Abstract
Little is known about Subgenomic RNA (sgRNA) dynamics in patients with Coronavirus diseases 2019 (COVID-19). We collected 147 throat swabs, 74 gut swabs and 46 plasma samples from 117 COVID-19 patients recruited in the LOTUS China trial (ChiCTR2000029308) and compared E and orf7a sgRNA load in patients with different illness duration, outcome, and comorbidities. Both sgRNAs were detected in all the three types of samples, with longest duration of 25, 13, and 17 days for E sgRNA, and 32, 28, and 17 days for orf7a sgRNA in throat, gut, and plasma, respectively. A total of 95% (57/60) of patients had no E sgRNA detected after 10 days post treatment, though 86% of them were still E RNA positive. High correlation on titer was observed between sgRNA encoding E and orf7a gene. sgRNA showed similar variation in the standard care and Lopinavir-Ritonavir group. Patients with diabetes and heart diseases showed higher pharyngeal E sgRNA at the first day (P = 0.016 and 0.013, respectively) but no difference at five days after treatment, compared with patients without such commodities. Patients with hypertension and cerebrovascular diseases showed no difference in the pharyngeal sgRNA levels at both one and five days after treatment, compared with patients without these two commodities. E sgRNA levels in the initial infection showed no correlation with the serum antibody against spike, nucleoprotein, and receptor binding domains at ten days later. sgRNA lasted a long period in COVID-19 patients and might have little effect on humoral response.
Keywords: SARS-CoV-2, Subgenomic RNA (sgRNA), Viral load, Antibody
Highlights
-
•
147 throat swabs, 74 gut swabs and 46 plasma samples were used for subgenomic RNA (sgRNA) E and orf7a detection and quantification.
-
•
sgRNA of E and orf7a gene were detected in throat swabs, gut swabs and plasmas and lasted long period.
-
•
E sgRNA levels in the initial infection showed no correlation with the serum antibody against SARS-CoV-2 ten days later.
1. Introduction
Human coronavirus disease 2019 (COVID-19) emerged in late 2019 and continued its spread around the world (Huang et al., 2020). A novel coronavirus subsequently named severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) was identified as the cause of COVID-19 (Zhu et al., 2020). The SARS-CoV-2 has higher reproduction rate (R0) than its sibling SARS-CoV, and thus is more infectious than SARS-CoV (Petersen et al., 2020). Moreover, COVID-19 patients shed infectious virus before clinical symptoms onset, which can cause silent transmissions and accelerate its spread (Lee et al., 2020). Up to Jan 4th, 2021, the virus has caused over 200 million cases, including over 4 million deaths. Though several vaccines have been approved recently, increasing the production capacity and mass vaccination of population still need times. An efficient control and prevention of COVID-19 is still the main challenge the world has to face in the near future.
Similar to other coronavirus, SARS-CoV-2 is an enveloped, positive sense, single-stranded RNA virus with a large genome of ∼ 30 kb (Lu et al., 2020). The 5′ end of the genome contains two large open reading frames (ORF1ab) with length of ∼ 20 kb, while the remaining region to the 3′ end encodes structure proteins such as spike (S), envelop (E), membrane (M), nucleocapsid (N) and several accessory proteins (3a, 6, 7a, 7b and 10, NC_045512.2) (Finkel et al., 2021). When an infectious SARS-CoV-2 entered the host cells, it initiated its replication cycle by translating ORF1ab into polyproteins, which directly formed the sense genome RNA, and started the infection (Kim et al., 2020). The translation of structure proteins in the 3′ end of genome requires the generation of subgenomic RNAs (sgRNAs) (Sola et al., 2015). Firstly, the virus-encoded RNA-dependent RNA polymerase (vRDRP) produced an anti-sense RNA from the 3′ end toward the 5′ end of the viral genome (Sawicki et al., 2007). During negative-strand synthesis, vRDRP paused when it encountered transcriptional regulatory sequences (TRS) upstream of each structure protein ORFs, and then switched the template to the TRS located at the 5′ UTR of virus genome joined to common 5′-leader sequence of approximately 70 bp (Kim et al., 2020; Lu et al., 2020). These antisense sgRNAs serve as templates for transcription of a series of mRNA containing the same 5′ leader sequence.
Since sgRNA is transcribed only in infected cells and is not packaged into virions, some studies use sgRNA levels to reflect virus replication status. Recent study on 35 mild COVID-19 patients showed moderate agreement between virus culture and sgRNA detection for respiratory specimens (Perera et al., 2020). However, another study got challenging results that detection of E gene sgRNA predicted poorly to positive virus culture (van Kampen et al., 2021). Previous studies showed patients with chronic cardiovascular disease, hypertension, chronic neurological diseases, and cerebrovascular diseases more often had high viral RNA load (Magleby et al., 2021; Maltezou et al., 2021), but we still know little about the sgRNA variation in patients with such comorbidities. To obtain a thorough understanding of the association between sgRNAs and clinical manifestation, we performed E gene sgRNA quantification of 268 samples including throat swabs, gut swabs, and plasma from severe COVID-19 patients recruited in the Lopinavir Trial for Suppression of SARS-CoV-2 in China (LOTUS China Trial, ChiCTR2000029308) (Cao et al., 2020). We also quantified another sgRNA, the orf7a in 88 samples, to provide another sgRNA dynamics in COVID-19 patients. Our study provided an overview of the relationship between SARS-CoV-2 sgRNA and duration of illness, treatment, commodities, and serum antibody.
2. Materials and methods
2.1. Study design and sample collection
In the LOTUS China Trial, throat swabs, gut swabs, and plasma samples were collected on day 1 (before Lopinavir-Ritonavir was administered), 5, 10,14, 21, 28 days after admission (Cao et al., 2020). Throat swabs and gut swabs were maintained in viral transport medium and stored in −80 °C. This study used samples collected from the first four timepoints as few samples collected on the last two timepoints were positive for virus RNA. Not all the patients have samples covered the total four timepoints due to limited sample storage. All the patients were positive for SARS-CoV-2 tested by real-time PCR targeting the SARS-CoV-2 ORF1b and N genes. Demographic and clinical information was shown in the original paper (Cao et al., 2020). The research was approved by the Institutional Review Board of Jin Yin-Tan Hospital (KY2020-02.01). Written informed consent was obtained from all the patients or their legal representatives for medical research.
2.2. sgRNA and genome RNA quantification
Total nucleic acids were extracted from 400 μL of throat swab, gut swab, and plasma using the MagNA Pure 96 system (Roche, Basel, Switzerland), as described previously (Cao et al., 2020). A final 80 μL of elution was obtained from each sample and stored at −80 °C until use. SARS-CoV-2 sgRNAs were detected with methods modified from Wolfel's study (Wölfel et al., 2020). Briefly, two set of primers and probe covering the leader sequence and 5′ end of ORF E and orf7a were used to detect sgRNA encoding E-gene and orf7a, respectively (Supplementary Table S1). SARS-CoV-2 RNA was detected using primers and probe targeting the coding region of E gene (Corman et al., 2020), which detected genomic and sgRNA of S, 3a and E gene. Since S and 3a sgRNA showed low abundance in the previous diagnostic samples (Alexandersen et al., 2020), we used the difference between total E RNA and sgRNA load as substitute of the genomic RNA (gRNA) load. Quantification of sgRNA and RNA was conducted on QX200 Droplet Digital PCR System (Bio-Rad, Hercules, USA) with 5 μL nucleic acids added as template. Briefly, 22 μL of reaction mix was prepared using 1-Step RT-ddPCR Advanced Kit for Probes (Cat.#1864021, Bio-Rad) and from which 20 μL was loaded into a DG8 Cartridge to generate droplets with the QX200 droplet generator (Bio-Rad). After generation, the droplets were carefully transferred into a 96-well plate, sealed and amplified in a C1000 Thermal Cycler (Bio-Rad) under the following thermal conditions described in the 1-Step RT-ddPCR Advanced Kit for Probes (Bio-Rad). After amplification, positive and negative droplets were read in the QX200 Droplet Reader (Bio-Rad). Vero E6 cells were infected with SARS-CoV-2 for 24 h and the cell lysate was served as positive control. A throat swab from an influenza patient (negative on SARS-CoV-2 RT-PCR) was used as negative control. All the primers and probes used in this study were listed on Supplementary Table S1.
2.3. Enzyme-linked immunosorbent assay (ELISA)
ELISA protocols were developed for detecting IgG and IgM against S, N, and receptor binding domain (RBD) of SARS-CoV-2 as described previously (Ren et al., 2021). The purified full-length N protein, extracellular domain of S proteins, and RBD protein (Cat: 40589-V08B1 and 40592-V08H, Sino Biological, Beijing, China) were used as coating antigens, respectively. Horseradish peroxidase-conjugated goat anti-human IgG (Cat: A0710, Sigma-Aldrich, St. Louis, MO, USA) was diluted to 1:60,000 working solution and used as the second antibody. The optimal coating concentration of antigen and optimal plasma dilutions were 10 ng/well and 1:400, respectively. The cutoff values of IgM and IgG were 0.1 and 0.3 for N, 0.13 and 0.21 for S, and 0.1 and 0.3 for RBD, respectively, as described in previous study (Ren et al., 2021).
2.4. Statistical analysis
Statistical analysis was conducted using the scipy module in the Python 3.8 version (Virtanen et al., 2020). Mann-Whitney U test and χ2 test were used for group comparison of continuous and categorical variables, respectively. Correlation of two variables was determined by Spearman's rank correlation test. A P < 0.05 was considered to be statistically significant.
3. Results
3.1. Overview of involved patients and samples
We collected 147 throat swabs, 74 gut swabs, and 46 plasma samples from 117 patients in our study, with median age of 60 [interquartile range (IQR) 52–71], 60 (IQR 51–71), 63 (IQR 53–68) years, respectively. All the 267 samples were subjected to E sgRNA quantification while orf7a sgRNA quantification was performed on 88 samples for limited sample storage. Hypertension was the most common comorbidity in the three groups (Table 1). Samples collected from the first two timepoints accounted for 76.9%, 66.2%, and 97.8% of the throat swabs, gut swabs, and plasma samples.
Table 1.
Characteristics of patients with samples tested in this study.
| Variable | Throat swab (n = 147) | Gut swab (n = 74) | Plasma (n = 46) | P | |
|---|---|---|---|---|---|
| Sex, n (%) | |||||
| Female | 52 (35.4) | 28 (37.8) | 15 (32.6) | 0.842 | |
| Male | 95 (64.6) | 46 (62.2) | 31 (67.4) | ||
| Age (y) | |||||
| Median (IQR) | 60 (52, 71) | 60 (51, 71) | 63 (53, 68) | 0.869 | |
| 20–40 | 10 (6.8) | 8 (10.8) | 4 (8.7) | ||
| 41–60 | 64 (43.5) | 30 (40.5) | 18 (39.1) | ||
| < 60 | 73 (49.7) | 36 (48.6) | 24 (52.2) | ||
| Comorbidity, n (%) | |||||
| Hypertension | 55 (37.4) | 17 (23) | 18 (39.1) | 0.42 | |
| Diabetes | 21 (14.3) | 9 (12.2) | 7 (15.2) | ||
| CVD | 17 (11.6) | 10 (13.5) | 5 (10.9) | ||
| Heart disease | 19 (12.9) | 6 (8.1) | 6 (13) | ||
| None | 53 (36.1) | 30 (40.5) | 9 (19.6) | ||
| Days after treatment, n (%) | |||||
| 1 | 83 (56.5) | 29 (39.2) | 35 (76.1) | 0.0002a | |
| 5 | 30 (20.4) | 20 (27.0) | 10 (21.7) | ||
| 10 | 23 (15.6) | 12 (16.2) | 1 (2.2) | ||
| 14 | 11 (7.5) | 13 (17.6) | 0 (0) | ||
| Outcome | |||||
| Death | 42 (28.6) | 23 (31.1) | 22 (47.8) | 0.049 | |
| Recovery | 105 (71.4) | 51 (68.9) | 24 (52.2) | ||
| Treatment | |||||
| Lopinavir-Ritonavir | 77 (52.4) | 35 (47.3) | 21 (45.7) | 0.639 | |
| Standard Care | 70 (47.6) | 39 (52.7) | 25 (54.3) | ||
IQR, interquartile range; CVD, Cerebrovascular diseases.
Fisher exact test. Chi-square test was used for the remaining variables comparison if not indicated.
3.2. sgRNA duration in the three types of samples
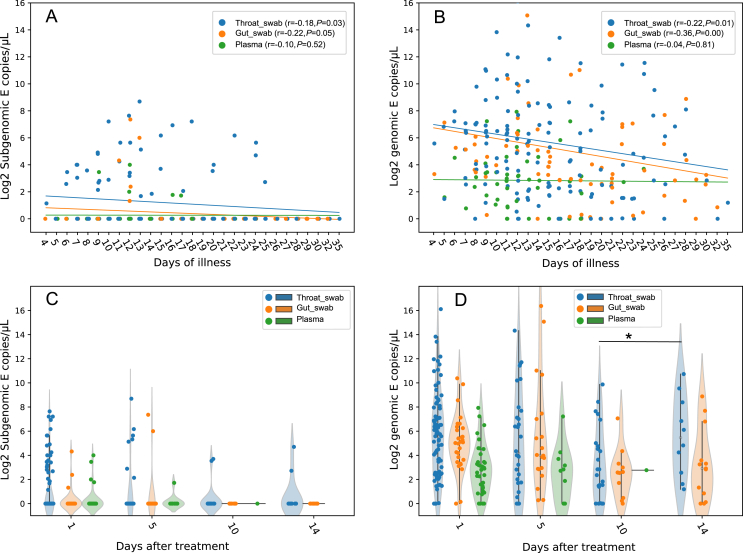
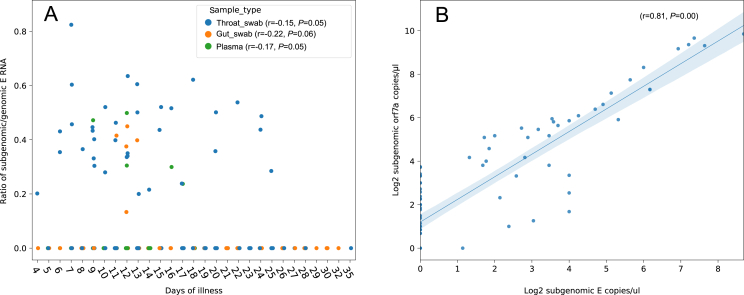
The E and orf7a sgRNA were detected in all the three types of samples. E sgRNA showed lower load than E gRNA in the same sample (Fig. 1A, Supplementary Fig. S1A); this is consistent with previous studies which detected E sgRNA in oropharyngeal swabs and stool samples (Moreira et al., 2021; Wölfel et al., 2020). Both E sgRNA and gRNA load decreased more slowly in the plasma than that in throat swabs and gut swabs (Fig. 1A and B), while orf7a sgRNA decreased slower in throat than the other two (Supplementary Fig. S1). E sgRNA lasted 25 days after illness onset in the throat swabs, followed by 17 days in the plasma and 13 days in gut swabs (Fig. 1A); orf7a sgRNA lasted even longer in the throat swabs (32 days) and gut swabs (28 days) (Supplementary Fig. S1). However, genome RNA can exist in the throat swabs and gut swabs for over 30 days post illness onset, followed by 24 days in plasma (Fig. 1B). On the individual level, E sgRNA reached peak titers about 15 days after disease onset and decreased since then (Supplementary Fig. S2). The ratio of gRNA to sgRNA E showed no association with the patient's illness duration (Fig. 2A). The sgRNA encoding orf7a showed high correlation with the E sgRNA load in the same sample (Fig. 2B). Few samples showed sgRNA positive 10 days after treatment, both in Lopinavir-Ritonavir treatment and Standard Care groups (Table 2, Fig. 1C, Supplementary S1B). Meanwhile, most of these samples still showed gRNA positive 14 days after Lopinavir-Ritonavir and standard treatment (Table 2, Fig. 1D). In all the four timepoints, the percentage of positive samples for E sgRNA or gRNA showed no difference between Lopinavir-Ritonavir and standard treatment group, according to the χ2/Fisher exact test (P = 0.216–1) on a total of 147, 60, 36, 24 samples 1, 5, 10, 14 days after treatment, respectively. However, patients with death outcome showed higher percentage of samples with positive sgRNA in day 1 post treatment than patients with recovery outcome (P = 0.046), while no significant difference was observed in the remaining 3 timepoints between the two groups, based on χ2/Fisher exact test (P = 0.167–1) (Table 2). Orf7a sgRNA also showed no difference in the frequency of positive samples between the two groups (Supplementary Table S2).
Fig. 1.
Scatterplots of the E gene sgRNA (A) and gRNA (B) load in the three sample types with different duration of illness (n = 264). Spearman's rank correlation coefficient (r) and P value were attached for each sample type. Sample types were indicated with different colors. Generalized estimating equations were used to fit the dynamics of sgRNA/gRNA load. Violin plot of sgRNA (C) and gRNA (D) load versus the duration of treatment (days). ∗P < 0.05 by Mann-Whitney U test. sgRNA, subgenomic RNA; gRNA, genomic RNA.
Fig. 2.
The ratio of subgenomic E titer to genomic E titer through the illness duration (n = 241) (A); high correlation between the titer of subgenomic E and subgenomic orf7a (n = 88) (B). Correlation coefficient (r) and P value was calculated by spearman's rank test.
Table 2.
Comparison of the number and frequency (%) of specimens that were positive for subgenomic and genomic E gene with different duration and clinical outcomes.
| Days after treatment | Subgenomic E |
Genomic E |
||||
|---|---|---|---|---|---|---|
| Treatment, n (%) |
P | Treatment, n (%) |
P | |||
| Standard Care | Lopinavir-Ritonavir | Standard Care | Lopinavir-Ritonavir | |||
| 1 | 14/79 (17.7) |
18/68 (26.5) | 0.280 | 74/79 (93.7) | 63/68 (92.6) | 1a |
| 5 | 4/29 (13.8) |
6/31 (19.4) | 0.732a | 25/29 (86.2) | 27/31 (87.1) | 1a |
| 10 | 0/19 (0) |
2/17 (11.8) | 0.216a | 18/19 (94.7) | 13/17 (76.5) | 0.167a |
| 14 |
0/7 (0) |
2/17 (11.8) |
1a |
7/7 (100) |
15/17 (88.2) |
1a |
| Outcome Death | Recovery | P | Outcome Death | Recovery | P | |
| 1 | 18/58 (31) |
14/89 (15.7) | 0.046 | 56/58 (96.6) |
81/89 (91) | 0.316 |
| 5 | 5/20 (25) |
5/40 (12.5) | 0.278a | 19/20 (95) |
33/40 (82.5) | 0.249 |
| 10 | 0/4 (0) |
2/32 (6.3) | 1a | 4/4 (100) |
27/32 (84.4) | 1a |
| 14 | 1/5 (20) |
1/19 (5.3) | 0.415a | 5/5 (100) |
17/19 (89.5) | 1a |
Fisher exact test. Chi-square test was used for the remaining variables comparison if not indicated, by chi2_contingency function in the scipy module in python 3.8.
3.3. sgRNA load in patients with different comorbidity
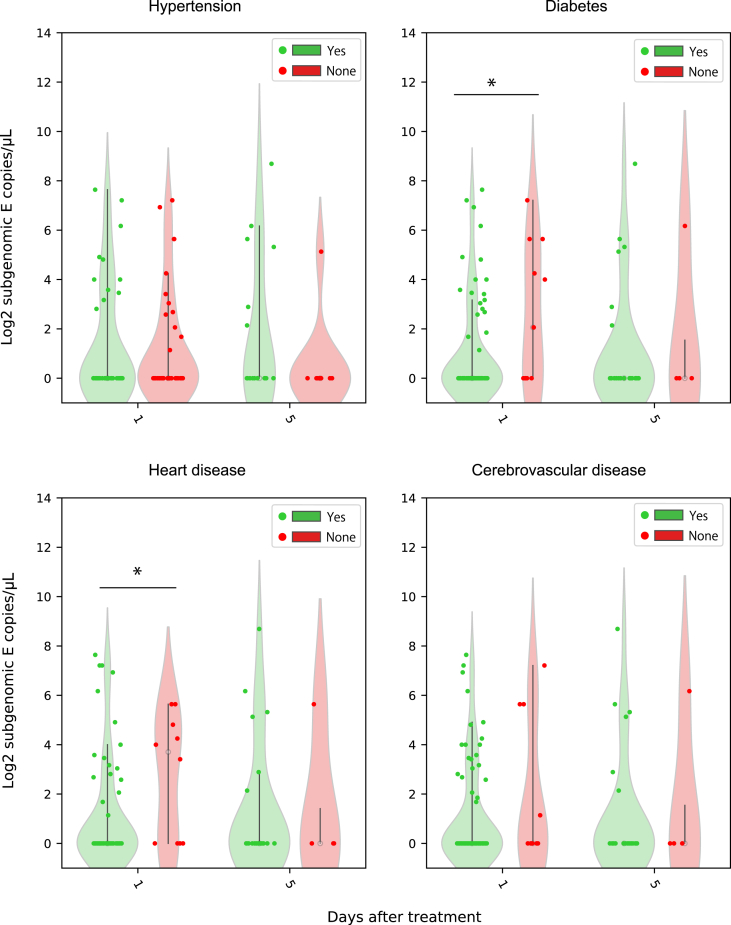
We collected 55, 21, 17, 19 throat swabs from patients with hypertension, diabetes, cerebrovascular disease, and heart disease, respectively. We compared the sgRNA load in patients with or without these comorbidities on day 1 and 5 after treatment (Fig. 3). E sgRNA load showed no difference in patients with or without hypertension and cerebrovascular diseases on day 1 and day 5 after treatment, and also no difference in patients with diabetes and heart diseases on day 5 after treatment. E sgRNA load was higher in patient with heart diseases than those without (P = 0.013), but lower in patients with diabetes than those without, in the samples of the first timepoint (P = 0.016). Similar results were observed on orf7a sgRNA load, that orf7a sgRNA load showed no difference between patients with the four comorbidities or without comorbidities (Supplementary Fig. S3).
Fig. 3.
Comparison of the subgenomic RNA load between patients with and without comorbidities on different duration of treatment (n = 113). Violin plots were generated for subgenomic RNA load in patients with (green) or without (red) hypertension, diabetes, heart diseases, cardiovascular diseases, after one or five days of treatment. ∗P < 0.05 by Mann-Whitney U test.
3.4. Throat sgRNA load at the time of detection correlated poorly with serum antibody 10 days later
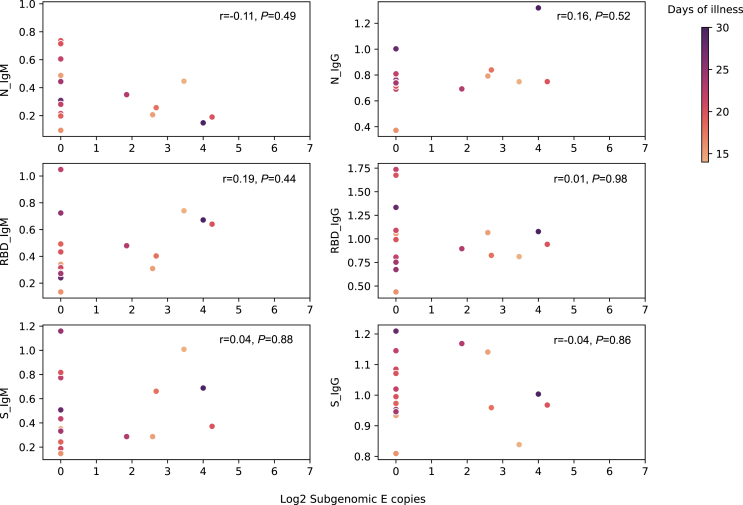
Higher sgRNA may indicate potent virus replication and thus induce efficient adaptive immune response. We measured the E sgRNA load in the throat swabs from 22 patients at the time of detection and also the antibody against N, S, and RBD 10 days later. The median duration of illness in these patients was 20.5 days (IQR 18–23). All the patients were positive for IgM and IgG against the three tested SARS-CoV-2 proteins 10 days after detection of infection, except one patient showed IgM negative against the N protein. However, E sgRNA load showed no correlation with the IgM and IgG titer 10 days after the sgRNA test (Fig. 4); both IgM and IgG titer showed no difference between patients with positive and negative E sgRNA (Supplementary Fig. S4).
Fig. 4.
Relationship between subgenomic RNA load in the throat and serum antibody against SARS-CoV-2 N, RBD, and S protein (n = 18). Duration of illness was mapped on the color bar. Spearman's rank correlation test was used to determine the correlation between subgenomic RNA loads and antibody titer. The P value resulted from Spearman's rank correlation test were shown in each subplot.
4. Discussion
As a coronavirus, SARS-CoV-2 carries a large genome of ∼ 30 kb which contains the common 5′ “leader” sequences of ∼ 70 bp. Up on mRNA transcription, this leader sequence fused to the structure protein ORF from the down-stream part of genome. Since only actively replicating SARS-CoV-2 initiates mRNA transcription, the sgRNA level may reflect the virus replication status in vivo and play a role on patients' clinical symptoms (Perera et al., 2020). We tested the E and orf7a sgRNA load in throat, gut and plasma samples from severe COVID-19 patients and found that sgRNA could be detected over 32 days after illness and poorly correlated with comorbidities and serum antibody response.
Viral load kinetics and duration of viral shedding are important determinant for disease transmission. A meta-analysis involved 79 studies showed mean duration of SARS-CoV-2 genome RNA shedding was 17.0 days in upper respiratory tract, 17.2 days in stool, and 16.6 days in serum samples (Cevik et al., 2021). However, detection of virus RNA did not indicate infectious virus presence. A couple of studies used sgRNA as an indicator of active virus replication or infectious virus shedding, though these studies had small sample size (Park et al., 2018; Wölfel et al., 2020). In our study, we detected E sgRNA up to 25, 17, and 13 days after illness onset in throat, plasma, and gut, respectively, as well as even longer duration of orf7a persistence in throat (Supplementary Fig. S1A). Similar to our results, two recent studies described the detection of SARS-CoV-2 sgRNA in upper respiratory tract to 17–22 days after initial detection of infection (Alexandersen et al., 2020; van Kampen et al., 2021); another study also reported sgRNA detection in stool samples from COVID-19 patients (Moreira et al., 2021). However, no previous study report sgRNA presence in plasma from COVID-19 patients. This is the first study which detects sgRNA in plasma sample after SARS-CoV-2 infection. We still do not know the origin of these sgRNA in plasma, as they may be produced in the vascular endothelial cells after SARS-CoV-2 infection or released from the infection sites (eg. lung), which need further exploration.
sgRNA from E gene was used to qualified SARS-CoV-2 sgRNA in several studies (van Kampen et al., 2021; Wölfel et al., 2020). However, sgRNA from other genes may show different dynamics in the same patients. Orf7a sgRNA showed the highest abundance in the previous meta-transcriptome study, and we also observed higher titer of orf7a sgRNA than E sgRNA in most samples (Fig. 2B) and lower decline rate in throat samples (Supplementary Fig. S1A). Moreover, the orf7a sgRNA lasted a longer duration and its titer highly correlated with that of E sgRNA (Fig. 2B, Supplementary Fig. S1A).
Currently, virus culture is commonly used to assess infectious virus shedding in samples. Several studies considered there was an association between positive virus culture and sgRNA presence in diagnostic samples (Perera et al., 2020), but studies involved hospitalized patients with larger sample size resulted in contradicting results (van Kampen et al., 2021). Soren et al. provided evidence that sgRNA was protected by cellular membranes and that the detection of sgRNA in diagnostic samples might not be a reliable indicator of active SARS-CoV-2 replication/infection (Alexandersen et al., 2020). In our study, we detected sgRNA up to 13–32 days after illness onset, which should not indicate infectious virus presence as low odds of live virus detection beyond 9–10 days of illness in respiratory sample (Cevik et al., 2021; Walsh et al., 2020) and feces samples. Moreover, no live virus isolated from plasma was reported so far, which underlined that the sgRNA detected in plasma in our study could not represent live virus presence. If sgRNA is just the leftover of mRNA produced during active virus replication and outlasts the detection of infectious virus (van Kampen et al., 2021), it is reasonable that sgRNA shows poor correlation with the patient's clinical features. Meanwhile, the E sgRNA load in the throat also poorly predicted the antibody titer 10 days later, which indicated that the sgRNA might have little effect on humoral response.
Our study observed significant difference on the E sgRNA load between patients with and without diabetes and heart disease at the first day after treatment. Type 2 diabetes mellitus reduced the expression of angiotensin-converting enzyme 2 (ACE2) which might play an anti-inflammatory role in the lung and further lead to a different sgRNA load in the patients with diabetes (Wu et al., 2020). Moreover, patients with preexisting pathologies involving renin-angiotensin-aldosterone system such as hypertension, chronic heart failure developed more severe clinical outcomes from COVID-19 (Nishiga et al., 2020), passably due to the upregulation of ACE2 receptors (which would facilitate viral entry) and increased the sgRNA load in their samples (Wu et al., 2020). However, the orf7a sgRNA showed no difference between patients with or without these four comorbidities, indicating different dynamics of different sgRNA in COVID-19 patients. SARS-CoV-2 lineage is also associated with sgRNA expression level. The samples in LOTUS China trial were collected from January 18, 2020 to February 3, 2020, which were most likely belonging to lineage A (Rambaut et al., 2020). Thus, for other variants such as Alpha or the latest Delta, sgRNA may show different dynamics.
Our study has some limitations. Firstly, only a small number of patients have samples of multiple timepoints for sgRNA test, which may lead to an underestimation of the duration of sgRNA in these patients. Secondly, we only tested the sgRNA load from E and orf7a gene of SARS-CoV-2, which were also used as sgRNA targets in a number of previous studies (Wölfel et al., 2020). However, sgRNA from other genes, such as N gene, may show higher abundance in the sample and achieve higher detection sensitivity (Nomburg et al., 2020). Finally, our study only included hospitalized patients with severe COVID-19 and lacked the sgRNA load data on mild COVID-19 patients. Thus, further studies including more genes of sgRNA are needed to determine the full picture of sgRNA dynamics in specific patient groups.
5. Conclusions
In conclusion, SARS-CoV-2 E and orf7a sgRNA presented in throat, plasma, and gut samples from the severe COVID-19 patients and lasted for a long duration. E sgRNA load in the throat showed poor correlation with patient's clinical features and antibody raised later. E and orf7a sgRNA might be not a good biomarker to predict patients' clinical outcome and infectious virus presence. We still need to develop a better tool to predict patient's clinical outcome and immune response.
Data availability
All the data from this study were attached in Supplementary Tables S3 and S4.
Ethics statement
The trial was approved by the institutional review board of Jin Yin-Tan Hospital. Written informed consent for clinical trial and medical research was obtained from all patients or from the patient's legal representative if the patient was too unwell to provide consent. The trial was conducted in accordance with the principles of the Declaration of Helsinki and the Good Clinical Practice guidelines of the International Conference on Harmonisation.
Author contributions
Zou Xiaohui: conceptualization, data curation, writing-original draft, writing-review, editing. Mu Shengrui: investigation, methodology, project administration, resources. Wang Yeming: investigation, methodology, project administration, resources. Guo Li: software, supervision, validation, visualization. Ren Lili: software, supervision, validation, visualization. Deng Xiaoyan: software, supervision, validation, visualization. Li Haibo: formal analysis, funding acquisition. Zhao Jiankang: formal analysis, funding acquisition. Zhang Yulin: formal analysis, funding acquisition. Li Hui: formal analysis, funding acquisition. Lu Binghuai: formal analysis, funding acquisition. Huang Chaolin: conceptualization, data curation, writing-original draft, writing-review, editing. Cao Bin: conceptualization, data curation, writing-original draft, writing-review, editing.
Conflict of interest
The authors declare that they have no conflict of interest.
Acknowledgement
Special thanks to Dr. Xiaolu Nie from the Beijing Children's Hospital, Capital Medical University, for her kindly help on data analysis. This work was supported by Natural Science Foundation of China (81900009 and 82041011/H0104), National Key Research and Development Program of China (2018YFC1200100 and 2018YFC1200102), and Chinese Academy of Medical Sciences Innovation Fund for Medical Sciences (CIFMS 2018-I2M-1–003, 2020-I2M-2–013, and 2020-I2M-CoV19-005). All funders had no role in the design, analysis or writing of this article.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.virs.2022.01.008.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Scatterplots of the orf7a subgenomic RNA load in the three sample types with different duration of illness (n = 88) (A). Spearman’s rank correlation coefficient (r) and P value was attached for each sample type. Sample types were indicated with different colors. Generalized estimating equations were used to fit the dynamics of sgRNA load. B Violin plot of orf7a subgenomic RNA load versus the duration of treatment (days). Mann-Whitney U test showed no difference between cohorts with different days after treatment.
E sgRNA dynamic on the individual level (n = 14) through the illness duration. Each patient included 3–4 timepoints data and labeled with different color. Throat swabs (n = 12) and gut swabs (n = 3) were labeled with circle and asterisk, respectively. No significant difference was detected.
Comparison of the orf7a subgenomic RNA load between patients with and without four comorbidities 1–5 days after treatment (n = 88). Mann-Whitney U test showed no statistical significance between patients with and without such diseases.
Comparison of antibody titer (at day 10) against SARS-CoV-2 N, RBD and S protein between patients with positive and negative E sgRNA in the throat swab at day 1 (n = 18), which showed no significant difference between them using the t-test.
References
- Alexandersen S., Chamings A., Bhatta T.R. SARS-CoV-2 genomic and subgenomic RNAs in diagnostic samples are not an indicator of active replication. Nat. Commun. 2020;11:1–13. doi: 10.1038/s41467-020-19883-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao B., Wang Y., Wen D., Liu W., Wang J., Fan G., Ruan L., Song B., Cai Y., Wei M., Li X., Xia J., Chen N., Xiang J., Yu T., Bai T., Xie X., Zhang L., Li C., Yuan Y., Chen H., Li H., Huang H., Tu S., Gong F., Liu Y., Wei Y., Dong C., Zhou F., Gu X., Xu J., Liu Z., Zhang Y., Li H., Shang L., Wang K., Li K., Zhou X., Dong X., Qu Z., Lu S., Hu X., Ruan S., Luo S., Wu J., Peng L., Cheng F., Pan L., Zou J., Jia C., Wang J., Liu X., Wang S., Wu X., Ge Q., He J., Zhan H., Qiu F., Guo L., Huang C., Jaki T., Hayden F.G., Horby P.W., Zhang D., Wang C. A trial of lopinavir–ritonavir in adults hospitalized with severe Covid-19. N. Engl. J. Med. 2020;382:1787–1799. doi: 10.1056/NEJMoa2001282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cevik M., Tate M., Lloyd O., Maraolo A.E., Schafers J., Ho A. SARS-CoV-2, SARS-CoV, and MERS-CoV viral load dynamics, duration of viral shedding, and infectiousness: a systematic review and meta-analysis. Lancet Microbe. 2021;2:e13–e22. doi: 10.1016/S2666-5247(20)30172-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K., Bleicker T., Brünink S., Schneider J., Schmidt M.L., Mulders D.G., Haagmans B.L., van der Veer B., van den Brink S., Wijsman L., Goderski G., Romette J.L., Ellis J., Zambon M., Peiris M., Goossens H., Reusken C., Koopmans M.P., Drosten C. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25:2000045. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkel Y., Mizrahi O., Nachshon A., Weingarten-Gabbay S., Morgenstern D., Yahalom-Ronen Y., Tamir H., Achdout H., Stein D., Israeli O., Beth-Din A., Melamed S., Weiss S., Israely T., Paran N., Schwartz M., Stern-Ginossar N. The coding capacity of SARS-CoV-2. Nature. 2021;589:125–130. doi: 10.1038/s41586-020-2739-1. [DOI] [PubMed] [Google Scholar]
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L., Xie J., Wang G., Jiang R., Gao Z., Jin Q., Wang J., Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. The lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Lee J.-Y., Yang J.-S., Kim J.W., Kim V.N., Chang H. The architecture of SARS-CoV-2 transcriptome. Cell. 2020;181:914–921. doi: 10.1016/j.cell.2020.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., Kim T., Lee E., Lee C., Kim H., Rhee H., Park S.Y., Son H.-J., Yu S., Park J.W., Park J.W., Choo E.J., Park S., Loeb M., Kim T.H. Clinical course and molecular viral shedding among asymptomatic and symptomatic patients with SARS-CoV-2 infection in a community treatment center in the Republic of Korea. JAMA Intern. Med. 2020;180:1447–1452. doi: 10.1001/jamainternmed.2020.3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N., Bi Y., Ma X., Zhan F., Wang L., Hu T., Zhou H., Hu Z., Zhou W., Zhao L., Chen J., Meng Y., Wang J., Lin Y., Yuan J., Xie Z., Ma J., Liu W.J., Wang D., Xu W., Holmes E.C., Gao G.F., Wu G., Chen W., Shi W., Tan W. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. The lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magleby R., Westblade L.F., Trzebucki A., Simon M.S., Rajan M., Park J., Goyal P., Safford M.M., Satlin M.J. Impact of severe acute respiratory syndrome coronavirus 2 viral load on risk of intubation and mortality among hospitalized patients with coronavirus disease 2019. Clin. Infect. Dis. 2021;73:e4197–e4205. doi: 10.1093/cid/ciaa851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maltezou H.C., Raftopoulos V., Vorou R., Papadima K., Mellou K., Spanakis N., Kossyvakis A., Gioula G., Exindari M., Froukala E., Martinez-Gonzalez B., Panayiotakopoulos G., Papa A., Mentis A., Tsakris A. Association between upper respiratory tract viral load, comorbidities, disease severity, and outcome of patients with SARS-CoV-2 infection. J. Infect. Dis. 2021;223:1132–1138. doi: 10.1093/infdis/jiaa804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira L.V.L., de Souza Luna L.K., Barbosa G.R., Perosa A.H., Chaves A.P.C., Conte D.D., Carvalho J.M.A., Bellei N. Test on stool samples improves the diagnosis of hospitalized patients: detection of SARS-CoV-2 genomic and subgenomic RNA. J. Infect. 2021;82:186–230. doi: 10.1016/j.jinf.2020.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiga M., Wang D.W., Han Y., Lewis D.B., Wu J.C. COVID-19 and cardiovascular disease: from basic mechanisms to clinical perspectives. Nat. Rev. Cardiol. 2020;17:543–558. doi: 10.1038/s41569-020-0413-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomburg J., Meyerson M., DeCaprio J.A. Pervasive generation of non-canonical subgenomic RNAs by SARS-CoV-2. Genome Med. 2020;12:1–14. doi: 10.1186/s13073-020-00802-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park W.B., Poon L.L., Choi S.-J., Choe P.G., Song K.-H., Bang J.H., Kim E.S., Kim H.B., Park S.W., Kim N.J., Peiris M., Oh M.D. Replicative virus shedding in the respiratory tract of patients with Middle East respiratory syndrome coronavirus infection. Int. J. Infect. Dis. 2018;72:8–10. doi: 10.1016/j.ijid.2018.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perera R.A., Tso E., Tsang O.T., Tsang D.N., Fung K., Leung Y.W., Chin A.W., Chu D.K., Cheng S.M., Poon L.L., Chuang V.W.M., Peiris M. SARS-CoV-2 virus culture and subgenomic RNA for respiratory specimens from patients with mild coronavirus disease. Emerg. Infect. Dis. 2020;26:2701. doi: 10.3201/eid2611.203219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen E., Koopmans M., Go U., Hamer D.H., Petrosillo N., Castelli F., Storgaard M., Al Khalili S., Simonsen L. Comparing SARS-CoV-2 with SARS-CoV and influenza pandemics. Lancet Infect. Dis. 2020;20:e238–e244. doi: 10.1016/S1473-3099(20)30484-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A., Holmes E.C., O'Toole Á., Hill V., McCrone J.T., Ruis C., du Plessis L., Pybus O.G. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat. Microbiol. 2020;5:1403–1407. doi: 10.1038/s41564-020-0770-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren L., Fan G., Wu W., Guo L., Wang Yeming, Li X., Wang C., Gu X., Li C., Wang Y., Wang G., Zhou F., Liu Z., Ge Q., Zhang Y., Li H., Zhang L., Xu J., Wang C., Wang J., Cao B. Antibody responses and clinical outcomes in adults hospitalized with severe coronavirus disease 2019 (COVID-19): a post hoc analysis of LOTUS China trial. Clin. Infect. Dis. 2021;72:e545–e551. doi: 10.1093/cid/ciaa1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawicki S.G., Sawicki D.L., Siddell S.G. A contemporary view of coronavirus transcription. J. Virol. 2007;81:20–29. doi: 10.1128/JVI.01358-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sola I., Almazan F., Zuniga S., Enjuanes L. Continuous and discontinuous RNA synthesis in coronaviruses. Annu. Rev. Virol. 2015;2:265–288. doi: 10.1146/annurev-virology-100114-055218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Kampen J.J., van de Vijver D.A., Fraaij P.L., Haagmans B.L., Lamers M.M., Okba N., van den Akker J.P., Endeman H., Gommers D.A., Cornelissen J.J., Hoek R.A.S., van der Eerden M.M., Hesselink D.A., Metselaar H.J., Verbon A., de Steenwinkel J.E.M., Aron G.I., van Gorp E.C.M., van Boheemen S., Voermans J.C., Boucher C.A.B., Molenkamp R., Koopmans M.P.G., Geurtsvankessel C., van der Eijk A.A. Duration and key determinants of infectious virus shedding in hospitalized patients with coronavirus disease-2019 (COVID-19) Nat. Commun. 2021;12:1–6. doi: 10.1038/s41467-020-20568-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virtanen P., Gommers R., Oliphant T.E., Haberland M., Reddy T., Cournapeau D., Burovski E., Peterson P., Weckesser W., Bright J., van der Walt S.J., Brett M., Wilson J., Millman K.J., Mayorov N., Nelson A.R.J., Jones E., Kern R., Larson E., Carey C.J., Polat İ., Feng Y., Moore E.W., VanderPlas J., Laxalde D., Perktold J., Cimrman R., Henriksen I., Quintero E.A., Harris C.R., Archibald A.M., Ribeiro A.H., Pedregosa F., van Mulbregt P. SciPy 1.0: fundamental algorithms for scientific computing in Python. Nat. Methods. 2020;17:261–272. doi: 10.1038/s41592-019-0686-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh K.A., Spillane S., Comber L., Cardwell K., Harrington P., Connell J., Teljeur C., Broderick N., de Gascun C.F., Smith S.M., Ryan M., O'Neill M. The duration of infectiousness of individuals infected with SARS-CoV-2. J. Infect. 2020;81:847–856. doi: 10.1016/j.jinf.2020.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wölfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Müller M.A., Niemeyer D., Jones T.C., Vollmar P., Rothe C., Hoelscher M., Bleicker T., Brünink S., Schneider J., Ehmann R., Zwirglmaier K., Drosten C., Wendtner C. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- Wu L., O'Kane A.M., Peng H., Bi Y., Motriuk-Smith D., Ren J. SARS-CoV-2 and cardiovascular complications: from molecular mechanisms to pharmaceutical management. Biochem. Pharmacol. 2020;178:114114. doi: 10.1016/j.bcp.2020.114114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G.F., Tan W. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Scatterplots of the orf7a subgenomic RNA load in the three sample types with different duration of illness (n = 88) (A). Spearman’s rank correlation coefficient (r) and P value was attached for each sample type. Sample types were indicated with different colors. Generalized estimating equations were used to fit the dynamics of sgRNA load. B Violin plot of orf7a subgenomic RNA load versus the duration of treatment (days). Mann-Whitney U test showed no difference between cohorts with different days after treatment.
E sgRNA dynamic on the individual level (n = 14) through the illness duration. Each patient included 3–4 timepoints data and labeled with different color. Throat swabs (n = 12) and gut swabs (n = 3) were labeled with circle and asterisk, respectively. No significant difference was detected.
Comparison of the orf7a subgenomic RNA load between patients with and without four comorbidities 1–5 days after treatment (n = 88). Mann-Whitney U test showed no statistical significance between patients with and without such diseases.
Comparison of antibody titer (at day 10) against SARS-CoV-2 N, RBD and S protein between patients with positive and negative E sgRNA in the throat swab at day 1 (n = 18), which showed no significant difference between them using the t-test.
Data Availability Statement
All the data from this study were attached in Supplementary Tables S3 and S4.