Abstract
Nucleocapsid protein (N protein) is the primary antigen of the virus for development of sensitive diagnostic assays of COVID-19. In this paper, we demonstrate the significant impact of dimerization of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) N-protein on sensitivity of enzyme-linked immunosorbent assay (ELISA) based diagnostics. The expressed purified protein from E. coli is composed of dimeric and monomeric forms, which have been further characterized using biophysical and immunological techniques. Indirect ELISA indicated elevated susceptibility of the dimeric form of the nucleocapsid protein for identification of protein-specific monoclonal antibody as compared to the monomeric form. This finding also confirmed with the modelled structure of monomeric and dimeric nucleocapsid protein via HHPred software and its solvent accessible surface area, which indicates higher stability and antigenicity of the dimeric type as compared to the monomeric form. The sensitivity and specificity of the ELISA at 95% CI are 99.0% (94.5–99.9) and 95.0% (83.0–99.4), respectively, for the highest purified dimeric form of the N protein. As a result, using the highest purified dimeric form will improve the sensitivity of the current nucleocapsid-dependent ELISA for COVID-19 diagnosis, and manufacturers should monitor and maintain the monomer-dimer composition for accurate and robust diagnostics.
Keywords: SARS-CoV-2, Nucleocapsid protein, Protein dimerization
Graphical abstract
1. Introduction
COVID-19 is a widespread global pandemic that has significantly damaged the financial stability and access to treatment for many, especially our most marginalized societies [1], [2], [3]. The development of vaccines and various therapeutic molecules has made a significant contribution to controlling the pandemic [4], [5]. Diagnostics has also played a major role, with most tests serving as an indicator of transmission at the time when the virus is in the upper respiratory tract [6]. Some advanced tests, based on duplex digital enzyme-linked immunosorbent assay (dELISA), have been employed for simultaneous detection of spike and nucleocapsid (N) proteins of SARS-CoV-2 [7]. However, detection of pathogen-specific antibodies that develop within days of infection is also a durable biomarker of prior exposure. The antibody-based assay is useful in identifying those who have been exposed to the virus and have antibodies to resist infection upon subsequent exposure of the SARS-CoV-2 [8], [9], [10].
The SARS-CoV-2 genome is composed of approximately 30,000 nucleotides, which encodes four structural proteins including spike (S) protein, envelope (E) protein, membrane (M) protein, and nucleocapsid (N) protein [11].
SARS-CoV-2 N protein is a ~45.6 kDa phosphoprotein, comprising of a N-terminal domain (NTD) and a C-terminal domain (CTD), connected by a loosely structured linkage region containing a serine/arginine-rich (SR) domain [12], [13]. N protein has two different oligomeric states that support its two functions: unmodified protein forms a structured oligomer suitable for nucleocapsid assembly, while phosphorylated protein forms a liquid-like compartment suitable for viral genome processing[14].
An assessment into the role of N protein similar to other coronaviruses, that SARS-CoV-2 N protein plays in the self-association, interactions with other proteins and RNA of virus thus represent extremely multivalent [15]. The residues from 45 to 181 of the NTD are responsible for the binding of viral RNA to the N protein. SR area linking the NTD and CTD is the site of phosphorylation which is assumed to control N protein performance [16]. Hydrophobic CTD of the N protein contains residues responsible for the homodimerization of the N protein [17], [18], [19], [20]. Homodimers of N protein are recorded to self-assemble into higher-order oligomeric complexes, possibly through cooperative interactions of homodimers [21]. Development of higher-order oligomeric complexes requires both dimerization domain and the expanded asymmetric moiety of the CTD [12], [22], [23]. Yeast two-hybrid analysis was used to demonstrate self-association of the full-length SARS-CoV N protein and the isolated C-terminal region [24], and the purified full-length protein was found to self-associate in solution mainly in dimer form [25]. Under physiological buffer conditions, N-protein condenses with specified RNA genomic regions, with the condensation enhanced at human body temperature (33 °C and 37 °C) and reduced at room temperature (22 °C) [26].
Upon SARS-CoV-2 infection, viral genomic RNA gets associated with the N protein to develop a ribonucleoprotein complex. This complex then packages itself into a helical conformation and combines itself with the M protein of the virion [13]. Despite being present within the viral particle and not very exposed to the surface, SARS-CoV-2 infected patients show elevated and earlier humoral response to the N protein rather than the spike [27]. This is the reason why the N protein is being widely used in vaccine development and serological assays [27], [28], [29]. It has been shown for SARS-CoV that the C-terminal region of the N protein is crucial for eliciting antibodies in immunological process [30], [31]. Most diagnostic assays are based on the antigenic proteins, either N or S protein, of the SARS-CoV-2 [32], [33], [34], [35], [36], [37], [38]. Several formats of ELISA have been developed to detect IgM/IgG antibodies in a patient's serum against the SARS-CoV-2 N protein [39], [40].
Structural study of the full-length coronavirus N protein expressed in Escherichia coli is complicated since the recombinant N protein is very susceptible to proteolysis [18]. As a result, minimal information exists on the structure of the SARS-CoV-2 N protein monomer and its assembly into higher-order complexes. In this study, full-length protein of SARS-CoV-2 was successfully expressed in E. coli BL21 (DE3) as aggregated inclusion bodies. Two major peaks of the N protein were identified as a monomeric and dimeric conformation via size exclusion chromatography coupled with multi-angle static light scattering (MALS), circular dichroism (CD), and fluorescence spectroscopy. Further, the antigenicity of these conformations was compared through a highly sensitive and precise ELISA-based antibody test. The epitope and solvent accessibility of the monomer and dimer forms of the N protein was also predicted using bioinformatics tools to study the structural stability and antigenicity of these conformations. It is evident that use of the dimeric form will increase the sensitivity of the current nucleocapsid dependent ELISA for rapid COVID-19 diagnostic. Further, the results indicate that monitoring and maintaining of the monomer-dimer composition is critical for accurate and robust diagnostics. To the best of our knowledge this is the first in-depth investigation into impact of dimerization of SARS-CoV-2 nucleocapsid protein on sensitivity of enzyme-linked immunosorbent assay (ELISA) based diagnostics of COVID-19.
2. Materials and methods
2.1. Construct and expression of SARS-CoV-2 N protein
Escherichia coli BL21 (DE3) purchased from Novagen – Merck Life Science Private Limited, India (Cat. No.69450-4), was used in the current study. The expression construct of SARS-CoV-2 N protein was commercially procured through Addgene in a pGBW-m4046785 vector with N protein gene insert of 1253 bp under the control of T7 promoter. The expression construct was transformed in E. coli BL21 (DE3) strain and bacterial culture was grown in terrific broth at 37.0 °C in the presence of 25 μg ml−1 chloramphenicol. When the O.D. at 600 of primary culture reached up to 1.0 ± 0.2, the secondary culture (100.0 ml) was inoculated with 5.0 ml of primary seed culture. Bacterial cultivations were carried out at 37.0 °C. 1.0 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) was used to induce the secondary culture in the mid-log phase. Cells were harvested after 12 h of induction and subjected to primary downstream processing steps to confirm protein expression.
2.2. Production of SARS-CoV-2 N protein in bioreactor
Protein expression was scaled-up in a 1.3 L bioreactor (Eppendorf, USA) with 0.5 L initial volume. Gas flow rate was maintained between 0.5 and 1.5 vvm (0.5–0.6 Lmin−1) by the mass flow controller. The pH of the media was monitored by a pH probe and maintained at 7.0 ± 0.2 by using 3 N phosphoric acid and 12.5% ammonia. Temperature was maintained at 37.0 °C. Dissolved oxygen of the batch was controlled at 30% saturation by cascading the stirrer speed between 300 and 900 rpm. Bioreactor was monitored and controlled by the Biocommand software (Eppendorf, USA). Fed batch media containing glycerol (200 g l−1, v/v) and yeast extract (1%, w/v) was continuously fed to the bacterial culture to enhance the biomass. Protein expression was induced with 1 mM IPTG for 8 h and cells were harvested by centrifugation at 8000 rpm for 15 min. Cell pellet was washed with 0.9% (w/v) NaCl, resuspended in lysis buffer (20 mM Tris-HCL, 150 mM NaCl, 0.5 mM EDTA, pH 8.0), lysed using an Ultrasonicator system (Oscar Ultrsonics Pvt., Ltd., India) for 30 min with 30s on/off (50% duty cycle). Lysed cells were then centrifuged at 7000 rpm for 15 min at 4 °C, supernatant was discarded, and the obtained IBs were washed twice with saline. Protein expression was analysed through SDS-PAGE.
2.3. Purification of SARS-CoV-2 N protein
IBs were solubilized in 100 mM Tris-HCl buffer containing 6 M urea (pH 8.0) for 2 h. The solution was then centrifuged, and supernatant was collected. To prepare CEX load, the pH was adjusted to 7.0 using acetic acid and conductivity was adjusted to <3.0 mS/cm using deionised water. The CEX column (SP Sepharose FF, Cytiva USA) was equilibrated with 20 mM phosphate buffer (pH 7.0) and the load was pumped on the CEX column at 5 min retention time. The bound N protein was eluted using 1 M NaCl in 20 mM phosphate buffer (pH 7.0). This purification step also doubled as an on-column refolding step for the N protein. The elute contained a mixture of monomer and dimer of the protein which was separated using preparative SEC (Superdex 200, Cytiva USA). Phosphate buffer (100 mM, pH 7.4) with 10% glycerol (v/v) was used to equilibrate the SEC column and CEX elute (1% of column volume) was injected at 45 min retention time. The SEC output was fractionated, and each fraction was analysed using analytical SEC. Purified monomer and dimer fractions were used for further analysis.
2.4. Immunoblotting
Purified SARS-CoV-2 N protein was electrophoresed on 4–10% SDS-PAGE (Bio-Rad) and stained with Coomassie Brilliant Blue G-250 (CBBG-250). For the detection of N protein through immunoblotting, the obtained protein bands were transferred onto a 0.22 μm nitrocellulose membrane (MDI) using a Trans-Blot Turbo Transfer System (Bio-Rad). The membrane was blocked with 5% (w/v) skimmed milk in Tris buffered saline-0.05% (v/v) Tween-20 (TBST) under gentle shaking at room temperature for 1 h. The membrane was then washed thrice with 1X-TBST and incubated with anti-SARS-CoV-2 N protein antibodies (catalogue No. ab272852, Abcam, 1:1000) in TBST with BSA (2%, w/v). The membrane was then washed thrice with 1X-TBST. Immunoblot was then incubated with HRP conjugated-goat anti-human IgG secondary antibodies (Millipore, AP309P) in a dilution of 1:10,000 for 1 h. The immunoblot was washed thrice with 1X TBST and was visualized on SuperSignal™ West Pico PLUS Chemiluminescent Substrate (Thermo Fischer Scientific), and chemiluminescent signals were captured using ImageQuant LAS 500 instrument (GE Healthcare).
2.5. Peptide mass fingerprinting
In-gel digestion with trypsin protocol was followed for mapping of purified recombinant SARS-CoV-2 N protein [41]. Gel band of interest was excised and transferred into microcentrifuge tube and destained by incubating for 30 min in 100 μl of 50 mM ammonium bicarbonate/acetonitrile (1:1, v/v) with vertexing. Then the gel pieces were incubated and vortexed in 200 μl of acetonitrile. Trypsin (Agilent Technologies, California, USA) was added and incubated at 37 °C for 12–14 h. Peptides were extracted by adding 100 lL of 1:2 solution of 5% formic acid and acetonitrile and incubated for 15 mins in a shaker at 37 °C. The liquid obtained was evaporated by Speed-Vac vacuum centrifuge and was reconstituted in the 0.1% formic acid for the LC-MS. Digested peptides were separated on a C18 column (Advance Bio Peptide mapping Plus C18, 2.7 μm, 2.1 × 150 mm) using Agilent 1260 HPLC with detector at 214 nm. Column temperature was maintained at 55 °C. The column was equilibrated with 98% solvent A (0.1% TFA in water) and 2% solvent B (0.1% TFA in acetonitrile) for 10 min with a flow rate of 0.5 mlmin−1. Elution was achieved with a linear gradient of 2–45% B for 45 min followed by 45–60% B for 10 min, then linear gradient to 100% B for 10 min. Column was cleaned with 100% B for 10 min followed by equilibration with 98% A for 10 min. LC was coupled with ESI-TOF (Agilent Technologies, California USA) and TIC were recorded for m/z 100–3200. The capillary was set at a temperature of 300 °C with a gas flow rate of 8 L/min and nebulizer at 35 psig in positive ion mode. MS spectrum was analysed with Agilent MassHunter Qualitative analysis software (B.07.00).
2.6. Molecular mass identification of monomer and dimer form of N protein
SEC was performed on Dionex Ultimate 3000 HPLC (Thermo Scientific, Sunnyvale, CA, USA) with Superdex 200 (Cytiva, Marlborough, USA) maintained at 25 °C. The column was equilibrated with 50 mM phosphate buffer of pH 6.8, 300 mM NaCl salt concentration and 0.02% sodium azide at a flow rate of 0.5 ml/min. Both the fractions of the purified N protein were injected and run for 50 min at a flow rate of 0.5 ml/min and detected at 280 nm. SEC was coupled with MALS from Wyatt technologies, CA, USA, to confirm the molecular mass of the monomer and dimer fractions. All the buffers were filtered through a 0.22 μm membrane (Pall Life Sciences, NY, USA).
2.7. Circular dichroism spectroscopy
Circular dichroism spectra were recorded with a Jasco J-1500 spectrophotometer (Jasco Inc., Maryland, U.S). Secondary structure was measured in the Far-UV range from 195 to 250 nm with a 1 nm step size. Data were normalized by subtracting the baseline with the buffer and smoothed with Savitzky–Golay smoothing filter.
2.8. Fluorescence spectroscopy
Fluorescence spectroscopy was performed on a Cary Eclipse Fluorescence Spectrophotometer (Agilent Technologies, Santa Clara, California, United States) using Costar 96-well black polystyrene plate. The tryptophan fluorescence was recorded with excitation at 285 nm and emission between 300 and 500 nm. Slits for both excitation and emission were 5 nm.
2.9. Enzyme linked immunosorbent assay
The fraction of dimeric form of purified N protein of SARS-CoV-2 was diluted at 10 ngμl−1 in 0.05 M carbonate-bicarbonate buffer, pH 9.6. The diluted protein was coated in an increasing gradient (25–200 ng well−1) on a 96-microtiter ELISA plate (Nunc, Thermo Fisher Scientific) overnight at 4 °C. On the subsequent day, unbound protein was removed, and wells were washed thrice with 1X TBST buffer. Wells were then blocked with 4% (w/v) skimmed milk prepared in 1X TBST buffer and incubated at 37 °C for 1 h. The anti-SARS-CoV-2 N protein antibodies (Abcam) were diluted in 1X TBST and 100 μl of the diluted antibodies were allowed to interact with the coated N protein in the ELISA wells at 37 °C for 1 h. Wells were then washed with 200 μl of 1X TBST buffer three times followed by incubation with 100 μl of goat IgG-HRP antibody (Thermo Fisher Scientific) prepared in 1X TBST buffer. The wells were then washed three times with 200 μl of 1X TBST buffer. One hundred microlitre 3,3′,5,5′-tetramethylbenzidine substrate (Thermo Fisher Scientific) was added to each well and incubated for 10–15 min. The reaction was stopped by adding 100 lL of 0.18 M sulphuric acid and the optical densities of the plate wells were measured using Biotek plate reader at 450 nm.
2.10. Monomeric/dimeric structure modeling
The sequence of SARS-CoV-2 N protein was collected from Uniprot database with uniport ID: PODTC9. Multi template approach was used to build the model of SARS-CoV-2 N protein. Modeling of the structure was performed using the online version of the HHpred tool [42]. This identified the most promising template for building the structure of the SARS-CoV-2 N protein. Final template-based modeling was performed using the modeller tool [43]. This resulted in monomeric structure of SARS-CoV-2 N protein sequence. Dimeric structure was built using the PDB template 6WZO structure [27]. Pymol tool was used to superimpose the structure of 6WZO and modelled monomeric structure to build its dimeric form.
2.11. Solvent accessible surface area (SASA) calculation
SASA was calculated for monomeric and dimeric form using the naccess tool (http://www.bioinf.manchester.ac.uk/naccess/nacdownload.html).
2.12. Epitopic prediction
Discontinuous epitopes were predicted using the 3D structure of a protein. Monomer and dimer were compared using several tools to identify these discontinuous fragments of the protein that can act as epitopes for antibody binding. Ellipro [44] was first used for this prediction. The starting residues of 1–48 in the monomer and dimer model protein structure did not appear as globular and were present at the terminal in extended conformation. They were not included in epitope prediction to avoid false positives. Later, a similar analysis was performed with the DiscoTope server. This method predicted the probability of each residue to be part of an epitope.
2.13. Ethical approval and patient's sera
The patients' sera used in this study has been approved by the Guru Teg Bahadur Hospital Ethics Committee in Dilshad Garden, Delhi East, India with a reference number GTBHEC 2021/P-149. The negative control sample was taken from 40 healthy subjects before the COVID-19 pandemic. The 100 RT-PCR positive samples used in this study were split into two groups. The first group has 70 RT-PCR positive samples, whereas the second group includes RT-PCR positive 30 confirmed COVID-19 associated mucormycosis patients. Table 1 lists the number and category of the patient's sample.
Table 1.
Patients sample used in the study.
| S. No. | Sample type | Category | Number |
|---|---|---|---|
| 1. | Pre-epidemic samples (before 2018) | Negative | 40 |
| 2. | Positive sample (RT-PCR Confirmed) | Group 1 (COVID-19) | 70 |
| 3. | COVID-19 associated mucormycosis (RT-PCR confirmed) |
Group 2 (COVID-19) | 30 |
| Total | 140 | ||
2.14. ELISA validation with patients' sample
The assay was validated using 140 samples collected during COVID-19 pandemic and pre pandemic time. Antibody geometric mean level with 95% confidence interval (CI) was reported in the Table 2 . Positive antibodies in SARS-CoV-2 samples were determined using the mean plus three standard deviations of absorbance values from 40 negative control. Table 2 summarizes the sensitivity and specificity of developed ELISA calculated based on cutoff of mean + 3SD and ROC curves. Our analysis examines all pre-pandemic samples clinically negative, and all PCR verified samples clinically positive.
Table 2.
The performance of nucleocapsid based IgG ELISA.
| Mean Negative OD + 3SD | ROC curve | |
|---|---|---|
| Cut Off | 0.36 | 0.35 |
| True Positive | 99/100 | 99/100 |
| True Negative | 38/40 | 36/40 |
| Sensitivity (95% CI) | 99.0% (94.5–99.9) | 99.0% (94.5–99.9) |
| Specificity (95% CI) | 95.0% (83.0–99.4) | 90.0% (76.3–97.2) |
| Accuracy (95% CI) | 97.8% (93.8–99.5) | 96.4% (91.8–98.8) |
| Positive Predictive Value (95% CI) | 98.02% (92.7–99.4) | 96.1% (90.7–98.4) |
| Negative Predictive Value (95% CI) | 97.44% (84.3–99.6) | 97.3% (83.6–99.6) |
2.15. Statistical analysis
Graph Pad Prism software was used to perform statistical analysis of the data. The significance level was set at 1% using student t-test.
3. Results
3.1. Expression and purification of SARS-CoV-2 N protein
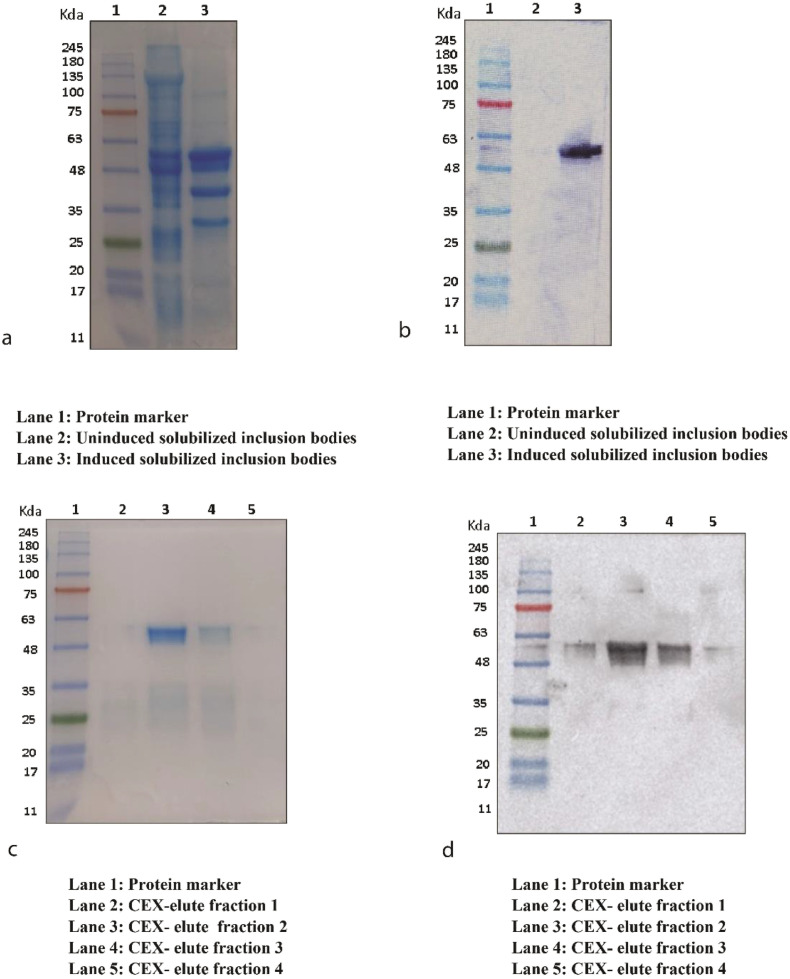
Full-length N protein gene construct was transformed into E. coli BL21 (DE3) cells. Robust expression of the full-length N protein was observed in 10% SDS-PAGE (Fig. 1a). The protein band with the molecular weight of about 51.38 kDa represents the full-length N protein expressed as IBs. The protein was further confirmed with immunoblotting using protein-specific antibody (Fig. 1b). Protein expression was later scaled-up in a bioreactor and a batch fermentation of transformed E. coli. BL21 (DE3) was performed with 10 g l−1 (v/v) of glycerol as a carbon source. Upon completion of batch, a DO shoot was observed (Supplementary Fig. 1) and feeding of 200 g l−1 (v/v) of the glycerol along with 1% (w/v) yeast extract was given to the bioreactor.
Fig. 1.
Nucleocapsid protein expression and purification. SDS-PAGE (10%) of expressed N protein (a). Immunoblotting of N protein (b). Coomassie staining of purified N protein fractions (c). Immunoblotting of purified N protein fraction using nucleocapsid specific antibody (d). CEX: Cation Exchange Chromatography.
Glycerol feeding resulted in attainment of higher cell density. Protein expression was induced by 1 mM IPTG at an optical density of 35 for 8 h. Biomass of about 20.3 g l−1 was generated in the fermentation batch of bioreactor. Due to overexpression of heterologous protein, product was accumulated in the form of IBs within the cytoplasm of the bacterial cell with a yield of about 6.25 g l−1. The titer of N protein was 2.17 mg per gram of inclusion bodies. The titer was not found to change from shake flask to bioreactor scale. However, the separation of monomer and dimer using size exclusion chromatography was found to be difficult to scale up.
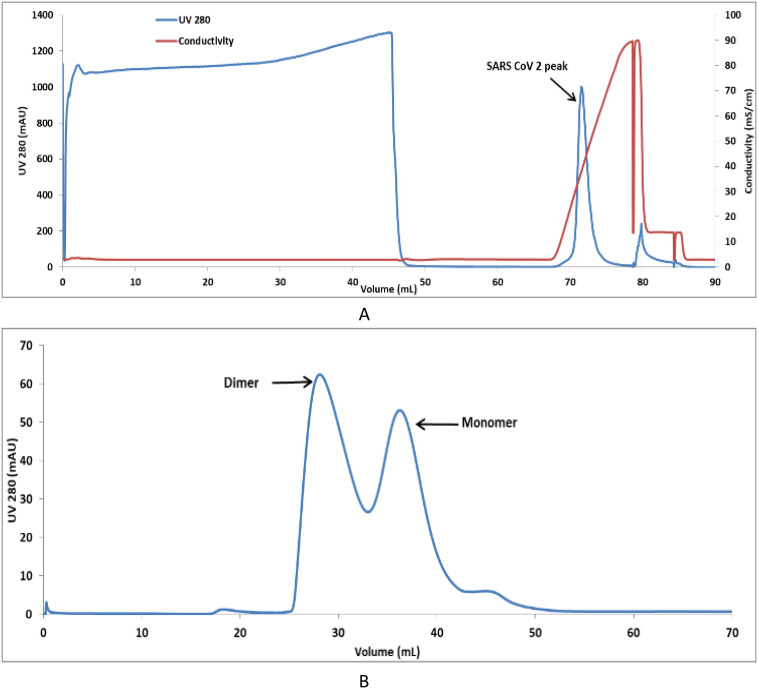
The inclusion bodies were solubilized, and the protein was captured using SP Sepharose FF resin and purified using CEX chromatography (Fig. 2 ). SARS-CoV-2 N protein of more than 95% purity was thus obtained (Fig. 1c) and confirmed with immunoblotting (Fig. 1d). The protein was conformed with in gel trypsin digestion followed by Liquid Chromatography with mass spectrometry (LC-MS) (Supplementary Fig. 2) and SEC-MALS (Supplementary Fig. 3) for protein molecular size determination.
Fig. 2.
Chromatogram of CEX chromatography (a) and preparative SEC (b) of Nucleocapsid protein of SARS-CoV-2.
Further, preparative SEC was performed to obtain fractions containing 5%, 10%, 25%, 55%, and 75% dimer (Fig. 3 ). Since it is impossible to distinguish the complete dimer from the monomer, fractions with the greatest possible dimer content were used in this analysis. N protein monomer and dimer rich pools were used for structural characterization and determination of ELISA sensitivity.
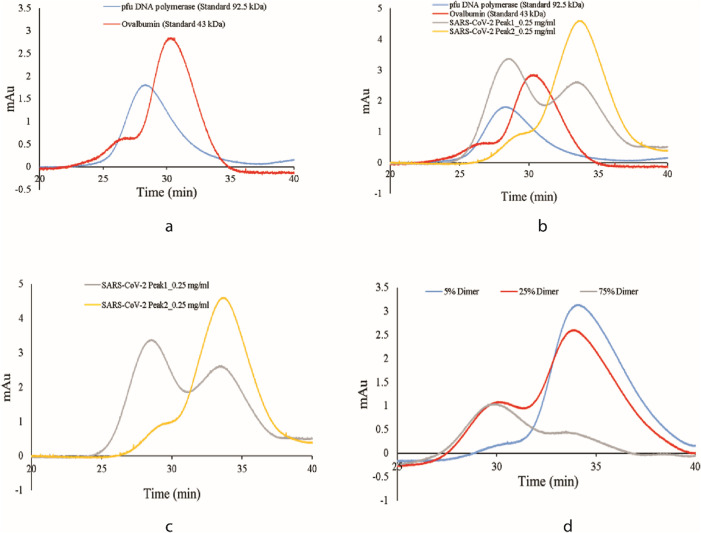
Fig. 3.
Chromatogram of analytical SEC (a) Standard protein of size 43 kDa and 92.5 kDa (b) overlay of purified N and standard protein (c) Purified N protein alone (d) Purified N protein of different fraction of dimer (10%, 25% and 75%).
3.2. Characterization of SARS-CoV-2 N protein
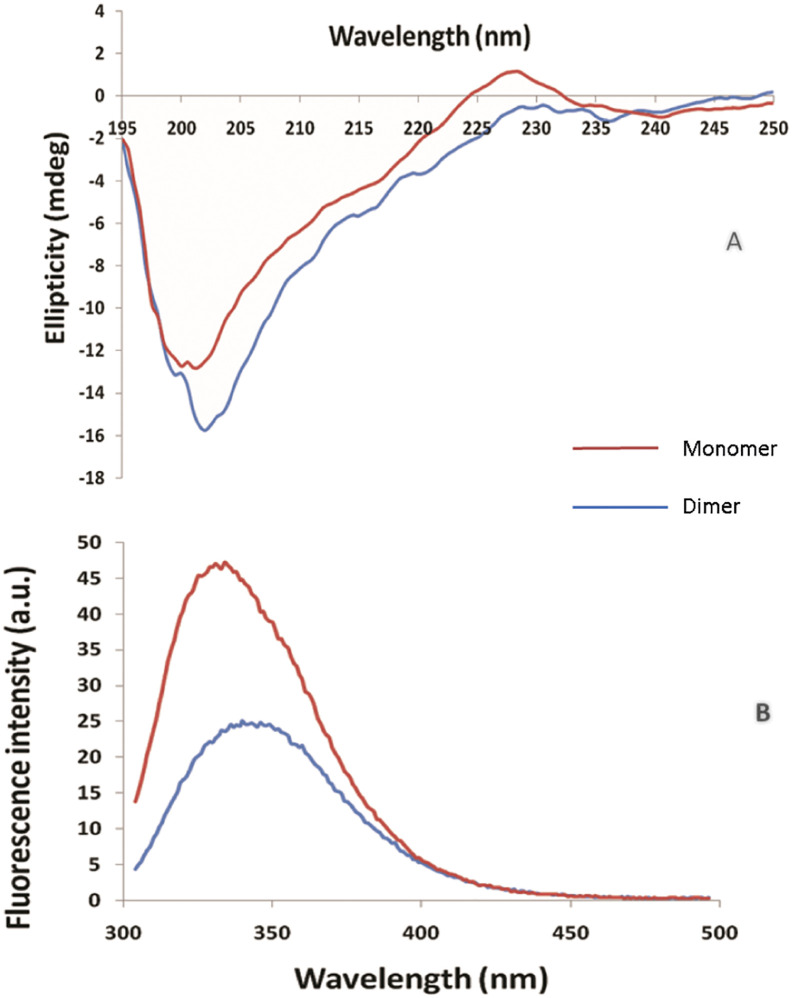
Purified SARS-CoV-2 N protein was analysed by SDS-PAGE, the band was excised and digested with trypsin. The proteolytic digested peptides were analysed using LC-MS. The sequence coverage of the digested protein showed 85.5% sequence coverage in comparison to in silico digested protein (Supplementary Fig. 2). SEC-MALS and analytical SEC of the N protein showed the monomer and dimer fractions to have molecular masses of 51.38 kDa and 108 kDa respectively (Supplementary Fig. 3). Further, purified SARS-CoV-2 protein was characterized for secondary structure by CD spectroscopy (Fig. 4a). It was observed that SARS-CoV-2 mainly consists of random coils as shown by the negative band at ~200 nm, which is consistent with reports in literature [45]. As is evident from data presented in Fig. 4b, both the monomer and dimer primarily consist of random coils. In dimer form, there is an increase in ellipticity at 218 nm, as well as a red shift in the negative band from 200 nm to 202 nm, suggesting an improved secondary structure due to oligomerization. Conformational state of SARS-CoV-2 was estimated by fluorescence spectroscopy with tryptophan excitation at 285 nm and emission in the range of 300–500 nm. Fluorescence spectra shows λmax of ~334 nm (Fig. 4b), indicating that the native structure of SARS-CoV-2 protein is similar to that reported for the SARS-CoV N protein [22]. Fluorescence spectra of dimer fraction exhibited a red shift in λmax from 334 nm to 340 nm, indicating exposure of the buried tryptophan and resulting in oligomerization of the protein.
Fig. 4.
Far-UV Circular Dichroism spectra of N protein of SARS-CoV-2 at 0.25 mg/ml concentration (a). Fluorescence spectra of N protein of SARS-CoV-2 at 0.25 mg/ml concentration (b).
3.3. Modeling of the structure of the monomeric/dimeric forms
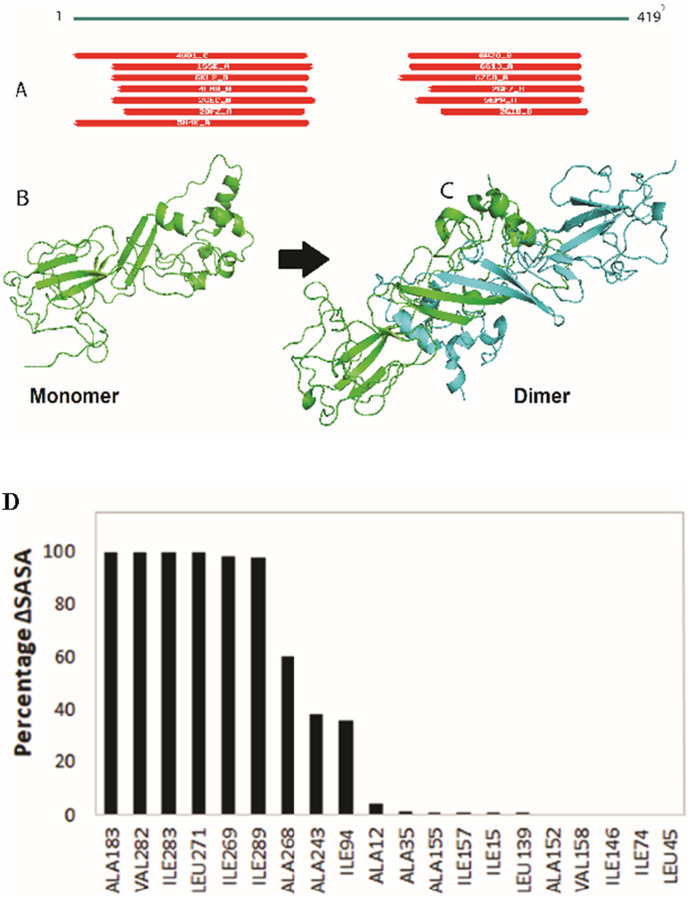
The SARS-CoV-2 N protein sequence was retrieved from the Uniprot database. It comprises of 419 amino acid residues. The current experimental structure has 30–40% of these residues, rendering it as the only structure for the viral N protein that has been uncovered by X-ray crystallography [46], [47], [48], [49]. Sequence alignment showed different potential templates covering the various segments of the protein. Fig. 5a shows the coverage of SARS-CoV-2 N protein sequence by different proteins in the sequence alignment. MERS CoV nucleocapsid (PDB: 4UD1) aligned best with the query sequence with 1e-58 E-value and covers 14-164 sequence. Fragment length 244-364 amino acids for SARS-CoV-2 N protein was experimentally solved and used as a second template (PDB: 6WZO). Selected templates (4UD1 and 6WZO) were used in modeller to build the model. This modelled the structure of SARS-CoV-2 N protein in its monomeric form as shown in Fig. 5b. Later, this monomeric form was superimposed with the partial experimental structure of the dimerization domain of SARS-CoV-2 N protein (6WZO). Transposing the two units of modelled monomers to the dimerization domain resulted in formation of the dimeric form of the protein as shown in Fig. 5c.
Fig. 5.
Structure modeling of SARS-CoV-2 N protein. (A) Sequence alignment of SARS-CoV-2 N protein with known structures. (B) monomer modelled structure built on 4UD1 and 6WZO templates. (C) dimeric form of protein built using 6WZO dimerization domain. (D) Percentage change in SASA for hydrophobic residues between monomer and dimer form of SARS-CoV-2 N protein.
3.4. Calculation of the solvent accessible surface area (SASA)
SASA was calculated for each residue in monomer and dimer form. This indicates the amount of area for a residue that is exposed to the solvent. Hydrophobic residues do not prefer polar environment and thus bury themselves in the native structure of the protein. Hydrophobic residues that show>50 Å2 SASA value in monomer indicate probable surface instability. These residues were marked and their corresponding percentage change in dimeric form was calculated. Fig. 5d shows the percentage change of these hydrophobic residues between the monomer and the dimer. Six such residues, namely A183, V182, I283, L271, I269, and I289, changed from completely exposed to buried state in dimeric form. Moreover, A268 buried by 60% while A243 and I194 buried by ≈ 35% compared to their monomeric conformation. This indicates that dimerization of protein helped to bury these hydrophobic residues at the interface that can stabilize the protein in the solution.
3.5. Prediction of epitopes
The 3D structure of a protein can be used to predict discontinuous epitopes. These epitopes are formed due to specific conformation of protein residues at the surface. To classify these discontinuous epitopes, many methods are used to evaluate monomer and dimer. Ellipro predicted 3 epitopic sites on the monomer surface and 6 epitopes for the dimer form. These 6 predicted epitopes for the dimer SARS-CoV-2 N are the duplicate of its corresponding monomer and hence improve the chance of antibody binding.
Among the predicted discontinuous 3D epitopes, patch 1:“R41, P42, Q43, G44, L45, P46, N47, N48, T49, A50, S51, W52, F53, T54, A55, E62, D63, L64, K65, F66, P67, G69, Q70, G71, V72, P73, I74, N75, T76, N77, S78, S79, P80, D81, D82, Q83, I84, Y112, L113, G114, T115, P122, Y123, G124, A125, V133, A134, T135, E136, G137, A138, L139, N140, T141, P142, K143, D144, H145, I146, G147, T148, R149, N150, P151, A152, N153, N154, A155, A156, I157, V158, L159, Q160, L161, P162, Q163, G164, T165, T166, L167, P168, K169, Y172, A173, E174, G175, Q176, T177, T257, P258, S259, G260” has the maximum score and it is present in duplicate for the dimeric form. A complete list of the epitopes predicted by Ellipro is given in Supplementary Table S1. Further, a similar analysis was performed with the DiscoTope server, which predicted the probability of each residue to be part of an epitope. It predicted 110 residues at the epitopic site at the DiscoTope threshold score ‘0’. However, dimer has 238 B-cell epitope residues out of 576 residues. A list of the predicted epitope residues is shown in Supplementary Table S2. Both the servers suggested that dimer has a greater number of structural epitopes and may have more affinity for antibodies.
3.6. Enzyme-linked immunosorbent assay
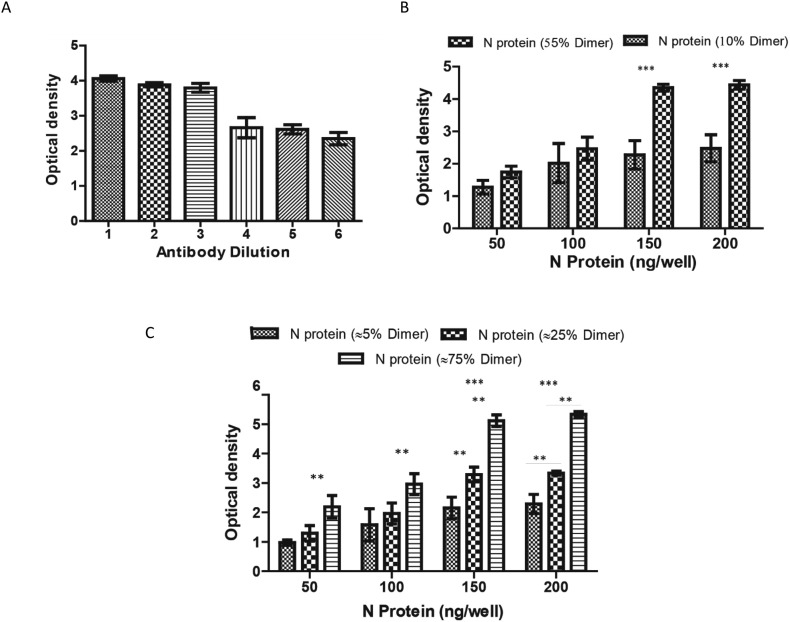
Antigenic response of the monomeric and dimeric form of the N protein was evaluated through indirect ELISA using monoclonal antibody targeted against nucleocapsid of SARS-CoV-2. In order to improve the assay performance, a systemic perusal of each step of the ELISA was performed. Variables such as antigen-coating amount and primary antibody dilution were optimised. Various quantities of N protein monomer and dimer fractions (50–200 ng well−1) were coated on the 96-well polystyrene plate. As illustrated in Fig. 6b, at a protein concentration of 150 ng well−1, dimeric fraction of the N protein was found highly sensitive for anti-N IgG detection. An optimum primary antibody dilution was investigated for N protein specific antibody after antigen coating. Six two-fold dilutions (1:1000, 1:2000, 1:3000, 1:4000, 1:5000 to 1:6000) of primary antibody were used for the detection of coated N-protein monomer and dimer. The detectability of the primary antibody was optimum at 1:3000 highest dilution as shown in Fig. 6a. This dilution will be kept for assay development. Owing to the superior antigenicity of the dimeric form of the N protein, impact of increasing percentage of dimer in the solution on its immunogenicity was further examined. Samples containing different percentages of N protein dimers (10–75%) were coated on the polystyrene plate and were detected by primary antibody (anti-N IgG, 1:3000). It can be inferred from Fig. 6b and c that as the percentage of the dimers increased in the solution, the sensitivity of the assay increased as well (p value <0.05).
Fig. 6.
SARS-CoV-2 N protein based indirect ELISA. (a) Development of an assay for dilution of commercial antibodies against the SARS-CoV-2 N protein (b) The effect of N protein fractions comprising 10% and 55% dimer on assay performance (c) Effect of different percentages of N protein dimer on assay performance, including 5%, 25%, and 75%.
3.7. Application in the diagnosis of COVID-19
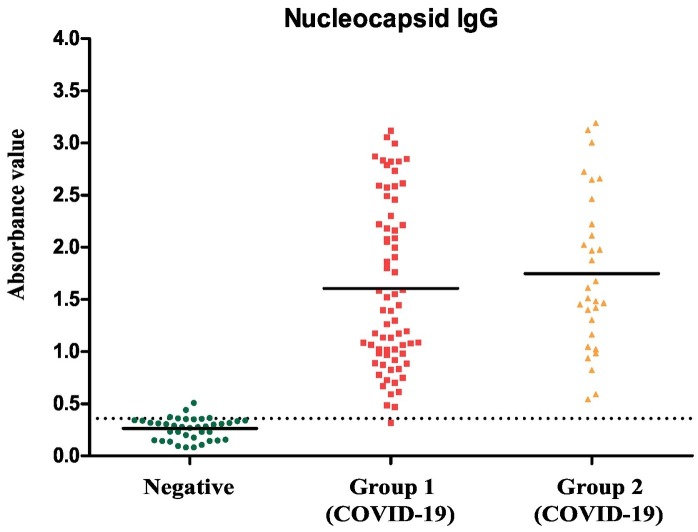
Anti-SARS-CoV-2 antibodies were evaluated in patient's samples using standardised ELISA (Fig. 7 ). The test accuracy as a clinical diagnostic tool was shown to have a high rate of sensitivity and specificity of 99.0% and 95%, respectively.
Fig. 7.
Indirect ELISA validation with human sample: The developed ELISA was used to detect IgG antibodies against SARS-CoV-2 N protein in healthy sample (n = 40) and COVID-19 patients collected in two groups, group 1 (n = 70) and group 2 (n = 30). The dotted lines show the assay cut-offs.
4. Discussion
A plausible approach to combat pandemic caused by SARS-CoV-2 is to improve the sensitivity of the existing diagnostics. N protein of the SARS-CoV-2 is an important candidate for the development of various diagnosis assays of the COVID-19. This work presents a reliable and sensitive ELISA based diagnostic assay for the detection of antibodies against N protein of SARS-CoV-2. The SARS-CoV-2 N protein antibody is more sensitive than the spike protein antibody for ELISA based identification of previous infections [50]. N protein is a highly immunogenic and generously expressed protein during infection of SARS-CoV-2. High levels of anti-N protein antibodies have been detected in sera in patients with prior infection of SARS-CoV-2. SARS-CoV-2 N protein is a highly basic protein with a pI of 10 [51]. The nucleocapsid is a multifunctional protein that interacts with RNA and other membrane proteins during virus assembly. Researchers have reported that homodimers of the full-length N protein are the fundamental unit of the ribonucleoprotein complex [52].
In this study, we purified monomeric and dimeric form of the nucleocapsid protein. The dimer and monomer ratio did not change in the concentration range under consideration. N proteins were subjected to denaturation using urea, so as to break any association that may be present between monomers of the protein. After purification using cation exchange chromatography, the N protein tends to dimerize at low concentration (1.5 mg/ml) and polymerize at higher concentration. It is the polymerization of monomeric protein through non-covalent bond (hydrophobic interaction between residues of dimerization domain) as SARS-CoV-2 N protein lacks the disulfide bond. Formation of the N protein dimer occurs in the solution after purification in the absence of genomic material. Samples rich in monomeric or dimeric forms were used to investigate the antigenic sensitivity to the SARS-CoV-2 nucleocapsid. The highest-grade dimer fraction of the N protein demonstrated high sensitivity and a wider dynamic range for antibody detection. Later, we investigated the phenomenon of high sensitivity of dimer using computational approach. Structures of N protein were modelled in their monomeric and dimeric forms. Solvent accessibility of hydrophobic residues was found to be lower in the dimeric form, thereby indicating better surface stability for dimer in polar solution. Additionally, 3D epitopes were predicted to find the potential of binding of the monomeric and the dimeric species with the antibody. Dimer species exhibited double the number of epitopes compared to the monomer, thereby enhancing the chances to interact with the antibody, an observation supported by experimental data. The IgG humoral response in COVID-19 and healthy individuals was assessed using this standardised ELISA, and a promising approach for improving sensitivity and specificity was developed. This is the first of its kind study, elucidating the impact of dimerization of SARS-CoV-2 nucleocapsid protein on sensitivity of enzyme-linked immunosorbent assay (ELISA) based diagnostics of COVID-19. The optical density calculation in an ELISA assay improved when a high proportion of the dimeric fraction was used as antigen. Thus, further modification of existing assays for the detection of SARS-CoV-2 antibodies and use of a high proportion of full-length nucleocapsid fragment dimer can further enhance the sensitivity of the existing rapid kit and ELISA assay.
CRediT authorship contribution statement
A.S.R., W.H.K., R.B. designed the study. W.H.K., N.K., S.G., V.B., and D.M performed the experiments. W.H.K., N.K., S.G, V.B., A.M., D.M. and R.B. analysed and interpreted data. A.M. performed the bioinformatics work. MAA and SD provide the patient's sample. W.H.K., N.K., S.G., V.B., and A.M., wrote the original manuscript. A.S.R. supervised the project and was the recipient of the funding. All authors were involved in the drafting, review, and approval of the report and the decision to submit for publication.
Declaration of competing interest
The authors declare that they do not have any competing interests.
Acknowledgments
This work was funded by a Centre of Excellence for Biopharmaceutical Technology grant from the Department of Biotechnology, Government of India (BT/COE/34/SP15097/2015) and CSR funding from Microsoft Corporation.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ijbiomac.2022.01.094.
Appendix A. Supplementary data
Supplementary material
Data availability
Data will be made available on request.
References
- 1.Iyer M., Jayaramayya K., Subramaniam M.D., Lee S.B., Dayem A.A., Cho S.G., Vellingiri B. COVID-19: an update on diagnostic and therapeutic approaches. BMB Rep, 2020;53(4):191–205. doi: 10.5483/BMBRep.2020.53.4.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kumar V., Doshi K.U., Khan W.H., Rathore A.S. COVID-19 pandemic: mechanism, diagnosis, and treatment. J. Chem. Technol. Biotechnol. 2021;96(2):299–308. [Google Scholar]
- 3.Rathore A.S. Covid 19 – pandemic in India. J. Chem. Technol. Biotechnol. 2020;95(7):1841. doi: 10.1002/jctb.6470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Khan W.H., Hashmi Z., Goel A., Ahmad R., Gupta K., Khan N., Alam I., Ahmed F., Ansari M.A. COVID-19 pandemic and vaccines update on challenges and resolutions. Front. Cell. Infect. Microbiol. 2021;11 doi: 10.3389/fcimb.2021.690621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mishra A., Khan W.H., Rathore A.S. Synergistic effects of natural compounds toward inhibition of SARS-CoV-2 3CL protease. J. Chem. Inf. Model. 2021;61(11):5708–5718. doi: 10.1021/acs.jcim.1c00994. [DOI] [PubMed] [Google Scholar]
- 6.Mahony J.B. Detection of respiratory viruses by molecular methods. Clin. Microbiol. Rev. 2008;21(4):716–747. doi: 10.1128/CMR.00037-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cai Q., Mu J., Lei Y., Ge J., Aryee A.A., Zhang X., Li Z. Simultaneous detection of the spike and nucleocapsid proteins from SARS-CoV-2 based on ultrasensitive single molecule assays. Anal. Bioanal. Chem. 2021;413(18):4645–4654. doi: 10.1007/s00216-021-03435-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tehrani Z.R., Saadat S., Saleh E., Ouyang X., Constantine N., DeVico A.L., Harris A.D., Lewis G.K., Kottilil S., Sajadi M.M. medRxiv; 2020. Specificity and Performance of Nucleocapsid and Spike-based SARS-CoV-2 Serologic Assays. 2020.08.05.20168476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yuen R., Steiner D., Pihl R., Chavez E., Olson A., Baird L., Korkmaz F., Urick P., Sagar M., Berrigan J., Gummuluru R., Corley R., Quillen K., Belkina A., Mostoslavsky G., Rifkin I., Kataria Y., Cappione A., Lin N., Bhadelia N., Snyder-Cappione J. medRxiv; 2020. SARS-CoV-2 Reactive Antibodies in Unexposed Individuals Revealed by a High Sensitivity, Low Noise Serologic Assay. 2020.09.15.20192765. [Google Scholar]
- 10.Yüce M., Filiztekin E., Özkaya K.G. COVID-19 diagnosis -a review of current methods. Biosens. Bioelectron. 2021;172 doi: 10.1016/j.bios.2020.112752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Satarker S., Nampoothiri M. Structural proteins in severe acute respiratory syndrome coronavirus-2. Arch. Med. Res. 2020;51(6):482–491. doi: 10.1016/j.arcmed.2020.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chang C.K., Hou M.H., Chang C.F., Hsiao C.D., Huang T.H. The SARS coronavirus nucleocapsid protein–forms and functions. Antivir. Res. 2014;103:39–50. doi: 10.1016/j.antiviral.2013.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Satarker S., Nampoothiri M. Structural proteins in severe acute respiratory syndrome coronavirus-2. Arch. Med. Res. 2020;51(6):482–491. doi: 10.1016/j.arcmed.2020.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carlson C.R., Asfaha J.B., Ghent C.M., Howard C.J., Hartooni N., Safari M., Frankel A.D., Morgan D.O. Phosphoregulation of phase separation by the SARS-CoV-2 N protein suggests a biophysical basis for its dual functions. Mol. Cell. 2020;80(6):1092–1103.e4. doi: 10.1016/j.molcel.2020.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cubuk J., Alston J.J., Incicco J.J., Singh S., Stuchell-Brereton M.D., Ward M.D., Zimmerman M.I., Vithani N., Griffith D., Wagoner J.A., Bowman G.R., Hall K.B., Soranno A., Holehouse A.S. The SARS-CoV-2 nucleocapsid protein is dynamic, disordered, and phase separates with RNA. Nat. Commun. 2021;12(1):1936. doi: 10.1038/s41467-021-21953-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu C., Qavi A.J., Hachim A., Kavian N., Cole A.R., Moyle A.B., Wagner N.D., Sweeney-Gibbons J., Rohrs H.W., Gross M.L., Peiris J.S.M., Basler C.F., Farnsworth C.W., Valkenburg S.A., Amarasinghe G.K., Leung D.W. bioRxiv; 2020. Characterization of SARS-CoV-2 N Protein Reveals Multiple Functional Consequences of the C-terminal Domain. 2020.11.30.404905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen C.Y., Chang C.K., Chang Y.W., Sue S.C., Bai H.I., Riang L., Hsiao C.D., Huang T.H. Structure of the SARS coronavirus nucleocapsid protein RNA-binding dimerization domain suggests a mechanism for helical packaging of viral RNA. J. Mol. Biol. 2007;368(4):1075–1086. doi: 10.1016/j.jmb.2007.02.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jayaram H., Fan H., Bowman B.R., Ooi A., Jayaram J., Collisson E.W., Lescar J., Prasad B.V.V. X-ray structures of the N- and C-terminal domains of a coronavirus nucleocapsid protein: implications for nucleocapsid formation. J. Virol. 2006;80(13):6612–6620. doi: 10.1128/JVI.00157-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Takeda M., Chang C.K., Ikeya T., Güntert P., Chang Y.H., Hsu Y.L., Huang T.H., Kainosho M. Solution structure of the c-terminal dimerization domain of SARS coronavirus nucleocapsid protein solved by the SAIL-NMR method. J. Mol. Biol. 2008;380(4):608–622. doi: 10.1016/j.jmb.2007.11.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu I.M., Oldham M.L., Zhang J., Chen J. Crystal structure of the severe acute respiratory syndrome (SARS) coronavirus nucleocapsid protein dimerization domain reveals evolutionary linkage between corona- and arteriviridae. J. Biol. Chem. 2006;281(25):17134–17139. doi: 10.1074/jbc.M602107200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nguyen T.H.V., Lichière J., Canard B., Papageorgiou N., Attoumani S., Ferron F., Coutard B. Structure and oligomerization state of the C-terminal region of the Middle East respiratory syndrome coronavirus nucleoprotein. acta crystallographicaSection D, Structural biology. 2019;75(Pt 1):8–15. doi: 10.1107/S2059798318014948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luo H., Chen J., Chen K., Shen X., Jiang H. Carboxyl terminus of severe acute respiratory syndrome coronavirus nucleocapsid protein: self-association analysis and nucleic acid binding characterization. Biochemistry. 2006;45(39):11827–11835. doi: 10.1021/bi0609319. [DOI] [PubMed] [Google Scholar]
- 23.Sabbih G.O., Korsah M.A., Jeevanandam J., Danquah M.K. Biophysical analysis of SARS-CoV-2 transmission and theranostic development via N protein computational characterization. Biotechnol. Prog. 2020;37(2):1–14. doi: 10.1002/btpr.3096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Surjit M., Liu B., Kumar P., Chow V.T.K., Lal S.K. The nucleocapsid protein of the SARS coronavirus is capable of self-association through a C-terminal 209 amino acid interaction domain. Biochem. Biophys. Res. Commun. 2004;317(4):1030–1036. doi: 10.1016/j.bbrc.2004.03.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luo H., Ye F., Sun T., Yue L., Peng S., Chen J., Li G., Du Y., Xie Y., Yang Y. In vitro biochemical and thermodynamic characterization of nucleocapsid protein of SARS. Biophys. Chem. 2004;112(1):15–25. doi: 10.1016/j.bpc.2004.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Iserman C., Roden C.A., Boerneke M.A., Sealfon R.S.G., McLaughlin G.A., Jungreis I., Fritch E.J., Hou Y.J., Ekena J., Weidmann C.A., Theesfeld C.L., Kellis M., Troyanskaya O.G., Baric R.S., Sheahan T.P., Weeks K.M., Gladfelter A.S. Genomic RNA elements drive phase separation of the SARS-CoV-2 nucleocapsid. Mol. Cell. 2020;80(6):1078–1091.e6. doi: 10.1016/j.molcel.2020.11.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ye Q., West A.M., Silletti S., Corbett K.D. Architecture and self-assembly of the SARS-CoV-2 nucleocapsid protein. Protein Sci. 2020;29(9):1890–1901. doi: 10.1002/pro.3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huergo L.F., Conzentino M.S., Gerhardt E.C., Santos A.R., de Oliveira Pedrosa F., Souza E.M., Nogueira M.B., Forchhammer K., Rego F.G., Raboni S.M. medRxiv; 2020. Magnetic Bead-based ELISA Allow Inexpensive, Rapid and Quantitative Detection of Human Antibodies Against SARS-CoV-2. [Google Scholar]
- 29.Timani K.A., Ye L., Ye L., Zhu Y., Wu Z., Gong Z. Cloning, sequencing, expression, and purification of SARS-associated coronavirus nucleocapsid protein for serodiagnosis of SARS. J. Clin. Virol. 2004;30(4):309–312. doi: 10.1016/j.jcv.2004.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dutta N.K., Mazumdar K., Lee B.H., Baek M.W., Kim D.J., Na Y.R., Park S.H., Lee H.K., Kariwa H., Mai le Q., Park J.H. Search for potential target site of nucleocapsid gene for the design of an epitope-based SARS DNA vaccine. Immunol. Lett. 2008;118(1):65–71. doi: 10.1016/j.imlet.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu C., Qavi A.J., Hachim A., Kavian N., Cole A.R., Moyle A.B., Wagner N.D., Sweeney-Gibbons J., Rohrs H.W., Gross M.L., Peiris J.S.M., Basler C.F., Farnsworth C.W., Valkenburg S.A., Amarasinghe G.K., Leung D.W. bioRxiv; 2020. Characterization of SARS-CoV-2 N Protein Reveals Multiple Functional Consequences of the C-terminal Domain. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bonelli F., Sarasini A., Zierold C., Calleri M., Bonetti A., Vismara C., Blocki F.A., Pallavicini L., Chinali A., Campisi D., Percivalle E., DiNapoli A.P., Perno C.F., Baldanti F. Clinical and analytical performance of an automated serological test that identifies S1/S2-neutralizing IgG in COVID-19 patients semiquantitatively. J. Clin. Microbiol. 2020;58(9) doi: 10.1128/JCM.01224-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Diego-Martin B., González B., Vazquez-Vilar M., Selma S., Mateos-Fernández R., Gianoglio S., Fernández-del-Carmen A., Orzáez D. bioRxiv; 2020. Pilot Production of SARS-CoV-2 Related Proteins in Plants: A Proof of Concept for Rapid Repurposing of Indoors Farms Into Biomanufacturing Facilities. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee H.K., Lee B.H., Dutta N.K., Seok S.H., Baek M.W., Lee H.Y., Kim D.J., Na Y.R., Noh K.J., Park S.H., Kariwa H., Nakauchi M., Mai le Q., Heo S.J., Park J.H. Detection of antibodies against SARS-Coronavirus using recombinant truncated nucleocapsid proteins by ELISA. J. Microbiol. Biotechnol. 2008;18(10):1717–1721. [PubMed] [Google Scholar]
- 35.Liu W., Liu L., Kou G., Zheng Y., Ding Y., Ni W., Wang Q., Tan L., Wu W., Tang S., Xiong Z., Zheng S. Evaluation of nucleocapsid and spike protein-based enzyme-linked immunosorbent assays for detecting antibodies against SARS-CoV-2. J. Clin. Microbiol. 2020;58(6) doi: 10.1128/JCM.00461-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Makatsa M.S., Tincho M.B., Wendoh J.M., Ismail S.D., Nesamari R., Pera F., de Beer S., David A., Jugwanth S., Gededzha M.P., Mampeule N., Sanne I., Stevens W., Scott L., Blackburn J., Mayne E.S., Keeton R.S., Burgers W.A. medRxiv; 2020. SARS-CoV-2 Antigens Expressed in Plants Detect Antibody Responses in COVID-19 Patients. 2020.08.04.20167940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mazzini L., Martinuzzi D., Hyseni I., Lapini G., Benincasa L., Piu P., Trombetta C.M., Marchi S., Razzano I., Manenti A., Montomoli E. bioRxiv; 2020. Comparative Analyses of SARS-CoV-2 Binding (IgG, IgM, IgA) and Neutralizing Antibodies From Human Serum Samples. 2020.08.10.243717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yokoyama R., Kurano M., Morita Y., Shimura T., Nakano Y., Qian C., Xia F., He F., Kishi Y., Okada J., Yoshikawa N., Nagura Y., Okazaki H., Moriya K., Seto Y., Kodama T., Yatomi Y. medRxiv; 2020. Validation of a New Automated Chemiluminescent Anti-SARS-CoV-2 IgM and IgG Antibody Assay System Detecting Both N and S Proteins in Japan. 2020.07.16.20155796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu L., Liu W., Zheng Y., Jiang X., Kou G., Ding J., Wang Q., Huang Q., Ding Y., Ni W., Wu W., Tang S., Tan L., Hu Z., Xu W., Zhang Y., Zhang B., Tang Z., Zhang X., Li H., Rao Z., Jiang H., Ren X., Wang S., Zheng S. A preliminary study on serological assay for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in 238 admitted hospital patients. Microbes Infect. 2020;22(4–5):206–211. doi: 10.1016/j.micinf.2020.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Prince H.E., Givens T.S., Lapé-Nixon M., Clarke N.J., Schwab D.A., Batterman H.J., Jones R.S., Meyer W.A., Kapoor H., Rowland C.M., Haji-Sheikhi F., Marlowe E.M. Detection of SARS-CoV-2 IgG targeting nucleocapsid or spike protein by four high-throughput immunoassays authorized for emergency use. J. Clin. Microbiol. 2020;58(11) doi: 10.1128/JCM.01742-20. e01742-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shevchenko A., Tomas H., Havliš J., Olsen J.V., Mann M. Nat. Protoc. 2007;1(6):2856. doi: 10.1038/nprot.2006.468. [DOI] [PubMed] [Google Scholar]
- 42.Söding J., Biegert A., Lupas A.N. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005;33(suppl_2):W244–W248. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Webb B., Sali A. Comparative protein structure modeling using MODELLER. Curr. Protoc. Bioinformatics. 2016;54(1):5.6.1–5.6.37. doi: 10.1002/cpbi.3. 2020.09.15.20192765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ponomarenko J., Bui H.-H., Li W., Fusseder N., Bourne P.E., Sette A., Peters B. ElliPro: a new structure-based tool for the prediction of antibody epitopes. BMC Bioinformatics. 2008;9(1):1–8. doi: 10.1186/1471-2105-9-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zeng W., Liu G., Ma H., Zhao D., Yang Y., Liu M., Mohammed A., Zhao C., Yang Y., Xie J. Biochemical characterization of SARS-CoV-2 nucleocapsid protein. Biochem. Biophys. Res. Commun. 2020;527(3):618–623. doi: 10.1016/j.bbrc.2020.04.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zinzula L., Basquin J., Bohn S., Beck F., Klumpe S., Pfeifer G., Nagy I., Bracher A., Hartl F.U., Baumeister W. High-resolution structure and biophysical characterization of the nucleocapsid phosphoprotein dimerization domain from the Covid-19 severe acute respiratory syndrome coronavirus 2. Biochem. Biophys. Res. Commun. 2021;538:54–62. doi: 10.1016/j.bbrc.2020.09.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang M., He S., Chen X., Huang Z., Zhou Z., Zhou Z., Chen Q., Chen S., Kang S. Structural insight into the SARS-CoV-2 nucleocapsid protein C-terminal domain reveals a novel recognition mechanism for viral transcriptional regulatory sequences. Front. Chem. 2020;8 doi: 10.3389/fchem.2020.624765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ye Q., West A.M.V., Silletti S., Corbett K.D. Architecture and self-assembly of the SARS-CoV-2 nucleocapsid protein. Protein Sci. 2020;29(9):1890–1901. doi: 10.1002/pro.3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhou R., Zeng R., von Brunn A., Lei J. Structural characterization of the C-terminal domain of SARS-CoV-2 nucleocapsid protein. Mol.Biomed. 2020;1(1):2. doi: 10.1186/s43556-020-00001-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Burbelo P.D., Riedo F.X., Morishima C., Rawlings S., Smith D., Das S., Strich J.R., Chertow D.S., Davey R.T., Jr., Cohen J.I., Jr. medRxiv; 2020. Detection of Nucleocapsid Antibody to SARS-CoV-2 Is More Sensitive Than Antibody to Spike Protein in COVID-19 Patients. [Google Scholar]
- 51.Gao T., Gao Y., Liu X., Nie Z., Sun H., Lin K., Peng H., Wang S. Identification and functional analysis of the SARS-COV-2 nucleocapsid protein. BMC Microbiol. 2021;21(1):58. doi: 10.1186/s12866-021-02107-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lutomski C.A., El-Baba T.J., Bolla J.R., Robinson C.V. bioRxiv; 2020. Autoproteolytic Products of the SARS-CoV-2 Nucleocapsid Protein Are Primed for Antibody Evasion and Virus Proliferation. 2020.10.06.328112. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Data Availability Statement
Data will be made available on request.