Abstract
Simulation models are increasingly being used to inform epidemiologic studies and health policy, yet there is great variation in their transparency and reproducibility. In this review, we provide an overview of applications of simulation models in health policy and epidemiology, analyze the use of best reporting practices, and assess the reproducibility of the models using predefined, categorical criteria. We identified and analyzed 1,613 applicable articles and found exponential growth in the number of studies over the past half century, with the highest growth in dynamic modeling approaches. The largest subset of studies focused on disease policy models (70%), within which pathological conditions, viral diseases, neoplasms, and cardiovascular diseases account for one-third of the articles. Model details were not reported in almost half of the studies. We also provide in-depth analysis of modeling best practices, reporting quality and reproducibility of models for a subset of 100 articles (50 highly cited and 50 randomly selected from the remaining articles). Only 7 of 26 in-depth evaluation criteria were satisfied by more than 80% of samples. We identify areas for increased application of simulation modeling and opportunities to enhance the rigor and documentation in the conduct and reporting of simulation modeling in epidemiology and health policy.
Keywords: health policy, reproducibility, simulation modeling
Abbreviations:
- MeSH
Medical Subject Headings
INTRODUCTION
Increasingly complex health systems have necessitated and advancements in computational tools have enabled the application of simulation models to inform epidemiology and health policy. Simulation models help analyze and interpret the complexity of a health issue, enabling the design, evaluation, and improvement of policies while minimizing unintended consequences (1, 2).
Simulation applications span a wide range of disciplines, such as health care reform (3), health care delivery (4, 5), cancer research (6, 7), chronic disease (8, 9), mental health (10, 11), and infectious diseases (12–17), among others. These models are often complex, follow different methods and formalizations, and are integrated into major decisions with potential for significant impact. Therefore, we need a more systematic look at which application areas are covered and how the methodological rigor and replicability of studies have evolved. However, although other studies have assessed transparency and reproducibility of simulation models of a single disease or a subset of modeling techniques (18–23), a broader systematic view that allows for comparison over application areas, methods, or time across health policy and epidemiology is lacking (24).
In this review, we address this gap through 3 interrelated aims. First, we provide a broad comparative view of the state of simulation-modeling research across various health domains and modeling approaches to provide context, inform current trends, and identify potential gaps. Second, we provide a systematic assessment of the modeling practices in existing research, focusing on rigor and quality in reporting of design, implementation, validation, and dissemination, which influence the credibility and impact of simulation-modeling research. Third, we assess the reproducibility of existing simulation-modeling research. Reproducibility is the cornerstone of scientific research, and the case for enhancing reproducibility has been made repeatedly and has given rise to various guidelines and methodologies for authors to produce reproducible simulation models (25–30). Reproducibility is key to building a cumulative science, revising problems in light of new data, and building confidence in the reliability of existing findings (31–33). Documenting the state of the field informs interpretation of present simulation-modeling approaches in health policy, particularly those that address health crises such as the COVID-19 pandemic (18, 22). Our work enables the measurement of progress in modeling reproducibility.
We pursued the first and second objectives by conducting a broad systematic review of 50 years of simulation-modeling research in health policy and epidemiology across modeling methodologies and research domains. The third objective was accomplished by sampling a subset of that body of literature for in-depth transparency evaluation. We do not summarize relevant substantive findings. Instead, our focus is on the trends in application areas, methods, modeling practices, and documentation and reproducibility.
METHODS
Search strategy and selection criteria
PubMed was the primary search database we searched for the terms “simulation,” “simulate,” “simulated,” “simulating,” “policy,” and “policies” in the title and abstract. Our definition of “simulation” was not limited to any discipline, and we included research from a wide range of fields. We complemented the results by searching 71 journals categorized in Health Policy and Services within Web of Science for articles containing the word “simulation” in the title or abstract. We limited the search in both databases to English-language, peer-reviewed articles. Our search included any articles indexed in these databases before March 2016 (the beginning of this project). We reviewed the abstracts of the resulting sample to identify articles about studies in which simulation was the main method of research to address an epidemiological or health policy research question. The full text was inspected in cases where the abstract did not establish the inclusion criteria. Articles in which simulation modeling and/or policy was mentioned but simulation modeling was not used as the main research method were excluded, as were reviews and meta-analyses. Figure 1 summarizes the search and the inclusion and exclusion process.
Figure 1.
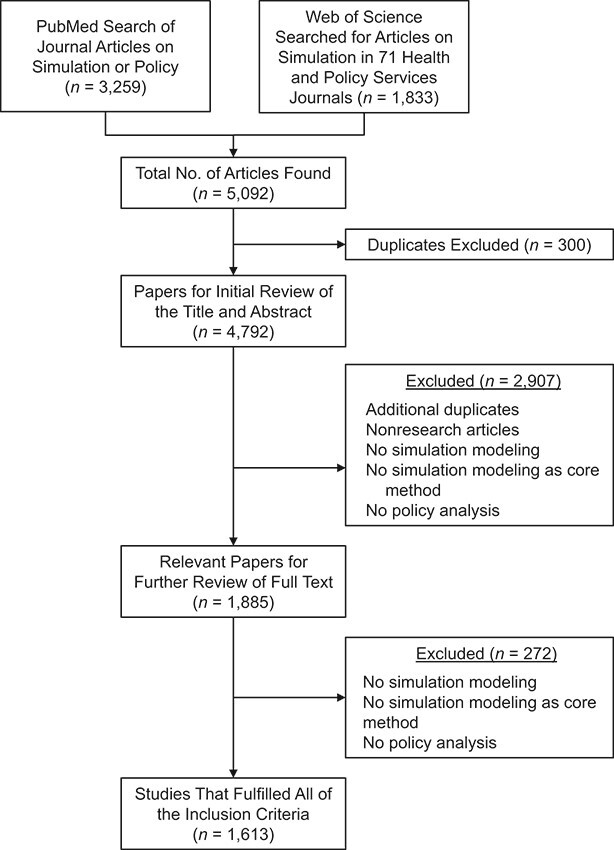
Study selection flowchart. Shown is the selection process for articles included in the review. Note that some articles met more than 1 criterion for exclusion.
Data extraction for full sample
We extracted the title, abstract, publication year, journal, and author information directly from PubMed and Web of Science. To extract model type and more detailed reporting characteristics, we obtained and examined the full text of each article. To access the associated Medical Subject Headings (MeSH) terms from the articles selected for analysis, we developed a web-scraping script in Python that we used on PubMed in July 2018. We used a Python package (scholar.py) to scrape citation data for each article, including number of citations and journal impact factor, from Google Scholar in October 2018.
Chronological, clustering, and trend analysis
We identified the categorizations of each MeSH term using the National Institute of Health’s MeSH Browser and determined the distribution of the full sample of articles across second-level categorizations (e.g., within Diseases [C], Neoplasms [C04]). Where we report the MeSH-term frequencies, we exclude the following categories because they were very general (and thus frequent) but not informative about policy areas of interest (we kept the articles containing them; each article may contain multiple second-level categorizations): Eukaryota; Amino Acids; Peptides and Proteins; and Hormones, Hormone Substitutes, and Hormone Antagonists.
Using the same MeSH-term frequency data, we present the distribution of all second-level MeSH terms across the articles. We use 4 first-level MeSH terms, color-coded in Figure 2, to represent high-level categories of topics: Anatomy, Chemicals and Drugs, Diseases, and Organisms.
Figure 2.
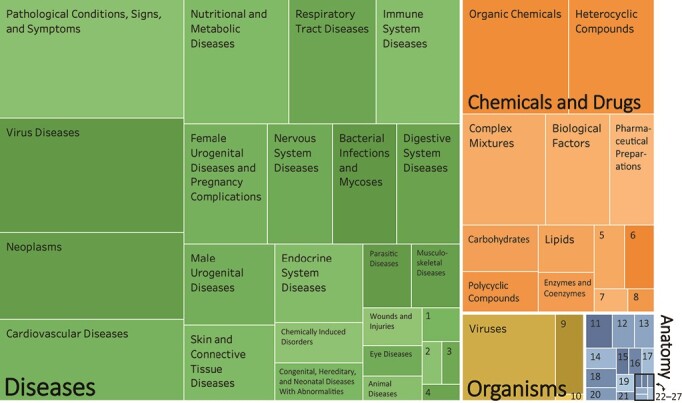
Medical Subject Heading–term categorization. Diseases (green), Chemicals and Drugs (orange), Organisms (yellow), and Anatomy (blue). For the boxes labeled with numbers, the key is as follows: 1. Hemic and Lymphatic Diseases; 2. Occupational Diseases; 3. Otorhinolaryngologic Diseases; 4. Stomatognathic Diseases; 5. Nucleic Acids, Nucleotides, and Nucleosides; 6. Inorganic Chemicals; 7. Biomedical and Dental Materials; 8. Macromolecular Substances; 9. Bacteria; 10. Organism Forms; 11. Body Regions; 12. Cells; 13. Fluids and Secretions; 14. Hemic and Immune Systems; 15. Musculoskeletal System; 16. Urogenital System; 17. Embryonic Structures; 18. Cardiovascular System; 19. Plant Structures; 20. Tissues; 21. Sense Organs; 22. Digestive System; 23. Respiratory System; 24. Nervous System; 25. Animal Structures; 26. Stomatognathic System; and 27. Integumentary System.
Finally, adopting from Adams and Gurney (34), we aggregated collaboration and location data for authors of each of the papers to determine a multilateral collaboration score and related this metric with the results of modeling rigor and reporting evaluation. See the Web Appendix (available at https://doi.org/10.1093/aje/mxab006) for more information.
Categorization based on modeling approach
The review of titles and abstracts across all articles began with an evaluation of 4 high-level properties (35) of models and their reporting: 1) static or dynamic (time dependency), 2) stochastic or deterministic, 3) event driven or continuous, and 4) model documentation (i.e., whether model equations are included in the article or its appendix, referenced in another paper, or not available (actual replication was not attempted)). The first 3 categorizations were used in lieu of reporting the simulation-modeling approach (e.g., Markov decision modeling, microsimulation, compartmental modeling, system dynamics, agent based approaches), because there is significant overlap among these categories and no uniform categorization exists. Categorical properties (35) were selected because they are mutually exclusive, informative (see Web Table 1 for definitions of categorical criteria), and represent specific modeling approaches. They also are useful for identifying the type of real-world mechanisms and focus areas (e.g., feedback mechanisms, stochasticity) tackled in different studies. For example, static and deterministic models may focus mostly on analytical derivations, whereas policy analyses may require feedback-rich models.
Evaluation of modeling rigor and reporting
After the broad overview and categorization of the full sample of articles, we selected 100 articles on which we performed an in-depth evaluation; we compared articles that received much attention (i.e., were highly cited) with typical articles (i.e., not highly cited) to determine if any measures of quality and transparency correlated with attention. Thus, we identified 50 highly cited articles and 50 articles randomly selected from all those not in the highly cited group. In identifying the 50 highly cited articles, we controlled for the recency of the articles, using a year fixed effect in a regression to predict expected citation; we then classified highly cited articles as those with the largest fractional deviation from expected. The 50 highly cited and 50 randomly selected articles are listed in Web Table 2 and Web Table 3, respectively.
Building upon best modeling practice guidelines recommended by the Professional Society for Health Economics and Outcomes Research (36, 37), the Agency for Healthcare Research and Quality (38), and other studies in the literature (24, 39, 40), we assembled a set of 26 concrete criteria for evaluating the quality and rigor of simulation-based models, as reported in the identified articles. These guidelines include criteria that we did not use in the present review (e.g., items targeted to assess economic evaluation studies). These previously validated, individual criteria spanned transparency in reporting of the model’s context, conceptualization, and formalization, along with analysis of the results and any external influences. To minimize potential bias, we considered each criterion to have equal weight. In assessing replication, we only determined whether a study replication could be attempted, not whether the final replication results would match those of the original (see the criteria and their definitions in Web Table 4 and additional detail on criteria assessment in the Web Appendix).
Two researchers, with the help of the first author, used an initial pilot test to evaluate assessment criteria and calibrate consistent answers over an initial sample of 10 articles. The uncalibrated agreement rate on this initial sample across 2 coders was 75%. Once consistency in evaluating criteria was established, the 2 researchers completed the coding and had a 90% agreement level. The remaining 10% of split coded items were discussed and resolved; reported findings are based on the resulting consensus.
Statistical analysis
For the subset of 100 sampled articles with additional information, logistic regression was performed to evaluate the relationship among article publication information (i.e., journal impact factor; collaboration; number of pages; time since publication; and location in United States or not), categorical modeling type designation (i.e., dynamic or static; continuous or event driven; stochastic or deterministic), and overall final evaluation score to identify potential correlates of reproducibility and formal quality measures. To compare the fulfillment of criteria by a sample of top-cited articles and a sample of randomly selected articles, a 2-sample test of proportions was performed.
RESULTS
Study identification and selection
We identified 5,092 articles from an initial search in the PubMed and Web of Science databases, of which 300 were duplicates found in both databases. We then reviewed the abstracts and titles of the resulting 4,792 articles. Of these, 1,855 articles met the initial criteria for full-text review. We excluded 272 articles after a the full-text inspection, which left 1,613 studies that contained simulation modeling as a core method, in addition to a policy analysis. This was the full sample for the study. Figure 1 presents a summary of study selection.
Chronological, clustering, and trend analysis
Figure 2 presents the breakdown of research areas. The research areas across articles were categorized as follows: Diseases (37.2%), followed by Chemicals and Drugs (34.0%), Organisms (27.5%), and Anatomy (1.2%). Each category was broken down into the most common subcategories. In Diseases, the most commonly occurring subcategories were Pathological Conditions, Signs and Symptoms, and Virus Diseases; in Chemicals and Drugs, Organic Chemicals was the most common; in Organisms, Viruses constituted the vast majority of terms; in Anatomy, Body Regions (n = 11 articles) and Cells (n = 9 articles) were most common. See Web Figure 1 for the number of articles in the top 20 research areas. It should be noted that 5% (n = 77 of 1,613) of the articles evaluated did not contain MeSH terms and were not included in Figure 2. In this sample, the average article had 12.4 (standard deviation, 4.8) second-level MeSH-term subcategories; thus, some articles may appear under multiple subheadings in the graph.
Categorization based on modeling approach
Figure 3 presents the trend in reporting of models from 1967 to 2016. Until 2007, the proportion of articles in which model equations were not reported at all was greater than the ones in which authors did report them. Between 2007 and 2016, model equations were reported in more studies than those that did not report model equations. The overall percentage of studies in which an earlier study was cited with its model equations (9%) increased from 1967 to 2016, with the largest increase occurring between 2007 and 2011.
Figure 3.
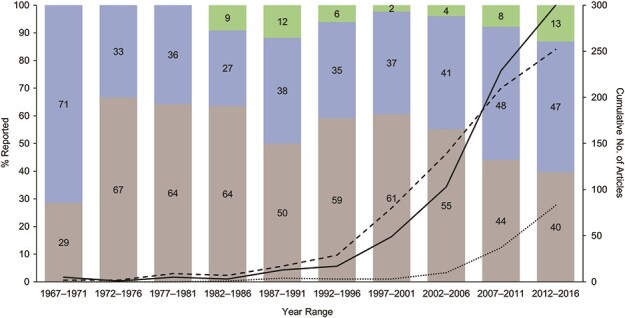
Model equation reporting trend over time. Articles were divided into 5-year publishing-date increments and then assessed by whether they contained model equations or citations of model equations. Gray represents the percentage of articles in which model equations were not reported; blue represents the percentage of articles in which model equations were reported; and green represents the percentage of articles in which reference was made to model equations published elsewhere. Dashed line represents the number of articles in which model equations were not reported; solid line represents the number of articles in which model equations were reported; and dotted line represents articles in which reference was made to model equations published elsewhere.
Model reporting patterns varied across deterministic and stochastic, static and dynamic, and event-driven and continuous permutations (see Web Figure 2). Of articles in which static and deterministic models were reported (n = 135), model equations were not reported in 79%, whereas model equations were reported on within the text of the article or appendix in 15%; and in 6%, another article was cited in which the model equations were referenced. Models that were both static and stochastic were reported on in 193 studies; the authors of 44% of these did not report their model equations, whereas model equations were given within the text of the article or appendix in 52%, and 4% cited another article in which the model equations were referenced.
More specifically, authors did not report their model equations in 68% of articles containing models that were dynamic, deterministic, and continuous (n = 207), whereas in 24% of these articles, model equations were presented in the text of the article of appendix, and in 8%, another article was cited in which the model equations were referenced. In 53% of articles on studies that used dynamic, deterministic, and event-driven models (n = 159), authors did not report their model equations; equations were reported within the text of the article or appendix of 38%, and in 8%, another article was cited in which the model equations were referenced. In addition, in 36% of articles in which models were used that were dynamic, stochastic, and continuous (n = 409), authors did not report their model equations; in 54%, equations were reported within the text of the article or appendix, and in 9% of these articles, another article was cited in which the model equations were referenced. In 31% of articles of studies in which dynamic, stochastic, and event-driven models were used (n = 451), authors did not report their model equations, whereas in 57% of these articles, equations were reported within the text of the article or appendix, and in 12%, another article was cited in which the model equations were referenced. Overall, dynamic, stochastic, and event-driven models had the highest rate of reporting model equations (see Web Figure 2 for details). Moreover, in approximately half of the articles, authors used event-driven simulations and continuous formalizations were used in the other half (see Web Figures 3 and 4 for breakdown over time of different model types).
Detailed evaluation of modeling rigor and reporting
Figure 4 presents our findings on whether the 100 sampled articles satisfied the 26 evaluation criteria. Of the 26 criteria, 3 were potentially better addressed in the random selection of articles than in the highly cited ones: generalizability discussion (z = 1.69; P = 0.09), high-level model visualization (z = 1.80; P = 0.07), and discussion about strategies or policies (z = 3.28; P < 0.001). Two criteria were potentially better addressed in the most-cited articles: quality of calibration fit (z = −1.78; P = 0.08) and model calibration (z = −2.52; P = 0.01) (Figure 4).
Figure 4.
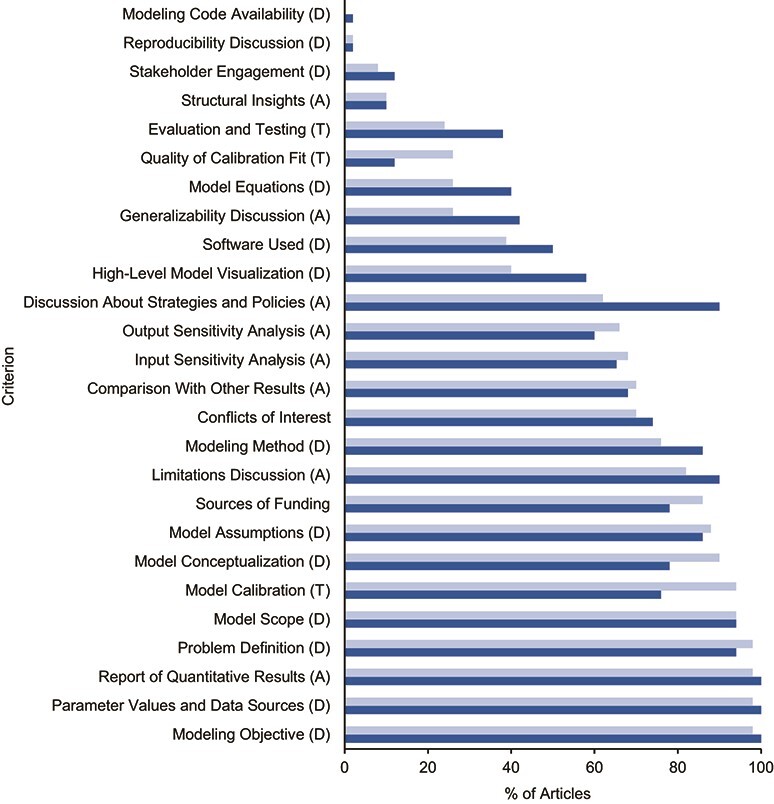
Percentage of articles satisfying 26 in-depth evaluation criteria in 4 areas. Each criterion was assessed for the 50 most-cited articles (light blue) and 50 articles randomly selected from the remaining articles (dark blue). The percentage of articles from each group meeting the criteria is presented. Five criteria (Quality of Calibration Fit; Generalizability Discussion; High-level Model Visualization; Discussion About Strategies or Policies; and Model Calibration) were significantly different between randomly selected and most-cited articles (P < 0.1). A, analysis; D, development; T, testing.
Modeling code availability and reproducibility discussion were the lowest in both groups (i.e., highly cited articles and randomly selected, not highly selected articles), with only approximately 2% of the articles from either group satisfying these criteria. Conversely, limitations discussion, assumptions, scope, objective, problem definition, parameter values and data sources, and the reporting of quantitative results were satisfied by the majority (>80%) of selected studies in both categories.
Overall, assessing average performance within the 3 categories of criteria revealed no significant differences between highly cited and random articles; on average, development criteria were fulfilled in 62% (95% confidence interval (CI): 52%, 72%) of randomly selected articles and 58% (95% CI: 47%, 69%) of highly cited articles. Testing criteria were fulfilled in 42% (95% CI: 33%, 51%) of randomly selected articles and 48% (95% CI: 37%, 59%) of highly cited articles. Analysis criteria were fulfilled in 66% (95% CI: 58%, 74%) of randomly selected articles and 60% (95% CI: 52%, 68%) of highly cited articles.
We found no strong predictor of aggregate quality measures. A linear regression was performed to analyze the overall evaluation score (maximum score = 26 points) of the 100 sampled articles on the basis of whether the model used in the reported study was static or dynamic, event driven or continuous, stochastic or deterministic; the journal impact factor; the number of citations; multilateral collaboration score; number of pages; time since publication; and the location of the authors’ affiliation (United States or not). The independent variables had no significant impact on the score; each 95% CI included zero. See Web Table 5 for the details of the regression analysis.
DISCUSSION
We conducted a broad review of published journal articles about studies in which authors used simulation modeling in epidemiology and health policy to identify trends, document application domains, and assess modeling rigor and reproducibility. Between 1967 and 2016, this line of research has become more common, and the types of models have shifted to better represent the complexity of real-world health issues, particularly within a dynamic and stochastic framework.
In our sample of simulation-modeling articles, the greatest proportion were on disease-focused studies. Areas such as pathological conditions and viral diseases receive a great deal of modeling attention and deservedly so; several infectious diseases, such as malaria, human immunodeficiency virus, and salmonellosis, spread within complex and varying systems, requiring insights to be uncovered through careful modeling studies rather than reactive policy-making, which may expend unnecessary resources. Simulation modeling is well suited to aid understanding of complex issues where resources are limited. Although it is difficult to assess the unmet potential in different application areas, on the basis of our analysis, we speculated about 3 areas with notable room for growth in simulation modeling: parasitic diseases (41, 42), chemically induced disorders (including opioid use disorder) (43–45), and congenital and hereditary diseases. These ailments all affect the population in increasingly complex ways (46, 47) but are not as prominent in current applications, potentially limiting the capability of policy makers to test the impact of interventions.
Although the best models are informative and transparent, opaque modeling studies can conceal underlying assumptions and errors. Simulation studies aid decision- and policy-making, but there must be a high level of trust in the methods and execution for their full potential to be realized. We found that in nearly half of the studies included in this review, authors did not report their model equations. This is consistent with findings of a recent study in which only 7.3% of simulation-modeling researchers responded when asked to post their codes to a research registry clearinghouse; ultimately, only 1.6% agreed to post these details (48). We encourage researchers to open their work to others; do so provides many opportunities to learn from others and enhances the work.
We also found that dynamic models tend to be better reported than static models, and the same is true for stochastic and event-driven versus deterministic and continuous models. Our broad review of the literature shows that, beginning in 2002, researchers have moved toward creating dynamic and stochastic models to match increasingly complex problems. During this period, improvements were made in reporting model equations. We hope this trend will continue to accelerate as simulation models become more frequently relied upon in health policy decisions.
Our analysis also highlights much room for improvement in terms of reproducibility and in rigor and quality in reporting of studies in all 3 reporting categories (i.e., development, testing, and analysis). An in-depth review of a subset of the sample revealed that models used in the studies reported in the most-cited articles performed no better than those reported in randomly selected articles when assessed against a host of modeling-process evaluation criteria. In addition, factors such as collaboration, number of pages, number of citations, and journal reputation had no significant impact on reproducibility or quality measures.
Overall, connecting a model’s structural qualities to the purported insights is at the heart of developing the readers’ intuition and thus having a lasting impact; on the basis of our review, much more can be done on that front (37, 49, 50). Although some of these suggestions may require some additional work for modelers, they are key to building confidence in models, gaining and maintaining the trust of decision makers, and the cumulative improvement of modeling research in general (51). We present these gaps in model reporting and rigor in the hope that by identifying common issues future publications will help improve model development and analysis and enhance reproducibility.
An important limitation of this study is our search strategy that focused on articles using the term “simulation” (and its variants) in the title or abstract. We expect a sizeable number of articles exist that use simulation but do not use the term or its variants in the title and abstract, instead referring to various modeling approaches. Expanding the search strategy to include mathematical, computational, economic, or other modeling approaches (indeed, there are at least 400 terms for these approaches (52)) would have enlarged the initial sample to tens of thousands of records, which would have been infeasible to review and analyze in depth. Considering this, we chose to review the large subset of the relevant literature that is explicit about using a simulation model, while acknowledging the undercount inherent in excluding nonexplicit simulation-modeling approaches. We cannot rule out that this balancing act might have somehow biased our results. Future research may include evaluating studies against various field-specific definitions of simulation modeling to increase the coverage of the sample and comparing those results with our findings to assess whether differences exist between simulation studies that name the use of a simulation method and those that do not.
This review may also be limited by our selection of only peer-reviewed studies published in English and indexed in PubMed and Web of Science by March 2016. We used PubMed’s MeSH-term categorization to identify the focus of each article; however, we were limited by potential overlap and errors in these categories. Our sampling frame stops in 2016; yet, given the rapid growth of the field, updates every few years to track new trends should add value. In addition, models of health policy may be developed privately, discussed in institutional reports, or remain unpublished. Thus, only including journal articles may omit some of the relevant results in simulation modeling.
For an in-depth analysis of the 100 articles, we devised a scoring system whereby we assigned 1 point to each binary criterion met, following other examples of binary assessment (36–38). This assignment only enumerates the existence of a set of criteria and does not offer a more nuanced understanding of quality. We also did not include differential weights for these criteria when assessing the overall quality. We acknowledge that those criteria vary in their importance to overall quality and impact of research; yet, in the absence of an objective method for aggregating them, we preferred to avoid imposing our own subjective weights on various criteria, instead providing the raw evaluation data for replication and extensions.
Additional reviews can be done to focus on ways to systematically analyze and improve the reproducibility of simulation modeling. For example, review authors could investigate reproducibility across application domains and levels of analysis (e.g., cell, individual, or society) to inform concrete suggestions for specific communities of research. Moreover, simulation models may play a major role in designing health policy in real time (e.g., as they have in response to COVID-19), and evaluating these models in near-real time is essential when conditions rapidly change; brief, yet timely, reviews over a narrow subject area can enhance policy makers’ trust in modeling results (18). Researchers also can compare our results on the reproducibility of simulation models in epidemiology and health policy with other application domains of simulation modeling. Tracking the same metrics over time provides another measure of progress. Researchers may also explore alternative weights for criteria depending on their importance for reproducibility and contribution to overall study quality. Finally, using our data set, machine-learning methods may be trained to identify reproducibility and quality metrics more efficiently and apply those criteria to larger bodies of research.
In summary, our analysis highlights the changes in simulation-modeling studies’ topics, methods, and quality over the past 50 years and suggests several areas for improvement. Regardless of the quality of the underlying model, lack of reproducibility is a major challenge that erodes confidence in the policies and decisions the studies using these models inform and their broader impact, and thus lack of reproducibility deserves more attention in simulation research. We hope this review facilitates conversations about research gaps that can benefit from simulation modeling and motivates modelers to collaborate in addressing those gaps and increasing the diversity of research areas while enhancing the rigor and reproducibility of simulation research in health policy.
Supplementary Material
ACKNOWLEDGMENTS
MGH Institute for Technology Assessment, Harvard Medical School, Boston, Massachusetts, United States (Mohammad S. Jalali, Catherine DiGennaro); and Sloan School of Management, Massachusetts Institute of Technology, Cambridge, Massachusetts, United States (Mohammad S. Jalali, Abby Guitar, Karen Lew, Hazhir Rahmandad).
This report has received funding from the European Union’s Horizon 2020 research and innovation programme under grant agreement no. 875171.We express our gratitude to Professors Donald Burke, John Sterman, and Gary King, who shared their suggestions and thoughts. We thank participants at Medical Decision Making and System Dynamics Conferences, who provided constructive feedback on initial versions of this report. We also thank Meera Gregerson, Yikang Qi, and Yuan, at Massachusetts Institute of Technology, who assisted in data extraction and analysis.
Data are reported in the Web material.
Conflict of interest: none declared.
REFERENCES
- 1. Sterman JD. Learning from evidence in a complex world. Am J Public Health. 2006;96(3):505–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hurd HS, Kaneene JB. The application of simulation models and systems analysis in epidemiology: a review. Prev Vet Med. 1993;15(2–3):81–99. [Google Scholar]
- 3. Glied S, Tilipman N. Simulation modeling of health care policy. Annu Rev Public Health. 2010;31:439–455. [DOI] [PubMed] [Google Scholar]
- 4. Fone D, Hollinghurst S, Temple M, et al. Systematic review of the use and value of computer simulation modelling in population health and health care delivery. J Public Health. 2003;25(4):325–335. [DOI] [PubMed] [Google Scholar]
- 5. Dean B, Barber N, Ackere A, et al. Can simulation be used to reduce errors in health care delivery? The hospital drug distribution system. J Health Serv Res Policy. 2001;6(1):32–37. [DOI] [PubMed] [Google Scholar]
- 6. Goldie SJ, Goldhaber-Fiebert JD, Garnett GP. Chapter 18: public health policy for cervical cancer prevention: the role of decision science, economic evaluation, and mathematical modeling. Vaccine. 2006;24(suppl 3):S155–S163. [DOI] [PubMed] [Google Scholar]
- 7. De Gelder R, Heijnsdijk EA, Van Ravesteyn NT, et al. Interpreting overdiagnosis estimates in population-based mammography screening. Epidemiol Rev. 2011;33(1):111–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Smith BT, Smith PM, Harper S, et al. Reducing social inequalities in health: the role of simulation modelling in chronic disease epidemiology to evaluate the impact of population health interventions. J Epidemiol Community Health. 2014;68(4):384–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Borg S, Ericsson Å, Wedzicha J, et al. A computer simulation model of the natural history and economic impact of chronic obstructive pulmonary disease. Value Health. 2004;7(2):153–167. [DOI] [PubMed] [Google Scholar]
- 10. Ghaffarzadegan N, Ebrahimvandi A, Jalali MS. A dynamic model of post-traumatic stress disorder for military personnel and veterans. PLoS One. 2016;11(10):e0161405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ghaffarzadegan N, Larson RC, Fingerhut H, et al. Model-based policy analysis to mitigate post-traumatic stress disorder. In: In: Gil-Garcia JR, Pardo T, Luna-Reyes LF, eds. Policy Analytics, Modelling, and Informatics. Berline, Germany: Springer; 2018:387–406. [Google Scholar]
- 12. Davies NG, Kucharski AJ, Eggo RM, et al. Effects of non-pharmaceutical interventions on COVID-19 cases, deaths, and demand for hospital services in the UK: a modelling study. Lancet Public Health. 2020;5(7):e375–e385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Li R, Pei S, Chen B, et al. Substantial undocumented infection facilitates the rapid dissemination of novel coronavirus (SARS-CoV-2). Science. 2020;368(6490):489–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Stehlé J, Voirin N, Barrat A, et al. Simulation of an SEIR infectious disease model on the dynamic contact network of conference attendees. BMC Med. 2011;9(1):87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Perez P, Dray A, Moore D, et al. SimAmph: an agent-based simulation model for exploring the use of psychostimulants and related harm amongst young Australians. Int J Drug Policy. 2012;23(1):62–71. [DOI] [PubMed] [Google Scholar]
- 16. Murray CJ. Forecasting the impact of the first wave of the COVID-19 pandemic on hospital demand and deaths for the USA and European economic area countries [preprint]. medRxiv. 2020.04.21.20074732. ( 10.1101/2020.04.21.20074732.2020.04.21.20074732). [DOI] [Google Scholar]
- 17. Fasina FO, Njage PM, Ali AM, et al. Development of disease-specific, context-specific surveillance models: avian influenza (H5N1)-related risks and behaviours in African countries. Zoonoses Public Health. 2016;63(1):20–33. [DOI] [PubMed] [Google Scholar]
- 18. Jalali MS, DiGennaro C, Sridhar D. Transparency assessment of COVID-19 models. Lancet Glob Health. 2020;8(12):e1459–e1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Smith H, Varshoei P, Boushey R, et al. Simulation modeling validity and utility in colorectal cancer screening delivery: a systematic review. J Am Med Inform Assoc. 2020;27(6):908–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nianogo RA, Arah OA. Agent-based modeling of noncommunicable diseases: a systematic review. Am J Public Health. 2015;105(3):e20–e31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Christodoulou E, Ma J, Collins GS, et al. A systematic review shows no performance benefit of machine learning over logistic regression for clinical prediction models. J Clin Epidemiol. 2019;110:12–22. [DOI] [PubMed] [Google Scholar]
- 22. Wynants L, Van Calster B, Collins GS, et al. Prediction models for diagnosis and prognosis of COVID-19: systematic review and critical appraisal. BMJ. 2020;369:m1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Unal B, Capewell S, Critchley JA. Coronary heart disease policy models: a systematic review. BMC Public Health. 2006;6(1):213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Grimm V, Augusiak J, Focks A, et al. Towards better modelling and decision support: documenting model development, testing, and analysis using TRACE. Ecol Model. 2014;280:129–139. [Google Scholar]
- 25. Goodman SN, Fanelli D, Ioannidis JPA. What does research reproducibility mean. Sci Transl Med. 2016;8(341):341ps12–41ps12. [DOI] [PubMed] [Google Scholar]
- 26. Begley CG, Ioannidis JPA. Reproducibility in science. Circ Res. 2015;116(1):116–126. [DOI] [PubMed] [Google Scholar]
- 27. Collins FS, Tabak LA. Policy: NIH plans to enhance reproducibility. Nature News. 2014;505(7485):612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lorscheid I, Heine B-O, Meyer M. Opening the ‘black box’ of simulations: increased transparency and effective communication through the systematic design of experiments. Comput Math Organ Theory. 2012;18(1):22–62. [Google Scholar]
- 29. Moons KG, Altman DG, Reitsma JB, et al. Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD): explanation and elaboration. Ann Intern Med. 2015;162(1):W1–W73. [DOI] [PubMed] [Google Scholar]
- 30. Alarid-Escudero F, Krijkamp EM, Pechlivanoglou P, et al. A need for change! A coding framework for improving transparency in decision modeling. Pharmacoeconomics. 2019;37(11):1329–1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rahmandad H, Sterman JD. Reporting guidelines for simulation-based research in social sciences. System Dynamics Review. 2012;28(4):396–411. [Google Scholar]
- 32. Galea S, Riddle M, Kaplan GA. Causal thinking and complex system approaches in epidemiology. Int J Epidemiol. 2010;39(1):97–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Speybroeck N, Van Malderen C, Harper S, et al. Simulation models for socioeconomic inequalities in health: a systematic review. Int J Environ Res Public Health. 2013;10(11):5750–5780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Adams J, Gurney KA. Bilateral and multilateral coauthorship and citation impact: patterns in UK and US international collaboration. Front Res Metr. 2018;3(12). 10.3389/frma.2018.00012. [DOI] [Google Scholar]
- 35. Müller M, Pfahl D. Simulation methods. In: Shull F, Singer J, DIK S, eds. Guide to Advanced Empirical Software Engineering. Berlin, Germany: Springer; 2008:117–152. [Google Scholar]
- 36. Caro JJ, Briggs AH, Siebert U, et al. Modeling good research practices--overview: a report of the ISPOR-SMDM Modeling Good Research Practices Task Force-1. Med Decis Making. 2012;32(5):667–677. [DOI] [PubMed] [Google Scholar]
- 37. Eddy DM, Hollingworth W, Caro JJ, et al. Model transparency and validation: a report of the ISPOR-SMDM Modeling good research practices task force–7. Med Decis Making. 2012;32(5):733–743. [DOI] [PubMed] [Google Scholar]
- 38. Dahabreh IJ, Trikalinos TA, Balk EM, et al. Guidance for the conduct and reporting of modeling and simulation studies in the context of health technology assessment. In: Methods Guide for Effectiveness and Comparative Effectiveness Reviews. Rockville, MD: Agency for Healthcare Research and Quality; 2008. [PubMed] [Google Scholar]
- 39. Philips Z, Ginnelly L, Sculpher M, et al. Review of guidelines for good practice in decision-analytic modelling in health technology assessment. Health Technol Assess. 2004;8(36) iii-iv, ix-xi:1–158. [DOI] [PubMed] [Google Scholar]
- 40. Dahabreh IJ, Trikalinos TA, Balk EM, et al. Recommendations for the conduct and reporting of modeling and simulation studies in health technology assessment. Ann Intern Med. 2016;165(8):575–581. [DOI] [PubMed] [Google Scholar]
- 41. Patz JA, Graczyk TK, Geller N, et al. Effects of environmental change on emerging parasitic diseases. Int J Parasitol. 2000;30(12):1395–1405. [DOI] [PubMed] [Google Scholar]
- 42. Trtanj J, Jantarasami L, Brunkard J, et al. Ch. 6: Climate impacts on water-related illness. In: Crimmins A, Balbus J, Gamble JL, et al (eds). The Impacts of Climate Change on Human Health in the United States: A Scientific Assessment. Washington, DC: US Global Change Research Program; 2016:157–188. [Google Scholar]
- 43. Hui D, Arthur J, Bruera E. Palliative care for patients with opioid misuse. JAMA. 2019;321(5):511. [DOI] [PubMed] [Google Scholar]
- 44. Wei YJ, Chen C, Sarayani A, et al. Performance of the Centers for Medicare & Medicaid Services' opioid overutilization criteria for classifying opioid use disorder or overdose. JAMA. 2019;321(6):609–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Thrul J, Pabst A, Kraus L. The impact of school nonresponse on substance use prevalence estimates - Germany as a case study. Int J Drug Policy. 2016;27:164–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Laurichesse Delmas H, Kohler M, Doray B, et al. Congenital unilateral renal agenesis: prevalence, prenatal diagnosis, associated anomalies. Data from two birth-defect registries. Birth Defects Res. 2017;109(15):1204–1211. [DOI] [PubMed] [Google Scholar]
- 47. Philippe P, Mansi O. Nonlinearity in the epidemiology of complex health and disease processes. Theor Med Bioeth. 1998;19(6):591–607. [DOI] [PubMed] [Google Scholar]
- 48. Emerson J, Bacon R, Kent A, et al. Publication of decision model source code: attitudes of health economics authors. Pharmacoeconomics. 2019;37(11):1409–1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Barlas Y. Multiple tests for validation of system dynamics type of simulation models. Eur J Oper Res. 1989;42(1):59–87. [Google Scholar]
- 50. Brailsford SC. System dynamics: what’s in it for healthcare simulation modelers. 2008 Winter Simulation Conference 2008. 2008:1478–1483.
- 51. McDougal RA, Bulanova AS, Lytton WW. Reproducibility in computational neuroscience models and simulations. IEEE Trans Biomed Eng. 2016;63(10):2021–2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ören T. A critical review of definitions and about 400 types of modeling and simulation. SCS M&S Magazine. 2011;2(3):142–151. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


