules, including glucose, methylhistidine, glutathione, C20 sphingosine, and oleic acid, with LOQs one order of magnitude greater in the single quadrupole enhanced in-source multiple fragment ion monitoring, indicating that certain chemicals can be more efficiently detected with the single quadrupole (Table S3). Thus, although there is a higher chemical noise baseline in the EIC produced by single quadrupole enhanced in-source fragmentation multiple fragment ion monitoring, the majority of the compounds can provide the same LOQs as those produced using MRM. Further, we found that ~90% of the molecules had the same or higher upper limit of the LDR in single quadrupole enhanced in-source fragmentation multiple fragment ion monitoring. This indicates that saturation was the same between the two methods for most ion signals and only 10% of molecules were saturated in enhanced in-source fragmentation multiple fragment ion monitoring. Overall, single quadrupole enhanced in-source fragmentation multiple fragment ion monitoring achieved an LDR of up to 5 orders of magnitude for over 90% of the analytes investigated.
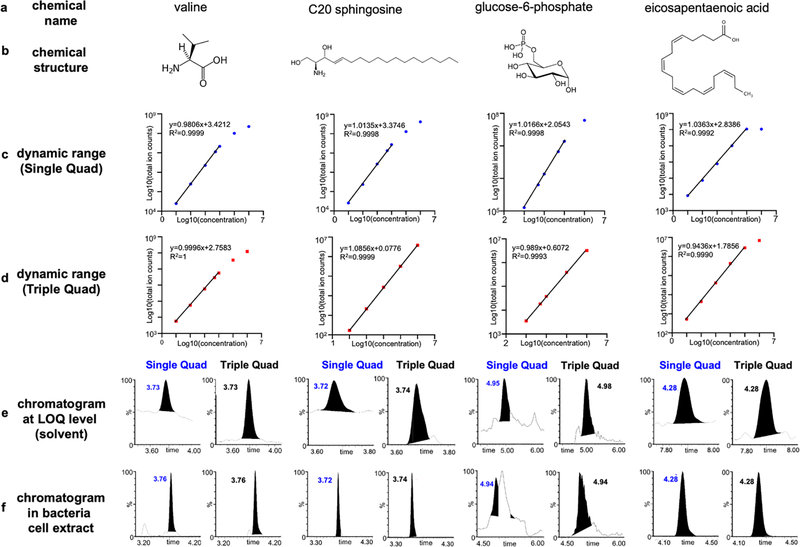
Four molecules with different physicochemical properties, including valine, C20 sphingosine, glucose-6-phosphate, and eicosapentaenoic acid (EPA), were selected to illustrate the differences in LDR between the two techniques (Figure 3a and 3b). As shown in Figure 3c and 3d, single quadrupole achieves similar or lower (C20 sphingosine) LOQs for the select compounds. Chromatograms of the four molecules at LOQ levels in solvent in both the single quadrupole enhanced in-source multiple fragment ion monitoring and triple quadrupole MRM modes are shown in Figure 3e. As illustrated, while single quadrupole monitoring comes with a greater level of chemical noise, the higher ion intensity enables comparable sensitivity. Of the four molecules, valine and EPA presented an identical dynamic range between the two modes; single quadrupole enhanced in-source multiple fragment ion monitoring analysis of C20 sphingosine proved more sensitive when compared to the triple quadrupole MRM allowing to extend the dynamic range towards lower analyte concentrations; for glucose-6-phosphate, detector saturation was observed at 1 mM in single quadrupole, while it was not observed in MRM mode. Chromatograms of the four molecules acquired using single quadrupole enhanced in-source fragmentation multiple fragment ion monitoring and triple quadrupole MRM modes in a bacteria cell extract are shown in Figure 3f. In summary, single quadrupole enhanced in-source multiple fragment ion monitoring performs equally well or in some cases better in terms of sensitivity and dynamic range for most metabolites investigated, while triple quadrupole MRM mode was less prone to detector saturation.
Figure 3.
Single quadrupole enhanced in-source multiple fragment ion monitoring and triple quadrupole MRM limit of quantitation (LOQ) and dynamic range. Chemical names (a) and structures (b), dynamic ranges (c, d), LC chromatograms at LOQ levels in solvent (e), and LC chromatograms in bacterial cell extracts (f) of four representative molecules with different physicochemical properties. Dynamic range data and LC chromatograms acquired in both single and triple quadrupoles are shown; the linearity was calculated based on the log10 transformed values; the ion used with enhanced in-source fragmentation multiple fragment ion monitoring and the transition used in MRM are shown next to the linear curve. Valine and glucose-6-phosphate were acquired using HILIC chromatography; C20 sphingosine and eicosapentaenoic acid were acquired using reverse phase chromatography.
Comparison of Single and Triple Quadrupoles of Cell Extracts and Plasma Samples
Additional analyses were performed using the above-mentioned calibration approaches to quantify these molecules from a bacteria cell extract using both single quadrupoles enhanced in-source fragmentation multiple fragment ion monitoring and triple quadrupole MRM. For those compounds which were not endogenously present, the bacteria cell extract was fortified with a defined amount of metabolite standards for quantification purposes. The concentrations calculated ranged from 1 nM to over 100 μM levels and good alignment was observed between the results calculated using the two techniques (Figure 2c; Table S4). We further analyzed these metabolites in a plasma extract and a mammalian cell extract using both single and triple quadrupoles, and concentrations of the detected metabolites are shown in Tables S5 and S6, respectively. The percent difference of the results calculated using the two techniques were below 7% for over 90% of the molecules measured in every matrix. Significantly co-eluting peaks from the matrix can interfere with the accuracy of peak integration and lead to a higher percent difference, e.g., uric acid in plasma.
Matrix Effect
A key advantage of MRM analysis of complex mixtures is the high selectivity achieved by the combination of two mass filters and a collision cell. As enhanced in-source fragmentation multiple fragment ion monitoring is performed with a single quadrupole, selectivity in complex mixture analysis is an important analytical feature that requires evaluation. Thus, we investigated three complex biological matrices, including a plasma extract, a mammalian cell extract, and a bacteria cell extract. As mentioned above, we first analyzed the three matrices and measured the concentrations of fifty metabolites in each of them. For those metabolites which were present in the matrices, we used the standard addition method to evaluate the matrix effects by spiking five to eight different concentration levels into the extract post-extraction19. For metabolites not endogenously present in the matrices, we spiked eight different concentration levels into each of the three different matrices post-extraction. Matrix effects were evaluated according to Matuszewski et al.19 and as described below, for all fifty metabolites in both MRM and enhanced in-source fragmentation selected ion monitoring modes. Overall, the two techniques showed similar matrix effect trends (suppression or enhancement) for the same metabolite and matrix. There were no significant differences of the matrix effects observed between enhanced in-source fragmentation multiple fragment ion monitoring and MRM. Five molecules, including phenylalanine, hypoxanthine, uridine, inosine, and guanosine, were selected to demonstrate the matrix effects of molecules in the three different matrices assessed using the standard addition method (Tables S7–S9). We established calibration lines for metabolites which were not present in the sample and compared their slopes in solvent and matrix as well as the standard deviations in replicate injections (n=5) with examples shown in Table 1. Overall, our results indicate that the difference of matrix effects between the two analytical modes, are compound and matrix dependent and independent of the analytical technique (single quadrupole multiple fragment ion monitoring versus MRM).
Table 1.
Matrix effects with the single quadrupole enhanced in-source multiple fragment ion monitoring and triple quadrupole MRM. The calibration lines for inosine in plasma extract and glucose-6-phosphate in bacteria cell extract as well as their calibrations from solvent using both single and triple quadrupole quantification techniques.
| Triple Quadrupole | Single Quadrupole | |||||
|---|---|---|---|---|---|---|
|
| ||||||
| solvent | plasma extract (inosine) | Mann-Whitney test | solvent | plasma extract (inosine) | Mann-Whitney test | |
| slopea | 195.4 | 197.1 | p > 0.01 | 2356.2 | 2389.8 | p > 0.01 |
| SDb | 2.4 | 6.5 | 46.8 | 109.3 | ||
| CV(%)c | 1.2 | 3.3 | 2 | 4.6 | ||
| solvent | bacteria cell (glucose-6-phosphate) | Mann-Whitney test | solvent | bacteria cell (glucose-6-phosphate) | Mann-Whitney test | |
| slope | 15 | 24.1 | p < 0.01 | 200.4 | 381.2 | p < 0.01 |
| SD | 0.6 | 1.2 | 2.4 | 5.2 | ||
| CV(%) | 3.9 | 4.8 | 1.2 | 1.4 | ||
slope, calculated using the linear calibration between total ion counts (y) and concentration (x) of the analyte
SD, standard deviation between slopes (n=5)
CV, coefficient of variation, calculated as the percent ratio between the standard deviation and mean value of the slope.
Accuracy/Precision of NIST Certified Plasma with Single and Triple Quadrupoles
The accuracy and reproducibility (Table 2) were examined with a quantitative isotope dilution-based analysis method using single quadrupole enhanced in-source multiple fragment ion monitoring via the analysis of 11 amino acids and 5 fatty acids in a certified NIST 1950 plasma sample. Calibration lines were prepared using the ratio between external calibration standards and the corresponding isotopically labelled internal standards versus the response-dependent concentration factor (concentration of the target analyte divided by the concentration of the internal standard). The results were compared to the certified or reference values provided by NIST for accuracy analysis. Precision was evaluated as intra- and inter-day CV as described above in the data analysis section.
Table 2.
Single quadrupole enhanced in-source multiple fragment ion monitoring of NIST SRM 1950 plasma, monitoring of 11 amino acids and 5 fatty acids.
| metabolite | certified/reference concentrationa (μM) | intraday | interday | ||||
|---|---|---|---|---|---|---|---|
|
| |||||||
| measured concentration (μM) | accuracyb (%) | CVc (%) | measured concentration (μM) | accuracyb (%) | CVc (%) | ||
| histidine | 72.6±3.6 | 67.9 | 93.6 | 1.1 | 66.2 | 91.2 | 2.1 |
| leucine/isoleucine | 155.9±9.7 | 164.2 | 105.3 | 5.6 | 160 | 102.6 | 8.7 |
| lysine | 140±14 | 134.5 | 96.1 | 0.4 | 136.7 | 97.6 | 0.8 |
| methionine | 22.3±1.8 | 22.2 | 99.7 | 1.4 | 21.3 | 95.5 | 2 |
| proline | 177±9 | 172.3 | 97.3 | 1.1 | 174.7 | 98.7 | 1.4 |
| threonine | 119.5±6.1 | 109.3 | 91.4 | 0.2 | 110.1 | 92.1 | 0.4 |
| tyrosine | 57.3±3.0 | 54.9 | 95.9 | 1.3 | 54.5 | 95.1 | 2.8 |
| valine | 182.2±10.4 | 181.3 | 99.5 | 0.6 | 181.8 | 99.8 | 0.7 |
| phenylalanine | 51±7 | 46.5 | 91.2 | 2.8 | 46.8 | 91.8 | 2.5 |
| arginine | 81.4±2.3 | 85.8 | 105.4 | 1.1 | 88 | 108.1 | 3.3 |
| eicosapentaenoic acid | 38.6±0.5 | 36.9 | 95.7 | 4.4 | 35.9 | 93 | 4.6 |
| linoleic acid | 2838±143 | 2081 | 73.3 | 4.9 | 2157.9 | 76 | 5.3 |
| oleic acid | 1614±154 | 1450 | 89.9 | 3.8 | 1456 | 90.2 | 5.3 |
| myristic acid | 80.1±17.0 | 98.6 | 123 | 3.6 | 98.3 | 122.7 | 3 |
| stearic acid | 644±41 | 822 | 127 | 1.3 | 829.7 | 128.8 | 1.4 |
reference concentrations for arginine and phenylalanine, certified values for all others
accuracy was calculated as the percent ratio between measured concentration and certified/reference concentrations (mean) in the sample
CV: coefficient of variation, calculated as the percent ratio between standard deviation and mean values of the measured concentrations (n=5).
As shown in Table 2, the results acquired using single quadrupole enhanced in-source fragmentation multiple fragment ion monitoring demonstrated very good accuracy (91–110% for amino acids; 76–129% for fatty acids) and inter-day precision (CV< 10%). The wider accuracy range for fatty acids may be partly ascribed to the different sample analysis protocol and instrument used by NIST (GC-MS). The LC (Agilent 1260 infinity) coupled with the single quadrupole mass spectrometer was not able to handle the high pressure, thus leucine and isoleucine were not distinguishable here. Therefore, a combined result for the two compounds was reported, which had a relative higher inter-day CV (8.7%).
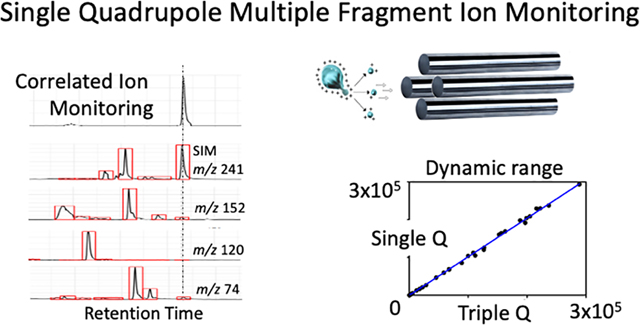
Correlated ion monitoring
Single quadrupole enhanced in-source fragmentation multiple fragment ion monitoring quantitative analysis as described above was validated using traditional selected ion monitoring technology. However, distinct from multi-SIM type approaches and to enhance selectivity, a correlated ion monitoring algorithm was developed to autonomously process, align, correlate, and filter SIM data. The correlated ion monitoring algorithm is designed to align and selectively compile multiple ions within one chromatogram and flag signals if they fall outside manually preset thresholds. The filtering was incorporated to help detect co-eluting molecules. The correlated ion monitoring chromatogram is created as a compilation of the individual ion signals only if each signal satisfies pre-set criteria. To accomplish this, the correlated ion monitoring algorithm analyzes the SIM traces within a pre-specified RT window from mzxml files. Correlated ion monitoring then performs peak picking, alignment, threshold, and ratio filtering with settings that can be adjusted to maximize performance.
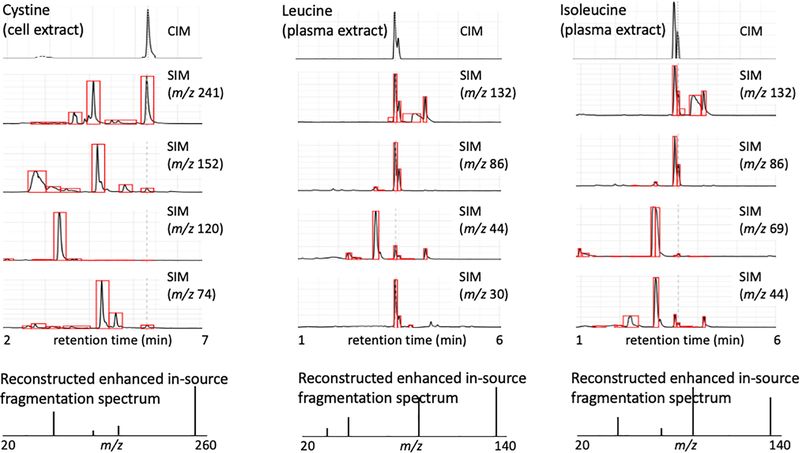
The function of monitoring the ratio between the precursor and the fragment ions was incorporated to identify co-eluting peaks that have either the same precursor or fragment ions. Thus, if a particular ion is found not to be within the pre-determined expected ratio parameters, the measured molecule is either flagged for quantitative removal, or the ion is eliminated from the quantification analysis. This approach could also be used for triple quadrupole since co-eluting peaks can also impact triple quadrupole quantitative accuracy. The correlated ion monitoring chromatogram represents a trace comparable to mass transitions obtained from an MRM experiment as demonstrated here with the analysis of cystine from a cell extract and leucine/isoleucine from a plasma extract (Figure 4). Additionally, correlated ion monitoring generates reconstructed single quadrupole enhanced in-source multiple fragment ion spectra allowing for the continuous monitoring of ion ratios and matching against authentic standard materials. In this way selectivity issues can autonomously be detected.
Figure 4.
Single quadrupole correlated ion monitoring data of cystine (cell extract), leucine (plasma extract), and isoleucine (plasma extract) correlates the precursor and multiple fragment ions to generate a correlated ion chromatogram. The data was peak picked, aligned and threshold filtered to provide deconvolution of the enhanced in-source multiple fragment ion monitoring data. Correlated ion monitoring also evaluates the ion ratios to monitor if they match the expected (p:f1:f2:f3) ratio, if not the molecule is flagged as being contaminated with co-eluting species. A possible correction for this would be to eliminate the contaminating ion as a quantifier. Additionally, reconstructed fragment spectra are generated allowing for continuous monitoring of ion ratios and matching against genuine standards. p: precursor ion; f1, f2 and f3: fragment ion 1, 2, and 3, respectively.
Applications and Limitations
Typically, additional MRM transitions must be monitored with a triple quadrupole instrument to enable the identification of quantified compounds, enhanced in-source multiple fragment ion monitoring adds this function to the single quadrupole mass spectrometer. Thus, single quadrupole enhanced in-source multiple fragment ion monitoring can significantly broaden the applications of single quadrupole mass spectrometers in quantitative analysis, especially those used for routine analysis. For example, single quadrupole enhanced in-source multiple fragment ion monitoring can be used for fragment ion monitoring which is performed daily in synthetic chemistry laboratories.29 While high resolution and high detection sensitivity are vital features for structure elucidation and trace analysis, fragment ion monitoring typically requires only unit mass resolution to provide verification of product formation and to track the comings and goings of reaction starting materials, products and intermediates.29 Further, due to high-maintenance equipment and complicated data analysis, a mass spectrometer (MS) is not widely used in quality control (QC) laboratories in the industry, e.g., pharmaceutical and food industry.29 However, single quadrupole enhanced in-source multiple fragment ion monitoring can help increase the application of MS-based methods in these QC laboratories, which are normally more sensitive and can substantially reduce time and cost. For example, single quadrupole enhanced in-source multiple fragment ion monitoring can be developed to monitor the post-translational modifications (PTMs) in complex protein drugs.29 Xu et al. have reported a single quadrupole method for PTMs detection and quantitation in a therapeutic monoclonal antibody.30 Single quadrupole enhanced in-source multiple fragment ion monitoring can also be used in the identification and quantitation of active ingredients, impurities, and degradation products in the development of chemical products such as pharmaceuticals and agrochemcials.29
However, in single quadrupole enhanced in-source multiple fragment ion monitoring analysis, highly co-eluted analytes with precursors and fragment ion masses like that of the target compound can result in ‘contaminated’ transitions, which can lead to false positive identifications or imprecise quantification. Thus, characteristic molecular ions need to be determined for the co-eluted target analytes using either tandem mass spectra and retention time databases such as METLIN or comparing the signal intensity (peak area) ratios across standards and samples as discussed earlier. Further, QC samples can be included to the sample sequence by adding a specified amount of analyte standard to the test sample. By comparing the peak intensity (area) ratio between QC and test samples, molecular ions specific to the target analytes can be determined. If these QC protocols do not perform with sufficient accuracy, an extended liquid chromatography gradient is needed to help single quadrupole enhanced in-source multiple fragment ion monitoring improve selectivity, especially for those closely eluted compounds with fragments or molecular ions sharing the same nominal masses.
Other ramifications of single quadrupole enhanced in-source multiple fragment ion monitoring are that it will expand the utility of related instrumentation (e.g., QTOF and ion mobility). QTOF instrumentation can now be used both as a quantitative tool, targeted identification, and for full scan analyses. For example, the TOF could be used for un-targeted analyses, the QTOF for identification, and the single quadrupole for targeted quantitative validation. The combination of which are unique to QTOF technology since Orbitrap instrumentation with parallel reaction monitoring (PRM) do not allow for high sensitivity quadrupole ion selection analyses. In addition, ion mobility technology (Figure S3) could be used as an alternative discriminating feature to decipher (for example) co-eluting species.
Conclusions
Even sixty years later, Wolfgang Paul’s original single quadrupole MS concept1 continues to provide interesting new capabilities, here we demonstrate that single quadrupoles can offer quantitative analyses using enhanced in-source multiple fragment ion generation and correlated ion monitoring (as illustrated in Figures 1 and 4) representing an alternative to traditional tandem MS approaches. The correlated ion monitoring algorithm enables these analyses by autonomously processing, aligning, filtering, and compiling multiple ions providing for both precursor and in-source fragment ions to be correlated within a single chromatogram, also enabling the detection of co-eluting species based on precursor and fragment ion ratios. The utilization of this existing platform facilitates high dynamic range, selectivity, accuracy, broad availability, and reproducibility, which can be deployed on complex matrices10 and on multiple mass spectrometer types (e.g., single quadrupole, triple quadrupole, QTOF, and Q-Orbitrap). This concept is potentially useful as one could utilize the same mass spectrometer (QTOF and Q-Orbitrap), without altering conditions, to perform both full scan and targeted quantitative analyses. Moreover, single quadrupole enhanced in-source multiple fragment ion monitoring can be coupled with other separation technologies including ultra-high resolution capillary electrophoresis, ion mobility (Figure S3), and gas chromatography to further enhance their sensitivity. Enhanced in-source multiple fragment ion monitoring could also provide for pseudo-MS3 and be particularly useful for the quantification of modified complex molecules such as oxidized phospholipids31 or closely related eicosanoids.32 Overall, single quadrupoles offer MRM level performance for quantitative analyses across a broad range of molecules on ubiquitous instrumentation; instruments that are generally inexpensive, easy to operate, and technically less complex.
Supplementary Material
Acknowledgements
The authors thank Dr. Dinghong Zhang at Olivia Osborn laboratory in the University of California San Diego and Dr. Wenjuan Li at James E. Voss laboratory in the Scripps Research Institute (La Jolla, CA) for cell sample preparation. This research was partially funded by National Institutes of Health grants R35 GM130385, P30 MH062261, P01 DA026146, and U01 CA235493 and by Ecosystems and Networks Integrated with Genes and Molecular Assemblies (ENIGMA), a Scientific Focus Area Program at Lawrence Berkeley National Laboratory for the U.S. Department of Energy, Office of Science, Office of Biological and Environmental Research, under contract number DE-AC02-05CH11231.
Footnotes
Supporting Information Available: Data and information for precursor and fragment ions investigated, limits of quantification, linear dynamic range, absolute concentration calculations, matrix effect, correlation curves between cone voltage and intensity of ions, fragment ion intensity comparisons, and ion mobility.
REFERENCES
- 1.Paul W Angew. Chem. 1990, 29 (7), 739–748. [Google Scholar]
- 2.Yost RA; Enke CG; McGilvery DC; Smith D; Morrison JD Int. J. Mass Spectrom. Ion Phys 1979, 30, 127–136. [Google Scholar]
- 3.Kondrat RW; McClusky GA; Cooks RG Anal. Chem. 1978, 50(14), 2017–2022. [Google Scholar]
- 4.Fenn JB; Mann M; Meng CK; Wong SF; Whitehouse CM Science 1989, 246, 64–71. [DOI] [PubMed] [Google Scholar]
- 5.Yuan M; Breitkopf SB; Yang X; Asara JM Nat. Protoc. 2012, 7, 872–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Addona TA; Abbatiello SE; Schilling B; Skates SJ; Mani DR; Bunk DM; Spiegelman CH; Zimmerman LJ; Ham AJL; Keshishian H; Hall SC; Allen S; Blackman RK; Borchers CH; Buck C; Cardasis HL; Cusack MP; Dodder NG; Gibson BW; Held JM; Hiltke T; Jackson A; Johansen EB; Kinsinger CR; Li J; Mesri M; Neubert TA; Niles RK; Pulsipher TC; Ransohoff D; Rodriguez H; Rudnick PA; Smith D; Tabb DL; Tegeler TJ; Variyath AM; Vega-Montoto LJ; Wahlander A; Waldemarson S; Wang M; Whiteaker JR; Zhao L; Anderson NL; Fisher SJ; Liebler DC; Paulovich AG; Regnier FE; Tempst P; Carr SA Nat. Biotechnol. 2009, 27, 633–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Venable JD; Dong MQ; Wohlschlegel J; Dillin A; Yates JR 3rd Nat. Methods 2004, 1, 39–45. [DOI] [PubMed] [Google Scholar]
- 8.Glish G; Vachet R Nat. Rev. Drug Discov. 2003, 2, 140–150. [DOI] [PubMed] [Google Scholar]
- 9.Mueller CA; Weinmann W; Dresen S; Schreiber A; Gergov M Rapid Commun. Mass Spectrom. 2005, 19, 1332–1338. [DOI] [PubMed] [Google Scholar]
- 10.Domingo-Almenara X; Montenegro-Burke JR; Ivanisevic J; Thomas A; Sidibé j.; Teav T; Guijas C; Aisporna AE; Rinehart D; Hoang L; Nordström A; Gómez-Romero M; Whiley L; Lewis MR; Nicholson JK; Benton HP; Siuzdak G Nat. Methods 2018, 15, 681–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Domingo-Almenara X; Montenegro-Burke JR; Guijas C; Majumder ELW; Benton HP; Siuzdak G Anal. Chem. 2019, 91, 3246–3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xue J; Domingo-Almenara X; Guijas C; Palermo A; Rinschen MM; Isbell J; Benton HP; Siuzdak G Anal. Chem. 2020, 92, 6051–6059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Seitzer PM; Searle BC J. Proteome Res. 2019, 18 (2), 791–796. [DOI] [PubMed] [Google Scholar]
- 14.Marquet P; Venisse N; Lacassie É; Lachâtre G Analusis 2000, 28 (10), 925–934. [Google Scholar]
- 15.Abranko L; Garcia-Reyes JF; Molina-Diaz AJ Mass Spectrom. 2011, 46 (5), 478–488. [DOI] [PubMed] [Google Scholar]
- 16.Xu YF; Lu W; Rabinowitz JD. Anal. Chem 2015, 87 (4), 2273–2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Müller C; Junker J; Bracher F; Giera M Nat. Protoc. 2019, 14, 2546–2570. [DOI] [PubMed] [Google Scholar]
- 18.Commission Decision (2002/657/EC) of 12 August 2002. Implementing Council Directive (96/23/EC) concerning the performance of analytical methods and the interpretation of results. Off J Eur Communities 2002, L221: 8–36. [Google Scholar]
- 19.United States Food and Drug Administration. Bioanalytical method validation guide for industry, 2018. https://www.fda.gov/media/70858/download (accessed on August 2020)
- 20.Matuszewski BK; Constanzer ML; Chavez-Eng CM Anal. Chem. 2003, 75, 3019–3030. [DOI] [PubMed] [Google Scholar]
- 21.Ivanisevic J; Elias D; Deguchi H; Averell PM; Kurczy M; Johnson CH; Tautenhahn R; Zhu Z; Watrous J; Jain M; Griffin J; Patti GJ; Siuzdak G Sci. Rep. 2015, 5, 12757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Serafim V; Tiugan DA; Andreescu N; Mihailescu A; Paul C; Velea I; Puiu M; Niculescu MD Molecules 2019, 24, 360–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Beyer BA; Fang M; Sadrian B; Montenegro-Burke JR; Plaisted WC; Kok BPC; Saez E; Kondo T; Siuzdak G; Lairson LL Nat. Chem. Biol. 2018, 14, 22–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Domingo-Almenara X; Guijas C; Billings E; Montenegro-Burke JR; Uritboonthai W; Aisporna AE; Chen E; Benton HP; Siuzdak G Nat. Commun. 2019, 10, 5811–5819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xue J; Guijas C; Benton HP; Warth B; Siuzdak G Nat. Methods 2020, 17, 953–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wishart DS; Jewison T; Guo AC; Wilson M; Knox C; Liu Y; Djoumbou Y; Mandal R; Aziat F; Dong E; Bouatra S; Sinelnikov I; Arndt D; Xia J; Liu P; Yallou F; Bjorndahl T; Perez-Pineiro R; Eisner R; Allen F; Neveu V; Greiner R; Scalbert A Nucleic Acids Res. 2013, 41 (Database issue), D801–D807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lisec J; Hoffmann F; Schmitt C; Jaeger C Anal. Chem. 2016, 88, 7487–7492. [DOI] [PubMed] [Google Scholar]
- 28.Liu H; Lam L; Dasgupta PK Talanta 2011, 87, 307–310. [DOI] [PubMed] [Google Scholar]
- 29.Bu X; Regalado EL; Hamilton SE; Welch CJ Trends Anal. Chem. 2016, 82, 22–34. [Google Scholar]
- 30.Xu W; Jimenez RB; Mowery R; Luo H; Cao M; Agarwal N; Ramos I; Wang X; Wang J MAbs 2017, 9 (7), 1186–1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jónasdóttir HS; Nicolardi S; Jonker W; Derks R; Palmblad M; Ioan-Facsinay A; Toes R; van der Burgt YEM; Deelder AM; Mayboroda OA; Gier M Anal. Chem. 2013, 85, 6003–6010. [DOI] [PubMed] [Google Scholar]
- 32.Chakraborty T; Thuer E; Heijink M; Tóth R, Bodai L; Vágvölgyi C; Giera M; Gabaldón T; Gácser A Virulence. 2018, 9 (1), 1019–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.