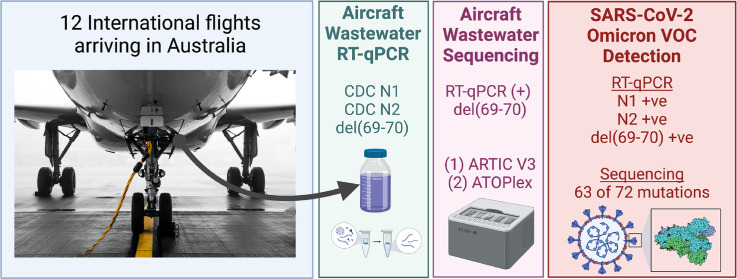
Abstract
On the 26th of November 2021, the World Health Organization (WHO) designated the newly detected B.1.1.529 lineage of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) the Omicron Variant of Concern (VOC). The genome of the Omicron VOC contains more than 50 mutations, many of which have been associated with increased transmissibility, differing disease severity, and potential to evade immune responses developed for previous VOCs such as Alpha and Delta. In the days since the designation of B.1.1.529 as a VOC, infections with the lineage have been reported in countries around the globe and many countries have implemented travel restrictions and increased border controls in response. We putatively detected the Omicron variant in an aircraft wastewater sample from a flight arriving to Darwin, Australia from Johannesburg, South Africa on the 25th of November 2021 via positive results on the CDC N1, CDC N2, and del(69–70) RT-qPCR assays per guidance from the WHO. The Australian Northern Territory Health Department detected one passenger onboard the flight who was infected with SARS-CoV-2, which was determined to be the Omicron VOC by sequencing of a nasopharyngeal swab sample. Subsequent sequencing of the aircraft wastewater sample using the ARTIC V3 protocol with Nanopore and ATOPlex confirmed the presence of the Omicron variant with a consensus genome that clustered with the B.1.1.529 BA.1 sub-lineage. Our detection and confirmation of a single onboard Omicron infection via aircraft wastewater further bolsters the important role that aircraft wastewater can play as an independent and unintrusive surveillance point for infectious diseases, particularly coronavirus disease 2019.
Keywords: SARS-CoV-2, COVID-19, Wastewater surveillance, Human health risks, Enveloped viruses, Aircraft
Graphical abstract
1. Introduction
On the 25th of November 2021, the South African National Institute for Communicable Diseases (NCID) announced the detection of a new lineage of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), B.1.1.529, in South Africa, based on genomic sequencing from 22 coronavirus disease 2019 (COVID-19) cases clustered in Gauteng province (National Institute for Communicable Diseases (NICD), 2021a). The following day the World Health Organization (WHO) Technical Advisory Group on SARS-CoV-2 Virus Evolution (TAG-VE) classified B.1.1.529 as a VOC named Omicron based on the presence of mutations associated with an increased transmissibility, virulence and immune escape and decreases in the effectiveness of public health measures for previous VOCs (WHO, 2021). The B.1.1.529 genome includes over 50 mutations, the highest number observed in a VOC to date, with more than 30 in the spike protein responsible for binding to host cell receptors (UK Health Security Agency, 2021). In South Africa between the 25th of November and 2nd of December, cases of COVID-19 and the percentage of positive tests increased rapidly in Gauteng province (National Institute for Communicable Diseases (NICD), 2021a). Per NICD, the reproductive number was greater than two in Gauteng province in late November with single day increases in the positive proportion as high as 22.4% (National Institute for Communicable Diseases (NICD), 2021b). A preliminary retrospective analysis of SARS-CoV-2 reinfections in South Africa indicated that Omicron is associated with a significant increase in the risk of reinfection (2.39×) compared to the Beta and Delta VOCs (Pulliam et al., 2021). The findings suggest Omicron may have an enhanced ability to evade immunity acquired during previous infections.
On the same day of the WHO VOC classification, travel-related infections with Omicron were identified in Belgium, Hong Kong, and Israel and the European Centre for Disease Prevention and Control classified the probability of introduction and local transmission in the European Union/European Economic Area as high to very high (European CDC, 2021). By the 27th of November, at least 115 Omicron infections had been identified in Botswana, South Africa, Hong Kong, Belgium, Israel, Germany, the Netherlands, and the United Kingdom (California Department of Public Health (CA DPH), 2021). In response, more than 50 countries, including the United States (US) and Australia implemented some form of travel restrictions or enhanced border controls to impede the transmission of Omicron (Mallapaty, 2021). Nonetheless by November 29th, Australia had reported its first case of Omicron in a person arriving on a repatriation flight from Johannesburg, South Africa on the 25th of November (https://www.abc.net.au/news/2021-11-29/nt-covid-outbreak-katherine-traveller-positive-for-omicron/100657690), and within days Australia began reporting cases of community transmission in New South Wales (NSW) and Australian Capital Territory (ACT).
The surveillance of wastewater from passenger aircraft may afford an efficient means of monitoring VOCs, especially in the context of emerging VOCs for which introduction via travel is a concern. Early in the COVID-19 pandemic, SARS-CoV-2 RNA was successfully detected in wastewater from passenger aircraft following international flights (Ahmed et al., 2020). More recently, aircraft wastewater surveillance demonstrated 83.7% accuracy in detecting SARS-CoV-2 infections among passengers of international flights all of whom had tested negative for COVID-19 prior to boarding (Ahmed et al., 2021). In the current study, we report the first successful detection of the Omicron VOC based on a consensus genome in aircraft wastewater from an international flight arriving in Australia.
2. Materials and methods
2.1. Wastewater sampling from the aircraft lavatory
Aggregated wastewater samples (~1 L in volume) were collected from the wastewater exiting lavatories of ten flights arriving to Australia in October to November 2021. Four repatriation flights landed at Darwin International Airport (DRW), Northern Territory, Australia, and six regular flights landed at Sydney International Airport (SYD), Sydney, NSW, Australia. These flights originated from O. R. Tambo International airport, Johannesburg (JNB) South Africa, Los Angeles International Airport (LAX), California, USA, Indira Gandhi International Airport (DEL), Delhi, India, and Istanbul Airport (IST), Istanbul, Turkey. These samples were supplemented by two additional wastewaters collected from flights from JNB to DRW in April and August 2021 for a total of twelve wastewater samples from aircrafts. Lavatory tanks full level on arrival ranged between 8 to 76%. For all these flights, wastewater was sampled using a specifically designed sample trap capable of retaining an aliquot of bulk wastewater sample exiting the aircraft lavatory tank (~200–2000 L capacity depending on the aircraft model) before entering a waste service truck system. The sample collection process has been described in detail in our previous study (Ahmed et al., 2021).
2.2. RT-qPCR inhibition test
Each 50 mL aircraft wastewater sample was seeded with ~105 gene copies (GC) of murine hepatitis virus (MHV) as an inhibition process control (Ahmed et al., 2021). The same concentration of MHV was also added to distilled water and subjected to RNA extraction. The quantification cycle (Cq) values obtained for the MHV-seeded distilled water (i.e., reference value) were compared with the Cq values of the MHV-seeded aircraft wastewater to determine RT-qPCR inhibition. If the Cq value of the wastewater RNA sample was >2 − Cq value difference compared to the reference Cq value for distilled water, the RNA sample was considered to have PCR inhibitors (Ahmed et al., 2021). The presence of PCR inhibition in nucleic acid samples extracted from wastewater was assessed using an MHV RT-PCR assay detailed elsewhere (Besselsen et al., 2002; Ahmed et al., 2021).
2.3. Sample concentration and RNA extraction
Viruses were concentrated from each wastewater sample (50 mL) using an automated, rapid concentrator instrument (Concentrating Pipette Select™, CP Select™, InnovaPrep, Drexel, MO) as described in our previous study (Ahmed et al., 2021).
Before concentration, the sample was first centrifuged at 4000 g for 5 min to pellet the suspended solids and toilet paper using a Beckman Coulter Avanti J-15R Centrifuge. The supernatant (∼45 mL) was then concentrated using an unirradiated 0.05 μm PS hollow fiber filter concentrating pipette tip (Cat. No. CC08004–200) with the CP Select™. Optimized wastewater application settings provided in the instrument instructions were used. Upon concentration, the tip was eluted twice using CP elution fluid (HC08001) containing 0.075% Tween 20-and 25-mM Tris (Cat. No. HC08001). The eluates (0.8–1 mL) were collected in sterile 15-mL polypropylene tubes. Immediately after virus concentration, RNA was extracted from 140 μL of the eluate using the QIAamp Viral RNA Mini Kit (Cat. No. 52905) (Qiagen) with a minor modification. In the final step, a 100 μL volume of buffer AVE was used to elute the RNA instead of 60 μL.
2.4. RT-qPCR analysis
Previously published RT-qPCR assays that target the N gene of SARS-CoV-2 (US CDC N1 and N2) (CDC, 2019) were used for SARS-CoV-2 RNA detection and quantification in wastewater samples. Sequencing results from over 100 specimens collected in South Africa indicate the Omicron genome contains deletion del(69–70) that allows for rapid putative identification via the S gene target failure (SGTF) previously associated with the Alpha VOC while other targets including the N and RdRp genes remain unaffected (https://www.nicd.ac.za/frequently-asked-questions-for-the-b-1-1-529-mutated-sars-cov-2-lineage-in-south-africa/). The WHO TAG-VE has suggested that SGTF can be used as a marker of Omicron pending confirmation by sequencing (https://www.who.int/news/item/26-11-2021-classification-of-omicron-(b.1.1.529)-sars-cov-2-variant-of-concern). Analysis of newly reported Omicron genomes has suggested dividing B.1.1.529 into two sub-lineages: BA.1, for which SGTF can be used for putative identification, and BA.2, which does not contain the del(69–70) mutation and cannot be detected via SGTF (https://github.com/cov-lineages/pango-designation/issues/361). For the detection of the S gene del(69–70) mutation (a known mutation of B.1.1.7 or alpha variant), an S.delH69V70 mutation assay (ThermoFisher Scientific) was used. The primer and probe sequences, reaction composition, and cycling parameters for all the RT-qPCR assays used, including MHV, are shown in Supplementary Table ST1. Gamma-irradiated SARS-CoV-2 hCoV 19/Australia/VIC01/2020 was measured with digital PCR (dPCR) and used as the RT-qPCR standard for SARS-CoV-2 US CDC N1 and N2 assays. Standard dilutions ranged from 1 × 106 to 1 GC/μL. For MHV, gBlocks gene fragments were purchased from Integrated DNA Technologies (Integrated DNA Technology Coralville, IA, US) and used as a positive control. For the del(69–70) mutation assay, AcroMetrix™ Ultra-Low positive RNA control (full length genomic RNA from SARS-CoV-2) was used. For each RT-qPCR run, triplicate negative controls were included. The RT-qPCR assays were performed using a Bio-Rad CFX96 or Opus thermal cyclers (Bio-Rad Laboratories, Richmond, CA). For each sample, the SARS-CoV-2 RNA concentration (GC/50 mL) was calculated from the standard curve and accounts for the difference in eluate volumes. All samples were analyzed alongside three non-template controls. For US CDC N1 and N2 RT-qPCR, the ALOD values were defined as the minimum GC number with a 95% probability of detection and determined as previously described (Verbyla et al., 2016).
2.5. RT-qPCR quality control
To minimize RT-qPCR contamination, RNA extraction and RT-qPCR setup were performed in separate laboratories. A method negative control was included for each batch of wastewater samples. A reagent negative control was also included during RNA extraction to account for any contamination during extraction. For each RT-qPCR run, triplicate negative controls were included. All method, reagent and RT-qPCR negative controls were negative for the targets analyzed.
2.6. Sequencing and bioinformatics
Confirmation of the RT-qPCR-detected variant strain was accomplished by two multiplex tiling PCR sequencing approaches; the ARTIC Network V3 scheme with Nanopore, and ATOPlex amplicon sequencing. For Nanopore sequencing, short amplicon fragments specific for SARS-CoV-2 spanning the full genome were generated using the nCOV-2019 primer set V3 designed by the ARTIC Network (Tyson et al., 2020). Libraries were constructed from the generated amplicon using the Oxford Nanopore Rapid sequencing kit and sequenced on the Oxford Nanopore GridION-X5. The real-time monitoring tool Rampart v1.2.0 was used to monitor the distribution of amplicon sequences being generated in parallel with the sequencing run. Passed reads from GridION-X5 using high-accuracy base calling (Qscore ≥ 9) read lengths were filtered from 250 to 500 based on ARTIC pipeline. Pangolin v3.1.16 with pangoLEARN (updated 2021–11-25) (https://github.com/cov-lineages/pangolin), scorpio 0.3.14, constellations v0.0.27 and pango-designation v1.2.106 were employed to assign lineage to the corresponding sequences. The presence or absence of defining mutations was supported by at least ≥20 of 45 defining mutations (GitHub - cov- lineages/constellations). Dropout or missing data at the locus was identified as ambiguous mutations. The ambiguous base (N) is assigned if there are fewer than 20 reads covered that position. Multiple Alignment using Fast Fourier Transform (MAFFT) v7.475 (Katoh et al., 2002) were applied to generate the multiple sequence alignment which was masked by in-house scripts. The regions subjected to masking were based on reported problematic sites (De Maio et al., 2020). Using the alignment, a maximum likelihood tree was generated by IQTREE2 (Minh et al., 2020). Tree visualization and plots were produced by ggplot2 and ggtree v3.1.0 (Yu, 2020) on R v4.1. Besides the jurisdiction specific data, the analysis utilized the public data from GISAID (www.gisaid.org) and services from https://nextstrain.org and https://austrakka.net.au.
For ATOPlex sequencing, the ATOPlex SARS-CoV-2 full-length genome panel (MGI, China) was used to construct libraries of short amplicons (240–333 bp) according to the manufacturer's instructions. The RNA sample was first converted to cDNA using reverse transcriptase (RT) with random hexamers (5′-NNNNNN-3′) (MGI, China). The 20-μL RT reaction mixture contained 10 μL of RNA template, 4 μL of N6 buffer, 5 μL of RT buffer, and 1 μL of RT enzyme mix. The RT reaction was performed in a C1000 thermal cycler (Bio-Rad, USA) using the program: 10 min at 25 °C, 30 min at 42 °C, 15 min at 70 °C. Lambda phage DNA (200 GC) was added into each sample as a spike-in control to ensure each sample generated sufficient amplification products for sequencing (>4 ng/μL). Lambda phage DNA and SARS-CoV-2 primers were co-amplified in the same reaction. DNA/cDNA samples were subjected to two rounds of PCR for target enrichment (first round) and addition of dual barcode (second round). In the first round the PCR amplification mixture contained 25 μL of PCR Enzyme Mix, 0.5 μL of PCR Clean Enzyme, 4 μL of PCR Primer Pool, and 20 μL of the wastewater-derived cDNA. The first-round PCR cycling parameters were 5 min at 37 °C, 10 min at 95 °C, 13 cycles of 95 °C for 10 s, 64 °C for 1 min, 60 °C for 1 min and 72 °C for 10 s, followed by a final extension step at 72 °C for 2 min performed on a C1000 thermal cycler. The first-round PCR products were then purified using clean magnetic beads. In the second round the PCR amplification mixture contained 25 μL of PCR Enzyme Mix, 0.5 μL of PCR Clean Enzyme, 1 μL of PCR additive, 2 μL of PCR block (259 sets of SARS-CoV-2 primers each targeting a different ~200 bp region to encompass the entire genome (accession MN908947.3) (Wu et al., 2020), 10 sets of Lambda Phage DNA primers; and four sets of primers targeting the human Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene for human DNA/RNA contamination control into the purified PCR products from the first round. The second-round PCR was performed under the same cycling parameters as that of the first-round PCR, except 27 PCR cycles were used. The second-round PCR products were also purified using clean magnetic beads. After bead-based purification, the second-round PCR products were quantified with the Qubit dsDNA High Sensitivity Assay kit (Thermo Fisher Scientific, Waltham, MA, USA) to confirm the required concentration of ≥4 ng/μL.
Short amplicons libraries from the RNA sample were equally pooled and subjected to single-stranded circular DNA (ssDNA) library preparation with the MGIEasy Dual Barcode Circularization kit (MGI, Shenzen, China) to obtain circularized DNA molecules. These molecules were subsequently digested to form circularized single strand DNA (ssCirDNA) and then subjected to rolling circle amplification to generate DNA nanoballs (DNBs) based libraries. The DNBs were added to a silicon slide that contains a grid-like pattern of binding sites, which enables the DNBs to self-assemble into a dense grid of spots for sequencing. The bases (A, C, T or G) of the DNBs were identified through digital imaging during each cycle of sequencing, when complementary and fluorophores-nucleotide containing nucleotides were ligated to the DNBs (Drmanac et al., 2010). The DNB libraries were sequenced on a DNBSEQ-G400 instrument at BGI Australia with pair-end 100 sequencing set (MGI, Shenzhen, China).
In this study, the read processing for ATOPlex was carried out by following the SARS-CoV-2_MultiPCR_v1.0 workflow (https://github.com/MGI-tech-bioinformatics/SARS-CoV-2_Multi-PCR_v1.0). With the primers trimmed, mapped reads were used to obtain the numbers of reads that map to either the SARS-CoV-2 genome, Lambda phage DNA, or GAPDH. The SARS-CoV-2 read numbers in each sample were normalized against Lambda phage read numbers. The variants were calculated by the Freebayes version v9.9.2–27-g5d5b8ac, based on the mapped result generated by the BWA mem version 0.7.13-r1126 (Li, 2013). Furthermore, consensus genomes from each sample were generated based on the variant information and the genome of the strain of SARS-CoV-2 (accession MN908947.3).
The consensus genomes of the aircraft wastewater sample (i.e., A12) positive for the del(69–70) mutation by RT-qPCR was compared to the latest SARS-CoV-2 genomes, including the 21 K(Omicron), through the clade assignment, mutation identification, consensus genome quality checking tools, and phylogenetic placement and visualization, provided by Nextstrain (https://clades.nextstrain.org) (Aksamentov et al., 2021).
2.7. Ethics approval
Low-risk approval as defined by the National Statement on Ethical Conduct in Human Research was obtained from the CSIRO Ethics Committee (reference number 2020_031_LR). Passenger information were de-identified.
2.8. Sequence data
Sequence is deposited in NCBI's SRA under the project number PRJNA789244; SRA # SRP350896. GenBank accession no. OL869974.
3. Results
All 12 RNA samples were free from PCR inhibition as determined by Cq values from the MHV RT-qPCR assay and were therefore used for downstream RT-qPCR analysis for SARS-CoV-2 RNA. RT-qPCR negative controls were negative in all RT-qPCR runs. The amplification efficiencies of US CDC N1 and CDC N2 assays were 97.2 and 98.0%, respectively, within acceptable bounds (Bustin et al., 2009). The correlation coefficient (R 2) values were between 0.98 and 0.99 for US CDC N1 and N2, respectively. The slopes of the standard curves were −3.37 (US CDC N1), and −3.43 (US CDC N2). The y-intercept values were 37.9 (US CDC N1) and 40.2 (US CDC N2). The ALODs for the RT-qPCR assays were 9.50 and 26.7 GC/reaction for US CDC N1 and N2, respectively. The recovery efficiency of the CP Select was 31.2 ± 6.23%.
Among the 12 aircraft wastewater samples, amplifications were observed in two samples (A3 and A12) by US CDC N1 and US CDC N2 assays and the mean SARS-CoV-2 RNA concentrations ranged from 2.98 ± 0.13 log10 GC/50 mL of wastewater (A3) to 4.30 ± 0.02 log10 GC/50 mL (A12) using the US CDC N1 assay. The mean concentrations for samples A3 and A12 were 3.01 ± 0.27 and 4.19 ± 0.03 log10 GC/50 mL using the US CDC N2 assay. All aircraft wastewater samples except A12 (flight from JNB-DRW) did not produce amplifications by the del(69–70) assay. For sample A12, the mean Cq value was 35.5 ± 0.17 for the del(69–70) assay suggesting the putative detection of the Omicron variant sub-lineage BA.1 in this sample. Allelic discrimination plot of the del(69–70) mutation assay also suggested the presence of del(69–70) mutation present in the sample. RT-qPCR/RT-PCR results are summarized in Table 1 .
Table 1.
Detection of SARS-CoV-2 and variant Omicron RNA in aircraft wastewater samples collected from flights landed at Darwin International Airport (DRW) and Sydney International Airport (SYD).
| Flight No. | Departure port – sampling port | Flight duration | Sampling date | Number of passengers | Mean ± SD log10 GC/50 mL of wastewater |
Mean Cq ± SD |
|
|---|---|---|---|---|---|---|---|
| US CDC N1 | US CDC N2 | del(69–70) | |||||
| A1 | JNB-DRW | ~14 h | 11/04/2021 | 193 | <ALOD | <ALOD | <ALOD |
| A2 | JNB-DRW | ~14 h | 19/08/2021 | 166 | <ALOD | <ALOD | <ALOD |
| A3 | IST-DRW | ~14 h 25 min | 2/10/2021 | 142 | 2.98 ± 0.13 | 3.01 ± 0.27 | <ALOD |
| A4 | LAX-SYD | ~15 h | 03/11/2021 | 108 | <ALOD | <ALOD | <ALOD |
| A5 | LAX-SYD | ~15 h | 04/11/2021 | Freighter service (no passengers) | <ALOD | <ALOD | <ALOD |
| A6 | DEL-DRW | ~11 h 40 min | 07/11/2021 | 37 | <ALOD | <ALOD | <ALOD |
| A7 | LAX-SYD | ~15 h | 08/11/2021 | 109 | <ALOD | <ALOD | <ALOD |
| A8 | LAX-SYD | ~15 h | 09/11/2021 | 147 | <ALOD | <ALOD | <ALOD |
| A9 | DEL-DRW | ~11 h 40 min | 10/11/2021 | 77 | <ALOD | <ALOD | <ALOD |
| A10 | LAX-SYD | ~15 h | 14/11/2021 | Freighter service (no passengers) | <ALOD | <ALOD | <ALOD |
| A11 | LAX-SYD | ~15 h | 15/11/2021 | 191 | <ALOD | <ALOD | <ALOD |
| A12 | JNB-DRW | ~14 h | 25/11/2021 | 20 | 4.30 ± 0.02 | 4.19 ± 0.03 | 35.5 ± 0.17 |
JNB: O.R. Tambo International airport.
DRW: Darwin International Airport.
LAX: Los Angeles International Airport.
SYD: Sydney International Airport.
DEL: Indira Gandhi International Airport.
IST: Istanbul Airport.
ALOD: Assay limit of detection.
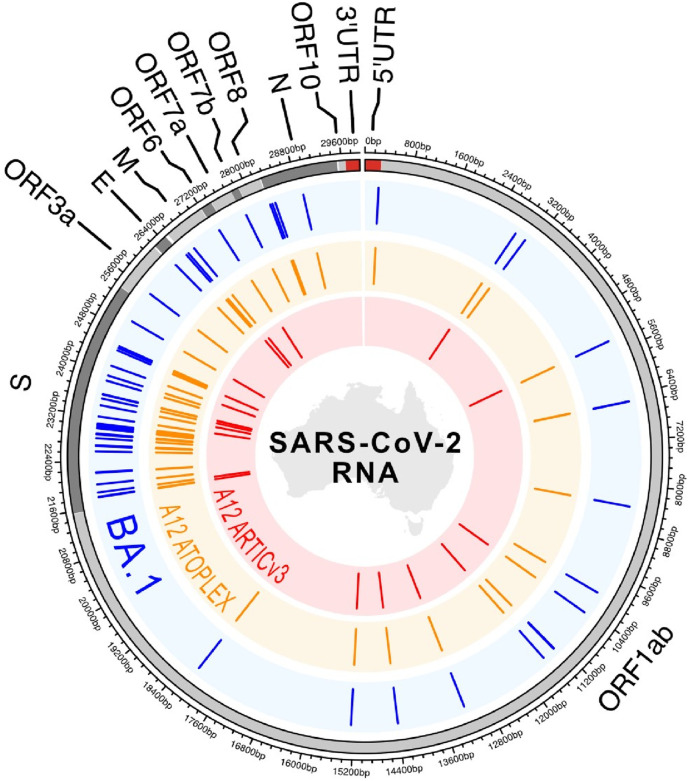
Sequence analysis was also consistent with the Omicron variant in aircraft wastewater sample A12 (Fig. 1 ). Targeted metagenomic sequencing with ARTIC V3 suggested the probable identification of VOC Omicron (B.1.1.529) lineage. Using Pangolin software, 17 of 45 defining mutations were detected in the consensus genome, whereas 20 of 45 defining mutations were detected by the local in-house computational pipelines (Fig. 2 ; Supplementary Table ST2). Pangolin software failed to detect three mutations due to a software glitch (i.e., multiple sequence alignment (MSA) missed some bases near the Ns). The local in-house pipeline use MAFFT for MSA and did not have this issue. Detection of ≥20 of 45 defining mutations is required to meet the SARS-CoV-2 lineage constellation definition (GitHub - cov- lineages/constellations). A total of 1,407,071 reads were obtained and 1,173,136 demultiplexed passed reads were used for the alignment. The read depth was normalized by 200× following the pipeline and resulted in a consensus sequence of ~61% coverage compared to the reference strain. The low genome coverage (i.e., missing data) and fragmented quality of the sequences detected in the sample resulted in limited ability to further confirm the lineage via the ARTIC protocol. Phylogenetic mapping indicated that wastewater sample A12 clustered reasonably well (20 of 45 defining mutations) with clinical isolates from Omicron Infections in Australia compared to the wild type (i.e., MN908947.3) (Fig. 2).
Fig. 1.
Variation map of BA.1 (Omicron) and aircraft wastewater sample A12 consensus sequences. Outermost ring represents the reference SARS-CoV-2 sequence (NCBI accession number NC_045512) with ORFs, 5′ and 3′ UTRs indicated. Blue ring with ticks indicates locations of mutations present in >90% of GISAID BA.1 sequence as of 9th of December 2021. Orange ring with ticks indicates sites of mutation represented in the consensus sequence retrieved from sample A12 using the ATOPlex; while red (innermost) ring with ticks represents mutations represented from the same sample but using the ARTIC V3 protocol.
Fig. 2.
Phylogenetic tree and constellations haplotyping of Australian Omicron sequences clustered with aircraft wastewater Omicron sequence (A12). Omicron mutations were highlighted in blue, ambiguous mutations were highlighted in yellow, and red is the reference strain (MN908947.3).
Further confirmation of Omicron detection was undertaken by ATOPlex sequencing of the A12 RNA sample. A total of 24,219,146 paired-end reads (100 bp) were generated from the sample A12 for a total of 4,843,829,200 bp of sequencing data. According to the pipeline, the sample was randomly subsampled to 20 million reads. In the resulting sample, 88.3% of the reads were above 99.9% accuracy (Phred score ≥ 30). Filtering of reads was conducted based on read quality, adaptor content and rate of unknown bases (“N”s), from which 96.1% of reads were used for downstream analysis. The SARS-CoV-2 consensus genome from ATOPlex sequencing detected 47 amino acid mutations and seven deletions (Supplementary Table ST3; https://covariants.org/variants/21K.Omicron). The mutations present in the sample consensus genome include all 36 mutations in the S gene along with mutations in the ORF1ab, ORF3a, ORF6, ORF7b, the E, M, and N genes used to define the Omicron VOC (Supplementary Table ST3). Phylogenetic clustering indicates the SARS-CoV-2 genome from the aircraft wastewater in sample A12 is most closely related to the Omicron variant (21K) of SARS-CoV-2 (Supplementary Fig. SF1). To better assess the coverage across different regions of the genome, a coverage analysis was performed using Python matplotdb function, showing that more than 99% of the genome regions were covered by the ATOPlex amplicons. Most of the mutations across the S gene have more than 100 reads of supporting coverage, and the highest coverage is up to 6630 reads (H655Y). Yet, some of the mutations had only 5 reads of coverage, including Q493R, G496S, Q498R and N501Y, indicating the high accuracy of ATOPlex could potentially identify these low-depth mutations with high sensitivity.
4. Discussion
In the current study, we demonstrate that wastewater surveillance can provide important information on the introduction of SARS-CoV-2 VOCs, such as Omicron, into a geographic region (in this case, Australia). To the best of our knowledge, this is the first report of the detection of the Omicron VOC in aircraft wastewater. The two aircraft samples positive for SARS-CoV-2 (A3, A12) in the current study were indicated by amplification across all technical replicates of both the US CDC N1 and N2 assays. Given the absence of amplification observed in all pertinent negative controls and the low Cq values, these detections were interpreted as true positives for SARS-CoV-2 RNA.
Since sample A12 was from an international flight arriving in DRW from JNB, South Africa on the 25th of November and this prompted us to screen for the presence of the Omicron variant. For the putative detection of Omicron, we used a del(69–70) assay which detects the mutation responsible for the S gene “dropout”. The mean Cq value for the A12 sample was 35.5 ± 0.17 suggesting the presence of the del(69–70) mutation and Omicron variant sub-lineage BA.1 in this sample. Two lineages (BA.1 and BA.2) of Omicron have been reported in Australia, Canada and South Africa that are genetically different. The proposed sub-lineage BA.2 does not have the del(69–70) mutation and will not be detected by SGTF. The del(69–70) mutation is found in the genomes of the Alpha (B.1.1.7; variant 501Y·V1) but is not commonly found in the Delta VOC (B.1.617) that was more prevalent in Australia at the time.
On the 29th of November, Northern Territory Health announced that a person who had arrived on a repatriation flight from South Africa on November 25 was infected with the Omicron variant after conducting genome sequencing. Wastewater sample A12 was collected from the same repatriation flight, strongly suggesting a link between the presence of Omicron in wastewater indicated by the positive del(69–70) RT-qPCR results and shedding by the infected individual. Detecting SARS-CoV-2/Omicron signal in aircraft A12 wastewater sample which received bodily fluids or feces from an infected individual is not surprising. In our previous study, we were able to achieve positive wastewater detection when only one infected passenger was on board on several occasions (Ahmed et al., 2021). This is due to the fact that unlike household toilets, which use several liters of water per flush, aircraft lavatories use a substantially reduced volume of water per flush, resulting in less dilution, increasing the likelihood for detecting small quantities of SARS-CoV-2 RNA.
To avoid potential misclassification, or rule out the presence of other SARS-CoV-2 strains from other passengers, the del(69–70) positive RNA sample was split and sequenced using both the ARTIC V3 protocol on a Nanopore platform and the ATOPlex sequencing. Both approaches have been previously used to detect VOCs in clinical and wastewater samples (Izquierdo-Lara et al., 2021; Lin et al., 2021; Ong et al., 2021). Sequencing results from each of these platforms indicated a consensus genome well aligned with the publicly available Omicron genomes. Thus, we conclude that the wastewater from the relevant flight contained RNA from the Omicron variant of SARS-CoV-2, possibly associated with the passenger identified through clinical testing and further genomic analysis during quarantine.
Importantly, because a passenger with COVID-19 may not be shedding via feces or may not use the lavatory for defecation during the flight, negative RT-qPCR results from aircraft wastewater cannot preclude the possibility of COVID-19 cases onboard. However, shedding from routes other than defecation, such as saliva and nasal fluids could be relevant for certain aircraft (Crank et al., 2022). For example, passengers could brush their teeth in the morning, rinse their mouth, and spit in the wash basin or deposit tissues with saliva and/or nasal fluid into the toilet. These shedding routes could be particularly relevant for aircraft (i.e., certain models) wastewater since the onboard fixtures greatly minimize the volume of water diluting any biological material. Furthermore, the aircraft wastewater holding tank is a closed system where homogenization of the biological material throughout the entire wastewater volume is possible and low ambient temperature at high altitude might preserve sample integrity. These characteristics would greatly increase the probability of detecting a small number of viruses in aircraft wastewater compared to wastewater surveillance at large municipal wastewater treatment plants where detection sensitivity may be compromised for various reasons (Ahmed et al., 2021).
Both sequencing approaches suggested the detection of Omicron VOC in sample A12. However, in this sample, the consensus genome obtained using the ARTIC V3 protocol was less complete (~61%) in comparison with that of ATOPlex (i.e., 99%). This resulted in comparatively fewer mutations detected by the ARTIC V3 protocol, particularly in the S gene (Supplementary Tables ST2 and ST3). There are several factors that may have contributed to the differences such as sub-sampling effects of the RNA aliquot that was shared with sequencing laboratories, the number of primers covering the entire length of the genomes, possible degradation of viral RNA in wastewater, differences in sequencing depth, and sequencing errors (Nemudryi et al., 2020; Ni et al., 2021). Notably, there are known mutations in the Omicron genome that cause primer dropout with certain amplicons in the ARTIC V3 primer scheme, such as the G142- mutation in the S gene interfering with primer 72_RIGHT (https://community.artic.network/t/sars-cov-2-v4-1-update-for-omicron-variant/342; https://community.artic.network/t/sars-cov-2-v4-1-update-for-omicron-variant/342; Davis et al., 2021). Hence there have been recent updates to the ARTIC protocol with the development of the V4.1 primer scheme that can improve genome coverage in problematic areas (https://community.artic.network/t/sars-cov-2-v4-1-update-for-omicron-variant/342).
We speculate that the low coverage observed within the ORF1ab and Spike region of the Omicron consensus genome generated with ARTIC V3 was likely caused by primer dropouts from known interfering Omicron mutations in those areas. In this case, the performance of the ATOPlex multiplex primer scheme did not appear to be as impacted by mutations present in Omicron, enabling a concordant consensus sequence. The identification of most lineage-defining mutations of Omicron at the consensus-level (e.g., they were the dominant mutations in the sequence data) in the aircraft wastewater sample using both ARTIC and ATOPlex would signify that either the SARS-CoV-2 genomic material originated from a single infected passenger, or that Omicron was the predominant variant among multiple infected passengers. Generating a consensus genome from the metagenomic data in this case was an effective way to identify the predominant VOC in the wastewater originating from one or few infected passengers; however, in the case of wastewater that has contributions from many infected persons with different genomic variants, the metagenomic data could be deconvoluted to infer the relative abundance of multiple VOCs by focusing on lineage-defining mutations (Lin et al., 2021). Linking wastewater genomic surveillance data combined with that of clinical patient testing in the post-arrival quarantine period could be a powerful approach to identify and verify VOC introductions and potential transmission chains.
One of the major limitations of targeted metagenomic sequencing is that rapid results cannot be obtained within the same timescale as RT-qPCR assays. While both require the same processing and extraction methodologies, sample preparation, sequencing and analysis extends the time to results by 72–96 h. Also, it is not clear how sensitive the sequencing approaches are for the trace detection of SARS-CoV-2 in wastewater, although in clinical samples with mixed variants detection levels from ~1.5% to 3% have been reported (Grubaugh et al., 2019). On the other hand, RT-qPCR assays may not be readily available for emerging variants, requiring the development of new assays and validation before application. Emerging variants may share a single nucleotide variant (SNV) with antecedent variants that are no longer prevalent. Thus, lead times in wastewater surveillance of an emerging variant might be significantly reduced by re-deploying previously validated RT-qPCR assays (Graber et al., 2021). For example, an allele-specific primer extension RT-qPCR assay has been deployed in Ontario, Canada that was originally intended to monitor C28311T allele frequency, a SNV specific for both Lambda and Omicron VOCs (pers. Comm. T. Graber). Furthermore, mutations at this location have the potential to affect the US CDC N1 assay performance. However, preliminary data indicate that there is no loss of sensitivity of the US CDC N1 assay on a C28311T RNA template above 10 GC (pers. Comm. Tyson Gerber). Indeed, the Omicron wastewater sample identified yielded amplification with the US CDC N1 assay. Here we demonstrate how RT-qPCR can be used to screen for putative VOCs, with sequencing used as a secondary confirmatory analysis.
5. Conclusions
-
•
We have demonstrated that surveillance of aircraft wastewater samples for SARS-CoV-2 provides valuable public health information regarding the global spread of emerging VOCs, such as Omicron. Pre-flight testing of clinical samples alone does not identify all COVID-19 cases among arriving passengers, thus post-flight testing of passengers may be beneficial when potentially more-infectious variants are predicted to be emerging globally, and quarantine measures may be necessary when new variants cause increase disease severity.
-
•
In a previous study, aircraft wastewater demonstrated 84% accuracy for COVID-19 cases onboard international flights (all of which were negative by pre-flight testing) (Ahmed et al., 2021). And in the current study, a single Omicron infection onboard an international flight was detected and corroborated by wastewater analysis with RT-qPCR and genomic sequences.
-
•
The application of routine monitoring of aircraft wastewater at airports requires further method optimization for trace detections and rapid on-site analysis rather than transporting samples to a centralized laboratory. Furthermore, collecting wastewater samples from aircraft is tedious and the success of this mode of surveillance largely dependent on the toileting behavior of passengers, the details of which cannot be easily surveyed or quantified.
-
•
Further research is needed to identify suitable sample matrices such as air sampling or surface swab sampling to determine whether these matrices are more appropriate than wastewater. A combination of sampling approaches and high-throughput analyses at major airport hubs may have the potential to provide rapid and scalable information about importations of potential variants to public health agencies.
Funding
The authors did not receive any funding for this project.
CRediT authorship contribution statement
Warish Ahmed: Methodology, Formal analysis, Investigation, Writing – original draft. Aaron Bivins: Formal analysis and writing – original draft. Wendy J.M. Smith: Methodology. Suzanne Metcalfe: Methodology. Mikayla Stephens: Methodology. Amy V. Jennison: Formal analysis, Data curation. Frederick A.J. Moore: Formal analysis, Data curation. Jayden Bourke: Methodology. Sanmarie Schlebusch: Formal analysis, Data curation. Jamie McMahon: Formal analysis, Data curation. Glen Hewitson: Formal analysis, Data curation. Son Nguyen: Formal analysis, Data curation. Jean Barcelon: Formal analysis, Data curation. Greg Jackson: Writing – original draft. Jochen F. Mueller: Conceptualization, Resources, Funding acquisition. John Ehret: Methodology, Resources. Ian Hosegood: Methodology, Resources. Wei Tian: Formal analysis, Data curation. Haofei Wang: Methodology. Lin Yang: Methodology. Paul M. Bertsch: Writing – original draft. Josh Tynan: Resources. Kevin Thomas: Writing – original draft. Kyle Bibby: Writing – original draft. Tyson E. Graber: Formal analysis, Writing – original draft. Ryan Ziels: Formal analysis, Writing – original draft. Stuart L. Simpson: Investigation, Funding acquisition, Writing – original draft.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We thank Qantas ground staff for collecting wastewater samples. Aaron Bivin's contribution was supported by NSF grant 2027752.
Editor: Damia Barcelo
References
- Ahmed W., Bertsch P.M., Angel N., Bibby K., Bivins A., Dierens L., Edson J., Ehret J., Gyawali P., Hamilton K.A., Hosegood I., Hugenholtz P., Jiang G., Kitajima M., Sichani H.T., Shi J., Shimko K.M., Simpson S.L., Smith W.J.M., Symonds E.M., Thomas K.V., Verhagen R., Zaugg J., Mueller J.F. Detection of SARS-CoV-2 RNA in commercial passenger aircraft and cruise ship wastewater: a surveillance tool for assessing the presence of COVID-19 infected travellers. J Travel Med. 2020;27 doi: 10.1093/jtm/taaa116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Simpson S.L., Bertsch P.M., Ehret J., Hosegood I., Metcalfe S.S., Smith W.J.M., Thomas K.V., Tynan J., Mueller J.F., Bivins Wastewater surveillance demonstrates high predictive value for COVID-19 infection on board repatriation flights to Australia. Environ. Int. 2021;158 doi: 10.1016/j.envint.2021.106938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aksamentov I., Roemer C., Hodcroft E.B., Neher R.A. Nextclade: clade assignment, mutation calling and quality control for viral genomes. J. Open Source Softw. 2021;6:3773. [Google Scholar]
- Besselsen D.G., Wagner A.M., Loganbill J.K. Detection of rodent coronaviruses by use of fluorogenic reverse transcriptase-polymerase chain reaction analysis. Comp Med. 2002;52:111–116. [PubMed] [Google Scholar]
- Bustin S.A., Benes V., Garson J.A., Hellemans J., Huggett J., Kubista M., Mueller R., Nolan T., Pfaffl M.W., Shipley G.L., Vandesompele J., Wittwer C.T. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009;55:611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- California Department of Public Health (CA DPH) Fact Sheet: Omicron Variant. 1 December 2021. 2021. https://www.cdph.ca.gov/Programs/CID/DCDC/Pages/COVID-19/Omicron-Variant-Fact-Sheet.aspx Accessed on 5 December 2021.
- CDC 2019-Novel Coronavirus (2019-nCoV) Real-Time rRT-PCR Panel Primers and Probes. 2019. https://www.cdc.gov/coronavirus/2019-ncov/downloads/rt-pcr-panel-primer-probes.pdf
- Crank K., Chen W., Bivins A., Lowry S., Bibby K. Contribution of SARS-CoV-2 RNA shedding routes to RNA loads in wastewater. Sci. Total Environ. 2022;806 doi: 10.1016/j.scitotenv.2021.150376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis J.J., Long S.W., Christensen P.A., Olsen R.J., Olson R., Shukla M., Subedi S., Stevens R., Musser J.M. Analysis of the ARTIC version 3 and version 4 SARS-CoV-2 primers and their impact on the detection of the G142D amino acid substitution in the spike protein. Microbiol. Spectr. 2021 doi: 10.1128/Spectrum.01803-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Maio N., Walker C., Borges R., Weilguny L., Slodkowicz G., Goldman N. Masking strategies for SARS-CoV-2 alignments. 2020. https://virological.org/t/issues-with-sars-cov-2-sequencing-data/473 Available from:
- Drmanac R., Sparks A.B., Callow M.J., Halpern A.L., Burns N.L., Kermani B.G., Carnevali P., Nazarenko I., Nilsen G.B., Yeung G., Dahl F., Fernandez A., Staker B., Pant K.P., Baccash J., Borcherding A.P., Brownley A., Cedeno R., Chen L., Chernikoff D., Cheung A., Chirita R., Curson B., Ebert J.C., Hacker C.R., Hauser B., Huang S., Jiang Y., Karpinchvk V., Koenig M., Kong C., Landers T., Le C., Liu J., Mcbride C.E., Morenzoni M., Morey R.E., Mutch K., Perazich H., Perry K., Peters B.A., Peterson J., Pethiyagoda C.L., Pothuraju K., Richter C., Rosenbaum A.M., Roy S., Shafto J., Sharanhovich U., Shannon K.W., Sheppy C.G., Sun M., Thakurja J.V., Tran A., Vu D., Zaranek A.W., Wu X., Crmanac S., Oliphant A.R., Banvaj W.C., Martin B., Ballinger D.G., hartlage R., Church G.M., Reid C.A. Human genome sequencing using unchained base reads on self-assembling DNA nanoarrays. Science. 2010;327(5961):78–81. doi: 10.1126/science.1181498. [DOI] [PubMed] [Google Scholar]
- European CDC Threat Assessment Brief: implicationsof the emergence and spread of the SARS-CoV-2 B.1.1.529 variant of concern (Omicron) for the EU/EEA. 26 November 2021. 2021. https://www.ecdc.europa.eu/en/publications-data/threat-assessment-brief-emergence-sars-cov-2-variant-b.1.1.529 Accessed on 5 December 2021.
- Graber T.E., Mercier É., Bhatnagar K., Fuzzen M., D'Aoust P.M., Hoang H.D., Tian X., Towhid S.T., Plaza-Diaz J., Eid W., Alain T., Butler A., Goodridge L., Servos M., Delatolla R. Near real-time determination of B.1.1.7 in proportion to total SARS-CoV-2 viral load in wastewater using an allele-specific primer extension PCR strategy. Water Res. 2021;15(205) doi: 10.1016/j.watres.2021.117681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grubaugh N.D., Gangavarapu K., Quick J., Matteson N.L., De Jesus J.G., Main B.J., Tan A.L., Paul L.M., Brackney D.E., Grewal S., Gurfield N., Van Rompay K., Isern S., Michael S.F., Coffey L.L., Loman N.J., Andersen K.G. An amplicon-based sequencing framework for accurately measuring intrahost virus diversity using PrimalSeq and iVar. Genome Biol. 2019;20:8. doi: 10.1186/s13059-018-1618-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izquierdo-Lara R., Elsinga G., Heijnen L., Munnink B., Schapendonk C., Nieuwenhuijse D., Kon M., Lu L., Aarestrup F.M., Lycett S., Medema G., Koopmans M., de Graaf M. Monitoring SARS-CoV-2 circulation and diversity through community wastewater sequencing, the Netherlands and Belgium. Emerg. Infect. Dis. 2021;27(5):1405–1415. doi: 10.3201/eid2705.204410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K., Misawa K., Kuma K., Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast fourier transform. Nucleic Acids Res. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. 2013. Aligning Sequence Reads, Clone Sequences and Assembly Contigs With BWA-MEM. arXiv:1303.3997v2 [q-bio.GN] [Google Scholar]
- Lin X., Glier M., Kuchinski K., Ross-Van Mierlo T., McVea D., Tyson J.R., Prystajecky N., Ziels R.M. Assessing multiplex tiling PCR sequencing approaches for detecting genomic variants of SARS-CoV-2 in municipal wastewater. mSystems. 2021;6 doi: 10.1128/mSystems.01068-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallapaty S. 2021. Omicron-variant Border Bans Ignore the Evidence, Say Scientists. [DOI] [PubMed] [Google Scholar]
- Minh B.Q., Schmidt H.A., Chernomor O., Schrempf D., Woodhams M.D., von Haeseler A., Lanfear R. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 2020;37:1530–1534. doi: 10.1093/molbev/msaa015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Institute for Communicable Diseases (NICD) New COVID-19 Variant Detected in South Africa. 25 November 2021. 2021. https://www.nicd.ac.za/new-covid-19-variant-detected-in-south-africa/ Accessed on 5 December 2021.
- National Institute for Communicable Diseases (NICD) Latest Confirmed Cases of COVID-19 in South Africa (2 December 2021). 2 December 2021. 2021. https://www.nicd.ac.za/latest-confirmed-cases-of-covid-19-in-south-africa-2-december-2021/ Accessed on 5 December 2021.
- Nemudryi A., Nemudraia A., Wiegand T., Surya K., Buyukyoruk M., Cicha C., Vanderwood K.K., Wilkinson R., Wiedenheft B. Temporal detection and phylogenetic assessment of SARS-CoV-2 in municipal wastewater. Cell Rep. Med. 2020;1 doi: 10.1016/j.xcrm.2020.100098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni G., Lu J., Maulani N., Tian W., Yang L., Harliwong I., Wang Z., Mueller J., Yang B., Yuan Z., Hu S., Guo J. Novel multiplexed amplicon-based sequencing to quantify SARS-CoV-2 RNA from wastewater. Environ. Sci. Technol. 2021;8:683–690. doi: 10.1021/acs.estlett.1c00408. [DOI] [PubMed] [Google Scholar]
- Ong S.W.X., Chiew C.J., Ang L.W., Mak T.-M., Cui L., Toh M.P.H.S., Lim Y.D., Lee P.H., Lee T.H., Chia P.Y., Maurer-Stroh S., Lin R.T.P., Lee Y.-S., Lee V.J., Lye D.C., Young B.E. Clinical and virological features of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variants of concern: a retrospective cohort study comparing B.1.1.7 (Alpha), B.1.351 (Beta), and B.1.617.2 (Delta) Clin. Infect. Dis. 2021 doi: 10.1093/cid/ciab721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulliam J.R.C., van Schalkwyk C., Govender N., von Gottberg A., Cohen C., Groome M.J., Dushoff J., Mlisana K., Moultrie H. medRXiv; 2021. Increased Risk of SARS-CoV-2 Reinfection Associated With Emergence of the Omicron Variants in South Africa. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyson J.R., James P., Stoddart D., Sparks N., Wickenhagen A., Hall G., Choi J.H., Lapointe H., Kamelian K., Smith A.D., Prystajecky N., Goodfellow I., Wilson S.J., Harrigan R., Snutch T.P., Loman N.J., Quick J. bioRxiv; 2020. Improvements to the ARTIC Multiplex PCR Method for SARS-CoV-2 Genome Sequencing Using Nanopore. [DOI] [Google Scholar]
- UK Health Security Agency SARS-CoV-2 variants of concern and variants under investigation in England: Variant of Concern: Omicron, VOC-21Nov-01 (B.1.1.529). 3 December 2021. 2021. https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/1038404/Technical_Briefing_30.pdf Accessed on 5 December 2021.
- Verbyla M.E., Symonds E.M., Kafle R.C., Cairns M.R., Iriarte M., Mercado Guzmán A., Coronado O., Breitbart M., Ledo C., Mihelcic J.R. Managing microbial risks from indirect wastewater reuse for irrigation in urbanizing watersheds. Environ. Sci. Technol. 2016;50(13):6803–6813. doi: 10.1021/acs.est.5b05398. [DOI] [PubMed] [Google Scholar]
- WHO Classification of Omicron (B.1.1.529): SARS-CoV-2 Variant of Concern, 26 November 2021. 2021. https://www.who.int/news/item/26-11-2021-classification-of-omicron-(b.1.1.529)-sars-cov-2-variant-of-concern Accessed on 5 December 2021.
- Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., Hu Y., Tao Z.W., Tian J.H., Pei Y.Y., Yuan M.L., Zhang Y.L., Dai F.H., Liu Y., Wang Q.M., Zheng J.J., Xu L., Holmes E.C., Zhang Y.Z. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G. Using ggtree to visualize data on tree-like structures. Curr. Prot. Bioinforma. 2020;69 doi: 10.1002/cpbi.96. [DOI] [PubMed] [Google Scholar]