Abstract
SARS-CoV-2 RNA can be detected in respiratory samples for weeks after onset of COVID-19 disease. Therefore, one of the diagnostic challenges of PCR positive cases is differentiating between acute COVID-19 disease and convalescent phase. The presence of SARS-CoV-2 nucleocapsid antigen in serum and plasma samples of COVID-19 patients has been demonstrated previously.
Our study aimed to characterize the analytical specificity and sensitivity of an enzyme-linked immunosorbent assay (Salocor SARS-CoV-2 Antigen Quantitative Assay Kit© (Salofa Ltd, Salo, Finland)) for the detection of SARS-CoV-2 nucleocapsid antigen in serum, and to characterize the kinetics of antigenemia. The evaluation material included a negative serum panel of 155 samples, and 126 serum samples from patients with PCR-confirmed COVID-19.
The specificity of the Salocor SARS-CoV-2 serum nucleocapsid antigen test was 98.0 %. In comparison with simultaneous positive PCR from upper respiratory tract (URT) specimens, the test sensitivity was 91.7 %. In a serum panel in which the earliest serum sample was collected two days before the collection of positive URT specimen, and the latest 48 days after (median 1 day post URT sample collection), the serum N antigen test sensitivity was 95.6 % within 14 days post onset of symptoms. The antigenemia resolved approximately two weeks after the onset of disease and diagnostic PCR.
The combination of simultaneous SARS-CoV-2 antigen and antibody testing appeared to provide useful information for timing of COVID-19. Our results suggest that SARS-CoV-2 N-antigenemia may be used as a diagnostic marker in acute COVID-19.
Keywords: SARS-CoV-2, COVID-19, Antigenemia, Enzyme immunoassay, Nucleocapsid
SARS-CoV-2 laboratory diagnostics rely primarily on molecular diagnostic techniques (Corman et al., 2020; Kubina and Dziedzic, 2020; Wiersinga et al., 2020). More recently, a variety of tests for SARS-CoV-2 antigen detection from upper respiratory tract (URT) specimens have established a complementary role (D’Cruz et al., 2020). While less sensitive, they benefit from being rapid, cheap, and performable outside of centralized laboratory facilities (Jääskeläinen et al., 2021).
Recent reports demonstrate SARS-CoV-2 nucleocapsid (N) antigenemia in COVID-19 patients (Deng et al., 2021; Hingrat et al., 2020; Lebedin et al., 2020; Shan et al., 2021; Thudium et al., 2021; Wang et al., 2021). While posing new questions on the pathophysiology of acute COVID-19, this may offer a novel diagnostic approach. Only limited information is available on the performance of serum antigen tests as a diagnostic method for SARS-CoV-2.
The aim of this study was to characterize the analytical specificity and sensitivity of an enzyme-linked immunosorbent assay (ELISA) for the detection of SARS-CoV-2 antigen in serum, namely Salocor SARS-CoV-2 Antigen Quantitative Assay Kit© (Salofa Ltd, Salo, Finland; later Salocor N-antigen ELISA) and to characterize the kinetics of antigenemia.
Serum samples were originally sent for diagnostic purposes to the Department of Virology and Immunology, Helsinki University Hospital Laboratory HUSLAB, Finland. Research permit HUS/157/2020 (Helsinki University Hospital, Finland) was obtained from the local review board.
The negative panel (N = 155; 148 cases) consisted of 144 serum samples collected in 2019, and 11 samples (positive for Aspergillus antigen) in 2020. Of the 155 specimens, 37 were sent for respiratory virus antibody testing; 32 samples were positive for anti-nuclear antibodies; 16 for phospholipase-A2-receptor antibodies; 8 for antineutrophil cytoplasmic (C-ANCA, P-ANCA and parallel C- and P-ANCA), and 4 for glomerular basement membrane antibodies. Two samples were from patients with Human coronavirus (HCoV) OC43 diagnosis (by PCR) and five with primary Epstein-Barr virus (EBV) infection. We also included serum specimens with a positive microbial antigen test as follows: 22 with Dengue virus NS1 antigen, 17 with hepatitis B virus surface antigen, 11 with Aspergillus antigen, and one with HIV p24 antigen. The median age of the negative panel cases was 53 years (range 2–89); 45 % were males (66/148).
There were two separate serum specimen panels from RT-PCR confirmed COVID-19 patients (panel A and panel B, both collected between 3–5/2020), who were previously SARS-CoV-2 RT-PCR positive from a URT specimen. They were also tested for SARS-CoV-2 IgG by both Abbott SARS-CoV-2 IgG (N antigen) and Euroimmun SARS-CoV-2 IgG (S1 antigen) according to manufacturer’s instructions.
Panel A comprised 70 serum samples from 62 cases (median age 54 years, range 24–86 years; 28/62 (45 %) males). The earliest serum sample was collected 2 days before the collection of the positive URT specimen, and the latest 48 days after (median 1 day post URT sample collection). The samples were previously tested with SARS-CoV-2 microneutralization (MNT) (Jääskeläinen et al., 2020). The date of onset of symptoms was available for 55/62 cases. Adjusted p-values for comparison of antigen and MNT result combinations were determined using Kruskal-Wallis test (GraphPad Prism 8.0.1).
Panel B comprised 56 serum samples from 27 cases (median age 50 years, range 27–65 years; 4/27 males); 2–4 consecutive serum samples from each. At least one serum from all 27 cases was SARS-CoV-2 IgG positive in both Abbott and Euroimmun tests. The earliest serum sample was collected 5 days before the collection of the positive URT specimen, and the latest 207 days after (median 101 days post collection). The date of onset of symptoms and MNT result were not available for this panel.
The PCR tests were carried out with one of the following methods: a laboratory-developed test based on Corman et al.; cobas® SARS-CoV-2 test kit on the cobas® 6800 system (Roche Diagnostics, Basel, Switzerland); and the Amplidiag® COVID-19 test on the Amplidiag® Easy platform (Mobidiag, Espoo, Finland). The performance of these tests in our laboratory is reported elsewhere (Mannonen et al., 2021).
The here-evaluated Salocor N-antigen ELISA (Salofa) is based on a double antibody sandwich ELISA test. The assay protocol is described in the Supplement. The assay cut-off is 2.97 pg/mL.
The receiver operating characteristic (ROC) curve and 95 % Clopper-Pearson confidence intervals were calculated for sensitivity and specificity (IBM SPSS statistical program package, version 25).
The specificity of the Salocor N antigen ELISA was 98.0 % (145/148) (Clopper-Pearson 95 % confidence interval 94.2–99.6 %) as determined by the negative panel. The three positive samples were collected in 2019 prior to circulation of SARS-CoV-2. One was from a patient with EBV primary infection (5 pg/mL). Two were originally sent for respiratory virus antibody screening; one with reported myocarditis (84.5 pg/mL) and one without any reported clinical outcome (4 pg/mL).
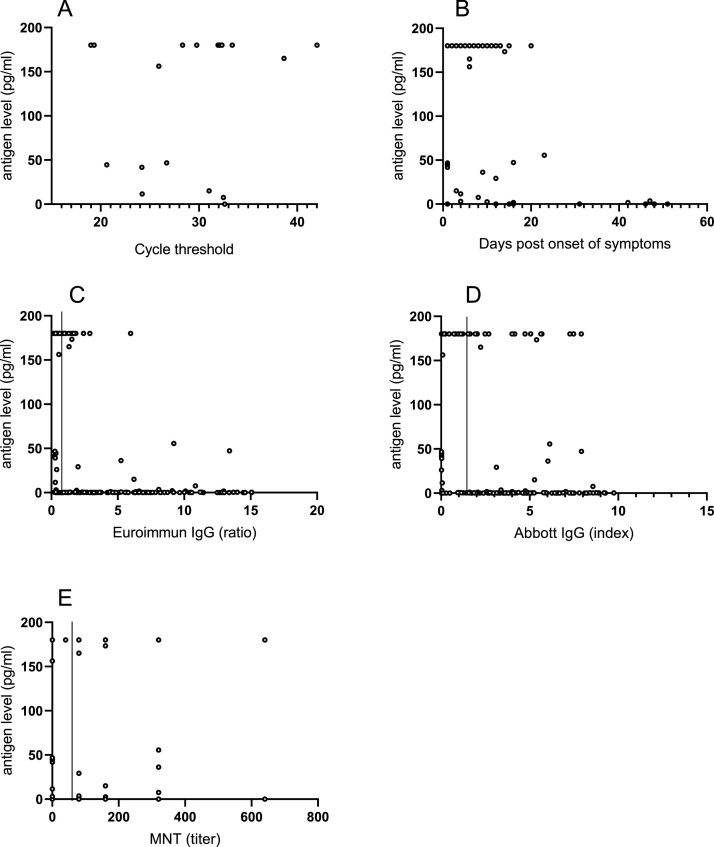
There was a simultaneous serum and PCR positive URT specimen available in 24 cases: N antigen was positive in 22/24, rendering 91.7 % sensitivity (Clopper-Pearson 95 % confidence interval 73.0−99.0%). The negative specimens were retrieved at one day and at 16 days post disease onset. The PCR cycle threshold values did not appear to associate with the N antigen concentrations (Fig. 1 a).
Fig. 1.
Serum antigen concentrations in relation to the timing of sampling and other tests. Serum antigen concentrations of >180 pg/mL are depicted as 180 pg/mL. A. Serum antigen concentrations compared to the Ct-value of a URT PCR sample obtained on the same day. B. Serum antigen concentrations relative to the timing of sampling after the onset of symptoms. C. Correlation of serum antigen concentration with the Euroimmun anti-S IgG assay results. The vertical line indicates the cut-off value of the Euroimmun assay D. Antigen concentrations in comparison with the. Abbott anti-N IgG assay results. The vertical line indicates the cut-off value of the Abbott assay E. Serum antigen concentrations relative to the microneutralization test titer. MNT titers of <40 (negative) are depicted as 0 and MNT titers of >2560 as 2560. Figure created using GraphPad Prism 8.0.1 software.
Using COVID-19 panel A, we calculated test sensitivity in relation to disease onset (Table 1 ). The sensitivity with specimens retrieved at ≤14 days post onset was 95.6 % and decreased to 50 % with specimens retrieved 15−21 days post onset (Table 1). The N antigen concentrations decreased over time from symptom onset (Fig. 1b). The furthest time point for a positive N antigen was observed at 47 days post disease onset (3.5 pg/mL) (Fig. 1b).
Table 1.
Sensitivity of serum antigen test in relation to time post symptom onset.
| Time post symptom onset (days) | Time post symptom onset (days, mean [SD]) | N | Sensitivity (% [CI 95 %]a) |
|---|---|---|---|
| ≤7 | 4.6 (2.3) | 26 | 96.2 (80.4−99.9) |
| 8−14 | 10.4 (1.7) | 24 | 91.7 (73.0−99.0) |
| ≤14 | 7.4 (3.5) | 45 | 95.6 (84.9−99.5) |
| 15−21 | 16.3 (1.9) | 6 | 50.0 (11.8−88.2) |
| ≤21 | 8.4 (4.4) | 51 | 90.2 (78.6−96.7) |
| ≥7 | 16.2 (12.5) | 41 | 75.6 (59.7−87.6) |
| ≥14 | 28.6 (14.9) | 14 | 42.9 (17.7−71.1) |
| ≥21 | 41.1 (10.3) | 7 | 28.6 (3.7−71.0) |
Clopper-Pearson confidence interval.
Area under the receiver operating characteristic (ROC) curve was 0.972 when samples collected ≤14 days post onset of symptoms were included in the analysis (data not shown).
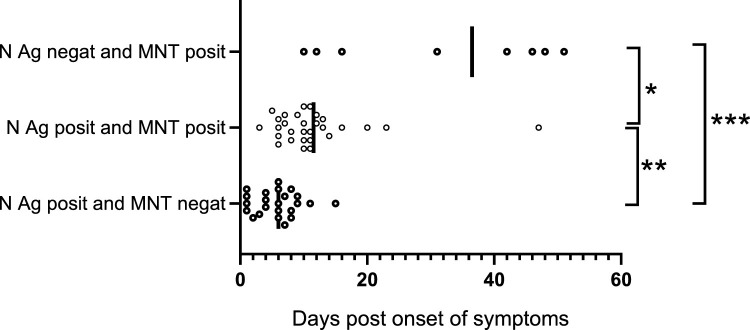
The median days from disease onset for subgroups in panel A were as follows: N antigen positive (≥2.97 pg/mL), MNT antibody negative (<40 titer) (6 days; 25 cases); N antigen positive, MNT antibody positive (10 days; 31 cases); N antigen negative, MNT antibody positive (37 days; 10 cases) (Fig. 2 ).
Fig. 2.
Simultaneous N antigen and MNT test results in relation to symptom onset. N antigen positive and MNT negative, median 6 days. N antigen positive, MNT positive, median 10 days. N antigen negative and MNT positive median. 37 days. Adjusted p-values (Kruskal-Wallis test): *p = 0.0317; **p 0.0015, and ***p=<0.0001. Figure created using GraphPad Prism 8.0.1 software.
There appeared to be a decreasing trend in N antigen concentration over increasing Euroimmun anti-S1 IgG test result (Fig. 1c), but no such trend was observed over increasing Abbott anti-N IgG test result (Fig. 1d), nor the increasing MNT titer (Fig. 1e).
Of the COVID-19 panel B, the only N antigen positive serum was retrieved on the date of positive PCR. All other 69 panel B serum specimens were N antigen negative; one was retrieved 5 days before positive PCR in URT, and the others between 12 and 207 days (median 107 days) after positive PCR in URT.
SARS-CoV-2 RNA can be detected for even 4–6 weeks post disease onset in URT samples (Hong et al., 2020; Kim et al., 2020). Therefore, one of the challenges in contact tracing of PCR positive cases is differentiating between acute COVID-19 disease and convalescent phase. The present study brings forward a potential tool to aid in timing of COVID-19 disease: detection of SARS-CoV-2N antigen in serum.
Some antigen tests for the detection of SARS-CoV have been described during and after the epidemic in 2002–2004 (Che et al., 2004; Cho et al., 2011). Previous reports on SARS-CoV-2 N antigenemia suggest a complementary role as a diagnostic test for COVID-19 during early infection (Shan et al., 2021; Thudium et al., 2021; Wang et al., 2021) and a prognostic role for disease severity has also been explored (Hingrat et al., 2020). We showed a very good specificity (98.0 %), and a reasonable sensitivity (91.7 %; in comparison with simultaneous positive PCR from URT specimens) for the Salocor N antigen test in line with previous reports. Higher sensitivity of up to 94–96 % can be achieved when patients are selected based on time of symptom onset, since SARS-CoV-2 N antigenemia appears to resolve within the first two weeks of illness. Particularly the combination of simultaneous SARS-CoV-2 antigen and antibody testing may provide useful information for timing of disease (Fig. 2). Both antibody and antigen tests can be performed from a single serum specimen in high throughput platforms. While this approach does not replace detection of SARS-CoV-2 RNA from URT, it may provide an additional aid.
Our study has limitations. Only a limited number of serum specimens was available with information on disease onset, particularly for the convalescent phase. This diminishes the ability of this study to provide a complete timeline for antigenemia in COVID-19. Information on disease severity was not available and could not be accounted for in our analysis. Potential cross-reactivity with seasonal coronaviruses was not assessed in our study. For the three false positive individuals, confirmatory tests for seasonal coronaviruses could not be performed, since no respiratory tract samples of these individuals were collected at the time of serum sampling.
More data are needed to clarify whether serum N antigen testing could be used to supplement current testing strategies for acute COVID-19. One interesting question is whether there is an association between antigenemia and viral shedding. Besides diagnostic differentiation between acute and convalescent COVID-19, N antigen testing from serum could potentially be deployed in epidemiological screening of asymptomatic infections, e.g. in recently vaccinated populations, which now becomes increasingly relevant.
Author contributions
MJA: Conceptualization, Investigation, Data curation, Writing – original draft, SKU: Conceptualization, Formal analysis, Resources, Writing – original draft, Writing – review and editing, Supervision, Project administration, SK: Investigation, Writing –review and editing, ML: Resources, Writing – review and editing, Supervision, Project administration HJ: Conceptualization, Formal analysis, Visualization, Resources, Writing – review and editing, Supervision, Project administration, AJJ: Conceptualization, Investigation, Formal analysis, Data curation, Writing – original draft, Supervision, Project administration.
Funding
This work has been supported by Helsinki University Hospital (TYH2019263 and TYH2021110).
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We would like to thank Marita Koivunen (HUS Diagnostic Center, HUSLAB, Helsinki, Finland), Eliisa Kekäläinen (Hus Diagnostic Center and University of Helsinki, Helsinki Finland) and Jesper Kivelä (Hus Diagnostic Center and University of Helsinki, Helsinki Finland).
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.jviromet.2022.114469.
Appendix A. Supplementary data
The following is Supplementary data to this article:
References
- Che X.-Y., Hao W., Wang Y., Di B., Yin K., Xu Y.-C., Feng C.-S., Wan Z.-Y., Cheng V.C.C., Yuen K.-Y. Nucleocapsid protein as early diagnostic marker for SARS. Emerg. Infect. Dis. 2004;10:1947–1949. doi: 10.3201/eid1011.040516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho S.-J., Woo H.-M., Kim K.-S., Oh J.-W., Jeong Y.-J. Novel system for detecting SARS coronavirus nucleocapsid protein using an ssDNA aptamer. J. Biosci. Bioeng. 2011;112:535–540. doi: 10.1016/j.jbiosc.2011.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K., Bleicker T., Brunink S., Schneider J., Schmidt M.L., Mulders D.G., Haagmans B.L., van der Veer B., van den Brink S., Wijsman L., Goderski G., Romette J.-L., Ellis J., Zambon M., Peiris M., Goossens H., Reusken C., Koopmans M.P., Drosten C. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. Bull. Eur. sur les Mal. Transm. = Eur. Commun. Dis. Bull. 2020;25:1. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Cruz R.J., Currier A.W., Sampson V.B. Laboratory testing methods for novel severe acute respiratory syndrome-coronavirus-2 (SARS-CoV-2) Front. Cell Dev. Biol. 2020;8:468. doi: 10.3389/fcell.2020.00468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Q., Ye G., Pan Y., Xie W., Yang G., Li Z., Li Y. High performance of SARS-Cov-2N protein antigen chemiluminescence immunoassay as frontline testing for acute phase COVID-19 diagnosis: a retrospective cohort study. Front. Med. 2021;8:1–8. doi: 10.3389/fmed.2021.676560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hingrat Q.L.E., Visseaux B., Laouenan C., Tubiana S., Bouadma L., Yazdanpanah Y., Duval X., Burdet C., Ichou H., Damond F., Bertine M., Benmalek N., Choquet C., Timsit J.-F., Ghosn J., Charpentier C., Descamps D., Houhou-Fidouh N., Diallo A., Le Mestre S., Mercier N., Paul C., Petrov-Sanchez V., Malvy D., Chirouze C., Andrejak C., Dubos F., Rossignol P., Picone O., Bompart F., Gigante T., Gilg M., Rossignol B., Levy-Marchal C., Beluze M., Hulot J.S., Bachelet D., Bhavsar K., Bouadma L., Chair A., Couffignal C., Da Silveira C., Debray M.P., Descamps D., Duval X., Eloy P., Esposito-Farese M., Ettalhaoui N., Gault N., Ghosn J., Gorenne I., Hoffmann I., Kafif O., Kali S., Khalil A., Laouénan C., Laribi S., Le M., Le Hingrat Q., Lescure F.-X., Lucet J.C., Mentré F., Mullaert J., Peiffer-Smadja N., Peytavin G., Roy C., Schneider M., Mohammed N.S., Tagherset L., Tardivon C., Tellier M.C., Timsit J.-F., Trioux T., Tubiana S., Visseaux B., Vanel N., Basmaci R., Angoulvant F., Kaguelidou F., Pages J., Tual C., Veislinger A., Couffin-Cardiergues S., Esperou H., Houas I., Jaafoura S., Papadopoulos A., Coelho A., Diouf A., Hoctin A., Mambert M., Bouscambert M., Gaymard A., Lina B., Rosa-Calatrava M., Terrier O., Benkerrou D., Dorival C., Meziane A., Téoulé F. Detection of SARS-CoV-2 N-antigen in blood during acute COVID-19 provides a sensitive new marker and new testing alternatives. Clin. Microbiol. Infect. 2020 doi: 10.1016/j.cmi.2020.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong K., Cao W., Liu Z., Lin L., Zhou X., Zeng Y., Wei Y., Chen L., Liu X., Han Y., Ruan L., Li T. Prolonged presence of viral nucleic acid in clinically recovered COVID-19 patients was not associated with effective infectiousness. Emerg. Microbes Infect. 2020;9:2315–2321. doi: 10.1080/22221751.2020.1827983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jääskeläinen A.J., Kuivanen S., Kekäläinen E., Ahava M.J., Loginov R., Kallio-Kokko H., Vapalahti O., Jarva H., Kurkela S., Lappalainen M. Performance of six SARS-CoV-2 immunoassays in comparison with microneutralisation. J. Clin. Virol. 2020;129 doi: 10.1016/j.jcv.2020.104512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jääskeläinen A.E., Ahava M.J., Jokela P., Szirovicza L., Pohjala S., Vapalahti O., Lappalainen M., Hepojoki J., Kurkela S. Evaluation of three rapid lateral flow antigen detection tests for the diagnosis of SARS-CoV-2 infection. J. Clin. Virol. 2021;137 doi: 10.1016/j.jcv.2021.104785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Hwang Y.J., Kwak Y. Prolonged SARS-CoV-2 detection and reversed RT-PCR results in mild or asymptomatic patients. Infect. Dis. (London, England) 2020:1–7. doi: 10.1080/23744235.2020.1820076. ahead-of-print. [DOI] [PubMed] [Google Scholar]
- Kubina R., Dziedzic A. Molecular and serological tests for COVID-19 a comparative review of SARS-CoV-2 coronavirus laboratory and point-of-care diagnostics. Diagnostics (Basel) 2020;10:434. doi: 10.3390/diagnostics10060434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebedin Y., Lyang O., Galstyan A., Belousov V., Rebrikov D. Serum SARS-CoV-2 nucleocapsid antigen detection is essential for primary diagnostics of SARS-CoV-2 associated pneumonia. medRxiv. 2020 doi: 10.1101/2020.09.24.20200303. [DOI] [Google Scholar]
- Mannonen L., Kallio-Kokko H., Loginov R., Jääskeläinen A., Jokela P., Antikainen J., Väre P., Kekäläinen E., Kurkela S., Jarva H., Lappalainen M. Comparison of two commercial platforms and a laboratory-developed test for detection of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) RNA. J. Mol. Diagn. 2021;23:407–416. doi: 10.1016/j.jmoldx.2021.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan D., Johnson J.M., Fernandes S.C., Suib H., Hwang S., Wuelfing D., Mendes M., Holdridge M., Burke E.M., Beauregard K., Zhang Y., Cleary M., Xu S., Yao X., Patel P.P., Plavina T., Wilson D.H., Chang L., Kaiser K.M., Nattermann J., Schmidt S.V., Latz E., Hrusovsky K., Mattoon D., Ball A.J. N-protein presents early in blood, dried blood and saliva during asymptomatic and symptomatic SARS-CoV-2 infection. Nat. Commun. 2021;12 doi: 10.1038/s41467-021-22072-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thudium R.F., Stoico M.P., Høgdall E., Høgh J., Krarup H.B., Larsen M.A.H., Madsen P.H., Nielsen S.D., Ostrowski S.R., Palombini A., Rasmussen D.B., Foged N.T. Early laboratory diagnosis of COVID-19 by antigen detection in blood samples of the SARS-CoV-2 nucleocapsid protein. J. Clin. Microbiol. 2021;59 doi: 10.1128/jcm.01001-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Hogan C.A., Verghese M., Solis D., Sibai M., Huang C., Röltgen K., Stevens B.A., Yamamoto F., Sahoo M.K., Zehnder J., Boyd S.D., Pinsky B.A. SARS-CoV-2 nucleocapsid plasma antigen for diagnosis and monitoring of COVID-19. Clin. Chem. 2021 doi: 10.1093/clinchem/hvab216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiersinga W.J., Rhodes A., Cheng A.C., Peacock S.J., Prescott H.C. Pathophysiology, transmission, diagnosis, and treatment of coronavirus disease 2019 (COVID-19): a review. JAMA. 2020;324:782–793. doi: 10.1001/jama.2020.12839. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.