Abstract
We sequenced a 396-bp region of the mitochondrial cytochrome b gene of the most common clinically important Candida species: Candida albicans, C. glabrata, C. parapsilosis, C. tropicalis, C. krusei, and C. lusitaniae. The recently described species of Candida, C. dubliniensis, associated with mucosal candidiasis in human immunodeficiency virus-infected individuals, was also included. Two to five strains of each species were examined. Some species represented intraspecies variation, which was not more than 1.8% (DNA). However, interspecies variations were more than 10 and 7%, respectively, for DNA and amino acid sequences. Multiple alignments of nucleotide and deduced amino acid sequences revealed species-specific nucleotides and amino acids. Nucleotide- and amino acid-based phylogenetic trees were constructed and are discussed. Using the database, it is possible to identify presumptive Candida species within a working day.
There has been a significant increase in the number of reports of systemic and mucosal infections caused by Candida species with the increase in the number of immunocompromised patients (3, 5, 9, 14, 21, 24, 25, 33). Candida albicans is the most frequently isolated causative agent of candidal infection in humans (more than 50%) and is generally accepted as the most pathogenic species of the genus Candida (3). However, in recent years, non-C. albicans Candida species, e.g., C. glabrata (formerly Torulopsis glabrata) (10 to 30%), C. parapsilosis (10 to 20%), C. tropicalis (10 to 20%), C. krusei, and C. lusitaniae, have been recovered with increasing frequency from cases of candidiasis (9, 16, 24). C. dubliniensis, phenotypically very similar to C. albicans and associated primarily with recurrent oral infections in human immunodeficiency virus-infected individuals, was first described in 1995 (23). The clinical significance of this species is under investigation.
Identification of Candida isolates to the species level in the clinical laboratory has become more important. Some Candida species, such as C. glabrata, are known to rapidly acquire decreased susceptibility to fluconazole; C. krusei is considered to be inherently resistant to fluconazole (20, 32). However, amphotericin B resistance is uncommon but appears to be most common among isolates of C. lusitaniae (27). C. parapsilosis can survive in the hospital environment, a fact which increases the chance of nosocomial transmission (5, 15). Therefore, species-specific identification is necessary for timely, targeted, and effective antifungal therapy and to facilitate hospital infection control measures.
The utility of a gene segment in accurate identification and phylogenetic analysis depends on the frequency of recombinational interchange of genetic material within the chosen gene segment. Mitochondria (mt) are self-replicating; in the budding yeast Saccharomyces cerevisiae, mt enter the bud immediately after the emergence of the bud and are equally distributed, although it is not clear how the mother cell maintains its own supply of mt (34). If this behavior is also true for the budding yeast Candida, the choice of a mitochondrial gene such as the cytochrome b gene is reasonable. Poulter et al. (17) reported a parasexual cycle for C. albicans; however, it is beyond our knowledge that parasexuality has any effect on genetic recombination of mitochondrial genes. Restriction fragment length polymorphisms of mitochondrial DNA have been shown to be useful genetic markers for estimating relationships among fungi (8, 11, 28). However, studies that investigate phylogenetic relationships based on the sequences of mitochondrial DNA (13, 29) are in their infancy. The utility of the sequences of the mitochondrial cytochrome b gene for identifying pathogenic Aspergillus species has been shown by Wang et al. (30, 31). However, similar techniques have not focused on yeasts.
In this study, we partially sequenced the mitochondrial cytochrome b genes of C. albicans, C. glabrata, C. parapsilosis, C. tropicalis, C. krusei, C. lusitaniae, and C. dubliniensis. Their identification and phylogenetic relationships are discussed.
MATERIALS AND METHODS
Yeast strains and isolation of DNA.
Two to five strains of each species of the most common pathogenic Candida species, including C. dubliniensis, and Filobasidiella neoformans were used in this study (Table 1). Total cellular DNA from these strains was extracted using a Gen Toru Kun kit (Takara Shuzo Co. Ltd., Otsu, Shiga, Japan) as recommended by the manufacturer, except that one-third of a loop of yeast cells from the YPD (1% [wt/vol] yeast extract, 2% [wt/vol] Polypeptone, 2% [wt/vol] glucose) slant was used to isolate DNA.
TABLE 1.
Fungal strains, strain origins, and accession numbers of cytochrome b genes sequenced in this studya
| Species | IFM no. | Origin | Accession no. |
|---|---|---|---|
| Candida albicans | 5728 | Clinical isolate | AB044909 |
| Candida albicans | 46907 | Clinical isolate | AB044910 |
| Candida albicans | 48311T | CBS 562 | AB044911 |
| Candida albicans | 48922 | Clinical isolate | AB044918 |
| Candida albicans | 49689 | Clinical isolate | AB044919 |
| Candida dubliniensis | 48184 | Clinical isolate | AB044912 |
| Candida dubliniensis | 48313T | CBS 7987 | AB044913 |
| Candida dubliniensis | 48314 | CBS 7988 | AB044914 |
| Candida glabrata | 5489 | Clinical isolate | AB044920 |
| Candida glabrata | 5768 | Clinical isolate | AB044921 |
| Candida glabrata | 40065 | Clinical isolate | AB044922 |
| Candida glabrata | 46843T | CBS 138 | AB044915 |
| Candida krusei (“Issatchenkia orientalis”) | 5798 | Clinical isolate | AB044924 |
| Candida krusei (“Issatchenkia orientalis”) | 40019 | Clinical isolate | AB044923 |
| Candida krusei (“Issatchenkia orientalis”) | 46834T | CBS 573 | AB044925 |
| Candida lusitaniae (“Clavispora lusitaniae”) | 49207T | IFO 1019 | AB044926 |
| Candida lusitaniae (“Clavispora lusitaniae”) | 49723 | Clinical isolate | AB044927 |
| Candida parapsilosis | 5464 | Clinical isolate | AB044928 |
| Candida parapsilosis | 40119 | Clinical isolate | AB044929 |
| Candida parapsilosis | 46829T | CBS 604 | AB044916 |
| Candida tropicalis | 40085 | Clinical isolate | AB044930 |
| Candida tropicalis | 46816T | CBS 94 | AB044917 |
| Candida tropicalis | 48776 | Clinical isolate | AB044931 |
| Candida tropicalis | 49042 | Clinical isolate | AB044932 |
| Candida tropicalis | 49101 | Clinical isolate | AB044933 |
| Filobasidiella neoformans | 48637T | CBS 132 | AB040656 |
CBS, Centraalbureau voor Schimmelcultures, Baarn, The Netherlands; IFM, Institute for Food Microbiology (at present, the Research Center for Pathogenic Fungi and Microbial Toxicoses, Chiba University), Chiba, Japan; IFO, Institute for Fermentation, Osaka, Japan.
PCR primers and amplification of the cytochrome b gene.
PCR primers, E1M4, E1M5, E2M4, and E2mr5 (Table 2), were designed by comparing previously published amino acid sequences for the mitochondrial cytochrome b genes of several organisms as described by Wang et al. (30). One microliter of extracted DNA was used to amplify the mitochondrial cytochrome b gene with a TaKaRa Ex Taq PCR amplification kit (Takara Shuzo). The reactions were performed in a final reaction mixture (50 μl) containing 10 pmol of each primer, 4 μl of 2.5 mM each deoxynucleoside triphosphate (dATP, dCTP, dGTP, and dTTP), 2.0 U of TaKaRa Ex Taq polymerase, and 5 μl of 10× reaction buffer (Takara Shuzo). The amplification reactions were performed with the following cycling parameters: 94°C for 2 min; 30 cycles of denaturation for 30 s at 94°C, annealing for 30 s at 50°C, and extension for 1 min at 72°C; and a final extension at 72°C for 10 min.
TABLE 2.
PCR primers used in this study
| Primer | Sequencea | Description |
|---|---|---|
| E1M4 | 5′-TGRGGWGCWACWGTTATTACTA-3′ | Forward |
| E1M5 | 5′-GTTATTACTAAYTTATTMTC-3′ | Forward |
| E2M4 | 5′-GGWATAGMWSKTAAWAYAGCATA-3′ | Reverse |
| E2mr5 | 5′-AGCACGTARWATTGCGTAGAATGG-3′ | Reverse |
R, A or G; W, A or T; Y, C or T; M, A or C; S, C or G; K, G or T.
Sequencing.
PCR products were purified using a QIAquick PCR purification kit (QIAGEN, Hilden, Germany) according to the manufacturer's instructions. Then, both strands of the PCR products were sequenced directly on an ABI prism 377 or 310 DNA sequencer using a Big Dye terminator cycle sequencing ready reaction kit (Applied Biosystems Japan Co. Ltd., Tokyo, Japan) as recommended by the kit manufacturer. From the DNA sequences, cytochrome b amino acid sequences were deduced using the yeast (Saccharomyces) mitochondrial genetic code.
Molecular phylogenetic analysis.
DNA and amino acid sequences were aligned using GENETYX-MAC genetic information processing software (Software Development Co., Ltd., Tokyo, Japan). Sequences were analyzed by the unweighted pair-group method with arithmetic mean (UPGMA) and neighbor joining (NJ) using GENETYX-MAC genetic information processing software or by maximum parsimony (MP) using the package Phylogenetic Analysis Using Parsimony, version 4.0b4a, for Macintosh (26). For the NJ analyses, the distance between the sequences was calculated using Kimura's two-parameter model (10).
Nucleotide sequence accession numbers.
Mitochondrial cytochrome b genes of Candida species partially sequenced in this study have been deposited in the DDBJ/EMBL/GenBank data library under the accession numbers shown in Table 1.
RESULTS
The mitochondrial cytochrome b genes of major pathogenic Candida species (Table 1), including C. dubliniensis, were amplified by PCR, and a 396-bp segment corresponding to positions 445 to 840 in the C. glabrata cytochrome b coding sequence (GenBank accession no. X53862) (2) was sequenced. Strains of C. albicans, C. glabrata, and C. tropicalis showed intraspecies variation (Table 3). Intraspecies variation was the highest for C. tropicalis (1.8%) and was divided into two DNA types.
TABLE 3.
Variation of cytochrome b sequences within different strains of a single Candida species
| Species and strain | % Variationa
|
|
|---|---|---|
| DNA | Amino acid | |
| C. albicans | ||
| IFM 48311T | ||
| IFM 5728 | 0.0 | 0.0 |
| IFM 46907 | 0.0 | 0.0 |
| IFM 48922 | 0.0 | 0.0 |
| IFM 49689 | 0.3 | 0.0 |
| C. glabrata | ||
| IFM 46843T | ||
| IFM 5489 | 0.0 | 0.0 |
| IFM 5768 | 0.0 | 0.0 |
| IFM 40065 | 0.0 | 0.0 |
| X53862 | 0.3 | 0.8 |
| C. tropicalis | ||
| IFM 46816T | ||
| IFM 40085 | 1.8 | 0.8 |
| IFM 48776 | 0.0 | 0.0 |
| IFM 49042 | 1.8 | 0.8 |
| IFM 49101 | 1.8 | 0.8 |
Compared to the type strain.
Although the intraspecies variations of these pathogenic yeasts were not higher than 1.8%, the interspecies variations were high. Table 4 (lower left portion) represents the pairwise nucleotide identities of these yeasts calculated from the nucleotide sequences of the cytochrome b genes. No pair of species represented more than 90% identical sequences. Even C. albicans and C. dubliniensis, the closest species, according to other DNA sequences, had differences in 40 nucleotides of the total of 396 nucleotides (89.9% identical).
TABLE 4.
Levels of cytochrome b nucleotide and amino acid sequence similarities for the most common pathogenic Candida species
| Species | % Cytochrome b sequence similaritya for:
|
||||||
|---|---|---|---|---|---|---|---|
| C. albicans | C. dubliniensis | C. parapsilosis | C. lusitaniae | C. krusei | C. tropicalis | C. glabrata | |
| C. albicans | 92.4 | 87.9 | 86.4 | 84.1 | 84.1 | 80.3 | |
| C. dubliniensis | 89.9 | 87.9 | 84.1 | 84.8 | 86.4 | 75.8 | |
| C. parapsilosis | 84.1 | 84.1 | 79.5 | 81.8 | 87.1 | 81.8 | |
| C. lusitaniae | 83.3 | 84.6 | 81.6 | 80.3 | 74.2 | 79.5 | |
| C. krusei | 81.8 | 84.1 | 81.6 | 84.3 | 75.8 | 79.5 | |
| C. tropicalis | 81.3 | 80.3 | 80.5 | 74.5 | 72.7 | 76.5 | |
| C. glabrata | 81.1 | 79.0 | 79.3 | 83.8 | 83.8 | 73.0 | |
Data in the lower left portion of the table indicate nucleotide sequence similarity, and data in the upper right portion indicate amino acid sequence similarity.
The Saccharomyces mitochondrial genetic code system was used to deduce amino acid sequences from cytochrome b gene sequences. In the yeast mitochondrial gene, UGA encodes tryptophan instead of polypeptide chain termination, AUA encodes methionine instead of isoleucine, and CUN encodes threonine instead of the leucine found in the universal codon system. C. glabrata has conserved the strong codon bias of S. cerevisiae (1); however, the CUN codon family codes for a leucine in C. parapsilosis mt (7). The reason for using the Saccharomyces mitochondrial genetic code system in the present study is that the system is accepted for yeasts by the DDBJ/EMBL/GenBank data library. Table 4 (upper right portion) represents the pairwise amino acid identities among different species. Based on amino acid sequences, except for C. albicans and C. dubliniensis (92.4% identical), no other pair had more than 90% identity. There were differences in 10 amino acids of the total of 132 amino acids between C. albicans and C. dubliniensis.
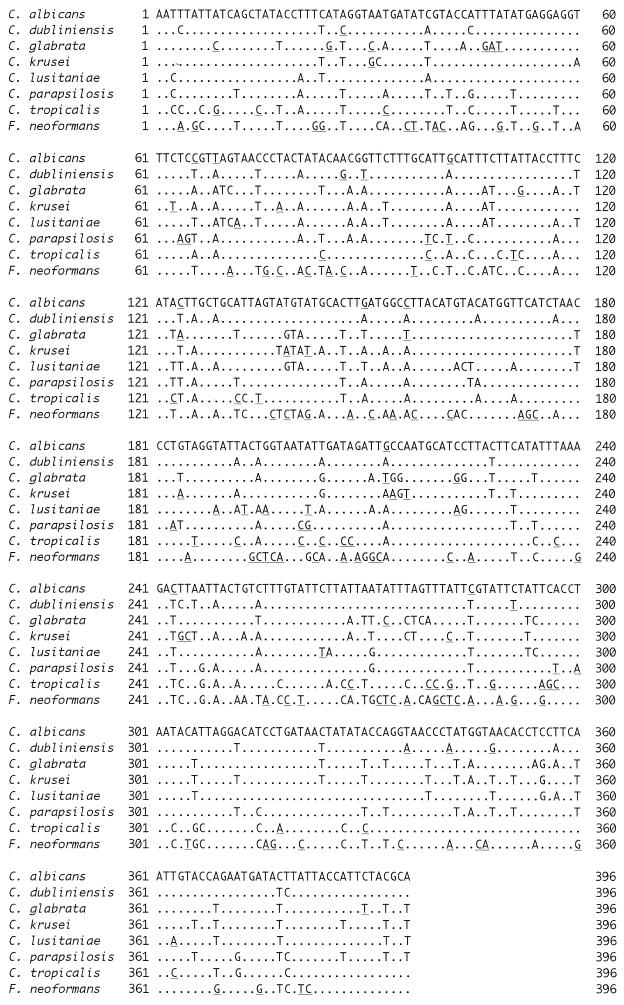
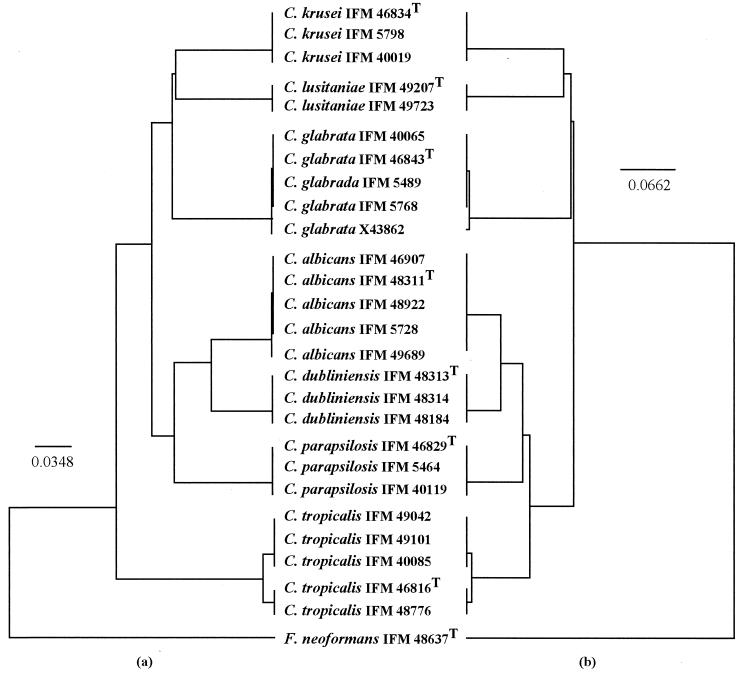
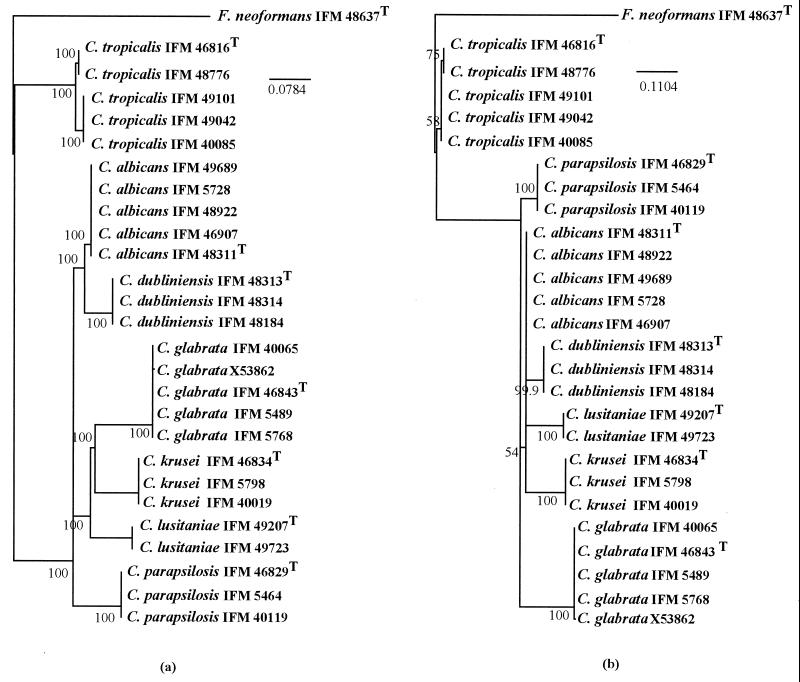
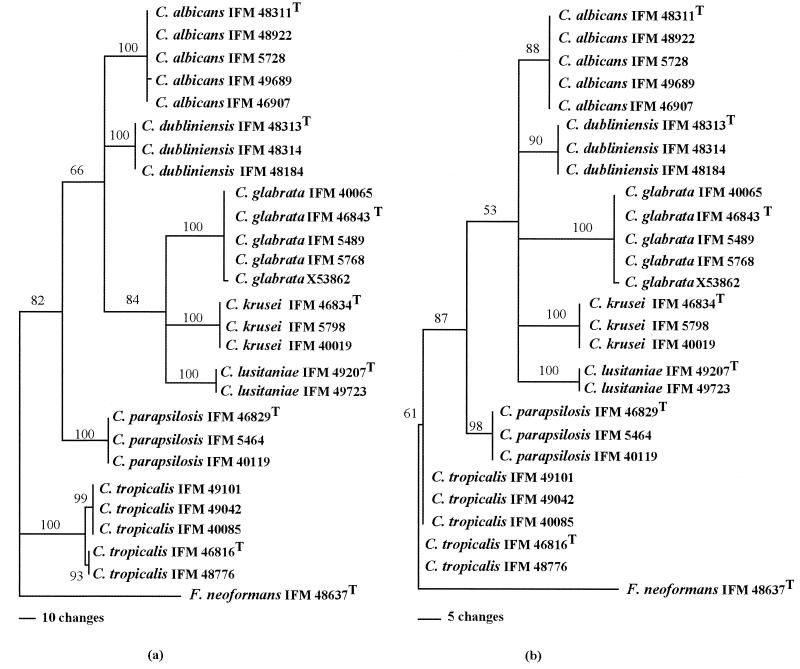
Multiple alignments of the nucleotide (Fig. 1) and amino acid (Fig. 2) sequences showed that individual species possessed characteristic sequences. The major pathogenic Candida species were positioned distinctly by any one of the phylogenetic trees constructed using UPGMA (Fig. 3), NJ (Fig. 4), or MP (Fig. 5) with basidiomycetous yeast F. neoformans as an outgroup. Although there was some difference in the degree of resolution when nucleotide and amino acid sequences were used, the topologies obtained by both sequence types and tree estimation algorithms used were consistent.
FIG. 1.
Comparison of nucleotide sequences of cytochrome b genes of the most common pathogenic Candida species (only type cultures) and F. neoformans. Dots indicate nucleotides identical to those in C. albicans. Species-specific nucleotides are underlined.
FIG. 2.
Comparison of deduced amino acid sequences for cytochrome b genes of the most common pathogenic Candida species (only type cultures) and F. neoformans. Dots indicate amino acids identical to those in C. albicans. Species-specific amino acids are underlined.
FIG. 3.
UPGMA trees of the most common Candida species, based on nucleotide (a) and deduced amino acid (b) sequences for the cytochrome b genes. The basidiomycetous yeast F. neoformans represents an outgroup. Bars represent the numbers of nucleotide and amino acid substitutions per nucleotide and amino acid sites.
FIG. 4.
NJ trees of the most common Candida species, based on nucleotide (a) and deduced amino acid (b) sequences for the cytochrome b genes. The basidiomycetous yeast F. neoformans represents an outgroup. The numerals given on the branches represent the confidence levels from 1,000 replicate bootstrap samplings (values lower than 50% are not shown). Bars represent the numbers of nucleotide and amino acid substitutions per nucleotide and amino acid sites, calculated using Kimura's two-parameter model (10).
FIG. 5.
Phylogenetic trees of the most common Candida species, based on nucleotide (a) and deduced amino acid (b) sequences for the cytochrome b genes. The trees were constructed by MP analysis (heuristic search, stepwise addition, tree-bisection-reconnection), where the basidiomycetous yeast F. neoformans represents an outgroup. Branch lengths are proportional to the numbers of nucleotide and amino acid changes, and the numerals given on the branches are the frequencies (>50%) with which a given branch appeared in 100 bootstrap replications.
DISCUSSION
This study reveals that the cytochrome b gene sequences of the major pathogenic Candida species differ from each other by more than 10%. Their amino acid sequences also show considerable differences. Except for C. albicans and C. dubliniensis (92.4% identical), no other pair showed more than 90% identity based on amino acid sequences. These results show the higher divergence of mitochondrial cytochrome b genes than of nuclear genes and its usefulness for investigating relationships of closely related species.
The sequence identity between C. dubliniensis and C. albicans is 89.9% based on the nucleotide sequences of the mitochondrial cytochrome b genes. However, the previously reported percentages of similarity between the organisms were 97.9% for the ACT1 gene (exon) (4), 97.52 to 97.75% for the V3 variable region of the large-subunit rRNA gene (23, 25), and 98.6% for the small-subunit rRNA gene (6). The predicted C. dubliniensis ACT1 protein sequence was identical to that of C. albicans, apart from a single conservative substitution, isoleucine to valine (4). On the other hand, the two organisms had 7.6% differences in the predicted protein sequences of the cytochrome b genes.
Wang et al. (30, 31) have shown that UPGMA is the best method for constructing phylogenetic trees based on cytochrome b sequences of fungi. In addition to UPGMA, sequences were analyzed using NJ and MP. There was very little difference among UPGMA (Fig. 3), NJ (Fig. 4), and MP (Fig. 5) phylogenetic trees based on the nucleotide or amino acid sequences for the cytochrome b genes of the most common pathogenic Candida species. This difference seems to be due to bootstrap values and differences in the methods of analysis. Moreover, different nucleotide codons may give rise to the same amino acid, a result which may produce differences in nucleotide- and amino acid-based phylogenetic trees. Although there was some difference in the degree of resolution when nucleotide and amino acid sequences were used, the topologies obtained with both sequence types and the tree estimation algorithms used were mutually consistent. The phylogenetic relationships of these pathogenic yeasts based on cytochrome b sequences were not 100% congruent with those based on rRNA; in addition, in the latter case, the relationships varied depending on the part of the sequences used to draw the phylograms, large or small-subunit rRNA (6, 23, 25).
This study represents the first phylogenetic investigation of the most common Candida species based on the sequences of the mitochondrial cytochrome b genes. The separation of the most common pathogenic Candida species, including the recently described species, C. dubliniensis, under investigation, was evident irrespective of the sequences, DNA or amino acid, and the methods of phylogenetic tree construction, UPGMA, NJ, or MP. The results supported the unique species designation of C. dubliniensis on the basis of its distinct phylogenetic position (supported by bootstrap analyses) and on the basis of the differences between its cytochrome b nucleotide and amino acid sequences and the corresponding sequences for C. albicans, 10.1 and 7.6%, respectively.
In a recent study, mitochondrial COX2 sequences revealed 2.1% intraspecies variability of C. glabrata isolates representing two types (22). These types correlated with the geographical origins of the strains, 90% of U.S. isolates belonging to type 1 and 82% of Brazilian isolates belonging to type 2. However, mitochondrial cytochrome b genes are more conserved at the intraspecies level (although the numbers of strains examined are limited). In this study, strains of the same species had identical sequences, except for C. albicans, C. glabrata, and C. tropicalis (Table 3). C. tropicalis had the highest variation at the intraspecies level—1.8%.
Rapid and accurate identification of pathogenic organisms is important clinically and epidemiologically (12, 18). Molecular methods provide accurate and rapid identification. Although a variety of methods, ranging from randomly amplified polymorphic DNA analysis to PCR-enzyme immunoassay, are used for DNA subtyping of Candida species, no “gold standard” exists (19). With an automated DNA sequencer, a DNA sequence can be obtained quite rapidly. Starting from DNA extraction from yeast cells, the sequence of a cytochrome b segment (396 bp) can be determined within a working day, and suspected Candida species can be identified correctly using a database.
This is the first report of the identification and phylogenetic analysis of the most common pathogenic Candida species by use of cytochrome b gene sequences. The 396-bp region of the cytochrome b gene examined in the present study has proved effective for identification and for studying phylogenetic relationships. High interspecies and low intraspecies divergences of cytochrome b gene sequences are attractive and may facilitate the design of probes for specific and rapid identification of pathogenic Candida species.
ACKNOWLEDGMENT
We thank the Ministry of Education, Tokyo, Japan, for providing a scholarship to S. K. Biswas during the study.
REFERENCES
- 1.Ainley W M, Macreadie I G, Butow R A. var1 gene on the mitochondrial genome of Torulopsis glabrata. J Mol Biol. 1985;184:565–576. doi: 10.1016/0022-2836(85)90303-1. [DOI] [PubMed] [Google Scholar]
- 2.Clark-Walker G D. Contrasting mutation rates in mitochondrial and nuclear genes of yeasts versus mammals. Curr Genet. 1991;20:195–198. doi: 10.1007/BF00326232. [DOI] [PubMed] [Google Scholar]
- 3.Coleman D C, Rinaldi M G, Haynes K A, Rex J H, Summerbell R C, Anaissie E J, Li A, Sullivan D J. Importance of Candida species other than Candida albicans as opportunistic pathogens. Med Mycol. 1998;36(Suppl. 1):156–165. [PubMed] [Google Scholar]
- 4.Donnelly S M, Sullivan D J, Shanley D B, Coleman D C. Phylogenetic analysis and rapid identification of Candida dubliniensis based on analysis of ACT1 intron and exon sequences. Microbiology. 1999;145:1871–1882. doi: 10.1099/13500872-145-8-1871. [DOI] [PubMed] [Google Scholar]
- 5.Fridkin S K, Jarvis W R. Epidemiology of nosocomial fungal infections. Clin Microbiol Rev. 1996;9:499–511. doi: 10.1128/cmr.9.4.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gilfillan G D, Sullivan D J, Haynes K, Parkinson T, Coleman D C, Gow N A. Candida dubliniensis: phylogeny and putative virulence factors. Microbiology. 1998;144:829–838. doi: 10.1099/00221287-144-4-829. [DOI] [PubMed] [Google Scholar]
- 7.Guelin E, Guerin M, Velours J. Isolation of the ATP synthase subunit 6 and sequence of the mitochondrial ATP6 gene of the yeast Candida parapsilosis. Eur J Biochem. 1991;197:105–111. doi: 10.1111/j.1432-1033.1991.tb15887.x. [DOI] [PubMed] [Google Scholar]
- 8.Hamari Z, Kevei F, Kovacs E, Varga J, Kozakiewicz Z, Croft J H. Molecular and phenotypic characterization of Aspergillus japonicus and Aspergillus aculeatus strains with special regard to their mitochondrial DNA polymorphisms. Antonie Leeuwenhoek. 1997;72:337–347. doi: 10.1023/a:1000578913759. [DOI] [PubMed] [Google Scholar]
- 9.Hazen K C. New and emerging yeast pathogens. Clin Microbiol Rev. 1995;8:462–478. doi: 10.1128/cmr.8.4.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kimura M. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 11.Kozlowski M, Stepien P P. Restriction enzyme analysis of mitochondrial DNA of members of the genus Aspergillus as an aid in taxonomy. J Gen Microbiol. 1982;128:471–476. doi: 10.1099/00221287-128-3-471. [DOI] [PubMed] [Google Scholar]
- 12.Nguyen M H, Peacock J E, Jr, Morris A J, Tanner D C, Nguyen M L, Snydman D R, Wagener M M, Rinaldi M G, Yu V L. The changing face of candidemia: emergence of non-Candida albicans species and antifungal resistance. Am J Med. 1996;100:617–623. doi: 10.1016/s0002-9343(95)00010-0. [DOI] [PubMed] [Google Scholar]
- 13.Paquin B, Forget L, Roewer I, Lang B F. Molecular phylogeny of Allomyces macrogynus: congruency between nuclear ribosomal RNA- and mitochondrial protein-based trees. J Mol Evol. 1995;41:657–665. doi: 10.1007/BF00175824. [DOI] [PubMed] [Google Scholar]
- 14.Perfect J R, Schell W A. The new fungal opportunists are coming. Clin Infect Dis. 1996;22(Suppl. 2):112–118. doi: 10.1093/clinids/22.supplement_2.s112. [DOI] [PubMed] [Google Scholar]
- 15.Pfaller M A. Nosocomial fungal infections: epidemiology of candidiasis. J Hosp Infect. 1995;30(Suppl.):329–338. doi: 10.1016/0195-6701(95)90036-5. [DOI] [PubMed] [Google Scholar]
- 16.Pfaller M A, Jones R N, Messer S A, Edmond M B, Wenzel R P. National surveillance of nosocomial blood stream infection due to species of Candida other than Candida albicans: frequency of occurrence and antifungal susceptibility in the SCOPE Program. Diagn Microbiol Infect Dis. 1998;31:327–332. doi: 10.1016/s0732-8893(97)00240-x. [DOI] [PubMed] [Google Scholar]
- 17.Poulter R, Jeffery K, Hubbard M J, Shepherd M G, Sullivan P A. Parasexual genetic analysis of Candida albicans by spheroplast fusion. J Bacteriol. 1981;146:833–840. doi: 10.1128/jb.146.3.833-840.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Price M F, LaRocco M T, Gentry L O. Fluconazole susceptibilities of Candida species and distribution of species recovered from blood cultures over a 5-year period. Antimicrob Agents Chemother. 1994;38:1422–1427. doi: 10.1128/aac.38.6.1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reiss E, Tanaka K, Bruker G, Chazalet V, Coleman D, Debeaupuis J P, Hanazawa R, Latge J P, Lortholary J, Makimura K, Morrison C J, Murayama S Y, Naoe S, Paris S, Sarfati J, Shibuya K, Sullivan D, Uchida K, Yamaguchi H. Molecular diagnosis and epidemiology of fungal infections. Med Mycol. 1998;36(Suppl. 1):249–257. [PubMed] [Google Scholar]
- 20.Rex J H, Rinaldi M G, Pfaller M A. Resistance of Candida species to fluconazole. Antimicrob Agents Chemother. 1995;39:1–8. doi: 10.1128/aac.39.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rex J H, Pfaller M A, Galgiani J N, Bartlett M S, Espinel-Ingroff A, Ghannoum M A, Lancaster M, Odds F C, Rinaldi M G, Walsh T J, Barry A L. Development of interpretive breakpoints for antifungal susceptibility testing: conceptual framework and analysis of in vitro-in vivo correlation data for fluconazole, itraconazole, and Candida infections. Subcommittee on Antifungal Susceptibility Testing of the National Committee for Clinical Laboratory Standards. Clin Infect Dis. 1997;24:235–247. doi: 10.1093/clinids/24.2.235. [DOI] [PubMed] [Google Scholar]
- 22.Sanson G F, Briones M R. Typing of Candida glabrata in clinical isolates by comparative sequence analysis of the cytochrome c oxidase subunit 2 gene distinguishes two clusters of strains associated with geographical sequence polymorphisms. J Clin Microbiol. 2000;38:227–235. doi: 10.1128/jcm.38.1.227-235.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sullivan D J, Westerneng T J, Haynes K A, Bennet D E, Coleman D C. Candida dubliniensis sp. nov.: phenotypic and molecular characterisation of a novel species associated with oral candidosis in HIV-infected individuals. Microbiology. 1995;141:1507–1521. doi: 10.1099/13500872-141-7-1507. [DOI] [PubMed] [Google Scholar]
- 24.Sullivan D J, Henman M C, Moran G P, O'Neill L C, Bennett D E, Shanley D B, Coleman D C. Molecular genetic approaches to identification, epidemiology and taxonomy of non-albicans Candida species. J Med Microbiol. 1996;44:399–408. doi: 10.1099/00222615-44-6-399. [DOI] [PubMed] [Google Scholar]
- 25.Sullivan D J, Haynes K, Bille J, Boerlin P, Rodero L, Lloyd S, Henman M, Coleman D. Widespread geographic distribution of oral Candida dubliniensis strains in human immunodeficiency virus-infected individuals. J Clin Microbiol. 1997;35:960–964. doi: 10.1128/jcm.35.4.960-964.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Swofford D L. PAUP*. Phylogenetic analysis using parsimony (and other methods). Version 4.0b4a for Macintosh. Sunderland, Mass: Sinauer Associates; 1998. [Google Scholar]
- 27.Vanden Bossche H, Dromer F, Improvisi I, Lozano-Chiu M, Rex J H, Sanglard D. Antifungal drug resistance in pathogenic fungi. Med Mycol. 1998;36(Suppl. 1):119–128. [PubMed] [Google Scholar]
- 28.Varga J, Kevei F, Vriesema A, Debets F, Kozakiewicz Z, Croft J H. Mitochondrial DNA restriction fragment length polymorphisms in field isolates of the Aspergillus niger aggregate. Can J Microbiol. 1994;40:612–621. doi: 10.1139/m94-098. [DOI] [PubMed] [Google Scholar]
- 29.Walker D J, Wakefield A E, Dohn M N, Miller R F, Baughman R P, Hossler P A, Bartlett M S, Smith J W, Kazanjian P, Meshnick S R. Sequence polymorphisms in the Pneumocystis carinii cytochrome b gene and their association with atovaquone prophylaxis failure. J Infect Dis. 1998;178:1767–1775. doi: 10.1086/314509. [DOI] [PubMed] [Google Scholar]
- 30.Wang L, Yokoyama K, Miyaji M, Nishimura K. The identification and phylogenetic relationship of pathogenic species of Aspergillus based on the mitochondrial cytochrome b gene. Med Mycol. 1998;36:153–164. [PubMed] [Google Scholar]
- 31.Wang L, Yokoyama K, Miyaji M, Nishimura K. Mitochondrial cytochrome b gene analysis of Aspergillus fumigatus and related species. J Clin Microbiol. 2000;38:1352–1358. doi: 10.1128/jcm.38.4.1352-1358.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wingard J R. Importance of Candida species other than C. albicans as pathogens in oncology patients. Clin Infect Dis. 1995;20:115–125. doi: 10.1093/clinids/20.1.115. [DOI] [PubMed] [Google Scholar]
- 33.Witek-Janusek L, Cusack C, Mathews H L. Candida albicans: an opportunistic threat to critically ill low birth weight infants. Dimens Crit Care Nurs. 1998;17:243–255. [PubMed] [Google Scholar]
- 34.Yang H C, Palazzo A, Swayne T C, Pon L A. A retention mechanism for distribution of mitochondria during cell division in budding yeast. Curr Biol. 1999;9:1111–1114. doi: 10.1016/s0960-9822(99)80480-1. [DOI] [PubMed] [Google Scholar]