In this Outlook, Grey and de Massy discuss a study by Pyatnitskaya et al. in this issue of Genes & Development that highlights the central role of the Saccharomyces cerevisiae ZMM protein Zip4 in how crossover formation and synapsis initiation are linked.
Keywords: aneuploidy, crossing over, homologous recombination, meiosis, chromosome segregation, DSB repair, protein–protein interactions, homologous synapsis
Abstract
During meiosis, a molecular program induces DNA double-strand breaks (DSBs) and their repair by homologous recombination. DSBs can be repaired with or without crossovers. ZMM proteins promote the repair toward crossover. The sites of DSB repair are also sites where the axes of homologous chromosomes are juxtaposed and stabilized, and where a structure called the synaptonemal complex initiates, providing further regulation of both DSB formation and repair. How crossover formation and synapsis initiation are linked has remained unknown. The study by Pyatnitskaya and colleagues (pp. 53–69) in this issue of Genes & Development highlights the central role of the Saccharomyces cerevisiae ZMM protein Zip4 in this process.
During meiosis, the proper segregation of chromosomes requires the establishment of at least one crossover (CO) between homolog pairs. COs result from the repair of programmed DNA double-strand breaks (DSBs) induced at meiotic prophase entry. Specific regulations, which occur early during DSB repair, control the channeling of repair toward the homolog rather than the sister chromatid, the frequency of COs, and the choice of repair toward CO or non-CO. CO spacing is regulated by a process called CO interference. In meiotic cells, an alternative minor pathway for COs, not subject to interference, is also active (Hunter 2015). Meiotic chromosomes are organized as an array of loops anchored to the chromosome axis that is formed by structural components and where DSB formation and repair take place. DSB repair by promoting interhomolog interactions brings the homolog axes into proximity. This pairing is further stabilized by the establishment of a tripartite structure called the synaptonemal complex (SC). The SC is composed of two lateral elements, the axes of each homolog, held together by a central region. Several studies have shown that synapsis initiates at DSB repair sites before extending along the entire chromosome length (Zickler and Kleckner 2015; Dubois et al. 2019).
A compelling feature of meiotic recombination is its regulation through an interplay of interactions between proteins that directly act at the DNA level and structural components of the chromosome axes and SC (Zickler and Kleckner 2015). The coordination of these events is crucial for CO control and for the stable interaction between homologs.
In recent years, it has been shown that a group of proteins called ZMM (for Zip, Msh, and Mer) are required for promoting the CO pathway regulated by interference (Pyatnitskaya et al. 2019). Cells harboring mutations of several of these proteins display a reduction in CO and also in SC formation and/or elongation, raising the question of the functional relationship between CO and SC formation. The analysis of mutants of SC structural components (Ecm11 and Gmc2) (Humphryes et al. 2013) and of a ZMM protein (Zip1) (Voelkel-Meiman et al. 2016) demonstrated that CO control and synapsis are functionally separated. In these mutants, CO formation is not or only slightly affected, whereas synapsis is deficient. These findings raised the question of how CO formation and synapsis formation are coordinated.
To gain insights into these questions (namely, the communication between proteins acting at the DNA level and proteins involved in SC structure), Borde's group (Pyatnitskaya et al. 2022) explored the interactions between candidate proteins in S. cerevisiae. They predicted that some ZMM proteins involved in DNA repair might interact with proteins involved in SC formation. They discovered that the ZMM component Zip4 interacts with the SC central element protein Ecm11. It was already known that ZIP4 is required for both CO and synapsis (Tsubouchi et al. 2006). Zip4 forms a complex with Zip2–Spo16, which binds to various branched DNA structures found in meiotic recombination intermediates (De Muyt et al. 2018; Arora and Corbett 2019). As it contains a tetratricopeptide repeat (TPR) motif, Zip4 might act as a platform for protein interactions. On the basis of primary sequence conservation and modeling data, Borde's group (Pyatnitskaya et al. 2022) convincingly identified residues in Zip4 and Ecm11 required for their interaction. The generation of mutants allowed elucidation of the function of this interaction. Borde's group (De Muyt et al. 2018) previously showed by ChIP analysis that Zip4 is recruited to DSB sites (and to axis sites). In the current study, they demonstrated that the SC component Ecm11 also is recruited to DSB sites. Interestingly, in mutants in which the Zip4–Ecm11 interaction is defective, Ecm11 recruitment to DSB sites was inefficient and Zip1 polymerization was deficient. Although reduced in some genomic regions, CO levels remained close to wild-type levels. Therefore, in these mutants, CO designation seems functional, including CO interference leading to high spore viability. Some of the observed variations in CO levels may be due to the downstream consequence of the synapsis defect in these mutants.
Then, to pinpoint Zip4's role in Ecm11 recruitment, Pyatnitskaya et al. (2022) designed a sophisticated experiment to induce the tethering of Ecm11 to Zip4. In this assay, upon induction of the interaction, synapsis formation was partially restored, thus showing that Ecm11 tethering to Zip4 is sufficient to initiate and promote SC elongation. Unexpectedly, COs, which were not supposed to be affected, were reduced. The investigators suggest that the kinetics of assembly of protein complexes in this artificial system may not reproduce the wild-type context. Kinetics of events and meiotic prophase progression are certainly important for coordinating DSB repair and synapsis. Indeed, the detailed analysis of ECM11 and GMC2 mutants at a specific DSB site (HIS4LEU2) revealed delays in recombination intermediate processing (Lee et al. 2021), underlying the importance of proper meiotic prophase progression for the coordination between DSB repair and synapsis.
Although the interfering CO pathway formation and SC formation are evolutionarily conserved, structural proteins such as yeast Zip4 and Ecm1 appear to be highly divergent, and the identification of orthologs among eucaryotes is therefore challenging. In this study, based on motif analysis and modeling, the investigators identified the mammalian TEX12 and SYCE2 as potential homologs of the central element proteins Ecm11 and Gmc2. Consistent with their hypothesis, they found that TEX12 interacts with TEX11 (the ortholog of Zip4) and with SYCE2.
Overall, this study highlights some of the components that control CO formation and its link to SC formation. Zip4 seems to be a major platform, linking the Zip2–Spo16 complex to Ecm1–Gmc2, and thus to SC initiation (see Fig. 1). Zip4 also interacts with Zip3, another essential protein involved in CO control through its SUMO ligase activity, and with Msh5 (De Muyt et al. 2018), which, with its partner Msh4, stabilizes recombination intermediates for CO. These important findings improve our understanding of the molecular interactions and functional regulations that mediate CO formation within the three-dimensional structure of the meiotic chromosome.
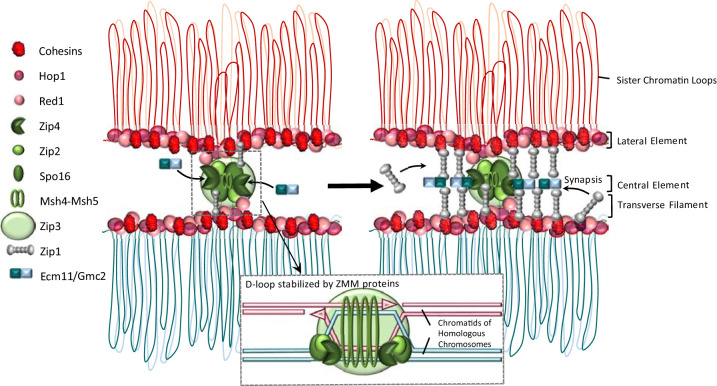
Figure 1.
Meiotic chromosomes are organized as an array of loops anchored to an axis (including cohesins, Hop1, and Red1). (Left panel) The ZMM proteins (Zip4, Zip2, Spo16, Msh4, Msh5, Zip3, Zip1, and Mer3) channel DSB repair toward the homolog and stabilize the D-loop intermediate. (Right panel) Zip4, by recruiting Ecm11 and Gmc2, is promoting synapsis initiation with the loading of Zip1 and other structural proteins, then elongating along the axis.
Acknowledgments
B.d.M. is funded by the European Research Council (ERC) under the European Union's Horizon 2020 research and innovation program (grant agreement no. 883605).
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.349286.121.
Freely available online through the Genes & Development Open Access option.
References
- Arora K, Corbett KD. 2019. The conserved XPF:ERCC1-like Zip2:Spo16 complex controls meiotic crossover formation through structure-specific DNA binding. Nucleic Acids Res 47: 2365–2376. 10.1093/nar/gky1273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Muyt A, Pyatnitskaya A, Andréani J, Ranjha L, Ramus C, Laureau R, Fernandez-Vega A, Holoch D, Girard E, Govin J, et al. 2018. A meiotic XPF–ERCC1-like complex recognizes joint molecule recombination intermediates to promote crossover formation. Genes Dev 32: 283–296. 10.1101/gad.308510.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois E, De Muyt A, Soyer JL, Budin K, Legras M, Piolot T, Debuchy R, Kleckner N, Zickler D, Espagne E. 2019. Building bridges to move recombination complexes. Proc Natl Acad Sci 116: 12400–12409. 10.1073/pnas.1901237116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphryes N, Leung WK, Argunhan B, Terentyev Y, Dvorackova M, Tsubouchi H. 2013. The Ecm11Gmc2 complex promotes synaptonemal complex formation through assembly of transverse filaments in budding yeast. PLoS Genet 9: e1003194. 10.1371/journal.pgen.1003194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter N. 2015. Meiotic recombination: the essence of heredity. Cold Spring Harb Perspect Biol 7: a016618. 10.1101/cshperspect.a016618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MS, Higashide MT, Choi H, Li K, Hong S, Lee K, Shinohara A, Shinohara M, Kim KP. 2021. The synaptonemal complex central region modulates crossover pathways and feedback control of meiotic double-strand break formation. Nucleic Acids Res 49: 7537–7553. 10.1093/nar/gkab566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyatnitskaya A, Borde V, De Muyt A. 2019. Crossing and zipping: molecular duties of the ZMM proteins in meiosis. Chromosoma 128: 181–198. 10.1007/s00412-019-00714-8 [DOI] [PubMed] [Google Scholar]
- Pyatnitskaya A, Andreani J, Guérois R, De Muyt A, Borde V. 2022. The Zip4 protein directly couples meiotic crossover formation to synaptonemal complex assembly. Genes Dev (this issue). 10.1101/gad.348973.121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsubouchi T, Zhao H, Roeder GS. 2006. The meiosis-specific zip4 protein regulates crossover distribution by promoting synaptonemal complex formation together with zip2. Dev Cell 10: 809–819. 10.1016/j.devcel.2006.04.003 [DOI] [PubMed] [Google Scholar]
- Voelkel-Meiman K, Cheng SY, Morehouse SJ, MacQueen AJ. 2016. Synaptonemal complex proteins of budding yeast define reciprocal roles in MutSγ-mediated crossover formation. Genetics 203: 1091–1103. 10.1534/genetics.115.182923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zickler D, Kleckner N. 2015. Recombination, pairing, and synapsis of homologs during meiosis. Cold Spring Harb Perspect Biol 7: a016626. 10.1101/cshperspect.a016626 [DOI] [PMC free article] [PubMed] [Google Scholar]