Abstract
Over the course of human evolution, shifts in dietary practices such as meat-eating and cooking, have resulted in reduced fiber intake, a trend that has been exaggerated more recently in industrialized populations. Reduced fiber consumption is associated with a loss of gut microbial taxa that degrade fiber, particularly butyrate. Therefore, this dietary shift in humans may have altered the abundance of microbial genes involved in butyrate production. This study uses a gene-targeted alignment approach to quantify the abundance of butyrate production pathway genes from published wild nonhuman primate and human gut metagenomes. Surprisingly, humans have higher diversity and relative abundances of butyrate production pathways compared with all groups of nonhuman primates except cercopithecoids. Industrialized populations of humans also differ only slightly in butyrate pathway abundance from nonindustrialized populations. This apparent resilience of butyrate production pathways to shifts in human diet across both evolutionary and modern populations may signal an evolutionary shift in host–microbe interactions in humans that increased SCFA production. Such a shift could have contributed to meeting the increased energy requirements of humans relative to nonhuman primates.
Keywords: gut microbiome, butyrate, metabolism, human evolution
Introduction
Microbially mediated short-chain fatty acid (SCFA) production from the fermentation of structural carbohydrates in the gut is a major source of energy for mammals and mediates host physiology and health (McNeil 1984; Cummings and Macfarlane 1997; Hooper et al. 2002; Tremaroli and Bäckhed 2012). In particular, butyrate production patterns have been linked to a range of health outcomes (Koh et al. 2016; Clavel et al. 2017) including immune function, mental health, neural development, inflammatory bowel disease, obesity, heart disease, diabetes, and colorectal cancer (Cho and Blaser 2012; Nicholson et al. 2012; Cani 2014; Selkrig et al. 2014; Sonnenburg and Bäckhed 2016; Zmora et al. 2019). Lower butyrate production and lower relative abundances of taxa associated with butyrate production correlate with a decline in gut bacterial diversity and adverse health impacts, including increases in inflammation (Qin et al. 2012; Le Chatelier et al. 2013; Ridaura et al. 2013; Kushugulova et al. 2018; Schirmer et al. 2018).
Production of butyrate in the gut is necessarily linked to the presence of fermentation substrates in the diet. Humans with diets that are higher in fiber and resistant starch have increased fecal concentrations of butyrate and/or more bacterial taxa that are known to produce butyrate (Phillips et al. 1995; Duncan et al. 2007; Brinkworth et al. 2009; Wu et al. 2011; Tremaroli and Bäckhed 2012; Yang et al. 2016). In contrast, low-fiber diets have been linked to reduced fecal butyrate and lower relative abundances of butyrate-producing microbes in humans, nonhuman primates, and mice (De Filippo et al. 2010; Amato et al. 2013; David et al. 2014; Nagpal et al. 2018), an effect which can compound across generations (Sonnenburg et al. 2016). Therefore, low-fiber diets may increase susceptibility to a range of health risks as a result of the associated altered butyrate production patterns. Indeed, modern industrialized human populations tend to be characterized by high fat, low-fiber diets, altered gut microbial communities, and increased prevalence of chronic, noncommunicable diseases (Sonnenburg ED and Sonnenburg JL 2019; Sonnenburg JL and Sonnenburg ED 2019).
Nevertheless, reduction in fiber consumption is not a recent phenomenon for humans. Since our divergence from other primates, humans have become more reliant on animal foods, expanded food processing techniques, including cooking, and have domesticated both plant and animal foods (Leonard and Robertson 1994; Teaford and Ungar 2000; Aiello and Wells 2002; Richards 2002; Wrangham and Conklin-Brittain 2003; Carmody and Wrangham 2009; DeCasien et al. 2017). Each of these dietary shifts has led to reductions in human fiber intake. Therefore, it is likely that SCFA production patterns in the human gut have shifted across evolution, not simply in response to industrialization over the past century. Although these changes may have influenced human susceptibility to certain diseases, they may also represent an evolutionary shift in human host–microbe interactions that support human’s unique energetic needs and life history traits (Amato 2016). Different SCFAs are used by different tissue types—butyrate is used primarily by the intestine, propionate is involved in gluconeogenesis in the liver, and acetate is used primarily by peripheral tissues, including the brain (Wong et al. 2006; Gao et al. 2009; van Eunen et al. 2013; Morrison and Preston 2016). There is evidence that diets higher in protein and lipids are associated with a gut microbiome that is more efficient at producing acetate (De Filippo et al. 2010; Wu et al. 2011; Tremaroli and Bäckhed 2012). Therefore, a decrease in butyrate production associated with changes in the human diet over evolution may have allowed a proportional increase in acetate production, nutritionally supporting the expansion in brain size in humans (Amato 2016).
Given that other primates have not experienced the same evolutionary diet shifts as humans, comparative data from humans and other primates can provide insight into this putative process. If humans exhibit reduced butyrate production patterns compared with all other primates, regardless of context, it suggests that our understanding of the interaction between diet, the gut microbiome, and health must reach beyond the modern human industrialized condition to consider a broader evolutionary context. Some evidence suggesting differences in human and nonhuman primate SCFA production patterns exists. One study reports that human gut microbiomes produce more SCFAs in vitro than gelada monkey gut microbiomes, but have a lower butyrate to propionate ratio (Frost et al. 2014). However, there is currently very little comparative data systematically describing SCFA production or production potential in humans and nonhuman primates.
In this study, we use a gene-targeted alignment approach to interrogate publicly available metagenomic data sets to test the hypothesis that butyrate production potential in humans from both nonindustrialized populations and industrialized populations is lower than that of wild nonhuman primates. Although multiple propionate, acetate, and butyrate production pathways have been well described in the literature (Wolin and Miller 1996; Duncan et al. 2002; Louis and Flint 2017), we chose to focus on butyrate due to the availability of a large reference database of butyrate production pathway genes (Vital et al. 2014, 2017). We analyze four major butyrate production pathways—one carbohydrate degradation pathway (acetyl-CoA), and three amino acid degradation pathways (4-aminobutyrate, glutarate, and lysine). Contrary to our expectations, we found that humans had higher abundances of the four pathways compared with most nonhuman primates. However, human pathway abundances are broadly similar to those of cercopithecoids. Additionally, we found differences in butyrate production pathways between humans in industrialized populations and humans in nonindustrialized populations. These data have important implications for the interactions between diet, the gut microbiome, and human health and allow us to situate an important metabolic function of the human gut microbiome within a broader evolutionary context.
Results
Pathway Abundance
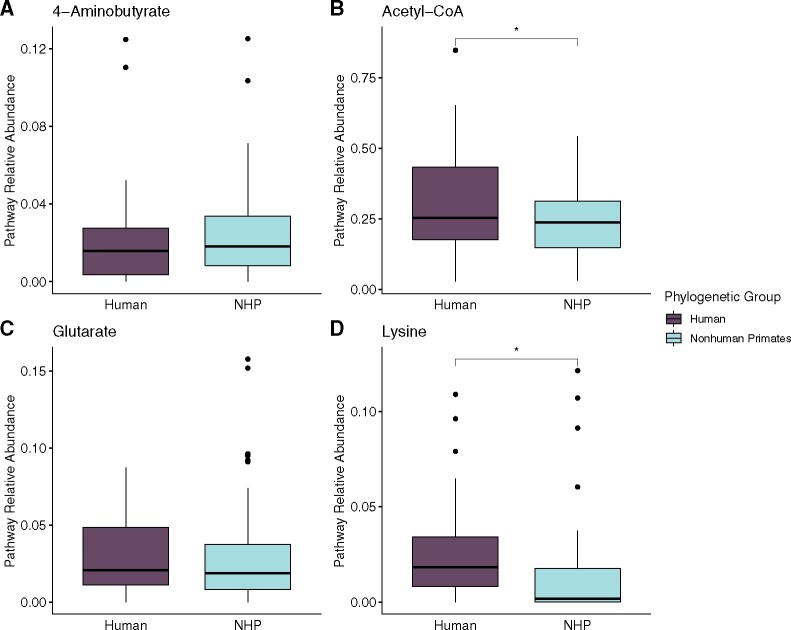
Shotgun metagenomic sequencing data sets from fecal samples from five major groups of primates—humans, apes, cercopithecoids (baboons, colobus monkeys, geladas, and guenons), platyrrhines (capuchins, howler monkeys, spider monkeys, and woolly monkeys), and lemurs and sifakas (see supplementary fig. S1 and table S1, Supplementary Material online, for details)—were queried using hidden Markov models (HMM) of genes from four butyrate production pathways: 4-aminobutyrate, acetyl-CoA, glutarate, and lysine. Although these pathways have different starting products, they share terminal enzymes (Louis and Flint 2017; Vital et al. 2017) (supplementary fig. S2, Supplementary Material online). We examined the effect of primate phylogeny—both using the broad taxonomic groups listed above and at the level of family—and the effect of dietary strategy (folivore, frugivore, grazer, and omnivore) on the abundance of all hits to each butyrate pathway relative to the abundance of housekeeping genes (pyrG, recA, and rplB) present in each sample. Due to very low detection of genes in the capuchin samples, they were only included in the family-level analysis. Humans had significantly higher acetyl-CoA pathway abundance and lysine pathway abundance (ANOVA, P = 0.022, P < 0.001) compared with nonhuman primates (fig. 1). Similarly, a significantly higher number of discrete pathways contributed to human butyrate production potential (ANOVA, P < 0.001), with humans having 0.696 more pathways compared with nonhuman primates (CI: 0.390–1.002).
Fig. 1.
The average abundance of four butyrate production pathways (A: 4-aminobutyrate, B: acetyl-CoA, C: glutarate, D: lysine) relative to the mean of housekeeping genes (pyrG, recA, and rplB) per sample in humans and nonhuman primates.
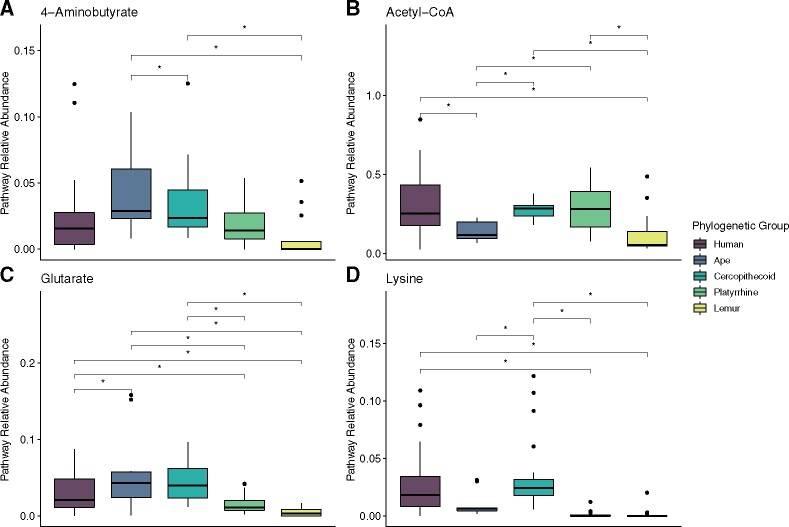
When comparing pathway abundance across major phylogenetic groups of primates, we found that the relative abundances of pathways in humans differed from most other groups of primates, with the exception of cercopithecoids (Tukey HSD, all Padj < 0.05) (fig. 2) (supplementary table S2, Supplementary Material online). We found a significant effect of phylogenetic group on all four individual butyrate production pathways (ANOVA, all P < 0.001) (fig. 2) (supplementary table S2, Supplementary Material online), though the magnitude of the differences in the abundance of the lysine pathway were slight. Humans had significantly higher relative abundances of the acetyl-CoA pathway compared with apes and lemurs, higher relative abundances of the glutarate pathway compared with platyrrhines and lemurs, and higher relative abundances of the lysine pathway compared with platyrrhines and lemurs (Tukey HSD, all Padj < 0.05) (fig. 2) (supplementary table S2, Supplementary Material online). Humans did not differ significantly from cercopithecoids in terms of relative pathway abundance for any of the pathways examined in this study (Tukey HSD, all Padj < 0.05) (fig. 2) (supplementary table S2, Supplementary Material online).
Fig. 2.
The average abundance of four butyrate production pathways (A: 4-aminobutyrate, B: acetyl-CoA, C: glutarate, D: lysine) relative to the mean of housekeeping genes (pyrG, recA, and rplB) in humans, nonhuman apes, cercopithecoids, platyrrhines, and lemurs.
Similarly, host phylogeny at the level of family also had a significant influence on the relative abundance of individual pathways (ANOVA, 4-aminobutyrate: P = 0.002, acetyl-CoA: P < 0.001, glutarate: P < 0.001, lysine: P < 0.001) (supplementary fig. S3 and table S2, Supplementary Material online). In particular, Cebidae and Indriidae generally had low pathway abundances, whereas Atelidae, Cercopithecidae, and Hominidae had elevated abundances of the acetyl-CoA pathway.
Within humans, lifestyle had a significant influence of the relative abundance of two pathways. Nonindustrialized populations had significantly higher relative pathway abundances of the glutarate pathway and significantly lower relative abundances of the lysine pathway compared with industrialized populations (ANOVA, both P < 0.05) (supplementary table S2, Supplementary Material online).
Surprisingly, dietary strategy had a moderate influence on relative pathway abundances. Glutarate and lysine pathway abundances were significantly associated with dietary strategy (ANOVA, glutarate: P = 0.003, lysine: P = 0.021). Pairwise comparisons revealed that omnivores did have higher relative abundances of the glutarate pathway compared with folivores and higher relative abundances of the lysine pathway compared with frugivores (Tukey HSD, both Padj < 0.05) (supplementary fig. S4 and table S2, Supplementary Material online). However, neither the relative abundance of the 4-aminobutyrate or acetyl-CoA pathways differed between primates with distinct dietary strategies (ANOVA, both P > 0.05) (supplementary fig. S4 and table S2, Supplementary Material online).
Functionally Complete Pathways within Single Microbial Taxa
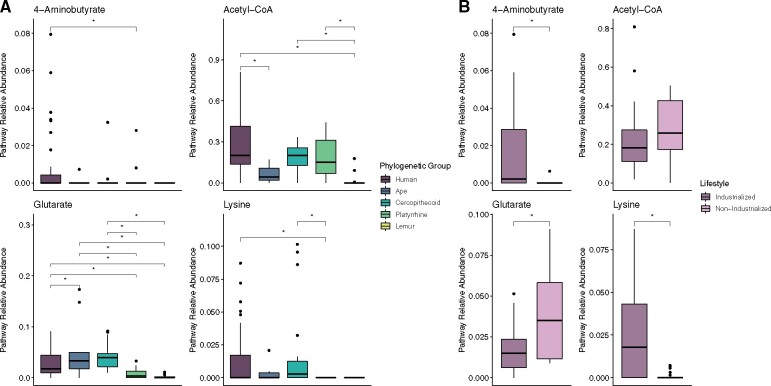
In order to examine only the abundance of butyrate production pathways conservatively believed to be functionally complete, we binned pathway hits by microbial genera and then included only those pathways where all pathway genes, including the terminal enzymes buk and/or but, were present within a single bacterial genus. Phylogenetic group had a significant effect on the relative abundance of all four complete pathways (ANOVA, all P < 0.05) (fig. 3 and supplementary table S3, Supplementary Material online), and, as with the analysis of all hits to butyrate pathways, humans were distinct from all clades of primates with the exception of cercopithecoids. Humans had significantly higher 4-aminobutyrate abundances than platyrrhines, higher acetyl-CoA abundances than apes and lemurs, higher glutarate abundances than apes, platyrrhines, and lemurs, and higher lysine abundances than platyrrhines (Tukey HSD, all Padj < 0.04) (fig. 3 and supplementary table S3, Supplementary Material online).
Fig. 3.
The relative abundance of functionally complete butyrate production pathways across all primates (A) and compared between industrialized and nonindustrialized populations of humans (B).
Relative abundances of complete pathways differed slightly between humans in industrialized populations (the United States and Great Britain) and nonindustrialized populations (Peru and Tanzania). Nonindustrialized populations had lower abundances of the 4-aminobutyrate and lysine pathways, but higher abundances of the glutarate pathway (ANOVA, all P < 0.05) (fig. 3 and supplementary table S3, Supplementary Material online). There were no significant differences in the relative abundance of the acetyl-CoA pathway between industrialized and nonindustrialized populations (ANOVA, P > 0.05).
Taxonomic Composition of Butyrate Production Pathways
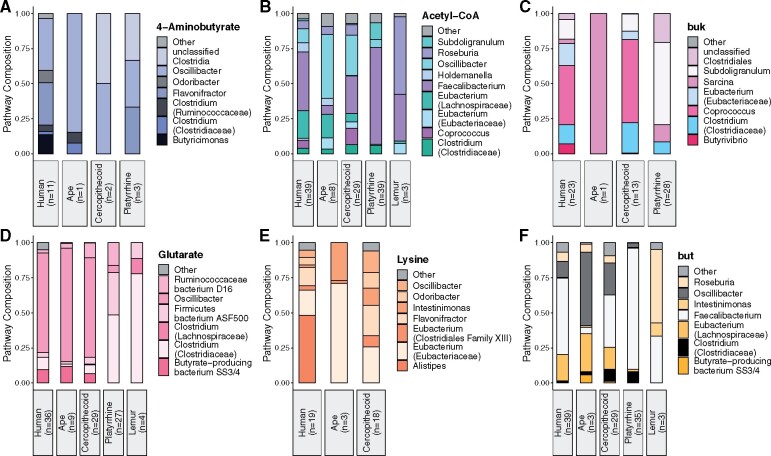
The taxonomic composition of butyrate production pathways was highly variable across phylogenetic groups (fig. 4 and supplementary table S4, Supplementary Material online). Three taxa distinguished humans from nonhuman primates—Butyricimonas and Odoribacter contributed to the 4-aminobutyrate pathway, and Alistipes contributed to the lysine pathway solely in humans. Taxonomic composition differed only slightly between industrialized and nonindustrialized populations of humans for the acetyl-CoA pathway and the but terminal enzyme. However, industrialized populations of humans had a greater number of taxa contributing to the 4-aminobutyrate, glutarate, and lysine pathways, and there were differences in the taxa contributing to the buk terminal enzyme (supplementary fig. S5, Supplementary Material online).
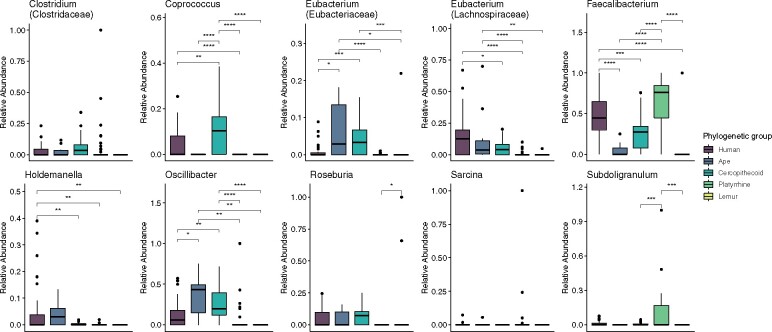
Fig. 4.
Taxonomic composition of 4-aminobutyrate (A), acetyl-CoA (B), glutarate (C), and lysine (D) butyrate producing pathways and the taxa with genes encoding the terminal enzymes buk (E) and but (F) in humans, nonhuman apes, cercopithecoids, platyrrhines, and lemurs.
Within the acetyl-CoA pathway, we compared the relative abundance of major taxa across phylogenetic groups (fig. 5 and supplementary table S5, Supplementary Material online). Humans were distinct from all other clades of primates, including cercopithecoids, in the relative abundance of taxa contributing to the acetyl-CoA pathway. Humans had higher relative abundances of Eubacterium (Lachnospiraceae) compared with cercopithecoids, platyrrhines, and lemurs (Tukey HSD, both P < 0.05), higher relative abundances of Faecalibacterium than apes, cercopithecoids, and lemurs (Tukey HSD, both P < 0.05), and higher relative abundances of Holdemanella compared with cercopithecoids, platyrrhines, and lemurs (Tukey HSD, both P < 0.05). Humans had lower relative abundances of Coprococcus compared with cercopithecoids and platyrrhines (Tukey HSD, both P < 0.05), lower relative abundances of Eubacterium (Eubacteriaceae) than apes or cercopithecoids (Tukey HSD, both P < 0.05), and lower relative abundances of Oscillibacter compared with apes or cercopithecoids (Tukey HSD, both P < 0.05). Within humans, nonindustrialized populations had higher relative abundances of Holdemanella and lower relative abundances of Eubacteria (Lachnospiraceae) (ANOVA, both P < 0.05) (supplementary table S5, Supplementary Material online).
Fig. 5.
Abundance of taxa contributing to the acetyl-CoA pathway in humans, nonhuman apes, cercopithecoids, platyrrhines, and lemurs.
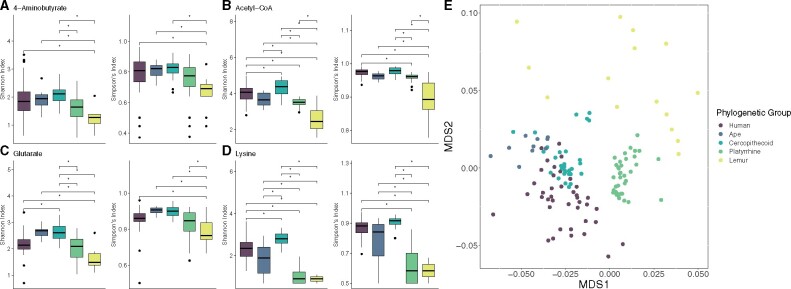
Alpha diversity—measured using both the Shannon diversity (SD: an index that accounts for both abundance and evenness of species) and Simpson (SI: an index that accounts only for evenness) indices—of taxa contributing to individual pathways differed significantly between phylogenetic groups for each butyrate production pathway (ANOVA, all P < 0.001) (fig. 6 and supplementary tables S6 and S7, Supplementary Material online). Alpha diversity values from a 16S analysis of the gut microbiome composition of a similar data set have been included as a reference point (supplementary table S7, Supplementary Material online) (Amato, Mallott, et al. 2019). Humans generally had within sample diversity values that were similar to apes, were slightly lower than cercopithecoids, and were significantly higher than platyrrhines and/or lemurs, depending on the pathway examined. Humans exhibited higher alpha diversity for the 4-aminobutyrate pathway compared with lemurs, higher alpha diversity than platyrrhines and lemurs for the acetyl-CoA pathway, higher alpha diversity than lemurs for the glutarate pathway, and higher alpha diversity than lemurs and platyrrhines for the lysine pathway (Tukey HSD, all P < 0.05). Humans had lower within sample diversity than cercopithecoids for all pathways except the 4-aminobutyrate pathway, but only when using the SI to calculate diversity (Tukey HSD, all P < 0.05).
Fig. 6.
Alpha (A–D) and beta (E) diversity of butyrate producing pathways in humans, nonhuman apes, cercopithecoids, platyrrhines, and lemurs.
The alpha diversity of butyrate production pathways did not differ between industrialized and nonindustrialized populations of humans, with the exception of the lysine pathway. Nonindustrialized human populations had lower SI values for the lysine pathway (ANOVA, P < 0.001).
The composition of taxa contributing to all butyrate production pathways was significantly influenced by phylogenetic group (PERMANOVA, Bray–Curtis distances, F = 11.497, R2 = 0.254, P < 0.001) (fig. 6 and supplementary table S8, Supplementary Material online). Pairwise comparisons indicated that the community composition was distinct for each phylogenetic group (pairwise PERMANOVA, all Padj = 0.001). Family similarly had a significant influence on taxonomic composition (F = 10.638, R2 = 0.284, P < 0.001). In contrast to the pathway abundance results, primates with different dietary strategies had significantly different butyrate production pathway taxonomic compositions (F = 7.010, R2 = 0.134, P < 0.001), though the strength of effect was lower than that for phylogenetic group or family.
Butyrate Production
In order to contextualize our computational analysis, we performed a literature search to find comparative data across primates on butyrate production. Although published values from nonhuman primates are limited and most describe the concentration of butyrate after a period of fermentation and not the butyrate production rate, humans do fall within the range of nonhuman primate variation (table 1). Butyrate concentrations—measured either from fecal metabolites or from in vitro fermentation—ranged from 3 to 26 mol% in humans and 3 to 29 mol% in nonhuman primates. Data from nonhuman primates are limited to eight species and within-species variation is high.
Table 1.
Butyrate Production Values from Published Studies of Humans and Nonhuman Primates.
| Species | Concentration | Proportion of SCFA | Method | Substrates | Study | Notes |
|---|---|---|---|---|---|---|
| Platyrrhine | ||||||
| Alouatta pigra | 1.7–2.8 mM | — | Fecal metabolite concentration | NA | Amato et al. (2015) | — |
| Cercopithecoid | ||||||
| Cercopithecus neglectus | 7.53 mM | 16.42 mol% | In vitro fermentation (12 h) | Blended species-specific diet | Lambert and Fellner (2012) | Captive individuals |
| Colobus guereza | 5.96 mM | 10.43 mol% | In vitro fermentation (12 h) | Blended species-specific diet | Lambert and Fellner (2012) | Captive individuals |
| Papio hamadryas | 9.04 mM | 10.86 mol% | In vitro fermentation (12 h) | Blended species-specific diet | Lambert and Fellner (2012) | Captive individuals |
| Papio hamadryas | 17.80 mM | 31.43 mol% | In vitro fermentation (24 h) | Blended species-specific diet | McKenney et al. (2014) | Captive individuals |
| Macaca fascicularis | — | 3 mol% | Fecal metabolite concentration | NA | Nagpal et al. (2018) | Captive individuals |
| Macaca mulatta | 4.41–4.57 nmol/g | — | Fecal metabolite concentration | NA | Hasegawa et al. (2018) | Captive individuals, juveniles only |
| Ape | ||||||
| Gorilla gorilla | 9.01 mM | 13.00 mol% | In vitro fermentation (12 h) | Blended species-specific diet | Lambert and Fellner (2012) | Captive individuals |
| Gorilla gorilla | 19.47 mM | 28.57 mol% | In vitro fermentation (24 h) | Blended species-specific diet | McKenney et al. (2014) | Captive individuals |
| Pan troglodytes | 7.30 mM | 9.20 mol% | In vitro fermentation (12 h) | Blended species-specific diet | Lambert and Fellner (2012) | Captive individuals |
| Pan troglodytes | 8.94 mM | 16.45 mol% | In vitro fermentation (24 h) | Blended species-specific diet | McKenney et al. (2014) | Captive individuals |
| Gorilla gorilla | — | 8.4–12 mol% | Fecal metabolite concentration | NA | Gomez et al. (2015) | — |
| Human | ||||||
| Humans—Industrialized | — | 8–26 mol% | In vitro fermentation (24 h) | Pectin, gum Arabic, oat bran, wheat bran, and cellulose | Bugaut and Bentéjac (1993) | — |
| Humans—Industrialized | 3.0–23.7 mM | 8.4–13.6 mol% | In vitro fermentation (7 days) | Amylopectin, pectin, inulin (dahlia), xylan (oat), and inulin (chicory) | Duncan et al. (2003) | — |
| Humans—Industrialized | 26.1 μmol/ml | — | Cecum metabolite concentration | NA | Bugaut and Bentéjac (1993) | — |
| Humans—Industrialized | 4.36–17.67 mM | 7–16 mol% | Fecal metabolite concentration | NA | Duncan et al. (2007) | — |
| Humans—Industrialized | — | 3 mol% | Fecal metabolite concentration | NA | Nagpal et al. (2018) | — |
| Humans—Industrialized | 0.675 ± 0.71 μmol/g | 16.60 ± 5.2% | Fecal metabolite concentration | NA | Schnorr et al. (2014) | — |
| Humans—Nonindustrialized | 0.601 ± 0.43 μmol/g | 12.16 ± 4.5% | Fecal metabolite concentration | NA | Schnorr et al. (2014) | — |
Discussion
This study aimed to situate human gut microbial butyrate production pathways in a broader evolutionary context than is currently common in the existing literature. We expected that the evolutionary changes in human diets toward decreased fiber consumption would be mirrored by functional changes in the human gut microbiome. Specifically, we predicted that butyrate production potential would be reduced in humans compared with nonhuman primates. Our gene-targeted alignment approach using shotgun metagenomic data from humans and nonhuman primates indicates this is not the case. Instead, compared with primates overall, humans had higher acetyl-CoA (a carbohydrate degradation pathway) and lysine (an amino acid degradation pathway) pathway abundances. When examining differences between humans and specific taxonomic groups of primates, humans also had similar or higher relative abundances of most butyrate production pathways compared with apes, platyrrhines, and lemurs, and similar butyrate production pathway relative abundances compared with cercopithecoids. Taxonomic analysis of butyrate production pathways revealed differences between humans and cercopithecoids, in terms of both the overall composition of taxa contributing to all butyrate production pathways and the relative abundances of major taxa contributing to the acetyl-CoA pathway. Additionally, our data suggest that humans in general have lost few taxa related to butyrate production compared with apes and cercopithecoids and do not show reduced diversity compared with nonhuman primates more generally. Individual humans on average have more butyrate production pathways present than nonhuman primate individuals. This is surprising, as a previous comparative study of gut microbial diversity across primates suggested that humans have lower overall gut microbial diversity compared with nonhuman primates (supplementary table S7, Supplementary Material online) (Amato, Mallott, et al. 2019).
Although we did not predict the observed similarities in butyrate production pathway abundances between humans and cercopithecoids, these findings mirror recent data describing patterns in overall gut microbiome composition and function among humans and nonhuman primates. Specifically, humans possess a gut microbiome that is more similar to that of cercopithecoids than that of phylogenetically more closely related apes (Amato, Mallott, et al. 2019; Gomez et al. 2019). This pattern is likely a result of humans and cercopithecoids occupying analogous ecological niches and experiencing a convergence in microbiome structure and function. Our data suggest a similar dynamic. Nevertheless, despite overall similarities in butyrate production potential between humans and cercopithecoids, we detected differences in the specific taxa contributing to the acetyl-CoA pathway between humans and cercopithecoids. As a result, humans and cercopithecoids are likely relying on different microbial taxa to achieve a similar functional output.
Despite the overall similarities that we detected in butyrate production potential between humans and cercopithecoids, our data indicate that humans have a higher potential compared with all other primates, as determined by multiple measures. The potentially increased capacity for butyrate production by the human microbiome may indicate an increase in the digestive efficiency of the gut microbiome over the course of human evolution (Amato 2016). Instead of shifting from butyrate production to acetate production as dietary fiber decreased, the human gut microbiome may have shifted toward a higher production of all SCFAs in response to a suite of human anatomical, behavioral, physiological, and dietary changes. Data describing the prevalence and relative abundances of production pathways for other SCFAs will provide further insight into these dynamics moving forward.
Increased butyrate production pathway diversity in humans compared with nonhuman primates may also signal increased resilience in SCFA functionality as a result of functional redundancy. Specifically, increased diversity makes it more likely that the most efficient strain (or pathway) for a given environmental context will emerge as an abundant member of the microbial community (Kettle et al. 2015). High rates of functional redundancy have been observed for other microbial pathways in the human colon (Egert et al. 2006; Kettle et al. 2014, 2015), and the gut microbiome is considered to be generally functionally robust compared with the microbiome of other body sites and other environments (Eng and Borenstein 2018). Further, functional redundancy has been shown to be particularly important in both butyrate and propionate production in humans (Mahowald et al. 2009; Reichardt et al. 2018). Experimental studies that can link the activity of specific production pathways to specific host diets should more rigorously test the extent to which these dynamics emerge.
As expected, we identified higher relative abundances of butyrate production pathways in nonindustrialized populations compared with industrialized populations. Similar patterns have been previously reported by studies examining both the taxonomic composition and function of the human microbiome (Schnorr et al. 2014; Smits et al. 2017; Jacobson et al. 2021). The gut microbiomes of nonindustrialized human populations generally have increased fiber degradation and SCFA production potential. For example, a study of variation in the terminal buk and but enzymes of the butyrate synthesis pathways reported lower relative abundances in industrialized populations (Jacobson et al. 2021). Given that industrialized diets typically incorporate much lower amounts of fiber than nonindustrialized populations, these findings are not surprising. However, what is surprising is that we did not detect any reductions in the taxonomic diversity of microbes that possess the butyrate production pathway. On the contrary, we found that more taxa contribute to amino acid degradation pathways in industrialized populations of humans compared with nonindustrialized population. This outcome was unexpected, as previous studies have found that industrialized populations have lower gut microbial diversity compared with nonindustrialized populations, likely due to differences in diet, antibiotic use, and environmental exposure (Blaser and Falkow 2009; Ou et al. 2013; Schnorr et al. 2014; Clemente et al. 2015; Smits et al. 2017; Blaser 2018). Additionally, this loss of diversity can be experimentally induced and increases across generations (Sonnenburg et al. 2016; Schulfer et al. 2018). However, decreases in the overall diversity of the gut microbiome do not necessarily translate to decreases in the diversity within a functional group (Smits et al. 2017) or of taxa contributing to a specific function. In fact, it has been previously reported that nonindustrialized Hadza hunter-gatherers have fewer bacterial taxa associated with butyrate production and lower fecal butyrate concentrations compared with industrialized Italian individuals (Schnorr et al. 2014). Similarly, a comparison of multiple industrialized, rural agricultural, pastoralist, and hunter-gatherer populations indicated that species-level richness, phylogenetic diversity, and evenness in butyrate synthesis genes is lower in nonindustrialized populations (Jacobson et al. 2021). Therefore, it appears that although industrialization causes declines in the overall diversity of the gut microbiome, other processes contribute to the maintenance of diversity in butyrate production pathways. For example, our data suggest that industrialized populations may have an increased capacity for producing butyrate from amino acids, specifically. This difference may reflect the fact that fewer carbohydrates and more amino acids are available as substrates for fermentation in lower-fiber, higher-fat, and higher-protein industrialized diets.
Overall, our results suggest that the human gut microbiome has an increased capacity for butyrate production that is resilient to dietary shifts across both evolutionary and modern timescales. Nevertheless, whether the observed increase in butyrate production potential in humans results in a higher rate or efficiency of butyrate production remains unclear. In a study of humans, the abundance of one butyrate-producing taxa, Faecalibacterium prausnitzii, was not correlated with transcriptional activity related to butyrate production—individuals with IBD had lower transcriptional activity but not lower absolute abundance of F. prausnitzii (Schirmer et al. 2018). Another study of healthy human subjects found low interindividual variation in butyrate production and uniformly high protein expression of butyrate-producing taxa (Tanca et al. 2017), although the authors note that enzyme abundance may not correlate with metabolite concentrations. The potential lack of relationship between pathway abundances and butyrate production may be particularly relevant for the amino acid degradation pathways, which exclusively use lysine and glutamate as substrates (Louis and Flint 2017) and would be limited by the amount of dietary protein that reaches the large intestine and contains these specific amino acids. In addition, although human gut microbiomes may or may not be producing butyrate at a higher rate than nonhuman primate gut microbiomes, shifts to a lower fiber diet across human populations or within an individual over time likely still decrease butyrate production through decreasing substrate availability (Phillips et al. 1995; Duncan et al. 2007; Brinkworth et al. 2009; De Filippo et al. 2010; Wu et al. 2011; Tremaroli and Bäckhed 2012; David et al. 2014; Yang et al. 2016). Our study, which relies on metagenomic data only, is unable to determine if humans have increased butyrate production compared with nonhuman primates in addition to having higher pathway gene abundances or to directly examine how differences in the consumption of specific nutrients influences butyrate production. Although published fecal butyrate concentrations in nonhuman primates suggest that humans and nonhuman primates have similar microbial-mediated butyrate production rates in the large intestine, the available data show a high level of variability influenced by host diet and/or fermentation substrates. They also primarily include captive individuals and are far from comprehensive in their coverage of nonhuman primate species. Captive primates have decreased fiber consumption, differ in their gut retention times, and have altered gut microbial communities relative to their wild counterparts (Amato et al. 2013; Clayton et al. 2016; Frankel et al. 2019; Houtz et al. 2021), all of which would influence butyrate production rates and capacity. The resulting available nonhuman primate fecal butyrate concentration data may be more similar to that of humans than fecal butyrate concentrations obtained from wild nonhuman primates would be. Additionally, the anatomy of the gastrointestinal tract varies across the Primate order (Lambert 1998), influencing retention times and fecal output. These differences are not often accounted for despite potential impacts the comparability of results.
It is also important to acknowledge that our analysis is necessarily simplified in order to best utilize the available reference sequences for butyrate production pathway genes. For example, we were not able to include the 3-methyasparate amino acid degradation pathway that results in butyrate production (Louis and Flint 2017), nor pathways by which lactate or acetate are fermented to butyrate (Duncan et al. 2002, 2004; Calder et al. 2004; Bourriaud et al. 2005). In addition, although the pathways analyzed in our study likely capture some cross-feeding of lactate or succinate as there is some overlap in the enzymatic pathways (Louis and Flint 2017), we are likely not fully accounting for the role of metabolic cross-feeding (Belenguer et al. 2006). Cross-feeding plays a role in SCFA production, as SCFAs and intermediate products are exchanged between bacterial taxa and contribute to butyrate formation (Duncan et al. 2004; Belenguer et al. 2006; Mahowald et al. 2009; George et al. 2018; Reichardt et al. 2018). High-quality genome scale metabolic models can be used to model these more complex associations contributing to butyrate production (van der Ark et al. 2017), but these models rely on well-characterized and well-annotated microbial reference genomes.
Finally, the development of database-independent approaches moving forward would avoid potential biases toward human-associated butyrate producers, as the reference database was developed primarily from bacterial genomes found in human gut microbiome samples from industrialized populations. We know that the human gut microbiome contains bacteria that are as yet uncultured or for which we do not have high-quality draft genomes (Bishara et al. 2018; Jacobson et al. 2021), particularly when examining nonindustrialized human populations who have been under sampled relative to industrialized populations (Jacobson et al. 2021). Nonhuman primate gut microbiomes are less well-characterized than human gut microbiomes (Amato, Sanders, et al. 2019; Manara et al. 2019). In our data set, humans from nonindustrialized populations had fewer taxa contributing to butyrate production pathways despite having higher pathway abundances, and Cebidae and Indriidae both had relatively few hits to our butyrate production pathway gene database. Although the gut microbiota of Cebidae are mainly characterized by taxa that are not known butyrate-producers, their gut microbiota includes a substantial fraction of unassigned or ambiguously assigned taxa (Mallott and Amato 2018; Mallott et al. 2018; Orkin et al. 2019). Similarly, the majority of taxa in the gut microbiome of Indriidae cannot be assigned to a genus (Perofsky et al. 2017; Springer et al. 2017). Despite potential biases toward human-associated microbiomes from industrialized populations in the database, we do see higher abundances of the glutarate pathway in apes compared with humans, and a higher diversity of taxa contributing to the acetyl-CoA, glutarate, and lysine pathways in cercopithecoids compared with humans. Additionally, we found five bacterial taxa containing butyrate production pathways that were only ever identified in nonhuman primate samples—Kineothrix, Anaerococcus, Nocardioides, Clostridiales bacterium NK3B98, and Lachnospiraceae bacterium NC2008 (supplementary table S4, Supplementary Material online). However, we may not be capturing the full diversity of taxa contributing to butyrate production in nonhuman primates or humans from nonindustrialized populations due to the biases in our reference database, which may contribute to our finding of a higher potential for butyrate production in humans and higher diversity of taxa contributing to butyrate production in industrialized populations of humans compared with nonindustrialized populations.
In conclusion, in spite of decreases in dietary fiber across both evolutionary and modern timescales, the human microbiome exhibits a capacity for butyrate production comparable to or higher than that of nonhuman primates. Human-associated gut microbiota have not lost butyrate production pathways, either in terms of abundance or diversity. Instead, the abundance of butyrate production pathways in human gut microbiomes appear similar to those of cercopithecoids, indicating that host ecology or physiology may be driving convergence of human and cercopithecoids gut microbiomes. We posit that higher abundances of butyrate-producing pathways as well as the observed redundancy and associated resilience in butyrate production pathways in humans may have resulted from an evolutionary shift toward a gut microbiome that produces more SCFAs to support increased in human energy requirements. However, comparative data across the primate order examining additional SCFAs and incorporating direct measures of SCFA production will be necessary to robustly test this hypothesis.
Materials and Methods
Data Sets
The data set used in this study included shotgun metagenomic sequencing data sets from the guts of 19 primate species (n = 100 samples) (Amato, Sanders, et al. 2019; Orkin et al. 2019), three industrialized populations of humans (n = 25) (Obregon-Tito et al. 2015; Mcdonald et al. 2018), one rural agricultural population of humans (n = 5) (Obregon-Tito et al. 2015), and two hunter-gatherer populations of humans (n = 15) (Obregon-Tito et al. 2015; Smits et al. 2017). For the human gut metagenomes, we selected adult individuals with a normal BMI (when possible). Details on the specific samples included and accession numbers for all samples are included (supplementary fig. S1 and table S1, Supplementary Material online).
We used an existing butyrate pathway gene reference library (Vital et al. 2014, 2017), updating it to include novel variants from the nonhuman primate metagenomes in our data set as described below. This database includes genes from known butyrate producers for the 4-aminobutyrate (4-hydroxybutyrate dehydrogenase: abfD, butyryl-CoA : 4-hydroxybutyrate CoA transferase: 4hbt, and 4-hydroxybutyryl-CoA dehydratase/vinylacetyl-CoA 3,2-isomerase: abfH), acetyl-CoA (acetyl-CoA transferase: thl; β-hydroxybutyryl-CoA dehydrogenase: bhbd; and crotonase: cro), glutarate (glutaconate-CoA transferase, α/β subunit: gctA/B; 2-hydroxyglutaryl-CoA dehydratase, α/β/γ subunit: hgCoAdA/B/C; glutaconyl-CoA decarboxylase, α/β subunit: gcdA/B), and lysine (lysine-2,3-aminomutase: kamA; β-lysine-5,6-aminomutase, α/β subunit: kamD/E; 3,5-diaminohexanoate dehydrogenase: kdd; 3-keto-5-aminohexanoate cleavage enzyme: kce; 3-aminobutyryl-CoA ammonia-lyase: kal; butyryl-CoA: acetoacetate CoA transferase: atoA/D) pathways, as well as genes shared by all pathways (butyryl-CoA dehydrogenase and electron transfer protein, α/β subunit: bcd-etfA/B) and terminal enzymes (butyrate kinase: buk; butyryl-CoA: acetate CoA transferase: but) (supplementary fig. S2, Supplementary Material online). Although the database does not include gene sequences for the phosphotransbutyrylase enzyme involved in the step just prior the terminal butyryl-phosphate to butyric acid reaction, the exclusion of this enzyme will not likely have a large effect on the estimates of individual butyrate pathway abundance using the methodology described below. Some of the enzymes noted above (thl, bhbd, cro, bcd-etfA/B) are also found in the pyruvate fermentation to butanol pathway (Jones and Woods 1986). Additionally, crotonyl-CoA can feed into the acetate production pathway (Duncan et al. 2002). However, the but and/or buk genes are absent in these other pyruvate fermentation pathways. Therefore, the functionally complete pathway analysis (see below) is restricted to bacterial genera in which these enzymes can result in butyrate production.
For samples where both a buk and a but gene was present within the same genera, we further screened the sequences. We first checked if the buk and but genes were found in the same species or strain of bacteria. If so, we then performed a translated nucleotide BLAST search against the GenBank. In two cases, we removed one of the two terminal enzymes from our database. For Coprococcus eutactus, only buk was retained, as the putative but sequences also matched acetyl-CoA hydrolase and 4hbt with high confidence and the published literature indicates that C. eutactus does not have a function but enzyme (Louis et al. 2007). Similarly, for Clostridium sp. L2-50, we only retained the buk sequences, as the but sequences had a high similarity to 4bht and the published literature was mixed (Duncan et al. 2002; Louis et al. 2004, 2007). However, for Eubacterium ventrosium, both buk and but were retained, as all high confidence hits for the buk sequences were to buk enzymes from other species.
Sequence Processing and Analysis
The metagenomic sequence analysis was based on a published workflow for analyzing butyrate pathways in gut microbial communities (Vital et al. 2014, 2017). Raw sequences from gut metagenome samples were trimmed and quality-filtered in Trimmomatic v0.33 (Bolger et al. 2014). Individual samples were aligned to protein HMMs for each butyrate pathway gene using Xander, a gene-targeted sequence aligner (Wang et al. 2015). Nucleotide sequences that aligned to each gene were dereplicated, chimeras were filtered, and FrameBot analysis was performed using USEARCH v8.1 (www.drive5.com/usearch, last accessed May 15, 2020) and RDPTools (github.com/rdpstaff/RDPTools). FrameBot corrected sequences were then aligned to the original HMMs using HMMER 3.0 (hmmer.org) and alignments were subject to complete linkage clustering at a 95% threshold in RDPTools. Clusters were then filtered to remove those that contained less than three sequences or contained sequences from the original reference database. Representative sequences from the remaining clusters were than blasted against the original reference database using BlastN v2.8.1, annotated with the top hit from the original reference database, and filtered for ≥80% alignment coverage and >90% identity to genes in the original database. These filtered representative sequences were then added to the reference database, along with sequences for three single-copy housekeeping genes—pyrG, recA, and rplB—obtained from FunGene (http://fungene.cme.msu.edu/, last accessed May 15, 2020). The updated database was dereplicated in USEARCH v.11.0.667, and quality-filtered gut metagenomes were aligned against the database using the very-sensitive option of Bowtie2 v2.2.3 (Langmead and Salzberg 2012). Resulting alignments were visualized using Samtools v0.1.19 (www.htslib.org, last accessed May 15, 2020).
Pathway Abundance, Presence, and Taxonomic Composition
Pathway abundance was calculated as the mean of all length-corrected unique pathway gene abundances, with the exception that the median of length-corrected gene abundances was used for 4-aminobutyrate pathway due to proportionally high numbers of the abfD gene. Genes that are shared across pathways (bcd-etfA/B) and the genes for the terminal enzymes (but/buk) were not included in pathway abundance calculations. Values are reported as abundances relative to the mean abundance of the three single-copy housekeeping genes within the same sample. A pathway was considered present if all genes for that pathway were found in a given sample.
In order to assess taxonomic composition of pathways and the abundance of functionally complete pathways, pathway gene hits were binned by genus, or higher taxonomic classifications if the identity of the hit was not resolved to the genus level. A pathway was considered complete for a specific bacterial taxon if all genes for that pathway were present and assigned to that taxon within a sample. The presence of gctA, atoA/D, and kal were not required to identify a pathway as complete, due to low detection rates across samples for those genes.
Statistical Analyses
Comparisons of pathway abundance and presence between humans and nonhuman primates, phylogenetic groups, and industrialized and nonindustrialized humans were performed using linear models in the mosaic package in R (Pruim et al. 2017). Tukey Honest Significant Differences were used for pairwise comparisons between groups. Alpha and beta diversity using all pathway hits were computed using the vegan package in R (Oksanen et al. 2020). Alpha diversity was calculated using Shannon–Weaver and Simpson’s indices. Beta diversity was calculated using Bray–Curtis dissimilarity indices. Differences in community composition between phylogenetic groups were tested using permutational multivariate analysis of variance (PERMANOVA) using the adonis function within the vegan package. Pairwise PERMANOVAs were performed using the pairwiseAdonis package in R.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
Thank you to Marius Vital for his advice on the computational methodology. This research was supported in part through the computational resources and staff contributions provided by the Genomics Compute Cluster which is jointly supported by the Feinberg School of Medicine, the Center for Genetic Medicine, and Feinberg’s Department of Biochemistry and Molecular Genetics, the Office of the Provost, the Office for Research, and Northwestern Information Technology. The Genomics Compute Cluster is part of Quest, Northwestern University’s high-performance computing facility, with the purpose to advance research in genomics. K.R.A. is supported as a Fellow in CIFAR’s “Humans and the Microbiome” program. E.K.M. contributed to conceptualization of the project, data acquisition and analysis, and writing the manuscript. K.R.A. contributed to conceptualization of the project and reviewing and editing the manuscript.
Data Availability
Sequencing data were drawn from published data sets available in the European Nucleotide Archive under accession numbers PRJEB22679, PRJEB11419, PRJNA268964, PRJNA485217, and PRJNA392180. All code for the analyses and the updated butyrate pathway gene database can be found at github.com/emallott/butyrate_pathway.
References
- Aiello LC, Wells JCK.. 2002. Energetics and the evolution of the genus Homo. Annu Rev Anthropol. 31(1):323–338. [Google Scholar]
- Amato KR. 2016. Incorporating the gut microbiota into models of human and non-human primate ecology and evolution. Am J Phys Anthropol. 159:S196–S215. [DOI] [PubMed] [Google Scholar]
- Amato KR, Leigh SR, Kent A, Mackie RI, Yeoman CJ, Stumpf RM, Wilson BA, Nelson KE, White BA, Garber PA.. 2015. The gut microbiota appears to compensate for seasonal diet variation in the wild black howler monkey (Alouatta pigra). Microb Ecol. 69(2):434–443. [DOI] [PubMed] [Google Scholar]
- Amato KR, Mallott EK, McDonald D, Dominy NJ, Goldberg T, Lambert JE, Swedell L, Metcalf JL, Gomez A, Britton GAO, et al. 2019. Convergence of human and Old World monkey gut microbiomes demonstrates the importance of human ecology over phylogeny. Genome Biol. 20(1):201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amato KR, Sanders J, Song SJ, Nute M, Metcalf JL, Thompson LR, Morton JT, Amir A, McKenzie V, Humphrey G, et al. 2019. Evolutionary trends in host physiology outweigh dietary niche in structuring primate gut microbiomes. ISME J. 13(3):576–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amato KR, Yeoman CJ, Kent A, Righini N, Carbonero F, Estrada A, Gaskins HR, Stumpf RM, Yildirim S, Torralba M, et al. 2013. Habitat degradation impacts black howler monkey (Alouatta pigra) gastrointestinal microbiomes. ISME J. 7(7):1344–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belenguer A, Duncan SH, Calder AG, Holtrop G, Louis P, Lobley GE, Flint HJ.. 2006. Two routes of metabolic cross-feeding between Bifidobacterium adolescentis and butyrate-producing anaerobes from the human gut. Appl Environ Microbiol. 72(5):3593–3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishara A, Moss EL, Kolmogorov M, Parada AE, Weng Z, Sidow A, Dekas AE, Batzoglou S, Bhatt AS.. 2018. High-quality genome sequences of uncultured microbes by assembly of read clouds. Nat Biotechnol. 36(11):1067–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaser MJ. 2018. The past and future biology of the human microbiome in an age of extinctions. Cell 172(6):1173–1177. [DOI] [PubMed] [Google Scholar]
- Blaser MJ, Falkow S.. 2009. What are the consequences of the disappearing human microbiota? Nat Rev Microbiol. 7(12):887–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B.. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30(15):2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourriaud C, Robins RJ, Martin L, Kozlowski F, Tenailleau E, Cherbut C, Michel C.. 2005. Lactate is mainly fermented to butyrate by human intestinal microfloras but inter-individual variation is evident. J Appl Microbiol. 99(1):201–212. [DOI] [PubMed] [Google Scholar]
- Brinkworth GD, Noakes M, Clifton PM, Bird AR.. 2009. Comparative effects of very low-carbohydrate, high-fat and high-carbohydrate, low-fat weight-loss diets on bowel habit and faecal short-chain fatty acids and bacterial populations. Br J Nutr. 101(10):1493–1502. [DOI] [PubMed] [Google Scholar]
- Bugaut M, Bentéjac M.. 1993. Biological effects of short-chain fatty acids in nonruminant mammals. Annu Rev Nutr. 13:217–241. [DOI] [PubMed] [Google Scholar]
- Calder AG, Stewart CS, Duncan SH, Lobley GE, Flint HJ, Holtrop G.. 2004. Contribution of acetate to butyrate formation by human faecal bacteria. Br J Nutr. 91:915. [DOI] [PubMed] [Google Scholar]
- Cani PD. 2014. The gut microbiota manages host metabolism. Nat Rev Endocrinol. 10(2):74–76. [DOI] [PubMed] [Google Scholar]
- Carmody RN, Wrangham RW.. 2009. The energetic significance of cooking. J Hum Evol. 57(4):379–391. [DOI] [PubMed] [Google Scholar]
- Cho I, Blaser MJ.. 2012. The human microbiome: at the interface of health and disease. Nat Rev Genet. 13(4):260–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clavel T, Gomes-Neto JC, Lagkouvardos I, Ramer-Tait AE.. 2017. Deciphering interactions between the gut microbiota and the immune system via microbial cultivation and minimal microbiomes. Immunol Rev. 279(1):8–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton JB, Vangay P, Huang H, Ward T, Hillmann BM, Al-Ghalith GA, Travis DA, Long HT, Tuan BV, Minh VV, et al. 2016. Captivity humanizes the primate microbiome. Proc Natl Acad Sci U S A. 113(37):10376–10381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemente JC, Pehrsson EC, Blaser MJ, Sandhu K, Gao Z, Wang B, Magris M, Hidalgo G, Contreras M, Noya-Alarcon O, et al. 2015. The microbiome of uncontacted Amerindians. Sci Adv. 1(3):e1500183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummings JH, Macfarlane GT.. 1997. Role of intestinal bacteria in nutrient metabolism. JPEN J Parenter Enteral Nutr. 21(6):357–365. [DOI] [PubMed] [Google Scholar]
- David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, Ling AV, Devlin AS, Varma Y, Fischbach MA, et al. 2014. Diet rapidly and reproducibly alters the human gut microbiome. Nature 505(7484):559–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeCasien AR, Williams SA, Higham JP.. 2017. Primate brain size is predicted by diet but not sociality. Nat Ecol Evol. 1(5):112. [DOI] [PubMed] [Google Scholar]
- De Filippo C, Cavalieri D, Di Paola M, Ramazzotti M, Poullet JB, Massart S, Collini S, Pieraccini G, Lionetti P.. 2010. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci U S A. 107(33):14691–14696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan SH, Barcenilla A, Stewart CS, Pryde SE, Flint HJ.. 2002. Acetate utilization and butyryl coenzyme A (CoA):acetate-CoA transferase in butyrate-producing bacteria from the human large intestine. Appl Environ Microbiol. 68(10):5186–5190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan SH, Belenguer A, Holtrop G, Johnstone AM, Flint HJ, Lobley GE.. 2007. Reduced dietary intake of carbohydrates by obese subjects results in decreased concentrations of butyrate and butyrate-producing bacteria in feces. Appl Environ Microbiol. 73(4):1073–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan SH, Louis P, Flint HJ.. 2004. Lactate-utilizing bacteria, isolated from human feces, that produce butyrate as a major fermentation product. Appl Environ Microbiol. 70(10):5810–5817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan SH, Scott KP, Ramsay AG, Harmsen HJM, Welling GW, Stewart CS, Flint HJ.. 2003. Effects of alternative dietary substrates on competition between human colonic bacteria in an anaerobic fermentor system. Appl Environ Microbiol. 69(2):1136–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egert M, de Graaf AA, Smidt H, de Vos WM, Venema K.. 2006. Beyond diversity: functional microbiomics of the human colon. Trends Microbiol. 14(2):86–91. [DOI] [PubMed] [Google Scholar]
- Eng A, Borenstein E.. 2018. Taxa-function robustness in microbial communities. Microbiome 6(1):45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankel JS, Mallott EK, Hopper LM, Ross SR, Amato KR.. 2019. The effect of captivity on the primate gut microbiome varies with host dietary niche. Am J Primatol. 81(12):e23061. [DOI] [PubMed] [Google Scholar]
- Frost GS, Walton GE, Swann JR, Psichas A, Costabile A, Johnson LP, Sponheimer M, Gibson GR, Barraclough TG.. 2014. Impacts of plant-based foods in ancestral hominin diets on the metabolism and function of gut microbiota in vitro. mBio 5(3):1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z, Yin J, Zhang J, Ward RE, Martin RJ, Lefevre M, Cefalu WT, Ye J.. 2009. Butyrate improves insulin sensitivity and increases energy expenditure in mice. Diabetes 58(7):1509–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George F, Daniel C, Thomas M, Singer E, Guilbaud A, Tessier FJ, Revol-Junelles A-M, Borges F, Foligné B.. 2018. Occurrence and dynamism of lactic acid bacteria in distinct ecological niches: a multifaceted functional health perspective. Front Microbiol. 9:2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez A, Petrzelkova K, Yeoman CJ, Vlckova K, Mrázek J, Koppova I, Carbonero F, Ulanov A, Modry D, Todd A, et al. 2015. Gut microbiome composition and metabolomic profiles of wild western lowland gorillas (Gorilla gorilla gorilla) reflect host ecology. Mol Ecol. 24(10):2551–2565. [DOI] [PubMed] [Google Scholar]
- Gomez A, Sharma AK, Mallott EK, Petrzelkova KJ, Jost Robinson CA, Yeoman CJ, Carbonero F, Pafco B, Rothman JM, Ulanov A, et al. 2019. Plasticity in the human gut microbiome defies evolutionary constraints. mSphere 4(4):e00271-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa Y, Curtis B, Yutuc V, Rulien M, Morrisroe K, Watkins K, Ferrier C, English C, Hewitson L, Slupsky CM.. 2018. Microbial structure and function in infant and juvenile rhesus macaques are primarily affected by age, not vaccination status. Sci Rep. 8(1):15867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper LV, Midtvedt T, Gordon JI.. 2002. How host-microbial interactions shape the nutrient environment of the mammalian intestine. Annu Rev Nutr. 22:283–307. [DOI] [PubMed] [Google Scholar]
- Houtz JL, Sanders JG, Denice A, Moeller AH.. 2021. Predictable and host-species specific humanization of the gut microbiota in captive primates. Mol Ecol. 30(15):3677–3687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson DK, Honap TP, Ozga AT, Meda N, Kagoné TS, Carabin H, Spicer P, Tito RY, Obregon-Tito AJ, Reyes LM, et al. 2021. Analysis of global human gut metagenomes shows that metabolic resilience potential for short-chain fatty acid production is strongly influenced by lifestyle. Sci Rep. 11(1):1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DT, Woods DR.. 1986. Acetone-butanol fermentation revisited. Microbiol Rev. 50(4):484–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kettle H, Donnelly R, Flint HJ, Marion G.. 2014. pH feedback and phenotypic diversity within bacterial functional groups of the human gut. J Theor Biol. 342:62–69. [DOI] [PubMed] [Google Scholar]
- Kettle H, Louis P, Holtrop G, Duncan SH, Flint HJ.. 2015. Modelling the emergent dynamics and major metabolites of the human colonic microbiota. Environ Microbiol. 17(5):1615–1630. [DOI] [PubMed] [Google Scholar]
- Koh A, De Vadder F, Kovatcheva-Datchary P, Bäckhed F.. 2016. From dietary fiber to host physiology: short-chain fatty acids as key bacterial metabolites. Cell 165(6):1332–1345. [DOI] [PubMed] [Google Scholar]
- Kushugulova A, Forslund SK, Costea PI, Kozhakhmetov S, Khassenbekova Z, Urazova M, Nurgozhin T, Zhumadilov Z, Benberin V, Driessen M, et al. 2018. Metagenomic analysis of gut microbial communities from a Central Asian population. BMJ Open. 8(7):e021682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert JE. 1998. Primate digestion: interactions among anatomy, physiology, and feeding ecology. Evol Anthropol. 7(1):8–20. [Google Scholar]
- Lambert JE, Fellner V.. 2012. In vitro fermentation of dietary carbohydrates consumed by African apes and monkeys: preliminary results for interpreting microbial and digestive strategy. Int J Primatol. 33(1):263–281. [Google Scholar]
- Langmead B, Salzberg SL.. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods. 9(4):357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Chatelier E, Nielsen T, Qin J, Prifti E, Hildebrand F, Falony G, Almeida M, Arumugam M, Batto JM, Kennedy S, et al. 2013. Richness of human gut microbiome correlates with metabolic markers. Nature 500(7464):541–546. [DOI] [PubMed] [Google Scholar]
- Leonard WR, Robertson ML.. 1994. Evolutionary perspectives on human nutrition: the influence of brain and body size on diet and metabolism. Am J Hum Biol. 6(1):77–88. [DOI] [PubMed] [Google Scholar]
- Louis P, Duncan SH, Mccrae SI, Jackson MS, Flint HJ, Millar J.. 2004. Restricted distribution of the butyrate kinase pathway among butyrate-producing bacteria from the human colon. J Bacteriol. 186(7):2099–2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis P, Flint HJ.. 2017. Formation of propionate and butyrate by the human colonic microbiota. Environ Microbiol. 19(1):29–41. [DOI] [PubMed] [Google Scholar]
- Louis P, McCrae SI, Charrier C, Flint HJ.. 2007. Organization of butyrate synthetic genes in human colonic bacteria: phylogenetic conservation and horizontal gene transfer. FEMS Microbiol Lett. 269(2):240–247. [DOI] [PubMed] [Google Scholar]
- Mahowald MA, Rey FE, Seedorf H, Turnbaugh PJ, Fulton RS, Wollam A, Shah N, Wang C, Magrini V, Wilson RK, et al. 2009. Characterizing a model human gut microbiota composed of members of its two dominant bacterial phyla. Proc Natl Acad Sci U S A. 106(14):5859–5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallott EK, Amato KR.. 2018. The microbial reproductive ecology of white-faced capuchins (Cebus capucinus). Am J Primatol. 80(8):e22896. [DOI] [PubMed] [Google Scholar]
- Mallott EK, Amato KR, Garber PA, Malhi RS.. 2018. Influence of fruit and invertebrate consumption on the gut microbiota of white-faced capuchins (Cebus capucinus). Am J Phys Anthropol. 165(3):576–588. [DOI] [PubMed] [Google Scholar]
- Manara S, Asnicar F, Beghini F, Bazzani D, Cumbo F, Zolfo M, Nigro E, Karcher N, Manghi P, Metzger MI, et al. 2019. Microbial genomes from gut metagenomes of non-human primates expand the tree-of-life with over 1,000 novel species. Genome Biol. 20(1):299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mcdonald D, Hyde E, Debelius JW, Morton JT, Gonzalez A, Ackermann G, Aksenov AA, Behsaz B, Brennan C, Chen Y, et al. 2018. American gut: an open platform for citizen science. mSystems 3(3):e00031-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenney EA, Ashwell M, Lambert JE, Fellner V.. 2014. Fecal microbial diversity and putative function in captive western lowland gorillas (Gorilla gorilla gorilla), common chimpanzees (Pan troglodytes), Hamadryas baboons (Papio hamadryas) and binturongs (Arctictis binturong). Integr Zool. 9(5):557–569. [DOI] [PubMed] [Google Scholar]
- McNeil NI. 1984. The contribution of the large-intestine to energy supplies in man. Am J Clin Nutr. 39(2):338–342. [DOI] [PubMed] [Google Scholar]
- Morrison DJ, Preston T.. 2016. Formation of short chain fatty acids by the gut microbiota and their impact on human metabolism. Gut Microbes. 7(3):189–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagpal R, Shively CA, Appt SA, Register TC, Michalson KT, Vitolins MZ, Yadav H.. 2018. Gut microbiome composition in non-human primates consuming a Western or Mediterranean diet. Front Nutr. 5:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson JK, Holmes E, Kinross J, Burcelin R, Gibson G, Jia W, Pettersson S.. 2012. Host-gut microbiota metabolic interactions. Science 336(6086):1262–1267. [DOI] [PubMed] [Google Scholar]
- Obregon-Tito AJ, Tito RY, Metcalf J, Sankaranarayanan K, Clemente JC, Ursell LK, Zech Xu Z, Van Treuren W, Knight R, Gaffney PM, et al. 2015. Subsistence strategies in traditional societies distinguish gut microbiomes. Nat Commun. 6:6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksanen J, Blanchet FG, Friendly M, Kindt R, Legendre P, McGlinn D, Minchin P R, O’Hara RB, Simpson GL, Solymos P, et al. 2020. vegan: Community Ecology Package. R package version 2.5–7. Available from: https://CRAN.R-project.org/package=vegan. [Google Scholar]
- Orkin JD, Campos FA, Guadamuz A, Melin AD, Myers MS, Hernandez SEC.. 2019. Seasonality of the gut microbiota of free-ranging white-faced capuchins in a tropical dry forest. ISME J. 13(1):183–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou J, Carbonero F, Zoetendal EG, DeLany JP, Wang M, Newton K, Gaskins HR, O’Keefe SJD.. 2013. Diet, microbiota, and microbial metabolites in colon cancer risk in rural Africans and African Americans. Am J Clin Nutr. 98(1):111–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perofsky AC, Lewis RJ, Abondano LA, Di Fiore A, Meyers LA.. 2017. Hierarchical social networks shape gut microbial composition in wild Verreaux’s sifaka. Proc R Soc B. 284(1868):20172274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips J, Muir JG, Birkett A, Lu ZX, Jones GP, O’Dea K, Young GP.. 1995. Effect of resistant starch on fecal bulk and fermentation-dependent events in humans. Am J Clin Nutr. 62(1):121–130. [DOI] [PubMed] [Google Scholar]
- Pruim R, Kaplan D, Horton N.. 2017. The mosaic Package: Helping Students to Think with Data Using R. R J. 9(1):77–102. [Google Scholar]
- Qin J, Li Y, Cai Z, Li S, Zhu J, Zhang F, Liang S, Zhang W, Guan Y, Shen D, et al. 2012. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 490(7418):55–60. [DOI] [PubMed] [Google Scholar]
- Reichardt N, Vollmer M, Holtrop G, Farquharson FM, Wefers D, Bunzel M, Duncan SH, Drew JE, Williams LM, Milligan G, et al. 2018. Specific substrate-driven changes in human faecal microbiota composition contrast with functional redundancy in short-chain fatty acid production. ISME J. 12(2):610–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards M. 2002. A brief review of the archaeological evidence for Palaeolithic and Neolithic subsistence. Eur J Clin Nutr. 56(12):1270–1278. [DOI] [PubMed] [Google Scholar]
- Ridaura VK, Faith JJ, Rey FE, Cheng J, Duncan AE, Kau AL, Griffin NW, Lombard V, Henrissat B, Bain JR, et al. 2013. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science 341(6150):1241214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirmer M, Franzosa EA, Lloyd-Price J, McIver LJ, Schwager R, Poon TW, Ananthakrishnan AN, Andrews E, Barron G, Lake K, et al. 2018. Dynamics of metatranscription in the inflammatory bowel disease gut microbiome. Nat Microbiol. 3(3):337–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnorr SL, Candela M, Rampelli S, Centanni M, Consolandi C, Basaglia G, Turroni S, Biagi E, Peano C, Severgnini M, et al. 2014. Gut microbiome of the Hadza hunter-gatherers. Nat Commun. 5:3654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulfer AF, Battaglia T, Alvarez Y, Bijnens L, Ruiz VE, Ho M, Robinson S, Ward T, Cox LM, Rogers AB, et al. 2018. Intergenerational transfer of antibiotic-perturbed microbiota enhances colitis in susceptible mice. Nat Microbiol. 3(2):234–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selkrig J, Wong P, Zhang X, Pettersson S.. 2014. Metabolic tinkering by the gut microbiome: implications for brain development and function. Gut Microbes. 5(3):369–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smits SA, Leach J, Sonnenburg ED, Gonzalez CG, Lichtman JS, Reid G, Knight R, Manjurano A, Changalucha J, Elias JE, et al. 2017. Seasonal cycling in the gut microbiome of the Hadza hunter-gatherers of Tanzania. Science 357(6353):802–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenburg ED, Smits SA, Tikhonov M, Higginbottom SK, Wingreen NS, Sonnenburg JL.. 2016. Diet-induced extinctions in the gut microbiota compound over generations. Nature 529(7585):212–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenburg ED, Sonnenburg JL.. 2019. The ancestral and industrialized gut microbiota and implications for human health. Nat Rev Microbiol. 17(6):383–390. [DOI] [PubMed] [Google Scholar]
- Sonnenburg JL, Bäckhed F.. 2016. Diet–microbiota interactions as moderators of human metabolism. Nature 535(7610):56–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenburg JL, Sonnenburg ED.. 2019. Vulnerability of the industrialized microbiota. Science 366(6464):eaaw9255. [DOI] [PubMed] [Google Scholar]
- Springer A, Fichtel C, Al-Ghalith GA, Koch F, Amato KR, Clayton JB, Knights D, Kappeler PM.. 2017. Patterns of seasonality and group membership characterize the gut microbiota in a longitudinal study of wild Verreaux’s sifakas (Propithecus verreauxi). Ecol Evol. 7(15):5732–5745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanca A, Abbondio M, Palomba A, Fraumene C, Manghina V, Cucca F, Fiorillo E, Uzzau S, Vizcaíno JA, Csordas A, et al. 2017. Potential and active functions in the gut microbiota of a healthy human cohort. Microbiome 5(1):79–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teaford MF, Ungar PS.. 2000. Diet and the evolution of the earliest human ancestors. Proc Natl Acad Sci U S A. 97(25):13506–13511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremaroli V, Bäckhed F.. 2012. Functional interactions between the gut microbiota and host metabolism. Nature 489(7415):242–249. [DOI] [PubMed] [Google Scholar]
- van der Ark KCH, van Heck RGA, Martin Dos Santos VAP, Belzer C, de Vos WM.. 2017. More than just a gut feeling: constraint-based genome-scale metabolic models for predicting functions of human intestinal microbes. Microbiome 5(1):78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Eunen K, Groen AK, Havinga R, Bakker BM, van Dijk TH, Lange K, Reijngoud D-J, Gerding A, den Besten G, Müller M, et al. 2013. Gut-derived short-chain fatty acids are vividly assimilated into host carbohydrates and lipids. Am J Physiol Gastrointest Liver Physiol. 305(12):G900–G910. [DOI] [PubMed] [Google Scholar]
- Vital M, Howe A, Tiedje J, Vital M, Adina Chuang Howe JMT.. 2014. Revealing the bacterial synthesis pathways by analyzing (meta)genomic data. mBio 5(2):e00889-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vital M, Karch A, Pieper D.. 2017. Colonic butyrate-producing communities in humans: an overview using omics data. mSystems 2:e00130-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Fish JA, Gilman M, Sun Y, Brown CT, Tiedje JM, Cole JR.. 2015. Xander: employing a novel method for efficient gene-targeted metagenomic assembly. Microbiome 3:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolin MJ, Miller TL.. 1996. Pathways of acetate, propionate, and butyrate formation by the human fecal microbial flora. Appl Environ Microbiol. 62(5):1589–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong JM, de Souza R, Kendall CW, Emam A, Jenkins DJ.. 2006. Colonic health: fermentation and short chain fatty acids. J Clin Gastroenterol. 40(3):235–243. [DOI] [PubMed] [Google Scholar]
- Wrangham R, Conklin-Brittain N.. 2003. Cooking as a biological trait. Comp Biochem Physiol A Mol Integr Physiol. 136(1):35–46. [DOI] [PubMed] [Google Scholar]
- Wu GD, Chen J, Hoffmann C, Bittinger K, Chen YY-Y, Keilbaugh SA, Bewtra M, Knights D, Walters WA, Knight R, et al. 2011. Linking long-term dietary patterns with gut microbial enterotypes. Science 334(6052):105–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Bindels LB, Munoz RRS, Martínez I, Walter J, Ramer-Tait AE, Rose DJ.. 2016. Disparate metabolic responses in mice fed a high-fat diet supplemented with maize-derived non-digestible feruloylated oligoand polysaccharides are linked to changes in the gut microbiota. PLoS One 11:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zmora N, Suez J, Elinav E.. 2019. You are what you eat: diet, health and the gut microbiota. Nat Rev Gastroenterol Hepatol. 16(1):35–56. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing data were drawn from published data sets available in the European Nucleotide Archive under accession numbers PRJEB22679, PRJEB11419, PRJNA268964, PRJNA485217, and PRJNA392180. All code for the analyses and the updated butyrate pathway gene database can be found at github.com/emallott/butyrate_pathway.