Abstract
Epigenetic modifications play important roles in genome evolution and innovation. However, most analyses have focused on the evolutionary role of DNA modifications, and little is understood about the influence of posttranscriptional RNA modifications on genome evolution. To explore the evolutionary significance of RNA modifications, we generated transcriptome-wide profiles of N6-methyladenosine (m6A), the most prevalent internal modification of mRNA, for 13 representative plant species spanning over half a billion years of evolution. These data reveal the evolutionary conservation and divergence of m6A methylomes in plants, uncover the preference of m6A modifications on ancient orthologous genes, and demonstrate less m6A divergence between orthologous gene pairs with earlier evolutionary origins. Further investigation revealed that the evolutionary divergence of m6A modifications is related to sequence variation between homologs from whole-genome duplication and gene family expansion from local-genome duplication. Unexpectedly, a significant negative correlation was found between the retention ratio of m6A modifications and the number of family members. Moreover, the divergence of m6A modifications is accompanied by variation in the expression level and translation efficiency of duplicated genes from whole- and local-genome duplication. Our work reveals new insights into evolutionary patterns of m6A methylomes in plant species and their implications, and provides a resource of plant m6A profiles for further studies of m6A regulation and function in an evolutionary context.
Keywords: RNA modification, bioinformatics, epitranscriptome, plant, epigenetic modifications, evolutionary genetics
Introduction
Epigenetics has informed hereditary and evolutionary theory since Conrad Waddington first defined the term in the 1940s (Waddington 1942; Jablonka and Lamb 2002). To date, epigenetic modifications such as methylation, which regulate many aspects of the genome including chromatin organization, structure, recombination, and gene expression (Kouzarides 2007; Colome-Tatche et al. 2012; Shi et al. 2019; Liang et al. 2020), have been recognized as important contributors to evolutionary novelties in eukaryote genomes (Diez et al. 2014; Vidalis et al. 2016; Wendel et al. 2016; Yi 2017). With the development of high-throughput epitranscriptome sequencing technologies, RNA modifications have also been identified as novel and dynamic regulators of gene expression. RNA N6-methyladenosine (m6A), the most abundant internal RNA modification, can affect many aspects of RNA metabolism (e.g., RNA splicing, localization, translation, stability) and regulates gene expression in many biological processes (Zhao, Roundtree, et al. 2017; Yang et al. 2018; Shen et al. 2019), including the DNA damage response (Xiang et al. 2017). The association of m6A modification with genome integrity and gene expression led to the question of whether the evolution of m6A modification interplays with genome evolution and innovation.
The evolutionary conservation of m6A was revealed through comparative analysis of the m6A methylomes of humans and mice (Dominissini et al. 2012; Song et al. 2021). Through the examination m6A methylomes from humans, chimpanzees, and rhesus macaques, the evolution of m6A was found to occur in parallel with the evolution of the gene sequence and mRNA abundance (Ma et al. 2017). Population genetics analysis showed that a proportion of m6A modifications occurred under positive selection pressure, and were associated with human traits or diseases (Zhang et al. 2020). Because the evolution of genome structures and the composition of m6A functional genes differ widely between humans and plants (Yue et al. 2019), the evolution of plant m6A warrants specific investigation. A single-species m6A analysis uncovered evolutionary conservation of m6A within Arabidopsis (Luo et al. 2014) and an association between m6A modification and the evolution of duplicate genes in maize (Zea mays) (Miao et al. 2020). However, virtually nothing is known about the global landscape and evolutionary dynamics of m6A methylomes in plant species or their potential implications for the evolution of plant genomes.
In this study, we first generated m6A profiles for 13 representative plant species spanning over half a billion years of evolution. We then performed comparative analysis of these m6A methylomes in order to address topics of conceptual importance to understanding the evolutionary implications of m6A methylomes in plant species. Specifically, we: 1) characterized the evolutionary patterns of conservation and diversity of m6A methylomes in plants species, 2) dissected the relationship between m6A modifications and gene origin and evolution, 3) revealed the evolutionary implications of m6A modifications on polyploidization in plant species, and 4) investigated the influence of m6A modifications on both gene family expansion and evolution triggered by local-genome duplications.
Results
Transcriptome-Wide Mapping of m6A Modifications for 13 Plant Species
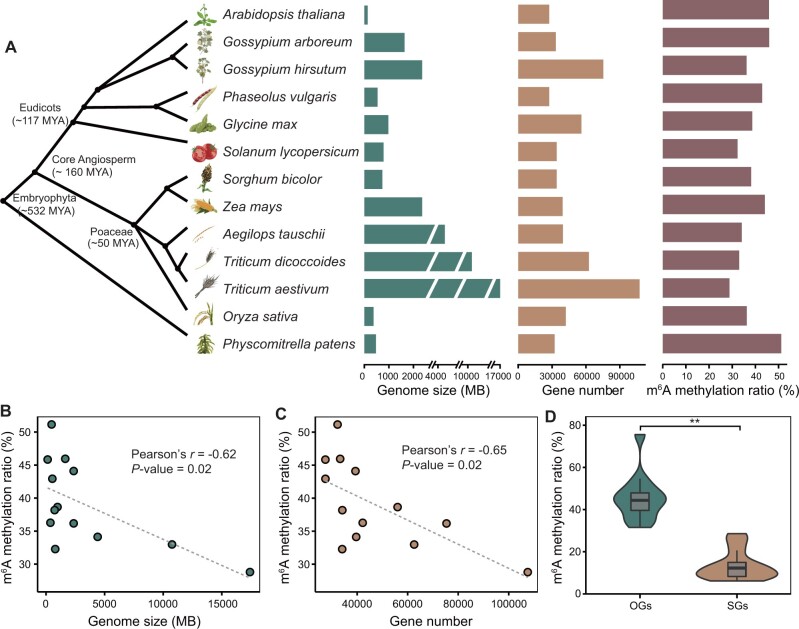
To explore the landscape of m6A modifications in the plant kingdom, we constructed and sequenced m6A-seq libraries from 12 plant species, namely A. thaliana, Gossypium arboreum, G. hirsutum, Glycine max, Phaseolus vulgaris, Sorghum bicolor, Z.mays, Aegilops tauschii, Triticum dicoccoides, T. aestivum, Oryza sativa, and Physcomitrella patens (supplementary data 1, Supplementary Material online). The sequencing reads are highly accumulated around the stop codon and within 3′-untranslated regions (3′-UTRs) in all m6A-seq libraries (supplementary fig. 1, Supplementary Material online). According to the previous study (Miao et al. 2020), we identified m6A peaks with high confidence for each species by intersecting peak regions in a pairwise fashion among all three replicates. Regions that overlap in at least two of three replicates were designated as high-confidence m6A peak regions (supplementary fig. 2, Supplementary Material online). Strong correlations were observed for the abundance of confident peaks between biological replicates (supplementary fig. 3, Supplementary Material online). After the integrated analysis of these data sets together with previously published m6A-seq data sets of four plant species (A. thaliana, O. sativa, Z. mays, and Solanum lycopersicum; supplementary data 2, Supplementary Material online), we obtained transcriptome-wide m6A profiles for 13 representative plant species (six dicots, six monocots, and one bryophyte; genome size 135 Mb–17 Gb; number of annotated protein-coding genes 27,420–107,545) representing over half a billion years phylogenetic timescale (fig. 1A). Among these 13 plant species, there were 3,743–31,719 high-confidence m6A peaks corresponding to 3,726–30,968 genes (supplementary data 3, Supplementary Material online). For each analyzed species, m6A peaks were preferentially enriched around the gene stop codon and 3′-UTR regions (supplementary fig. 4, Supplementary Material online), consistent with previous observations in both animal and plant species (Fu et al. 2014; Liang et al. 2020).
Fig. 1.
Characterization of m6A methylomes in 13 plant species. (A) Overview of genome size, gene number, and transcriptome-wide m6A methylation ratios in 13 plant species. (B) The m6A methylation ratio is negatively correlated with genome size. (C) The m6A methylation ratio is negatively correlated with gene number. (D) Comparison of m6A methylation ratios between OGs and SGs. Statistical analysis conducted using the Student’s t-test. **P<0.001.
These transcriptome-wide m6A profiles provide a new valuable resource for researchers interested in investigating the evolutionary dynamics of m6A in plants. The statistical analysis showed that the m6A methylation ratio for all genes was significantly negatively correlated with the genome size (Pearson’s r=−0.62, P = 0.02; fig. 1B) and the gene number (Pearson’s r=−0.65, P = 0.02; fig. 1C). To enable interspecies comparison, we identified orthologous genes (OGs) and species-specific genes (SGs) through comparative analysis of protein-coding genes from the 13 species (see Materials and Methods). We observed that the m6A methylation ratio for OGs was significantly higher than that for SGs (fig. 1D and supplementary data 4, Supplementary Material online). Of the one-to-one OGs among these species, the number of m6A OGs shared by different species were significantly more than expected by chance (supplementary data 5, Supplementary Material online). Gene ontology (GO) enrichment analysis showed that OGs with high-conservation of m6A methylation were mainly enriched in protein transport and RNA processing, whereas OGs with species-specific m6A methylation were mainly enriched in signal transduction and response to stress (supplementary fig. 5, Supplementary Material online). These results revealed evolutionary dynamics of m6A methylation, that should be related to the evolution of gene function. Then, we identified putative functional counterparts of the m6A methyltransferases (MT-A70, FIP37, VIRILIZER, and HAKAI), and reader proteins (YTH-domain proteins) in 13 species (supplementary data 6, Supplementary Material online). Statistical analysis showed that the global m6A levels for these species were positively correlated with the number of writer and reader genes (supplementary fig. 6, Supplementary Material online); such correlation would be indicative of coevolution between m6A methylome and m6A functional counterpart genes. Together, these results suggested that species differentiation and genome evolution are accompanied by conservation and variation in the m6A methylome.
Divergence of m6A Modification in OGs with Different Evolutionary Origins
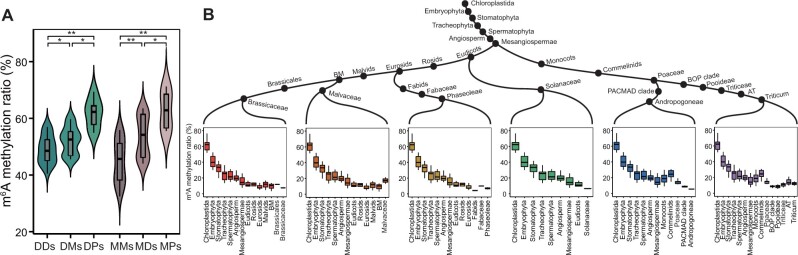
Previous phylostratigraphy analysis showed that OGs between early diverged species were older than those between recently diverged species (Domazet-Loso et al. 2007). Here, we broadly classified OGs with diverse evolutionary origins into six categories: dicot genes orthologous to dicot genes (DDs), dicot genes orthologous to monocot genes (DMs), dicot genes orthologous to Phy. patens genes (DPs), monocot genes orthologous to monocot genes (MMs), monocot genes orthologous to dicot genes (MDs), and monocot genes orthologous to Phy. patens genes (MPs). Therefore, of the dicot genes, DPs had the oldest evolutionary origins, followed by DMs, and DDs. Likewise, of the monocot genes, MPs had the oldest evolutionary origins, followed by MDs, and MMs. For both dicot and monocot genes, the mean m6A methylation ratio increased with evolutionary age (fig. 2A): DDs ([49.1 ± 4.8]%) < DMs ([52.2 ± 4.7]%) < DPs ([61.8 ± 5.3]%), MMs ([44.9 ± 7.5]%) < MDs ([53.5 ± 7.2]%) < MPs ([62.9 ± 6.9]%). Similarly, for each individual species of dicot and monocot, m6A methylation ratios for OGs between dicot and monocot were much higher than within dicot and monocot (supplementary fig. 7, Supplementary Material online). Then, we analyzed the number of shared m6A OGs among these six categories, and found that the intersections between DDs and DPs were smaller than those between DDs and DMs in dicot species, and the intersections between MMs and MPs were smaller than those between MMs and MDs in monocot species (supplementary fig. 8, Supplementary Material online).
Fig. 2.
m6A modifications were preferentially maintained in ancient genes. (A) Comparison of m6A methylation ratios between interspecific OGs and intraspecific OGs. DDs, dicot genes orthologous to dicot genes; DMs, dicot genes orthologous to monocot genes; DP, dicot genes orthologous to Physcomitrella patens genes; MMs, monocot genes orthologous to monocot genes; MDs, monocot genes orthologous to dicot genes; MPs, monocot genes orthologous to Phy. patens genes. Statistical analysis conducted using the Student’s t-test. *P<0.05; **P<0.001. (B) The m6A methylation ratios decreased from ancient PSs to young PSs.
We further investigated the changes in m6A methylation ratios for OGs with different evolutionary origins. These are defined in supplementary figure 9A, Supplementary Material online, a phylostratigraphic tree of genes built using 34 plants species with clear taxonomic classification. In total, 543,853 OGs were assigned to 29 internodes on the phylostratigraphic tree (named as phylostratigraphic stages [PSs]) according to the emergence of their founders in the phylogeny (supplementary fig. 9B, Supplementary Material online). The evolutionary origins of the OGs were identified by tracing each set to its earliest common ancestor and assessing the age of the OGs in the corresponding internodes. OGs at the Chloroplastida internode have the oldest evolutionary origins, and the evolutionary origins of OGs in the subsequent PSs gradually get newer as they advance along the phylostratigraphic paths. Figure 2B shows six phylostratigraphic paths for the 13 species. We found that for each of these six phylostratigraphic paths, the m6A methylation ratio for OGs decreased gradually from old PSs to young PSs (fig. 2B). These results indicated that m6A modifications were maintained in evolutionarily ancient genes.
Evolutionary Dynamics of m6A Modification between OGs
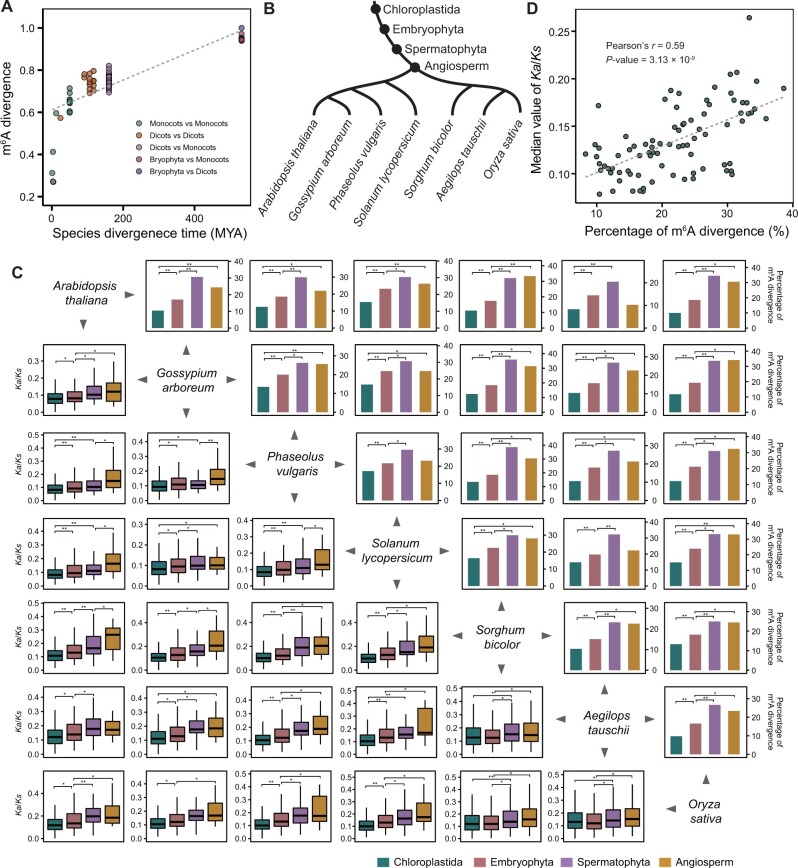
Previous studies demonstrated that ancient genes have been under strong purifying selection because of their fundamental role in plant survival (Guo 2013; Arendsee et al. 2014). This prompted us to ask whether dynamic selection forces may act on plant genomes to shape the diversity of m6A modifications throughout plant evolution. To answer this question, we first analyzed the correlation between m6A methylome divergence and species differentiation time. We used the Spearman correlation coefficient (SCC) to examine the similarity of m6A abundances between paired OGs in any two given species, and the divergence of m6A methylomes between these two species was measured as 1−SCC. We found that the divergence of m6A methylomes was positively correlated with species differentiation time (fig. 3A). We then investigated the relationship between the evolutionary rate (ω) and the divergence of m6A status for pairs of OGs among these species. Here, the evolutionary rate (ω) was defined as the ratio of nonsynonymous substitutions (Ka)/synonymous sites (Ks). The divergence of m6A status indicated the coexistence or loss of m6A modifications within paired OGs. In order to eliminate the ambiguity resulting from genomic duplication events, we only used single-copy OGs expressed (TPM>1) in seven diploid species: A. thaliana, G. arboreum, P. vulgaris, Sol. lycopersicum, S. bicolor, Aeg. tauschii, and O. sativa. In the four PSs shared by these seven diploid species (fig. 3B), both the frequency of sequence mutation (supplementary fig. 10, Supplementary Material online) and evolutionary rate (fig. 3C) of OGs increased from older to younger PSs, meaning that OGs originating from old PSs experienced a higher intensity of purifying selection and slower rates of evolution than those originating from young PSs. A similar increase was also observed for the divergence-m6A ratio of OGs originating from Chloroplastida to Spermatophyta PSs (fig. 3C), suggesting that evolutionarily ancient OG pairs had less divergence in m6A methylation than younger OG pairs. Further Pearson correlation analysis revealed a positive correlation between the divergence-m6A ratio and the evolutionary rate of paired OGs (Pearson’s r = 0.59, P = 3.13 × 10−9; fig. 3D). These results together indicate evolutionary constraints on m6A divergence of OG pairs such that the earlier an OG pair originated, the less m6A modification diverged.
Fig. 3.
Evolutionary patterns of m6A methylation divergence. (A) m6A methylome divergence is positively correlated with species differentiation time. (B) The four PSs shared by seven diploid species with m6A profiling in this study. (C) Comparison of the divergence ratio of m6A methylation and the evolutionary rate (Ka/Ks) of m6A-modified OGs among different PSs from old to young. Statistical analysis conducted using the Student’s t-test and χ2 test. *P<0.05; **P<0.001. (D) m6A divergence is positively correlated with the evolutionary rate.
Divergence of m6A Modification between Homologous Genes in the Context of Polyploidization in Plants
Polyploidization (whole-genome duplication [WGD]) is a common phenomenon in plants (Otto 2007; Soltis et al. 2015; Cheng et al. 2018) and an essential component of plant genome evolution and innovation (Freeling 2009; Van de Peer et al. 2009). For example, maize and soybean (Gly. max) have undergone recent autopolyploidization and subsequent rediploidization, and upland cotton (G. hirsutum), emmer wheat (T. dicoccon), and common wheat (T. aestivum) have undergone allopolyploidization. Our recent study, moreover, revealed an association between m6A modification and duplicate gene evolution in maize (Miao et al. 2020). By sequencing multiple polyploidy plant species with m6A methylomes in this study, we further explored the evolutionary implications of m6A modification in the context of plant genome polyploidization, with a special emphasis on the evolutionary divergence of m6A modifications triggered by autopolyploidization and allopolyploidization.
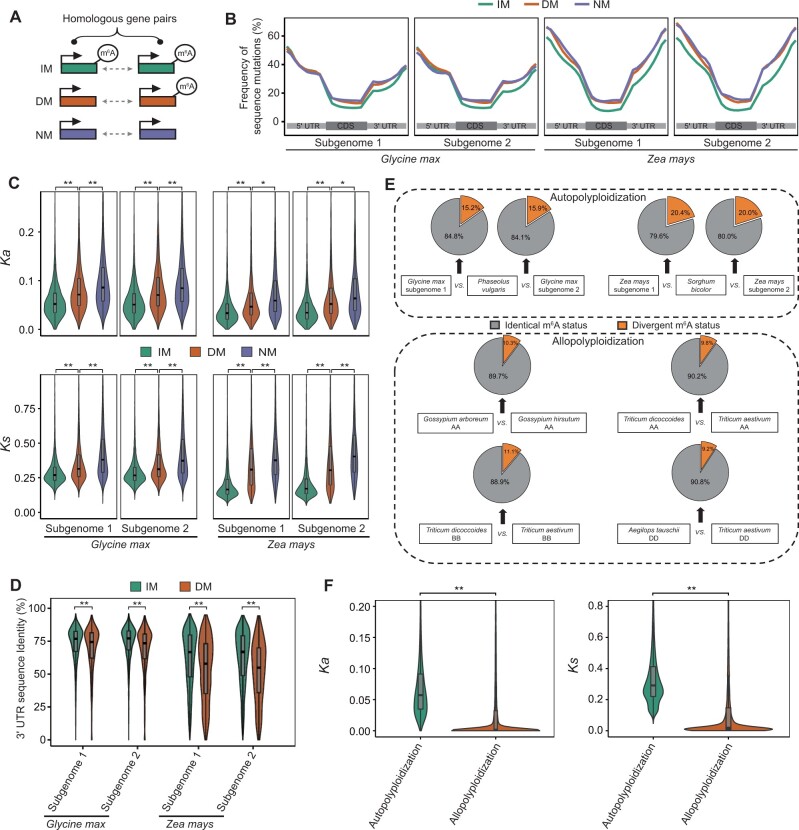
First, we identified subgenomes of each sequenced polyploid species by analyzing syntenic relationships between homologous genes in polyploids and their diploid relatives (e.g., Gly. max vs. P. vulgaris, Z. mays vs. S. bicolor; see Materials and Methods). Then, we classified homologous gene pairs (both partners expressed with TPM>1) into three groups (fig. 4A): IM pairs (both of the paired genes modified with m6A), DM pairs (only one of the paired genes modified with m6A), and NM pairs (none of the paired genes containing m6A peaks). Many sequence variations were found in the duplicated genes after polyploidization (Lockton and Gaut 2005; Freeling and Thomas 2006; Li et al. 2021). Next, we compared the sequence differences in IM, DM, and NM pairs. For both the autopolyploid species and the allopolyploid species, we found less sequence variation in IM than in DM and NM pairs (fig. 4B and supplementary fig. 11, Supplementary Material online). In addition, both Ka and Ks from IM pairs were significantly lower than from DM pairs. Notably, NM pairs had the largest Ka and Ks values (fig. 4C and supplementary fig. 12, Supplementary Material online). Focusing on 3′-UTR, where m6A peaks were mainly enriched, we found that sequence identity within IM pairs was significantly higher than that within DM pairs (fig. 4D and supplementary fig. 13, Supplementary Material online). In addition, we compared the length of 3′-UTR sequences between homologous gene pairs. As shown in supplementary figure 14A, Supplementary Material online, the differences of 3′-UTR length between homologous gene pairs were mainly distributed into peaks that span −500 to 500 nucleotides (zero indicates the same length of two partners of homologous gene pairs). Focusing on the highest part of these peaks, around zero, the peak of IM pairs is significantly higher than that of DM pairs (supplementary fig. 14A, Supplementary Material online), meaning less difference of 3′-UTR length between IM pairs compared with DM pairs (supplementary fig. 14B, Supplementary Material online). These results suggest that divergence of m6A modifications may be related to the occurrence of spontaneous mutations in homologous genes during the evolutionary process postpolyploidization.
Fig. 4.
Evolutionary characterization of m6A modification in the context of polyploidization in plants. (A) Model of three types of homologous gene pairs. IM, pairs in which both genes contained m6A peaks; DM, pairs in which one gene contained an m6A peak, whereas the other did not; NM, pairs in which neither gene contained an m6A peak. (B) Frequency of mutations in full-length transcript sequences of different types of OG pairs. (C) Comparison of nonsynonymous (Ka) and synonymous substitution sites (Ks) in different types of OG pairs. (D) Comparison of the 3′-UTR sequence identity of different types of OG pairs between polyploids (Glycine max, Z. mays) and their diploid relatives (Phaseolus vulgaris, Sorghum bicolor), respectively. (E) Fractions of m6A divergence between the two partners in OG pairs from species that experienced recent polyploidization and their respective diploid relatives. (F) Comparison of the frequency of sequence substitutions (Ka and Ks) of homologous genes between autopolyploid species and allopolyploid species. In (C), (D), and (F), statistical analyses were conducted using the Student’s t-test. *P<0.05; **P<0.001.
In addition to these similarities, we also found differences between autopolyploid and allopolyploid species. As shown in figure 4E, the divergence-m6A ratio of homologous genes (i.e., DM/[IM+DM+NM]) in autopolyploid species (15.2–20.4%) was significantly higher than in allopolyploid species (9.2–11.1%; Wilcoxon test, P < 0.05). This may be the result of a higher frequency of sequence substitutions (Ka and Ks) in homologous genes between autopolyploid species and their diploid relatives than between allopolyploid species and their diploid relatives (fig. 4F). Alternatively, this may be the result of higher m6A methylation ratios for duplicates than for singletons in autopolyploid species (soybean and maize; supplementary fig. 15, Supplementary Material online), That is, the autopolyploidization–rediploidization cycle included the loss of one copy of many duplicate gene pairs (i.e., gene fractionation) (Lockton and Gaut 2005; Freeling and Thomas 2006). The m6A modifications were retained on duplicate genes during the autopolyploidization–rediploidization cycle.
Based on these observations, we inferred that postpolyploidization, sequence variations may be important factors affecting the evolutionary divergence of m6A modifications. The asymmetric retention of m6A modifications between duplicates and singletons during the autopolyploidization–rediploidization cycle may provide for divergence of m6A modification in homologous gene pairs between autopolyploid species and their diploid relatives.
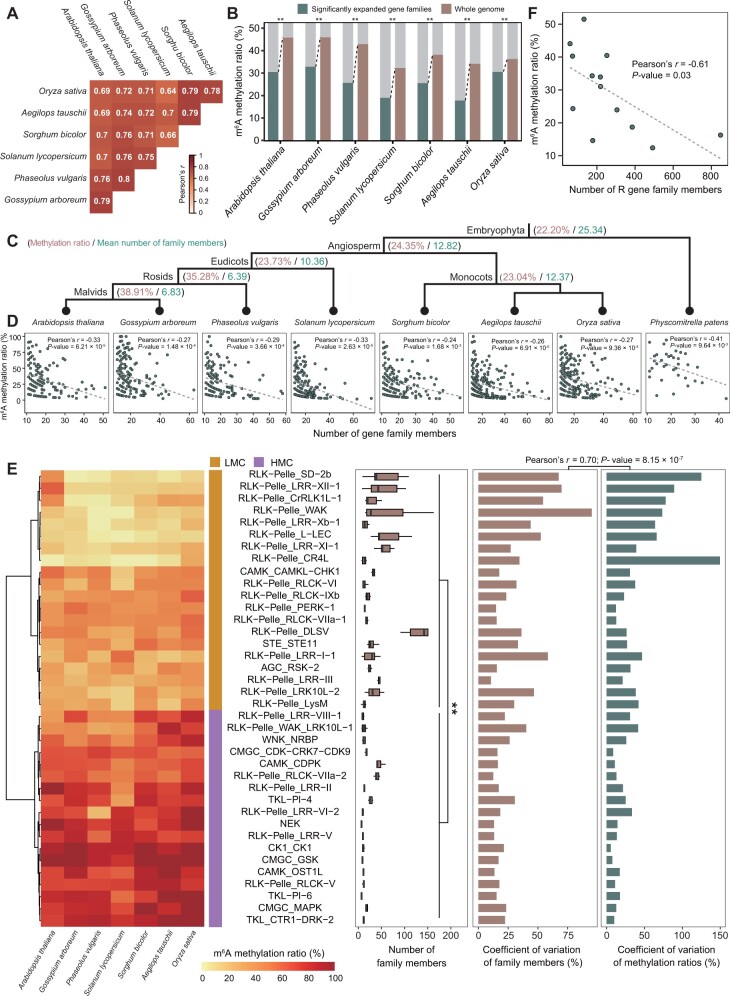
Number of Family Members Was Correlated with m6A Methylation Ratio during Gene Family Expansion
Unlike animals, plants cannot move away from unexpected extreme and harsh environments, thus long-term selective pressures leave genetic and genomic signatures in plant endurance. In addition to WGD, local-genome duplication, which results in significant expansions of specific gene families, is also an important factor in shaping natural variation for adaptation in plants (Rizzon et al. 2006; Freeling 2009; Wang, Yang, et al. 2019). To pave the way for understanding the evolutionary consequences of m6A modification in the context of gene family expansion, we constructed a phylogenetic tree of local-genome duplication events using genes from 27 diploid species that were reported to have not experienced WGD events since they split from their close relatives (supplementary fig. 16, Supplementary Material online). Our results showed that the gene family either expands or contracts over time at a rate of approximately 0.0011 per gene every million years. This is comparable to the rate previously estimated for Drosophila (0.0012) (Hahn et al. 2007), mammals (0.0016) (Demuth et al. 2006), and plants (0.0014) (Guo 2013). We identified 308 significantly expanded gene families shared by seven angiosperm species with m6A profiling in this study (supplementary data 7, Supplementary Material online). The methylation ratios of these significantly expanded gene families were highly positively correlated (fig. 5A), implying interspecific conservation of evolutionary patterns of m6A modification in the context of gene family expansion. For each angiosperm species, the m6A methylation ratios for genes within significantly expanded gene families were lower than those for genes of the whole genome (fig. 5B). In addition, the m6A methylation ratio increased and the mean number of gene family members decreased along phylogenetic timescale from old PSs to young PSs (fig. 5C). Moreover, the m6A methylation ratio was negatively correlated with the number of gene family members in each PS (supplementary fig. 17, Supplementary Material online) and each species (fig. 5D). These results suggested that elimination of m6A-modified genes during the evolutionary process of gene family expansion may be triggered by local-genome duplications.
Fig. 5.
Evolutionary consequences of m6A modification in the context of gene family expansion. (A) The pairwise correlation of methylation levels of significantly expanded gene families among seven diploid species. (B) Decreased m6A methylation ratios for genes within significantly expanded gene families. (C) The m6A methylation ratio increased and the mean number of gene family members decreased along the phylogenetic timescale from ancient PSs to young PSs. (D) The m6A methylation ratio was negatively correlated with the number of gene family members in each species. (E) For PK families in seven diploid species, the number of family members was significantly larger in LMC than in HMC, and the coefficient of variation in family members was positively correlated with the m6A methylation ratios. LMC, clusters with LMC ratio; HMC, clusters with HMC ratio. (F) For R gene families, the number of family members was negatively correlated with the m6A methylation ratio. In (B), statistical analysis conducted using the χ2 test. **P<0.001. In (E), statistical analysis conducted using the Wilcoxon test. **P<0.001.
The evolutionary correlation between methylation ratio and number of family members was also observed in different types of gene families (e.g., protein kinase [PK], transcription factor [TF], and disease-resistance protein [R]). The PK families are enzymes that can regulate the biological activity of proteins through phosphorylation of specific amino acids. Thirty-eight PK families with more than five family members could be divided into two clusters (low m6A methylation ratio [LMC] and high m6A methylation ratio [HMC]; fig. 5E). The number of family members was significantly larger in LMC than in HMC, and the coefficient of variation in the number of family members was positively correlated with that in the m6A methylation ratio across species (fig. 5E). Our results were similar in the TF families (supplementary fig. 18, Supplementary Material online). The R families are plant immune receptors that guard the plant against pathogens. Members of these families were identified based on the presence of nucleotide-binding site (NBS) and leucine-rich repeat (LRR) domains. Phylogenetic analysis showed that NBS-LRR genes could be classified as two subfamilies (supplementary fig. 19, Supplementary Material online): Toll/interleukin-1 receptor in the amino-terminal domain (TNL) and coiled-coil motifs in the amino-terminal domain (CNL). Proportions of CNLs were grouped into clades dominated by TNLs, suggesting that homologous recombination or homogenization occurred between some members of the two subfamilies (supplementary fig. 19, Supplementary Material online). Figure 5F depicts the negative correlation between the number of R family members and the m6A methylation ratio. This negative correlation was also present in subfamilies with distinct species compositions (supplementary fig. 20 and data 8, Supplementary Material online).
Together, these results illustrate evolutionary correlation between m6A modification and gene family expansion. That is, ancient gene families have experienced numerous expansion events that led to an increase in the number of family members accompanied by a decrease in m6A methylation ratio for family members, resulting in asymmetric expansion between m6A-modified members and non-m6A members.
Effects of m6A Modification on Gene Expression and Translational Efficiency in Genomic Duplications
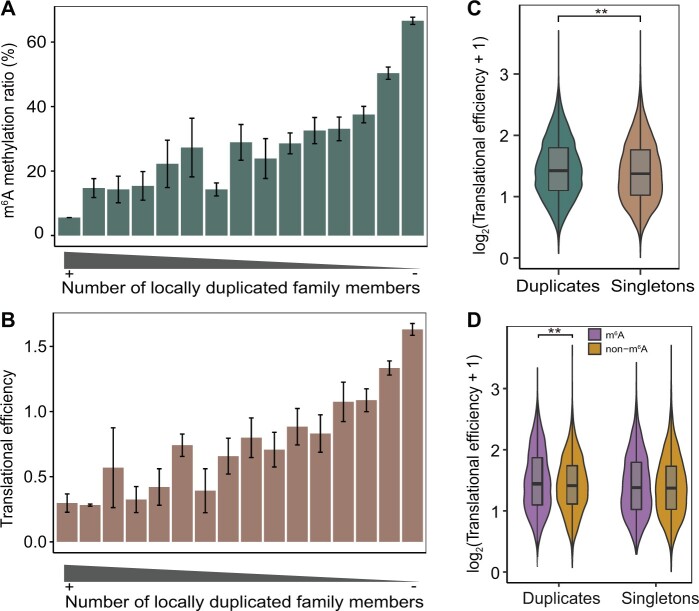
Previous studies have demonstrated that a variety of gene dosage compensation mechanisms at both the transcriptional and translational levels have evolved to alleviate harmful stoichiometric imbalances caused by genomic duplications (Veitia et al. 2008; Edger and Pires 2009; Song et al. 2020). For example, DNA methylation could rebalance gene dosage after gene duplication by inhibiting the initiation of transcription of duplicate genes (Chang and Liao 2012). Given that the effect of m6A methylation on translational efficiency is multifaceted in its strength and genic location (Luo et al. 2020), we wondered what the evolutionary effects of m6A methylation on gene expression and translational efficiency are in the context of genomic duplications.
Integrative analysis of the m6A-seq and ribosome profiling data sets (Luo et al. 2020) showed that both the expression level and translational efficiency of m6A-modified genes were significantly higher than those of non-m6A genes in maize (supplementary fig. 21, Supplementary Material online), therefore, we focused on duplicated genes identified in the maize genome. We identified 7,536 locally duplicated genes from 2,566 gene families. Members of gene families with the largest numbers of members had the lowest m6A methylation ratio (fig. 6A), average translational efficiency (fig. 6B) and expression level (supplementary fig. 22A, Supplementary Material online). These results indicated that, with gene family expansion, the reduced level of m6A modification may attenuate the expression level and translational efficiency of gene duplicates caused by local-genome duplication (supplementary figs. 23 and 24, Supplementary Material online). In addition, the translational efficiency of gene duplicates retained after WGD was significantly higher than that of singletons (fig. 6C). Notably, for duplicate genes, the translational efficiency of m6A-modified genes was significantly higher than that of non-m6A genes, but this difference was not observed in singletons (fig. 6D), implying biased impact of m6A modification on translational efficiency in WGD-duplicated genes. In addition, the expression level of m6A-modified genes was significantly higher than that of non-m6A genes for both duplicate and singleton genes (supplementary fig. 22B, Supplementary Material online).
Fig. 6.
Evolutionary effects of m6A modification on translational efficiency. (A) Gene families with more copies had lower m6A methylation levels. (B) Gene families with more copies had lower average translational efficiency. (C) Comparison of translational efficiency between duplicate genes and singletons in the context of a WGD event. (D) Comparison of translational efficiency between m6A-modified genes and non-m6A genes in duplicates and singletons, respectively. In (C) and (D), statistical analyses were conducted using the Student’s t-test. **P<0.001.
Collectively, our results showed that the evolutionary consequences of m6A modification and the influence on translational efficiency of duplicated genes varied depending on the type of duplication event.
Discussion
Although the progress in dissecting the role of m6A modification in gene regulation is expedited in the plant kingdom (Shen et al. 2019), our understanding of the mechanism of evolution of m6A modification remains limited. Some biological functions of m6A modification appear to be evolutionarily conserved in both mammals and plants (Zhao, Roundtree, et al. 2017), however, many previous observations of m6A modification in mammals have been challenged in plant species (Yue et al. 2019). Furthermore, some components of m6A modification have been present since early plant evolution, but some components were lost during plant evolution and speciation (Liang et al. 2020). Thus, our view of the evolutionary conservation and divergence of m6A modification across plants species has remained blurry.
In this study, we systematically compared m6A methylomes among 13 representative plant species, and provided novel insights into the patterns, processes, and influences of m6A evolution. As an ancient chemical modification, m6A modification may undertake basic molecular functions such as RNA metabolism and export, and therefore be widespread and conserved to a certain extent in plants. Transcriptome-wide methylation levels differed little between evolutionarily adjacent species (fig. 1). Physcomitrellapatens was an outlier, with the smallest genome and the highest methylation level. These results suggest that variation in the m6A methylome was relatively stable at short evolutionary timescales, whereas at long evolutionary timescales, the methylation levels varied as a result of species differentiation and genome evolution.
Natural selection can drive the evolution of plants through gene sequence mutation and copy number variation. m6A modifications were mainly enriched in the 3′-UTR gene region such that mutations in this region may affect the evolutionary divergence of the m6A methylome. In this study, by comparing OG pairs between polyploids and their diploid relatives, we found that m6A modifications were maintained in ancient genes, and 3′-UTR sequences were more conserved in IM pairs than in DM pairs. We speculated that throughout the long evolution of the plant kingdom, ancient genes have been under strong purifying selection, since ancient genes are mostly fundamental for plant survival, and this resulted in slower evolution and higher conservation of these sequences, which could in turn favor the retention of m6A modification in ancient OGs. The m6A modifications in turn affected the evolutionary fate of genes following duplication events by changing gene dosage. Consistent with these ideas, recent studies in mammals have shown that m6A modifications gained during evolution were subject to positive selection, whereas conserved m6A peaks were under purifying selection (Ma et al. 2017). Comparative study of m6A methylomes between humans and mice found that ∼40% of the m6A peaks in the 3′-UTR were subject to purifying selection, and the m6A sites gained by humans were subject to positive selection and associated with disease (Zhang et al. 2020). These results preliminarily illustrated the conservation of m6A modification, which contributed the evolutionary conservation and novelty of some genes key to speciation and species differentiation.
Changes in the number of copies of genes can enhance a species’ ability to adapt to change in its environment. Our results indicate that throughout the process of gene family expansion, m6A modification influenced the gene expression and translational efficiency of family members differently, possibly due to differences in duplication events. Gene families with many copies can be divided into two categories. One includes small-scale local duplication events, which resulted from expansion in short evolutionary time leading to gene dosage proliferation. According to the Gene Balance Hypothesis, duplicates of dosage-sensitive genes tend to be underretained or eliminated in order to maintain proper balance (Freeling 2009). The association between m6A modification and copy number variation and the influence of m6A modification on gene expression and translational efficiency may be a mechanism of dosage compensation that has evolved to alleviate harmful dosage imbalances. The other includes WGD events that increased the dosage of all genes simultaneously. The positive contribution of m6A modification to gene expression and translational efficiency of duplicate genes appears to support the Increased Gene Dosage Hypothesis, which predicts that the WGD and diploidization cycle yields some highly expressed genes that may be beneficial and in turn result in purifying selection to retain both gene duplicates (Conant and Wolfe 2008).
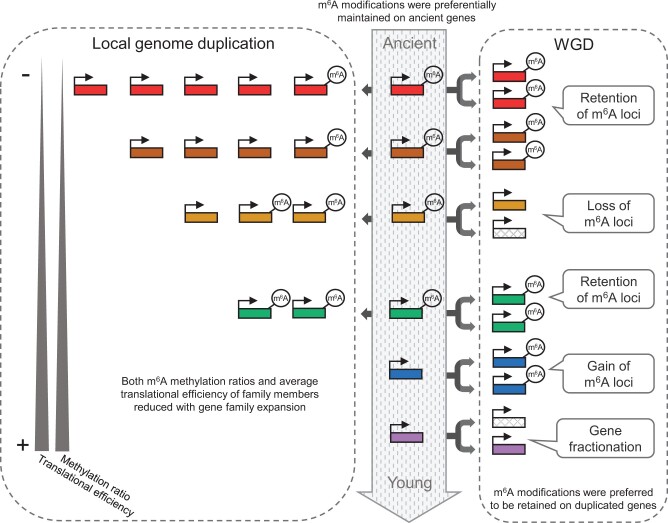
Based on the results in this study, we propose a conceptual model to explain the evolutionary role of the m6A methylome in plants (fig. 7). The process of plant speciation and genome evolution was accompanied by the divergence of m6A modification in OGs, and m6A modifications were maintained in evolutionarily ancient genes, increasing the diversity of m6A methylomes across plants. In the context of the WGD-diploidization cycle, variations in gene sequences affected the gain and loss of m6A loci, and m6A-modified genes were preferentially retained as duplicates, potentially leading to substantial effects on gene fractionation. In addition, local-genome duplication triggered variation in copy number among family members that was accompanied by the divergence of m6A modification. With gene family expansion, the reduced level of m6A modification attenuated the expression level and translational efficiency of duplicated genes, and this could be a mechanism of evolutionary reaction to gene dosage imbalances caused by local-genome duplication.
Fig. 7.
Graphic illustration of the evolutionary implications of m6A methylome in plants. The evolutionary origin of genes from ancient to young is denoted by arrow in the middle. m6A methylations are preferentially maintained on evolutionally ancient genes. The right panel shows the WGD-diploidization cycle. The variations of m6A methylation are labeled at the corresponding positions. The left panel shows local-genome duplication. The copy number increased from young genes to ancient genes in with decreased m6A methylation levels and translational efficiency.
Materials and Methods
Plant Material and Growth Conditions
Arabidopsis thaliana (Col-0) was sown in nutrient soil and cultivated in a greenhouse at 22 °C under a 16 h light/8 h dark cycle. On the 14th day after germination, all the above-ground parts of the seedlings of A. thaliana were collected and frozen in liquid nitrogen immediately and stored at −80 °C. Gossypium arboreum (Shixiya1, AA genome) and G. hirsutum (TM-1, AADD genome) were grown at 28 °C, and P. vulgaris (G19833) and Gly. max (Williams 82) were grown at 20 °C under a 16 h light/8 h dark cycle. The above-ground parts of the seedlings of G. arboretum, G. hirsutum, P. vulgaris, and Gly. max were collected on the 18th day after germination. Sorghum bicolor (BTx623) and Z. mays (B73) were sown in nutrient soil and cultivated in a greenhouse at 25 °C under a 16 h light/8 h dark cycle. Aegilopstauschii (AL8/78, DD genome), T. dicoccoides (Zavitan, AABB genome), and T. aestivum (Chinese Spring, AABBDD genome) were germinated in petri dishes and transplanted into soil under a 16 h light/8 h dark cycle at 22 °C. Oryza sativa (Nipponbare) was grown at 28 °C under a 14 h light/10 h dark cycle. When seedlings of these six monocot species developed three fully expanded leaves, the above-ground parts were collected and frozen at −80 °C. The single shoot tip of Phy. patens (Gransden 2004) was taken from the top of the fronds and inserted into BCD medium without ammonium tartrate at 25 °C and under a 16 h light/8 h dark cycle. After 4 weeks, the stems and leaves were mixed together and frozen in liquid nitrogen immediately and stored at −80 °C. For each species, three biological replicates were used for the experiments described below.
RNA Isolation and High-Throughput m6A-Seq
Total RNA was extracted and purified using TRIzol reagent (Invitrogen) according to the manufacturer’s instructions, and NanoDrop ND-1000 (NanoDrop) was used to quantify its amount and purity. RNA integrity was assessed using Bioanalyzer 2100 (Agilent, CA) with RIN number >7.0 and confirmed using denaturing agarose gel electrophoresis. Polyadenylated (PolyA+) mRNA selection was performed using Dynabeads Oligo (dT) 25-61005 (Thermo Fisher) with two rounds of purification from 30 μg total RNA. The poly(A) RNA was fragmented into small pieces using Magnesium RNA Fragmentation Module (NEB, Cat. e6150) under 86 °C for 7 min. The fragmented RNA was then used to construct the input library directly following conventional RNA-seq and the immunoprecipitation (IP) library was enriched using m6A-specific antibodies (No. 202003, Synaptic Systems, Germany) in IP buffer (50 mM Tris–HCl, 750 mM NaCl, and 0.5% Igepal CA-630) for 2 h at 4 °C. The Input and IP were reverse-transcribed using SuperScript II Reverse Transcriptase (Invitrogen, Cat. 1896649). U-labeled second-stranded DNAs were synthesized with Escherichiacoli DNA polymerase I (NEB, Cat. m0209), RNase H (NEB, Cat. m0297), and dUTP Solution (Thermo Fisher, Cat. R0133).
An A-base was added to the blunt ends of each strand. Adapters containing a T-base overhang were used for ligating the A-tailed fragmented DNA. Single- or dual-index adapters were ligated to the fragments, and size selection was performed with AMPureXP beads. After heat-labile UDG enzyme (NEB, Cat. m0280) treatment of the U-labeled second-stranded DNAs, the ligated products were amplified with PCR under the following conditions: initial denaturation at 95 °C for 3 min; eight cycles of denaturation at 98 °C for 15 s, annealing at 60 °C for 15 s, extension at 72 °C for 30 s, and final extension at 72 °C for 5 min. The average insert size for the final cDNA library was 300 ± 50 bp. Sequencing was carried out on the Illumina NovaSeq 6000 platform (LC-Bio Technology Co., Ltd., Hangzhou, China) with a 2 × 150-bp paired-end strategy (PE150).
Bioinformatic Analysis of m6A-Seq Data
Raw reads from 13 species were preprocessed by trimming the adapter sequences, low-quality bases, and undetermined bases using fastp v0.20.0 (Chen et al. 2018) with default parameters. Clean reads were mapped against their corresponding reference genomes using STAR v2.7.3a (Dobin et al. 2013) with the following settings: outFilterMismatchNmax, 0; outSAMattrIHstart, 0; outFilterMultimapNmax, 1; alignIntronMin, 10; alignIntronMax, an estimated value that covers 99.9% of all intron lengths in the analyzed species. The reference genomes and gene annotations in GFF3 format of A. thaliana, Z. mays, Aeg. tauschii, T. dicoccoides, and T. aestivum were obtained from Ensembl Plants (v43) (Howe et al. 2020); G. arboreum from Wang, Wang, et al. (2019); G. hirsutum from CottonGen database (Yu et al. 2014); Sol. lycopersicum from Sol Genomics Network (Lin et al. 2005); P. vulgaris, Gly. max, O. sativa, and S. bicolor from JGI (phytozome v12) (Goodstein et al. 2012) (supplementary data 9, Supplementary Material online). For strand-specific sequencing data, bam files were split into two sets based on samflags: forward strand set (-f 80 [first in pair] and -f 128 -F 16 [second in pair]) and reverse strand set (-f 64 -F 16 [first in pair] and -f 144 [second in pair]). SAMtools v1.9 (Li et al. 2009) was used to obtain reads with high-mapping quality (samtools view -F 1,804 -f 2 -q 30), sort bam files (samtools sort), and fix mate-pair information (samtools fixmate). Paired-end reads were converted to BED format using bedtools v2.29.0 (bedtools bamtobed -bedpe -mate1). Reads within genic regions were retained for subsequent analysis (bedtools intersect -wa -f 0.5). Mapped reads of input and IP were input into R package PEA (Zhai et al. 2018) to perform peak calling using the SlidingWindow method with default options. Peaks that overlapped in at least two of three replicates were merged as confidence m6A peaks using the slice function in R package IRanges (Lawrence et al. 2013). Stringtie v2.0 (Pertea et al. 2015) was used to estimate gene expression abundances from input libraries by calculating transcripts per million (TPM) (stringtie -e -j 10; –rf for strand-specific parameter). m6A peak abundance was calculated as (mapped fragments IP × Total mapped fragments Input)/(mapped fragments Input × Total mapped fragments IP) using DiffBind package (Ross-Innes et al. 2012). The called peaks with low m6A abundance (<2-fold enrichment) or within lowly expressed genes (TPM<1) were discarded.
Identification of OGs and SGs among 13 Species
The protein sequences of 611,220 protein-coding genes from 13 species were used to construct orthogroups using OrthoFinder v.2.4.0 (Emms and Kelly 2019) with default parameters. Briefly, 1) only the longest protein was selected for identification when several isoforms were available for one gene; 2) DIAMOND v.0.9.24 (Buchfink et al. 2015) was used for obtaining similarity relationship between protein sequences; and 3) clustering of genes into orthogroups was performed using MCL graph clustering algorithm. A total of 42,580 orthogroups were identified. We further defined genes appearing in nonspecies-specific orthogroups and presenting in at least two species as OGs, whereas genes other than OGs were defined as SGs.
GO Enrichment Analysis
The GO annotations were retrieved from Ensembl Plant Biomart (Kinsella et al. 2011), and CottonGen (Yu et al. 2014). GO enrichment analysis was performed using the enricher function from clusterProfiler R-package for the hypergeometric test (Yu et al. 2012).
Phylostratigraphic Analysis
The phylostratigraphic tree of genes or gene families was constructed using TimeTree (Kumar et al. 2017) according to methods detailed in previous studies (Domazet-Loso et al. 2007; Guo 2013; Lei et al. 2017). To obtain high-quality phylostratigraphic results, we analyzed 34 representative species having clear taxonomic and phylogenetic relationships (Wu et al. 2020). The orthogroups were assigned to different phylostrata representing different evolutionary ages. If there was an unnamed node in phylostrata, it was renamed using the combination of first letters of the child node names (i.e., AT represented Aeg. tauschii and Triticum, BM represented Brassicales and Malvaceae).
Divergence of m6A Methylomes
The species tree and divergence time of internal nodes were obtained from the TimeTree database (http://timetree.org/) using “BUILD A TIMETREE” function (Kumar et al. 2017). The m6A abundance divergence was defined as 1−ρ, where ρ was the Spearman’s correlation coefficient between m6A methylomes in two species.
Identification of Subgenomes in Species Having Undergone Recent Polyploidization
The paralogous gene pairs between subgenomes in Z. mays, Gly. max, and T. aestivum, and their syntenic counterparts in sister species S. bicolor/Z. mays and P. vulgaris/Gly. max were obtained from previously published data sets (Zhao, Zhang, et al. 2017; Brohammer et al. 2018; Ramirez-Gonzalez et al. 2018). The subgenomes of G. hirsutum were classified based on chromosomes (A and D subgenomes). The syntenic counterparts in G. arboreum/G. hirsutum (A subgenome), Aeg. tauschii/T. aestivum (D subgenome), and T. dicoccoides (A subgenome)/T. aestivum (A subgenome), and T. dicoccoides (B subgenome)/T. aestivum (B subgenome) were detected using SynMap within CoGe with a 1:1 quota-align ratio and default parameters (Haug-Baltzell et al. 2017). In maize, genes not included in duplicates formed by WGD were further classified into four duplication types using DupGen_finder (Qiao et al. 2019) with default parameters: tandem duplicates, proximal duplicates, transposed duplicates, and dispersed duplicates.
Analysis of Synonymous and Nonsynonymous Substitutions in OGs
The one-to-one OG pairs from either eight diploid species (A. thaliana, G. arboreum, P. vulgaris, Sol. lycopersicum, S. bicolor, Aeg. tauschii, O. sativa, and Phy. patens) or their sister species (G. arboreum/G. hirsutum, P. vulgaris/Gly. max, S. bicolor/Z. mays, Aeg. tauschii/T. dicoccoides/T. aestivum) were extracted from OrthoFinder, and their protein sequences were aligned using paraAT v2.0 (Zhang et al. 2012). The synonymous and nonsynonymous substitutions, and their resulting Ka/Ks (ω) values were calculated using KaKs_calculator v2.0 (Wang et al. 2010) with the Model Averaging method.
Analysis of Sequence Divergence
To estimate the sequence divergence between OG pairs, nucleotide sequences for gene pairs were aligned using needle in EMBL analysis tools (Chojnacki et al. 2017) with parameters -gapopen=10 and -gapextend=0.5. The sequence divergence between OG pairs from sister species genomes was calculated by ortholog comparisons. For example, sequence divergence in Z. mays genes was estimated by comparison with their orthologous in S. bicolor. For the aligned sequences of paired genes, gaps in the gene from the species that had not experienced recent polyploidy events were removed. For example, in S. bicolor/Z. mays gene pairs, the gaps in S. bicolor genes were removed. Aligned sequences were further divided into three nonoverlapping parts: 5′-UTR, coding sequence, and 3′-UTR. Only gene pairs with all three parts and each part longer than 100 bp were used in subsequent analysis. Each part was divided into ten bins and the mutation frequency for each bin was calculated using “inconsistent base/base number.” The sequence identity of 3′-UTR was calculated using “identical base/base number.”
Likelihood Analysis of Gene Gain and Loss
The program CAFE (computational analysis of gene family evolution) v4.2.1 (De Bie et al. 2006) was used to analyze the evolution of gene families based on a probabilistic graphical model. To reduce the influence from plant polyploidization events, 27 diploid species were selected to identify expansion and contraction of gene families. The number of genes in gene families was obtained from OrthoFinder results. Gene families with more than 100 gene copies in one or more species were removed from this analysis. The dated species tree was downloaded from the TimeTree database. We set the birth–death parameter to one and the P value to 0.01.
Identification of TFs, Kinases, and R Genes
Transcription factors and kinases were obtained by scanning consensus rules and HMM profiles using iTAK v1.7 (Zheng et al. 2016). The identification of R genes was performed using HMMER v3 (Potter et al. 2018) and CD-search (Marchler-Bauer and Bryant 2004). The protein sequences from 13 species were scanned using HMMER and Hidden Markov Model of NB-ARC (Pfam accession: PF00931) (El-Gebali et al. 2019). Genes with NB-ARC domain (E-value <1e-5) were further authenticated. The amino-terminal domain (TIR/coiled-coil/other) and carboxyl-terminal domain (LRR) were recognized by CD-search. Only genes with both NBS and LRR domains were defined as R genes.
Sequence Alignment and Estimation of the Phylogenetic Tree
Multiple sequence alignment of R genes was performed using muscle v3.8.1551 (Edgar 2004) with default parameters. The maximum-likelihood phylogenetic tree was then inferred with IQ-Tree v2.0.3 (Minh et al. 2020) with parameters -m VT+F+R9, -bb 1,000, and -T AUTO. All trees were rooted using P25941 from streptomyces as the outgroup.
Calculation of Translational Efficiency
The translational level and corresponding transcriptional level of maize genes were obtained from a previous study (Luo et al. 2020). The translational efficiency of each gene was calculated by “TPM (translational level)/TPM (transcriptional level)” as previously described (Lei et al. 2015; Luo et al. 2020).
Statistical Analysis
The Wilcox test, the χ2 test, and the Student’s t-test were respectively performed using the wilcox.test, chisq.test, and t.test functions in R package.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
We thank Professor Yikun He (Capital Normal University, China) for providing Physcomitrella patens material, Professor Lianjun Sun (China Agricultural University, China) for providing seeds of Glycine max, Professor Chunbao Zhang (Jilin Academy of Agricultural Sciences, China) for providing seeds of Phaseolus vulgaris, Dr Lei Ma (Institute of Cotton Research of CAAS, China) for providing seeds of Gossypium arboreum and G. hirsutum, the Wheat Genetics and Genomics Center (China Agricultural University, China) for providing seeds of Aegilops tauschii, Triticum dicoccoides, and Triticum aestivum, Professor Kunming Chen (Northwest A&F University, China) for providing seeds of Oryza sativa. We also thank High-Performance Computing (HPC) of Northwest A&F University for providing computing resources. This work was supported by the Youth 1000-Talent Program of China, the National Natural Science Foundation of China (Grant No. 32000410), the Hundred Talents Program of Shaanxi Province of China, the Fund of Northwest A&F University (Grant No. Z111021603), and the Fundamental Research Funds for the Central Universities (Grant No. 2452020041).
Author Contributions
C.M. and Z.M. conceived the project. T.Z., B.X., and Z.M. performed the data analysis. Y.Q. prepared plant materials. Z.M., T.Z., B.X., and C.M. wrote the article.
Data Availability
The raw-sequencing data generated in this study have been deposited in the Genome Sequence Archive (Wang et al. 2017) in National Genomics Data Center (National Genomics Data Center Members and Partners 2020), Beijing Institute of Genomics (BIG), Chinese Academy of Sciences, under accession number CRA004114 that are publicly accessible at https://bigd.big.ac.cn/gsa. The analysis pipeline and codes used in this study have been deposited in GitHub (https://github.com/cma2015/m6A_Evolution).
References
- Arendsee ZW, Li L, Wurtele ES.. 2014. Coming of age: orphan genes in plants. Trends Plant Sci. 19(11):698–708. [DOI] [PubMed] [Google Scholar]
- Brohammer AB, Kono TJY, Springer NM, McGaugh SE, Hirsch CN.. 2018. The limited role of differential fractionation in genome content variation and function in maize (Zea mays L.) inbred lines. Plant J. 93(1):131–141. [DOI] [PubMed] [Google Scholar]
- Buchfink B, Xie C, Huson DH.. 2015. Fast and sensitive protein alignment using DIAMOND. Nat Methods. 12(1):59–60. [DOI] [PubMed] [Google Scholar]
- Chang AY, Liao BY.. 2012. DNA methylation rebalances gene dosage after mammalian gene duplications. Mol Biol Evol. 29(1):133–144. [DOI] [PubMed] [Google Scholar]
- Chen S, Zhou Y, Chen Y, Gu J.. 2018. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34(17):i884–i890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng F, Wu J, Cai X, Liang J, Freeling M, Wang X.. 2018. Gene retention, fractionation and subgenome differences in polyploid plants. Nat Plants. 4(5):258–268. [DOI] [PubMed] [Google Scholar]
- Chojnacki S, Cowley A, Lee J, Foix A, Lopez R.. 2017. Programmatic access to bioinformatics tools from EMBL-EBI update: 2017. Nucleic Acids Res. 45(W1):W550–W553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colome-Tatche M, Cortijo S, Wardenaar R, Morgado L, Lahouze B, Sarazin A, Etcheverry M, Martin A, Feng S, Duvernois-Berthet E, et al. 2012. Features of the Arabidopsis recombination landscape resulting from the combined loss of sequence variation and DNA methylation. Proc Natl Acad Sci U S A. 109(40):16240–16245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conant GC, Wolfe KH.. 2008. Turning a hobby into a job: how duplicated genes find new functions. Nat Rev Genet. 9(12):938–950. [DOI] [PubMed] [Google Scholar]
- De Bie T, Cristianini N, Demuth JP, Hahn MW.. 2006. CAFE: a computational tool for the study of gene family evolution. Bioinformatics 22(10):1269–1271. [DOI] [PubMed] [Google Scholar]
- Demuth JP, De Bie T, Stajich JE, Cristianini N, Hahn MW.. 2006. The evolution of mammalian gene families. PLoS One 1:e85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diez CM, Roessler K, Gaut BS.. 2014. Epigenetics and plant genome evolution. Curr Opin Plant Biol. 18:1–8. [DOI] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR.. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29(1):15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domazet-Loso T, Brajkovic J, Tautz D.. 2007. A phylostratigraphy approach to uncover the genomic history of major adaptations in metazoan lineages. Trends Genet. 23(11):533–539. [DOI] [PubMed] [Google Scholar]
- Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, Cesarkas K, Jacob-Hirsch J, Amariglio N, Kupiec M, et al. 2012. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485(7397):201–206. [DOI] [PubMed] [Google Scholar]
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32(5):1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edger PP, Pires JC.. 2009. Gene and genome duplications: the impact of dosage-sensitivity on the fate of nuclear genes. Chromosome Res. 17(5):699–717. [DOI] [PubMed] [Google Scholar]
- El-Gebali S, Mistry J, Bateman A, Eddy SR, Luciani A, Potter SC, Qureshi M, Richardson LJ, Salazar GA, Smart A, et al. 2019. The Pfam protein families database in 2019. Nucleic Acids Res. 47(D1):D427–D432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emms DM, Kelly S.. 2019. OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol. 20(1):238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeling M. 2009. Bias in plant gene content following different sorts of duplication: tandem, whole-genome, segmental, or by transposition. Annu Rev Plant Biol. 60:433–453. [DOI] [PubMed] [Google Scholar]
- Freeling M, Thomas BC.. 2006. Gene-balanced duplications, like tetraploidy, provide predictable drive to increase morphological complexity. Genome Res. 16(7):805–814. [DOI] [PubMed] [Google Scholar]
- Fu Y, Dominissini D, Rechavi G, He C.. 2014. Gene expression regulation mediated through reversible m(6)A RNA methylation. Nat Rev Genet. 15(5):293–306. [DOI] [PubMed] [Google Scholar]
- Goodstein DM, Shu S, Howson R, Neupane R, Hayes RD, Fazo J, Mitros T, Dirks W, Hellsten U, Putnam N, et al. 2012. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 40(Database issue):D1178–D1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo YL. 2013. Gene family evolution in green plants with emphasis on the origination and evolution of Arabidopsis thaliana genes. Plant J. 73(6):941–951. [DOI] [PubMed] [Google Scholar]
- Hahn MW, Han MV, Han SG.. 2007. Gene family evolution across 12 Drosophila genomes. PLoS Genet. 3(11):e197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haug-Baltzell A, Stephens SA, Davey S, Scheidegger CE, Lyons E.. 2017. SynMap2 and SynMap3D: web-based whole-genome synteny browsers. Bioinformatics 33(14):2197–2198. [DOI] [PubMed] [Google Scholar]
- Howe KL, Contreras-Moreira B, De Silva N, Maslen G, Akanni W, Allen J, Alvarez-Jarreta J, Barba M, Bolser DM, Cambell L, et al. 2020. Ensembl Genomes 2020-enabling non-vertebrate genomic research. Nucleic Acids Res. 48(D1):D689–D695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jablonka E, Lamb MJ.. 2002. The changing concept of epigenetics. Ann N Y Acad Sci. 981:82–96. [DOI] [PubMed] [Google Scholar]
- Kinsella RJ, Kahari A, Haider S, Zamora J, Proctor G, Spudich G, Almeida-King J, Staines D, Derwent P, Kerhornou A, et al. 2011. Ensembl BioMarts: a hub for data retrieval across taxonomic space. Database (Oxford) 2011:bar030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T. 2007. Chromatin modifications and their function. Cell 128(4):693–705. [DOI] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Suleski M, Hedges SB.. 2017. TimeTree: a resource for timelines, timetrees, and divergence times. Mol Biol Evol. 34(7):1812–1819. [DOI] [PubMed] [Google Scholar]
- Lawrence M, Huber W, Pages H, Aboyoun P, Carlson M, Gentleman R, Morgan MT, Carey VJ.. 2013. Software for computing and annotating genomic ranges. PLoS Comput Biol. 9(8):e1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei L, Shi J, Chen J, Zhang M, Sun S, Xie S, Li X, Zeng B, Peng L, Hauck A, et al. 2015. Ribosome profiling reveals dynamic translational landscape in maize seedlings under drought stress. Plant J. 84(6):1206–1218. [DOI] [PubMed] [Google Scholar]
- Lei L, Steffen JG, Osborne EJ, Toomajian C.. 2017. Plant organ evolution revealed by phylotranscriptomics in Arabidopsis thaliana. Sci Rep. 7(1):7567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup. 2009. Genome Project Data Processing S 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25(16):2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, McKibben MTW, Finch GS, Blischak PD, Sutherland BL, Barker MS.. 2021. Patterns and processes of diploidization in land plants. Annu Rev Plant Biol. 72:1–24. [DOI] [PubMed] [Google Scholar]
- Liang Z, Riaz A, Chachar S, Ding Y, Du H, Gu X.. 2020. Epigenetic modifications of mRNA and DNA in plants. Mol Plant. 13(1):14–30. [DOI] [PubMed] [Google Scholar]
- Lin C, Mueller LA, Mc Carthy J, Crouzillat D, Petiard V, Tanksley SD.. 2005. Coffee and tomato share common gene repertoires as revealed by deep sequencing of seed and cherry transcripts. Theor Appl Genet. 112(1):114–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockton S, Gaut BS.. 2005. Plant conserved non-coding sequences and paralogue evolution. Trends Genet. 21(1):60–65. [DOI] [PubMed] [Google Scholar]
- Luo GZ, MacQueen A, Zheng G, Duan H, Dore LC, Lu Z, Liu J, Chen K, Jia G, Bergelson J, et al. 2014. Unique features of the m6A methylome in Arabidopsis thaliana. Nat Commun. 5:5630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo JH, Wang Y, Wang M, Zhang LY, Peng HR, Zhou YY, Jia GF, He Y.. 2020. Natural variation in RNA m(6)A methylation and its relationship with translational status. Plant Physiol. 182(1):332–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Zhao B, Chen K, Thomas A, Tuteja JH, He X, He C, White KP.. 2017. Evolution of transcript modification by N(6)-methyladenosine in primates. Genome Res. 27(3):385–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A, Bryant SH.. 2004. CD-Search: protein domain annotations on the fly. Nucleic Acids Res. 32(Web Server issue):W327–W331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao Z, Zhang T, Qi Y, Song J, Han Z, Ma C.. 2020. Evolution of the RNA N6-methyladenosine methylome mediated by genomic duplication. Plant Physiol. 182(1):345–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, von Haeseler A, Lanfear R.. 2020. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol. 37(5):1530–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Genomics Data Center Members and Partners. 2020. Database resources of the National Genomics Data Center in 2020. Nucleic Acids Res. 48:D24–D33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto SP. 2007. The evolutionary consequences of polyploidy. Cell 131(3):452–462. [DOI] [PubMed] [Google Scholar]
- Pertea M, Pertea GM, Antonescu CM, Chang TC, Mendell JT, Salzberg SL.. 2015. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol. 33(3):290–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter SC, Luciani A, Eddy SR, Park Y, Lopez R, Finn RD.. 2018. HMMER web server: 2018 update. Nucleic Acids Res. 46(W1):W200–W204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao X, Li Q, Yin H, Qi K, Li L, Wang R, Zhang S, Paterson AH.. 2019. Gene duplication and evolution in recurring polyploidization-diploidization cycles in plants. Genome Biol. 20(1):38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez-Gonzalez RH, Borrill P, Lang D, Harrington SA, Brinton J, Venturini L, Davey M, Jacobs J, van Ex F, Pasha A, et al. 2018. The transcriptional landscape of polyploid wheat. Science 361(6403):eaar6089. [DOI] [PubMed] [Google Scholar]
- Rizzon C, Ponger L, Gaut BS.. 2006. Striking similarities in the genomic distribution of tandemly arrayed genes in Arabidopsis and rice. PLoS Comput Biol. 2(9):e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross-Innes CS, Stark R, Teschendorff AE, Holmes KA, Ali HR, Dunning MJ, Brown GD, Gojis O, Ellis IO, Green AR, et al. 2012. Differential oestrogen receptor binding is associated with clinical outcome in breast cancer. Nature 481(7381):389–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L, Liang Z, Wong CE, Yu H.. 2019. Messenger RNA modifications in plants. Trends Plant Sci. 24(4):328–341. [DOI] [PubMed] [Google Scholar]
- Shi H, Wei J, He C.. 2019. Where, when, and how: context-dependent functions of RNA methylation writers, readers, and erasers. Mol Cell. 74(4):640–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltis PS, Marchant DB, Van de Peer Y, Soltis DE.. 2015. Polyploidy and genome evolution in plants. Curr Opin Genet Dev. 35:119–125. [DOI] [PubMed] [Google Scholar]
- Song B, Chen K, Tang Y, Wei Z, Su J, de Magalhaes JP, Rigden DJ, Meng J.. 2021. ConsRM: collection and large-scale prediction of the evolutionarily conserved RNA methylation sites, with implications for the functional epitranscriptome. Brief Bioinform. bbab088:1–17. [DOI] [PubMed] [Google Scholar]
- Song MJ, Potter BI, Doyle JJ, Coate JE.. 2020. Gene balance predicts transcriptional responses immediately following ploidy change in Arabidopsis thaliana. Plant Cell 32(5):1434–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Peer Y, Maere S, Meyer A.. 2009. The evolutionary significance of ancient genome duplications. Nat Rev Genet. 10(10):725–732. [DOI] [PubMed] [Google Scholar]
- Veitia RA, Bottani S, Birchler JA.. 2008. Cellular reactions to gene dosage imbalance: genomic, transcriptomic and proteomic effects. Trends Genet. 24(8):390–397. [DOI] [PubMed] [Google Scholar]
- Vidalis A, Zivkovic D, Wardenaar R, Roquis D, Tellier A, Johannes F.. 2016. Methylome evolution in plants. Genome Biol. 17(1):264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddington CH. 1942. The epigenotype. Endeavour 1:18–20. [Google Scholar]
- Wang D, Zhang Y, Zhang Z, Zhu J, Yu J.. 2010. KaKs_Calculator 2.0: a toolkit incorporating gamma-series methods and sliding window strategies. Genomics Proteomics Bioinformatics. 8(1):77–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Wang D, Zheng X, Qin A, Zhou J, Guo B, Chen Y, Wen X, Ye W, Zhou Y, et al. 2019. Multi-strategic RNA-seq analysis reveals a high-resolution transcriptional landscape in cotton. Nat Commun. 10(1):4714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N, Yang Y, Moore MJ, Brockington SF, Walker JF, Brown JW, Liang B, Feng T, Edwards C, Mikenas J, et al. 2019. Evolution of portulacineae marked by gene tree conflict and gene family expansion associated with adaptation to harsh environments. Mol Biol Evol. 36(1):112–126. [DOI] [PubMed] [Google Scholar]
- Wang Y, Song F, Zhu J, Zhang S, Yang Y, Chen T, Tang B, Dong L, Ding N, Zhang Q, et al. 2017. GSA: genome Sequence Archive. Genomics Proteomics Bioinformatics. 15(1):14–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendel JF, Jackson SA, Meyers BC, Wing RA.. 2016. Evolution of plant genome architecture. Genome Biol. 17:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Han B, Jiao Y.. 2020. Genetic contribution of paleopolyploidy to adaptive evolution in angiosperms. Mol Plant. 13(1):59–71. [DOI] [PubMed] [Google Scholar]
- Xiang Y, Laurent B, Hsu CH, Nachtergaele S, Lu Z, Sheng W, Xu C, Chen H, Ouyang J, Wang S, et al. 2017. RNA m(6)A methylation regulates the ultraviolet-induced DNA damage response. Nature 543(7646):573–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Hsu PJ, Chen YS, Yang YG.. 2018. Dynamic transcriptomic m(6)A decoration: writers, erasers, readers and functions in RNA metabolism. Cell Res. 28(6):616–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi SV. 2017. Insights into epigenome evolution from animal and plant methylomes. Genome Biol Evol. 9(11):3189–3201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G, Wang LG, Han Y, He QY.. 2012. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16(5):284–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Jung S, Cheng CH, Ficklin SP, Lee T, Zheng P, Jones D, Percy RG, Main D.. 2014. CottonGen: a genomics, genetics and breeding database for cotton research. Nucleic Acids Res. 42(Database issue):D1229–D1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue H, Nie X, Yan Z, Weining S.. 2019. N6-methyladenosine regulatory machinery in plants: composition, function and evolution. Plant Biotechnol J. 17(7):1194–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai J, Song J, Cheng Q, Tang Y, Ma C.. 2018. PEA: an integrated R toolkit for plant epitranscriptome analysis. Bioinformatics 34(21):3747–3749. [DOI] [PubMed] [Google Scholar]
- Zhang H, Shi X, Huang T, Zhao X, Chen W, Gu N, Zhang R.. 2020. Dynamic landscape and evolution of m6A methylation in human. Nucleic Acids Res. 48(11):6251–6264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Xiao J, Wu J, Zhang H, Liu G, Wang X, Dai L.. 2012. ParaAT: a parallel tool for constructing multiple protein-coding DNA alignments. Biochem Biophys Res Commun. 419(4):779–781. [DOI] [PubMed] [Google Scholar]
- Zhao BS, Roundtree IA, He C.. 2017. Post-transcriptional gene regulation by mRNA modifications. Nat Rev Mol Cell Biol. 18(1):31–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M, Zhang B, Lisch D, Ma J.. 2017. Patterns and consequences of subgenome differentiation provide insights into the nature of paleopolyploidy in plants. Plant Cell 29(12):2974–2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Jiao C, Sun H, Rosli HG, Pombo MA, Zhang P, Banf M, Dai X, Martin GB, Giovannoni JJ, et al. 2016. iTAK: a program for genome-wide prediction and classification of plant transcription factors, transcriptional regulators, and protein kinases. Mol Plant. 9(12):1667–1670. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw-sequencing data generated in this study have been deposited in the Genome Sequence Archive (Wang et al. 2017) in National Genomics Data Center (National Genomics Data Center Members and Partners 2020), Beijing Institute of Genomics (BIG), Chinese Academy of Sciences, under accession number CRA004114 that are publicly accessible at https://bigd.big.ac.cn/gsa. The analysis pipeline and codes used in this study have been deposited in GitHub (https://github.com/cma2015/m6A_Evolution).