Abstract
A coronavirus was isolated from feces of a diarrheic foal and serially propagated in human rectal adenocarcinoma (HRT-18) cells. Antigenic and genomic characterizations of the virus (isolate NC99) were based on serological comparison with other avian and mammalian coronaviruses and sequence analysis of the nucleocapsid (N) protein gene. Indirect fluorescent-antibody assay procedures and virus neutralization assays demonstrated a close antigenic relationship with bovine coronavirus (BCV) and porcine hemagglutinating encephalomyelitis virus (mammalian group 2 coronaviruses). Using previously described BCV primers, the N protein gene of isolate NC99 was amplified by a reverse transcriptase PCR (RT-PCR) procedure. The RT-PCR product was cloned into pUC19 and sequenced; the complete N protein of NC99 (446 amino acids) was then compared with published N protein sequences of other avian and mammalian coronaviruses. A high degree of identity (89.0 to 90.1%) was observed between the N protein sequence of NC99 and published sequences of BCV (Mebus and F15 strains) and human coronavirus (strain OC43); only limited identity (<25%) was observed with group 1 and group 3 coronaviruses. Based on these findings, the virus has been tentatively identified as equine coronavirus (ECV). ECV NC99 was determined to have close antigenic and/or genetic relationships with mammalian group 2 coronaviruses, thus identifying it as a member of this coronavirus antigenic group.
The Coronaviridae are a large group of RNA-containing viruses that infect a wide variety of avian and mammalian species (22, 29). The family is comprised of two genera, Coronavirus and Torovirus, which share similarities in morphology, genome organization, and genome expression (26). The coronavirus genome consists of a positive-sense, single-stranded RNA molecule that is 20 to 30 kb in size (19, 26). Virions are enveloped, pleomorphic, and 80 to 220 nm in diameter, and they have club-shaped peplomers approximately 20 nm in length (19, 26). Three major structural proteins, the surface glycoprotein (90 to 180 kDa), an integral membrane protein (20 to 35 kDa), and a nucleocapsid (N) protein (50 to 60 kDa), are known (19, 26). Additionally, some coronaviruses also contain a fourth major structural protein, the hemagglutinin-esterase protein (120 to 140 kDa) (12, 26).
Coronaviruses have been subdivided into three major antigenic groups based on antigenic differences identified by serological analyses, and these findings have been substantiated by nucleotide sequence analyses (21, 22, 29). Human coronavirus (HCV) strain 229E, porcine transmissible gastroenteritis virus (TGEV), canine coronavirus, and feline infectious peritonitis virus are members of group 1. HCV strain OC43, murine hepatitis virus (MHV), porcine hemagglutinating encephalomyelitis virus (HEV), and bovine coronavirus (BCV) are members of group 2. Infectious bronchitis virus (IBV) and turkey coronavirus (TCV) comprise group 3 (4).
Coronaviruslike viruses previously have been identified by electron microscopy in feces of foals and adult horses with enteric disease; however, isolation and subsequent characterization of these viruses have not been reported (1, 8, 13, 17). A coronavirus antigenically related to BCV was identified in feces and intestinal tissues of a diarrheic foal, based on immunohistochemistry and an antigen-capture enzyme-linked immunosorbent assay, but virus isolation attempts were unsuccessful (7). The purpose of the present report is to describe the isolation and characterization of a coronavirus from feces of a diarrheic foal.
MATERIALS AND METHODS
Clinical history.
A 2-week-old Arabian foal was presented to the North Carolina State University College of Veterinary Medicine with profuse, watery diarrhea and fever. Diarrhea and fever persisted for approximately 6 days after which the foal recovered. Feces were collected and examined by negative-stain electron microscopy as previously described (9).
Virus isolation, propagation, and purification.
Feces obtained from the diarrheic foal were prepared as a 10% suspension in RPMI 1640 (Sigma Chemical Co., St. Louis, Mo.), clarified by low-speed centrifugation, and filtered sequentially through 0.45- and 0.22-μm filters (Millipore Corp., Bedford, Mass.). The filtrate was inoculated onto confluent monolayers of human rectal adenocarcinoma (HRT-18) cells in 25-cm2 flasks and incubated for 60 min at 37°C. Inoculated cell cultures were maintained with RPMI 1640 supplemented with 1% fetal bovine serum (Gibco BRL, Grand Island, N.Y.) or serum-free RPMI 1640 supplemented with 0.25 μg of trypsin (type IX; Sigma Chemical Co.) per ml. Inoculated cell cultures were examined daily for cytopathic effects; they were passed at 4- to 5-day intervals to fresh monolayers of HRT-18 cells. Cell culture supernatant fluids were examined for the presence of virus by electron microscopy (9).
Isolate NC99 was harvested from infected cells and purified as previously described (11). The virus was located in fractionated sucrose gradients by hemagglutinating activity; hemagglutination assays were done using rat erythrocytes as previously described (25).
Cells, viruses, and antisera.
HRT-18 cells were obtained from D. A. Brian, University of Tennessee, Knoxville, Tenn. BCV strain Mebus and HEV strain Mengeling were obtained from the National Veterinary Services Laboratory, Ames, Iowa. BCV Mebus and HEV Mengeling were propagated in HRT-18 cells.
Antisera specific for isolate NC99 were prepared by immunization of guinea pigs. One-half milliliter of purified virus was mixed with an equal volume of incomplete Freund's adjuvant and inoculated subcutaneously into guinea pigs. This was repeated 4 and 7 weeks later, and serum was collected 10 days after the last inoculation.
Antisera specific for BCV Mebus, HEV Mengeling, and TGEV Purdue were obtained from the National Veterinary Services Laboratory. Antisera specific for IBV Massachusetts was obtained from SPAFAS, Inc., Norwich, Conn.
IFAT.
The indirect fluorescent-antibody test (IFAT) was performed as described before (10).
Virus neutralization.
Serial twofold dilutions of heat-inactivated antiserum (1/16 to 1/8,192) were prepared in serum-free RPMI 1640. Serum-free RPMI 1640 (0.5 ml) containing approximately 1,000 50% cell culture infective dose (CCID50) of virus was added to an equal volume of diluted antisera and mixed well. These mixtures then were incubated at 37°C for 60 min. The growth medium was decanted from monolayers of HRT-18 cells in 24-well tissue culture plates. Monolayers were rinsed twice with serum-free RPMI 1640, and 0.2 ml of each mixture was placed on a duplicate monolayer. After a 60-min incubation at 37°C in a humidified CO2 incubator, the inoculum was replaced with 1 ml of RPMI 1640 containing 0.05 mg of gentamicin per ml and 0.25 μg of trypsin (Type IX; Sigma Chemical Co.) per ml; incubation was continued for 3 days. Cells then were examined for hemadsorption using rat erythrocytes as previously described (25). The reciprocal of the highest dilution of antiserum that resulted in complete neutralization, as evidenced by the absence of hemadsorption, was considered to be the antibody titer.
RT-PCR.
Viral RNA was extracted from sucrose gradient-purified virus, and cDNA synthesis was accomplished by reverse transcriptase (RT)-PCR as previously described (3). PCR primers ECVf and ECVr were based on previously published N gene sequences of BCV Mebus (16, 18). ECVmid was designed from preliminary nucleotide sequence data to amplify the internal region of the gene when used with ECVr. Primers possessed EcoRI restriction sites to facilitate cloning of cDNA into pUC19. Primer sequences are as follows: ECVf, 5′-TGAATTCTCTGGCATGGACACCGCATT-3′; ECVr, 5′-TGAATTCCCAGGTGCCGACATAAGGTT-3′; and ECVmid, 5′-TGAATTCGTGATGAGGCTATTCCGACTA-3′.
Cloning.
RT-PCR products were cloned into pUC19 and transformed into competent Escherichia coli strain DH5α (Gibco-BRL, Grand Island, N.Y.) as described elsewhere (23).
Sequence analyses.
DNA was sequenced at the University of North Carolina, Chapel Hill, automated DNA sequencing facility, on a model 373A DNA sequencer (Applied Biosystems, Foster City, Calif.) using the Taq DyeDeoxy Terminator Cycle sequencing kit (Applied Biosystems). All sequences were confirmed by sequencing both strands.
Comparative analyses of N protein amino acid sequences were performed with the GeneStream Align program and the CLUSTAL W Multiple Sequence alignment program, version 1.7 (28). Phylogenetic trees were constructed for the N gene region using the MegAlign application of the Lasergene software package (DNASTAR, Madison, Wis.). Phylogenetic-tree construction was based on the neighbor-joining method. Coronavirus sequences were obtained from the GenBank database. These included BCV Mebus and F15, MHV A59, HCV OC43 and 229E, TGEV Purdue, IBV Beaudette, and TCV NC95 (2, 4, 6, 14–16, 20, 24).
Nucleotide sequence accession number.
The GenBank accession number for sequences in this study is AF251144.
RESULTS
Coronaviruslike particles were identified in feces of a diarrheic foal by negative-stain electron microscopy. Particles were pleomorphic and approximately 80 to 160 nm in diameter, and they had large club-shaped surface projections. Initial attempts to propagate virus from feces of the foal were unsuccessful with maintenance medium without trypsin supplementation. However, subsequent virus isolation attempts were successful when inoculated cells were maintained in serum-free medium containing 0.25 μg of trypsin per ml. No cytopathic effects were evident during the first two passages; however, cytopathic effects were detected by 4 days postinoculation at the third passage. Cytopathic effects were characterized by the presence of round refractile cells associated with the HRT-18 cell monolayer and by small, floating syncytia.
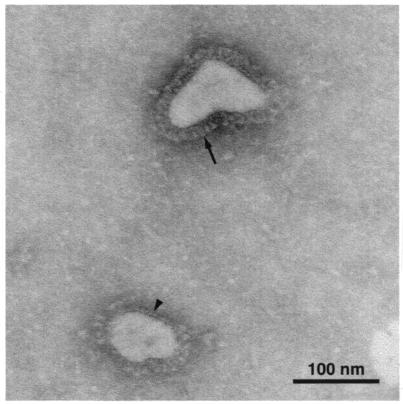
Electron-microscopic examination of cell culture supernatant fluid revealed the presence of typical coronavirus particles (Fig. 1). The observed coronavirus particles were 80 to 120 nm in diameter, and they appeared to possess a double row of peplomers: an outer row of large, club-shaped peplomers compatible with surface glycoprotein and an inner row of smaller peplomers compatible with hemagglutinin-esterase protein (5).
FIG. 1.
Coronavirus particles (80 to 120 nm in diameter) identified in infected HRT cell supernatant fluids. Note the presence on virion surfaces of an outer layer of large club-shaped peplomers (arrow) and an inner layer of short peplomers (arrowhead).
Antigenic and genomic characterization of the isolate, hereafter referred to as isolate NC99, were based on IFAT, serum-virus neutralization assays, and sequence analyses. Antisera specific for BCV and HEV (group 2 coronaviruses) reacted strongly against NC99 by IFAT (results not shown); positive fluorescence was not detected using TGEV-specific antisera (group 1) or IBV-specific antisera (group 3).
The isolate was compared by cross-neutralization studies with BCV, HEV, and TGEV (Table 1). A two-way antigenic relationship was demonstrated between NC99 and BCV Mebus; however, homologous reactions were substantially greater than heterologous reactions. No cross-neutralization was observed between NC99, HEV, and TGEV.
TABLE 1.
Antigenic relationship of ECV NC99 to other mammalian coronaviruses (BCV, porcine HEV, and porcine TGEV) as determined by virus neutralization assays
| Virus (strain) | Response to antiserum (titers)a
|
|||
|---|---|---|---|---|
| Guinea pig anti-ECV | Bovine anti-BCV | Porcine anti-HEV | Porcine anti-TGEV | |
| ECV (NC99) | 4,196 | 32 | <16 | <16 |
| BCV (Mebus) | 128 | 512 | <16 | <16 |
| HEV (Mengeling) | <16 | <16 | 256 | <16 |
Titers, highest dilution of antiserum that resulted in complete neutralization of virus.
RNA obtained from purified NC99 was amplified in an RT-PCR assay using synthetic primers that were based on N gene sequences of BCV Mebus. The PCR products were approximately 1.5 kb in size and identical in size with the product obtained using BCV Mebus RNA as template (data not shown). No PCR product was observed when RNA was harvested from uninfected HRT-18 cells and amplified by RT-PCR or when RT-PCR was run without RT (data not shown).
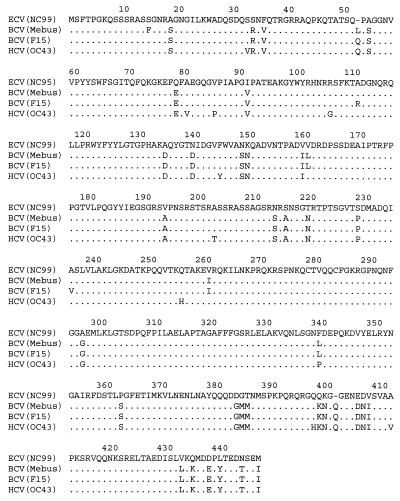
The deduced amino acid sequence of the N protein of NC99 is shown in Fig. 2; NC99 is compared with published sequences of two strains of BCV (Mebus and F15) and one of HCV (OC43). A comparison of the percent identity of NC99 N protein amino acid sequences with published sequences of BCV Mebus and F15, HCV OC43 and 229E, MHV A59, TGEV Purdue, and IBV Beaudette is presented in Table 2. The N protein sequence of NC99 had 89.0 to 90.1% identity with BCV strains Mebus and F15 and with HCV strain OC43. In contrast, N protein sequences of BCV strains Mebus and F15 had 99.1% identity with each other and 96.9% identity with HCV OC43. The N protein sequence of NC99 had <25% identity with HCV 229E, TGEV Purdue, and IBV Beaudette.
FIG. 2.
Comparison of N protein amino acid sequences of ECV strain NC99 and published sequences of BCV Mebus, BCV F15, and HCV OC43 (14–16). Amino acid sequence differences are shown for BCV strains and HCV OC43. –, position where an amino acid is missing; …, similar amino acids.
TABLE 2.
Percent sequence identity of N proteins of ECV NC99 and published sequences of BCV (Mebus, F15), HCV (OC43, 229E), MHV (A59), TGEV (Purdue), and IBV (Beaudette)a
| Virus (strain) | Sequence identity (%) with virus (strain)
|
||||||
|---|---|---|---|---|---|---|---|
| ECV (NC99) | BCV (Mebus) | BCV (F15) | HCV (OC43) | MHV (A59) | HCV (229E) | TGEV (Purdue) | |
| BCV (Mebus) | 90.1 | ||||||
| BCV (F15) | 89.9 | 99.1 | |||||
| HCV (OC43) | 89.0 | 96.9 | 96.9 | ||||
| MHV (A59) | 66.7 | 66.7 | 66.7 | 66.3 | |||
| HCV (229E) | 19.5 | 19.5 | 19.8 | 19.5 | 22.9 | ||
| TGEV (Purdue) | 20.2 | 20.2 | 20.4 | 19.9 | 22.3 | 19.5 | |
| IBV (Beaudette) | 21.8 | 20.3 | 20.3 | 18.1 | 21.3 | 18.1 | 20.4 |
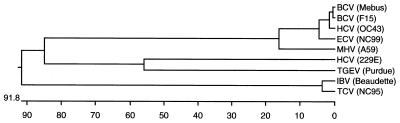
A phylogenetic tree was prepared to further examine relationships between NC99 and other coronaviruses based on a comparison of N protein amino acid sequences (Fig. 3). The phylogenetic tree was prepared based on N protein sequences of NC99 and published sequence data for selected avian and mammalian coronaviruses (Fig. 3). Phylogenetic analysis showed that NC99 was more closely related to group 2 coronaviruses (BCV, HCV OC43, and MHV) than to members of group 1 (TGEV and HCV 229E) and group 3 (IBV and TCV) coronaviruses. In addition, phylogenetic analyses based on N protein sequences demonstrated that BCV strains Mebus and F15 and HCV strain OC43 are more closely related to each other than they are to NC99.
FIG. 3.
Phylogenetic relationship of ECV NC99 and other avian and mammalian coronaviruses based on a comparison of N protein sequences (2, 3, 6, 14, 15, 16, 20, 24). Amino acid sequences were aligned using the CLUSTAL method, and phylogenetic trees were constructed using the neighbor-joining method. Analyses were done using the MegAlign application of the Lasergene software package.
DISCUSSION
Coronaviruslike viruses previously have been identified in feces of diarrheic foals and adult horses (1, 7, 8, 13, 17). In the present study, a coronavirus associated with diarrhea in a young foal was serially propagated in cell culture and partially characterized. The virus was identified as a coronavirus based on (i) virion size and morphology, (ii) antigenic relatedness to BCV and HEV as determined by serological procedures, and (iii) genetic relatedness to BCV, HCV strain OC43, and MHV as determined by N gene sequence analysis. The virus tentatively is identified as equine coronavirus (ECV) based on the origin of the virus.
The coronavirus N protein has been shown to be highly variable in amino acid composition between the viruses that comprise the three coronavirus antigenic groups but highly conserved within these groups (27, 30). In the present study, a high degree of identity (66.7 to 90.1%) was observed between the N protein sequences of ECV strain NC99 and N protein sequences of group 2 coronaviruses (BCV, HCV OC43, and MHV). In contrast, the ECV NC99 N protein had significantly lower identity (<25%) with N protein sequences of group 1 (TGEV and MHV strain 229E) and group 3 (IBV) coronaviruses. These findings indicate the identification of ECV NC99 as a member of the group 2 mammalian coronaviruses and support the findings based on serological analyses.
Sequence analyses also indicated that ECV NC99 was genetically distinct from other previously characterized group 2 coronaviruses. Based on N protein sequences, BCV strains Mebus and F15 shared 99% identity with each other, and these viruses shared 96% identity with HCV strain OC43. In contrast, ECV NC99 had 89 to 90.1% identity with BCV strains Mebus and F15 and HCV strain OC43. These findings, along with phylogenetic analyses, demonstrate that BCV strains Mebus and F15 and HCV OC43 are more closely related to each other than they are to ECV (NC99). These findings also suggest that ECV NC99 is a distinct coronavirus species and that it should be recognized as a new member of the group 2 mammalian coronaviruses. However, as the present study was based on a single isolate and only a relatively small portion of the coronavirus genome (1.5 kb), additional studies are needed to confirm these findings.
The findings of the present study are supported by a recent report in which a coronavirus antigenically related to BCV was identified in a foal with enteritis (7). The virus was identified in intestinal tissues of the foal by immunohistochemistry using BCV-specific monoclonal antibodies and in feces using an antigen-capture enzyme-linked immunosorbent assay designed for BCV detection; however, virus isolation was not successful, and further characterization of the virus was not done.
Although this study and those of others have identified coronaviruses or coronaviruslike viruses in foals and adult horses with enteric disease, the pathogenicity of these viruses and their etiologic role in enteric disease have not been examined. Additional studies are needed to determine the prevalence of ECV infection and the relative importance of ECV as a cause of enteric disease in horses.
ACKNOWLEDGMENT
This work was supported by the State of North Carolina.
REFERENCES
- 1.Bass E P, Sharpee R L. Coronavirus and gastroenteritis in foals. Lancet. 1975;ii:822. doi: 10.1016/S0140-6736(75)80058-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boursnell M E, Binns M M, Foulds I J, Brown T D. Sequences of the nucleocapsid genes from two strains of avian infectious bronchitis virus. J Gen Virol. 1985;66:573–580. doi: 10.1099/0022-1317-66-3-573. [DOI] [PubMed] [Google Scholar]
- 3.Breslin J J, Smith L G, Fuller F J, Guy J S. Sequence analysis of the matrix/nucleocapsid gene region of turkey coronavirus. Intervirology. 1999;42:22–29. doi: 10.1159/000024956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Breslin J J, Smith L G, Fuller F J, Guy J S. Sequence analysis of the turkey coronavirus nucleocapsid gene and 3′ untranslated region identifies the virus as a close relative of infectious bronchitis virus. Virus Res. 1999;65:187–193. doi: 10.1016/S0168-1702(99)00117-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brian D A, Hogue B G, Kienzle T E. The coronavirus hemagglutinin esterase glycoprotein. In: Siddell S G, editor. The coronaviridae. New York, N.Y: Plenum Press; 1995. pp. 165–179. [Google Scholar]
- 6.Cruciere C, Laporte J. Sequence analysis of bovine enteritic coronavirus (F15) genome. I. Sequence of the gene coding the nucleocapsid protein; analysis of the predicted protein. Ann Inst Pasteur. 1988;139:123–138. doi: 10.1016/S0769-2617(88)80012-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davis E, Rush B R, Cox J, DeBey B, Kapil S. Neonatal enterocolitis associated with coronavirus infection in a foal: a case report. J Vet Diagn Investig. 2000;12:153–156. doi: 10.1177/104063870001200210. [DOI] [PubMed] [Google Scholar]
- 8.Durham P J K, Stevenson B J, Farquhanson B C. Rotavirus and coronavirus associated diarrhea in domestic animals. N Z Vet J. 1979;27:30–32. doi: 10.1080/00480169.1979.34595. [DOI] [PubMed] [Google Scholar]
- 9.Guy J S, Barnes H J. Partial characterization of a turkey enterovirus-like virus. Avian Dis. 1991;35:197–203. [PubMed] [Google Scholar]
- 10.Guy J S, Barnes H J, Smith L G. Rapid diagnosis of infectious laryngotracheitis using a monoclonal antibody-based immunoperoxidase procedure. Avian Pathol. 1992;21:77–86. doi: 10.1080/03079459208418820. [DOI] [PubMed] [Google Scholar]
- 11.Guy J S, Brian D A. Bovine coronavirus genome. J Virol. 1979;29:293–300. doi: 10.1128/jvi.29.1.293-300.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holmes K V, Lai M M C. Coronaviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fundamental virology. 3rd ed. Vol. 1. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 1075–1093. [Google Scholar]
- 13.Huang J C M, Wright S L, Shipley W D. Isolation of coronavirus-like agent from horses suffering from acute equine diarrhoea syndrome. Vet Rec. 1983;113:262–263. doi: 10.1136/vr.113.12.262. [DOI] [PubMed] [Google Scholar]
- 14.Kamahora T, Soe L H, Lai M M. Sequence analysis of nucleocapsid gene and leader RNA of human coronavirus OC43. Virus Res. 1989;12:1–9. doi: 10.1016/0168-1702(89)90048-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kapke P A, Brian D A. Sequence analysis of the porcine transmissible gastroenteritis coronavirus nucleocapsid protein gene. Virology. 1986;151:41–49. doi: 10.1016/0042-6822(86)90102-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lapps W, Hogue B G, Brian D A. Sequence analysis of the bovine coronavirus nucleocapsid and matrix protein genes. Virology. 1987;157:47–57. doi: 10.1016/0042-6822(87)90312-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mair T S, Taylor F G R, Harbour D A, Pearson G R. Concurrent cryptosporidium and coronavirus infections in an Arabian foal with immunodeficiency syndrome. Vet Rec. 1990;126:127–130. [PubMed] [Google Scholar]
- 18.Majhdi F, Mocha H C, Kapil S. Isolation and characterization of a coronavirus from elk calves with diarrhea. J Clin Microbiol. 1997;35:2937–2942. doi: 10.1128/jcm.35.11.2937-2942.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murphy F A. Virus taxonomy. In: Fields B N, Knipe D M, Howley P M, editors. Fundamental virology. 3rd ed. Vol. 1. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 15–57. [Google Scholar]
- 20.Parker M M, Masters P S. Sequence comparison of the N genes of five strains of the coronavirus mouse hepatitis virus suggests a three domain structure for the nucleocapsid protein. Virology. 1990;179:463–468. doi: 10.1016/0042-6822(90)90316-J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pedersen N C. Antigenic relationship of feline infectious peritonitis virus to coronaviruses of other species. Arch Virol. 1978;58:45–53. doi: 10.1007/BF01315534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Robb J A, Bond C W. Coronaviridae. In: Fraenkel-Conrat H, Wagner R R, editors. Comprehensive virology. Vol. 14. New York, N.Y: Plenum Press; 1979. pp. 193–247. [Google Scholar]
- 23.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 24.Schreibner S S, Kamahora T, Lai M M. Sequence analysis of the nucleocapsid protein of human coronavirus 229E. Virology. 1989;169:142–151. doi: 10.1016/0042-6822(89)90050-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sharpee R L, Mebus C A, Bass E P. Characterization of a calf diarrheal coronavirus. Am J Vet Res. 1976;37:1031–1041. [PubMed] [Google Scholar]
- 26.Siddell S G. The Coronaviridae: an introduction. In: Siddell S G, editor. Coronaviridae. New York, N.Y: Plenum Press; 1995. pp. 1–9. [Google Scholar]
- 27.Siddell S G, Anderson R, Cavanagh D, Fujiwara K, Klenk H D, Macnaughton M R, Pensaert M, Stohlman S A, Sturman L, Zeijst B A M V D. Coronaviridae. Intervirology. 1993;20:181–189. doi: 10.1159/000149390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thompson J D, Gibson T J, Plewniak F, Jeanmougin F, Higgins D G. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wege H, Siddel S, ter Meulen V. The biology and pathogenesis of coronaviruses. Curr Top Microbiol Immunol. 1982;99:165–200. doi: 10.1007/978-3-642-68528-6_5. [DOI] [PubMed] [Google Scholar]
- 30.Williams A K, Wang L, Sneed L W, Collisson E W. Comparative analyses of the nucleocapsid genes of several strains of infectious bronchitis virus and other coronaviruses. Virus Res. 1992;25:213–222. doi: 10.1016/0168-1702(92)90135-V. [DOI] [PMC free article] [PubMed] [Google Scholar]