Abstract
Medications having the unwanted side effect of inhibiting 7-dehydrocholesterol reductase (DHCR7), one of the last enzymes in the cholesterol biosynthesis pathway, account for about 300 million yearly prescriptions in the United States. Many of these drugs are currently prescribed to pregnant women. Many DHCR7-inhibiting medications share chemical similarities, which can be the active substructure responsible for the medication affinity to the enzyme. This work highlights a computational strategy to identify enriched fragments in a set of DHCR7-inhibiting medications. The computational approach used here involves systematic fragmentation of molecules using the molBLOCKS tool, followed by enrichment analysis. The results of this approach highlight putative pharmacophores that might be responsible for the DHCR7-inhibiting activity of some of these medications. The identification of DHCR7-inhibiting substructures is an important step toward knowledge-based drug development and can improve the neurodevelopmental safety of medications.
Keywords: cholesterol metabolism, 7-dehydrocholesterol, pharmacophores, Smith-Lemli-Opitz Syndrome, molBLOCKS, DHCR7
Recent reports revealed that some highly prescribed pharmaceuticals have a marked effect on sterol metabolism, inhibiting different enzymes in the cholesterol biosynthesis pathway.1−7 High-throughput screening studies showed that 5% of U.S. Food and Drug Administration (FDA)-approved medications have the unwanted side effect of inhibiting 7-dehydrocholesterol reductase (DHCR7), leading to an increase in 7-dehydrocholesterol (7-DHC).1 The accumulation of 7-DHC, especially during neurodevelopment, has adverse effects on the brain.8−10 Previous studies demonstrated that 7-DHC is highly oxidizable and it generates dozens of oxysterols, species that affect neuronal differentiation and viability.11,12 Interestingly, cell culture experiments revealed that 7-DHC levels in cells treated with some DHCR7 inhibiting medications were comparable to those found in cells from patients with Smith-Lemli-Opitz Syndrome (SLOS),4 a neurodevelopmental disorder caused by genetic mutations in DHCR7.10,13−15 These observations have a public health relevance, as medications with this side effect of elevating 7-DHC account for over 200 million yearly prescriptions in the United States, and many are currently prescribed to pregnant women.16,17 When used during pregnancy, these medications can reach the brain of the developing fetus and inhibit DHCR7, creating what is essentially a chemically induced SLOS biochemical phenotype.2,3,18,19 Indeed, scholars have suggested that a higher incidence of fetal malformations, spontaneous abortions, and intrauterine death was associated with the use of several DHCR7 inhibitors taken during the first trimester of pregnancy, a critical period when a decrease in cholesterol has been linked to teratogenicity.20
With the increased use and demand of new medications, the FDA has approved new drugs that bear striking structural similarities to known inhibitors of DHCR7. Given the parallel cellular biochemistry found in SLOS and in human exposures to DHCR7 inhibitors, there has been increased interest in describing the set of DHCR7-active compounds that constitute potential exposure threats during neurodevelopment. We set out to determine whether the list of FDA-approved medications that lead to an increase in 7-DHC share specific enriched fragments that could potentially explain their DHCR7 inhibiting activity.
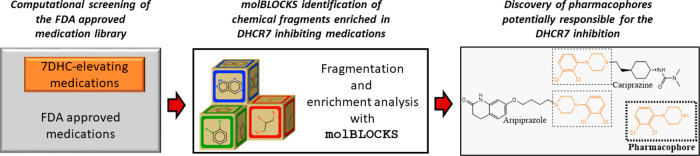
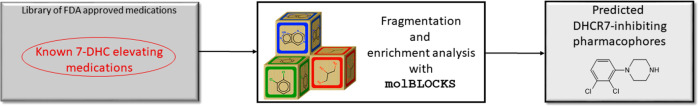
One of the classical strategies to develop medications is bioisosterism, an approach that relies on chemical substitutions of certain functional groups around a core chemical structure to improve affinity to the target enzyme.21 This core chemical structure, known as a pharmacophore, is perhaps the most important substructure in medications, as it drives the interaction between the ligand and receptor.21 Interestingly, drugs that are dissimilar overall but that share specific fragments often display nearly identical effects (or side effects),22 providing the rationale for our fragment-based approach. To answer the question of whether DHCR7-active compounds share enriched fragments that could be responsible for their activity, we used molBLOCKS.23 molBLOCKS is a suite of computational tools that uses a graph theoretical approach for breaking down small molecules into fragments based on a customizable list of chemical bonds. After fragmentation, molBLOCKS performs enrichment analysis to identify statistically overrepresented fragments in a list of active compounds compared to a background set. Figure 1 shows an overview of the approach.
Figure 1.
Experimental approach used to identify statistically overrepresented fragments in a list of active DHCR7 inhibiting medications. molBLOCKS was used to fragment known 7-DHC-elevating medications within the FDA-approved medications and isolate chemical structures present in each compound.
Results and Discussion
Previous studies used Neuro2a cells to evaluate the DHCR7-inhibiting capabilities of 970 compounds from the FDA-approved medication library.1 Cells were incubated with each compound and the levels of 7-DHC were assessed by LC-MS/MS. The authors of that study reported that ∼5% of the medications increased cellular levels of 7-DHC, acting as DHCR7 inhibitors. Antipsychotics, antidepressants, and antihypertensive medications were among the most potent DHCR7 inhibitors.
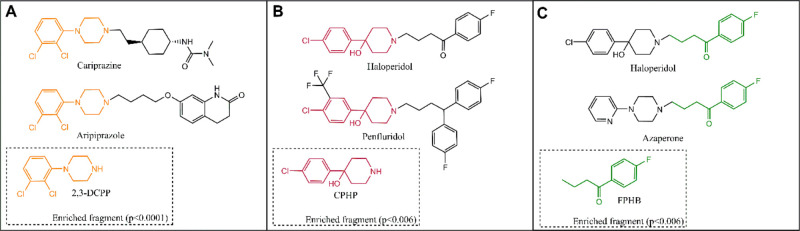
Figure 2 shows the results of the enrichment analysis, highlighting the three significantly (p < 0.05) enriched fragments and the parent drugs that contain them. Figure 2A depicts the identification of 2,3-dichlorophenyl piperazine (2,3-DCPP) as an enriched fragment that is present in aripiprazole and cariprazine, two potent DHCR7 inhibitors. Noteworthy, previous studies demonstrated that 2,3-DCPP alone is a potent DHCR7 inhibitor, elevating 7-DHC in cultured cells.4 Considering that 2,3-DCPP is itself an active compound and that molBLOCKS also identified it as a significantly enriched fragment and potential DHCR7 inhibiting pharmacophore, we interpret this as a validation of this approach as a tool to identify novel compounds. Figure 2B depicts the identification of 4-chlorophenyl-4-hydroxypiperidine (CPHP) as a significant fragment found in both haloperidol and penfluridol, two antipsychotics also known to elevate 7-DHC. Although 2,3-DCPP and CPHP are distinct chemical structures, they share similarities that give us clues of the essential chemical properties that can be associated with their 7-DHC-elevating abilities.
Figure 2.
Enriched fragments in the DHCR7 inhibiting set. Panels (A)–(C) highlight the three statistically enriched fragments in the set of top 30 DHCR7 inhibiting molecules. The enriched fragments are color matched and depicted within the dotted rectangles. The parent molecules containing the identified fragments are also shown.
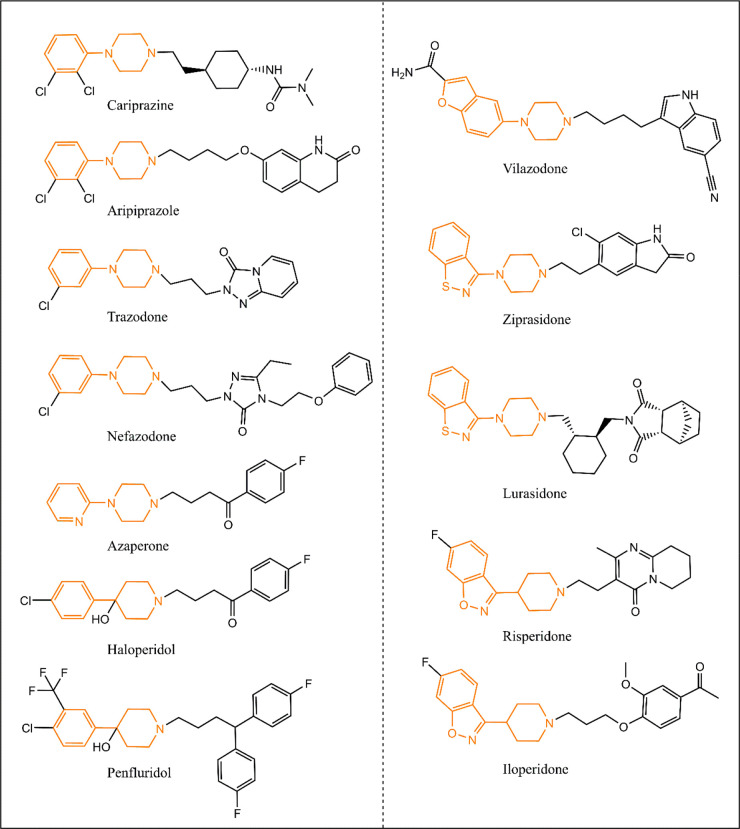
A side-by-side comparison between the chemical structures of 2,3-DCPP and CPHP reveals that both compounds have terminal modified phenyl rings linked to either a piperidine or piperazine ring. The hydroxyl group on the piperidine C-4 position on CPHP adds a heteroatom that makes the electron density in the piperidine ring resemble those of piperazines. These chemical features play a critical role in binding affinities to enzyme pockets, driving the interaction between the compound and the enzyme. Indeed, phenylpiperazines and 4-phenylpiperidines have been extensively used as one of the pharmacophores in many antipsychotics, antidepressant, and anxiolytics, many of which are active DHCR7 inhibitors (Figure 3).
Figure 3.
Chemical structures of several known DHCR7 inhibiting medications. Highlighted in brown are similar fragments among the different medications, which can be responsible for their DHCR7 inhibiting activities. Note that all medications contain either a piperidine or a piperazine ring.
A closer look at the chemical structures of DHCR7 active molecules depicted in Figure 3 shows that other structures besides phenylpiperazines and phenylpiperidines might lead to this side effect. Heterocyclic compounds linked to piperazines or piperidines are also fragments present in other DHCR7 inhibiting medications (e.g., ziprasidone, lurasidone, iloperidone, vilazodone, and risperidone). It seems that the presence of modified piperidines and piperazines contribute to the DHCR7 inhibiting properties of these medications. molBLOCKS also identified 4-(p-fluorophenyl)-4-hydroxybutane (FPHB) as an enriched fragment present in haloperidol and azaperone (Figure 2C). It is still unclear whether FPHB is a DHCR7 inhibiting fragment, which will be tested in subsequent studies. However, it is worth noting that azaperone contains a pyridylpiperazine, a structure that resembles those of the enriched fragments shown in Figure 2A and B. Thus, we hypothesize that this fragment is likely the chemical structure that contributes to azaperone’s DHCR7 inhibiting activities.
In order to identify pharmacophores responsible for the DHCR7 inhibition, we have successfully used moLBLOCKS to screen the FDA-approved medication library for common chemical structures in DHCR7 inhibiting molecules. The identification of 2,3-DCPP and CPHP illustrates the capabilities of the software to separate potentially active fragments from a pool of diverse molecules. However, there are key chemical properties not yet detectable by molBLOCKS. Given the overall chemical similarities seen in many DHCR7 inhibiting molecules (highlighted in brown in Figure 3), molBLOCKS could be further modified to allow the incorporation of additional features. These modifications would potentially allow the clustering of similar fragments highlighted based on molecular symmetry, electron density, hydrophobicity and molecular size, allowing for an improved identification of novel pharmacophores. Finally, the approach presented herein can be used as an initial step toward the identification of compounds responsible for (un)wanted biological effects, serving as a fast and inexpensive tool to identify the safety of novel pharmacophores.
Methods
SDF files for the 970 FDA-approved medications for which we have DHCR7 inhibition data were downloaded from PubChem.24 All molecules were converted to SMILES strings using OpenBabel.25 All molecules were fragmented with the fragment tool in molBLOCKS, using the default RECAP list of bonds that comes standard with the tool,26 with the extensive fragmentation flag turned on and a minimum fragment size equal to 12 atoms. After fragmentation, we performed fragment enrichment analysis with the molBLOCKS analyze tool, using all 970 FDA-approved medications as background and the top 30 DHCR7 inhibiting molecules as the target set. We used a False Discovery Rate (FDR) equal to 0.05 to determine the statistical significance of the enriched fragments.
Acknowledgments
T.G.-M. has been funded in part by The National Institutes of Health, NIMH MH110636 (KM) and NIMH MH067234 (KM). T.G.-M. would like to thank the support of Drs. Karoly Mirnics and Zeljka Korade at UNMC.
Author Contributions
§ D.G. and T.G.-M. contributed equally to this study.
The authors declare no competing financial interest.
References
- Kim H. Y.; Korade Z.; Tallman K. A.; Liu W.; Weaver C. D.; Mirnics K.; Porter N. A. Inhibitors of 7-Dehydrocholesterol Reductase: Screening of a Collection of Pharmacologically Active Compounds in Neuro2a Cells. Chem. Res. Toxicol. 2016, 29 (5), 892–900. 10.1021/acs.chemrestox.6b00054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korade Z.; Heffer M.; Mirnics K. Medication effects on developmental sterol biosynthesis. Mol. Psychiatry 2021, 10.1038/s41380-021-01074-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genaro-Mattos T. C.; Allen L. B.; Anderson A.; Tallman K. A.; Porter N. A.; Korade Z.; Mirnics K. Maternal aripiprazole exposure interacts with 7-dehydrocholesterol reductase mutations and alters embryonic neurodevelopment. Mol. Psychiatry 2019, 24 (4), 491–500. 10.1038/s41380-019-0368-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genaro-Mattos T. C.; Tallman K. A.; Allen L. B.; Anderson A.; Mirnics K.; Korade Z.; Porter N. A. Dichlorophenyl piperazines, including a recently-approved atypical antipsychotic, are potent inhibitors of DHCR7, the last enzyme in cholesterol biosynthesis. Toxicol. Appl. Pharmacol. 2018, 349, 21–28. 10.1016/j.taap.2018.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wages P. A.; Kim H. H.; Korade Z.; Porter N. A. Identification and characterization of prescription drugs that change levels of 7-dehydrocholesterol and desmosterol. J. Lipid Res. 2018, 59 (10), 1916–1926. 10.1194/jlr.M086991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall P.; Michels V.; Gavrilov D.; Matern D.; Oglesbee D.; Raymond K.; Rinaldo P.; Tortorelli S. Aripiprazole and trazodone cause elevations of 7-dehydrocholesterol in the absence of Smith-Lemli-Opitz Syndrome. Mol. Genet. Metab. 2013, 110 (1–2), 176–178. 10.1016/j.ymgme.2013.04.004. [DOI] [PubMed] [Google Scholar]
- Canfran-Duque A.; Casado M. E.; Pastor O.; Sanchez-Wandelmer J.; de la Pena G.; Lerma M.; Mariscal P.; Bracher F.; Lasuncion M. A.; Busto R. Atypical antipsychotics alter cholesterol and fatty acid metabolism in vitro. J. Lipid Res. 2013, 54 (2), 310–324. 10.1194/jlr.M026948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korade Z.; Xu L.; Shelton R.; Porter N. A. Biological activities of 7-dehydrocholesterol-derived oxysterols: implications for Smith-Lemli-Opitz syndrome. J. Lipid Res. 2010, 51 (11), 3259–3269. 10.1194/jlr.M009365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korade Z.; Xu L.; Mirnics K.; Porter N. A. Lipid biomarkers of oxidative stress in a genetic mouse model of Smith-Lemli-Opitz syndrome. J. Inherit Metab Dis 2013, 36 (1), 113–122. 10.1007/s10545-012-9504-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter F. D. RSH/Smith-Lemli-Opitz syndrome: a multiple congenital anomaly/mental retardation syndrome due to an inborn error of cholesterol biosynthesis. Mol. Genet. Metab. 2000, 71 (1–2), 163–174. 10.1006/mgme.2000.3069. [DOI] [PubMed] [Google Scholar]
- Xu L.; Korade Z.; Porter N. A. Oxysterols from free radical chain oxidation of 7-dehydrocholesterol: product and mechanistic studies. J. Am. Chem. Soc. 2010, 132 (7), 2222–2232. 10.1021/ja9080265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L.; Mirnics K.; Bowman A. B.; Liu W.; Da J.; Porter N. A.; Korade Z. DHCEO accumulation is a critical mediator of pathophysiology in a Smith-Lemli-Opitz syndrome model. Neurobiol Dis 2012, 45 (3), 923–929. 10.1016/j.nbd.2011.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter F. D. Smith-Lemli-Opitz syndrome: pathogenesis, diagnosis and management. Eur. J. Hum Genet 2008, 16 (5), 535–541. 10.1038/ejhg.2008.10. [DOI] [PubMed] [Google Scholar]
- Porter F. D.; Herman G. E. Malformation syndromes caused by disorders of cholesterol synthesis. J. Lipid Res. 2011, 52 (1), 6–34. 10.1194/jlr.R009548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith D. W.; Lemli L.; Opitz J. M. A Newly Recognized Syndrome of Multiple Congenital Anomalies. J. Pediatr 1964, 64, 210–217. 10.1016/S0022-3476(64)80264-X. [DOI] [PubMed] [Google Scholar]
- Genaro-Mattos T. C.; Klingelsmith K. B.; Allen L. B.; Anderson A.; Tallman K. A.; Porter N. A.; Korade Z.; Mirnics K. Sterol Biosynthesis Inhibition in Pregnant Women Taking Prescription Medications. ACS Pharmacol Transl Sci. 2021, 4 (2), 848–857. 10.1021/acsptsci.1c00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentile S.; Tofani S.; Bellantuono C. Aripiprazole and pregnancy: a case report and literature review. J. Clin Psychopharmacol 2011, 31 (4), 531–532. 10.1097/JCP.0b013e318222bc65. [DOI] [PubMed] [Google Scholar]
- Genaro-Mattos T. C.; Anderson A.; Allen L. B.; Tallman K. A.; Porter N. A.; Korade Z.; Mirnics K. Maternal cariprazine exposure inhibits embryonic and postnatal brain cholesterol biosynthesis. Mol. Psychiatry 2020, 25 (11), 2685–2694. 10.1038/s41380-020-0801-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korade Z.; Allen L. B.; Anderson A.; Tallman K. A.; Genaro-Mattos T. C.; Porter N. A.; Mirnics K. Trazodone effects on developing brain. Transl Psychiatry 2021, 11 (1), 85. 10.1038/s41398-021-01217-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boland M. R.; Tatonetti N. P. Investigation of 7-dehydrocholesterol reductase pathway to elucidate off-target prenatal effects of pharmaceuticals: a systematic review. Pharmacogenomics J. 2016, 16 (5), 411–429. 10.1038/tpj.2016.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patani G. A.; LaVoie E. J. Bioisosterism: A Rational Approach in Drug Design. Chem. Rev. 1996, 96 (8), 3147–3176. 10.1021/cr950066q. [DOI] [PubMed] [Google Scholar]
- Ahmed J.; Worth C. L.; Thaben P.; Matzig C.; Blasse C.; Dunkel M.; Preissner R. FragmentStore--a comprehensive database of fragments linking metabolites, toxic molecules and drugs. Nucleic Acids Res. 2011, 39 (Database), D1049–D1054. 10.1093/nar/gkq969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghersi D.; Singh M. molBLOCKS: decomposing small molecule sets and uncovering enriched fragments. Bioinformatics 2014, 30 (14), 2081–2083. 10.1093/bioinformatics/btu173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.; Chen J.; Cheng T.; Gindulyte A.; He J.; He S.; Li Q.; Shoemaker B. A.; Thiessen P. A.; Yu B.; Zaslavsky L.; Zhang J.; Bolton E. E. PubChem in 2021: new data content and improved web interfaces. Nucleic Acids Res. 2021, 49 (D1), D1388–D1395. 10.1093/nar/gkaa971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Boyle N. M.; Banck M.; James C. A.; Morley C.; Vandermeersch T.; Hutchison G. R. Open Babel: An open chemical toolbox. J. Cheminform 2011, 3, 33. 10.1186/1758-2946-3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewell X. Q.; Judd D. B.; Watson S. P.; Hann M. M. RECAP--retrosynthetic combinatorial analysis procedure: a powerful new technique for identifying privileged molecular fragments with useful applications in combinatorial chemistry. J. Chem. Inf. Comput. Sci. 1998, 38 (3), 511–522. 10.1021/ci970429i. [DOI] [PubMed] [Google Scholar]