Abstract
It is conventionally assumed that conserved pathways evolve slowly with little participation of gene evolution. Nevertheless, it has been recently observed that young genes can take over fundamental functions in essential biological processes, for example, development and reproduction. It is unclear how newly duplicated genes are integrated into ancestral networks and reshape the conserved pathways of important functions. Here, we investigated origination and function of two autosomal genes that evolved recently in Drosophila: Poseidon and Zeus, which were created by RNA-based duplications from the X-linked CAF40, a subunit of the conserved CCR4–NOT deadenylase complex involved in posttranscriptional and translational regulation. Knockdown and knockout assays show that the two genes quickly evolved critically important functions in viability and male fertility. Moreover, our transcriptome analysis demonstrates that the three genes have a broad and distinct effect in the expression of hundreds of genes, with almost half of the differentially expressed genes being perturbed exclusively by one paralog, but not the others. Co-immunoprecipitation and tethering assays show that the CAF40 paralog Poseidon maintains the ability to interact with the CCR4–NOT deadenylase complex and might act in posttranscriptional mRNA regulation. The rapid gene evolution in the ancient posttranscriptional and translational regulatory system may be driven by evolution of sex chromosomes to compensate for the meiotic X chromosomal inactivation (MXCI) in Drosophila.
Keywords: CAF40, Poseidon, Zeus, CCR4–NOT, MXCI, MSCI, new gene function
Introduction
The complex regulation of gene expression is essential for a proper cell function. Accurate spatial and temporal transcriptional activation and repression, as well as posttranscriptional regulation, is determined by intricate regulatory networks, often involving extensive signaling processes and the recruitment of multiprotein regulatory complexes. Thus, describing the components of such regulatory circuits, as well as understanding their role in the evolution of gene regulation can extend our comprehension of how organisms adapt and diversify over time (Erwin and Davidson 2009; Halfon 2017). Fundamental cellular functions, including basic regulatory processes common to distantly related organisms, are often assumed to be carried out by old conserved elements, whereas evolutionary young genes would be involved in more restrict, even dispensable, activities (Miklos and Rubin 1996; Kondrashov 2012). However, recent case studies have challenged this view, showing that young genes can be incorporated into ancestral regulatory networks, with major impact in the expression of numerous genes (Matsuno et al. 2009; Ding et al. 2010; Chen et al. 2012). The importance of such integration of new elements into fundamental cellular processes is illustrated by dramatic examples of young genes which were experimentally shown to have acquired indispensable roles in development or reproduction in a short evolutionary time, even when present in a single or a small clade of species (Chen et al. 2010; Saleem et al. 2012; Ross et al. 2013; VanKuren and Long 2018; Lee et al. 2019; Kasinathan et al. 2020).
The incorporation of new genetic elements, in particular evolutionary young genes, into ancestral regulatory networks remains elusive and underexplored (Abrusan 2013; Zhang et al. 2015). In particular, little is known about the molecular mechanisms involved in the evolution of regulatory networks driven by recently evolved gene duplicates. For instance, it is not clear to what extent the regulatory role of a young gene diverges from that of the parental copy, as well as what specific cellular processes and phenotypes they impact. In this context, the detailed comparison of old genes and their young duplicated paralogs can shed light on the mechanisms leading to the integration of new elements into preexisting cellular processes.
Extensive comparative genomic analyses have revealed an intriguing evolutionary pattern of gene traffic: there is a strong excess of parental genes on the X chromosome that produced autosomal duplicated genes with specific expression in the male germline (Betran et al. 2002; Kaessmann et al. 2009; Vibranovski, Lopes, et al. 2009). This pattern was confirmed for various organisms such as flies (Betran et al. 2002; Dai et al. 2006; Bai et al. 2007; Zhang et al. 2010), mosquitos (Toups and Hahn 2010), and mammals (Emerson et al. 2004; Carelli et al. 2016). The preferential fixation of male-biased duplicated copies into autosomes likely reflects the fact that, during spermatogenesis, the silencing of X chromosome in meiosis and later stages, as observed in mammals (Richler et al. 1992) and Drosophila (Vibranovski, Zhang, et al. 2009; Mahadevaraju et al. 2021) provided a selective mechanism to drive the X to A gene traffic in new gene duplication (Vibranovski, Lopes, et al. 2009; Jiang et al. 2017; Long and Emerson 2017). Consequently, natural selection favors the fixation of autosomal duplicate copies that escape the X chromosome and compensate for the expression of its parental gene (Betran et al. 2002; Vibranovski, Lopes, et al. 2009, 2012; Casola and Betran 2017). In addition, other factors were proposed to interpret the gene traffic out of the X. It was considered that the testis was a rapidly evolving organ, prone to the accumulation of new elements, consistent with intense sexual selection (Harrison et al. 2015). It was also shown that during late spermatogenesis stages, the transcription of new gene copies is facilitated by the permissive chromatin state, which may facilitate the promiscuous transcription, insertion, and subsequent evolution of newly arisen genes (Kaessmann 2010; Witt et al. 2019).
Consistent with the pattern described above, previous studies have reported that male reproductive tissues express specific versions of several housekeeping genes involved in basic cellular processes, such as the proteasomal (Zhong and Belote 2007), transcriptional (Hiller 2004), and translational machineries (Baker and Fuller 2007). It was suggested that these duplicated copies may represent specialized versions of their parental genes, required to accomplish the intense and coordinated changes in gene expression observed during spermatogenesis (White-Cooper 2010). It is not clear, however, why the duplicated, specific copies occur so frequently, or to what extent they diverge in function from their parental ones (Belote and Zhong 2009).
In this study, we investigate evolutionary and functional impacts of Poseidon and Zeus, two autosomal young genes expressed in the testes, which independently retroposed from X-linked CAF40, and are only found in some Drosophila species (Zhang et al. 2010). CAF40 is an ancient gene, broadly expressed in all fly tissues. The locus encodes a highly conserved protein in eukaryotes, with orthologs identified and experimentally studied from mammals (e.g., mouse and human) to insects (e.g., Drosophila) to fungi (e.g., yeast) (Collart and Panasenko 2017). CAF40, also known as Rcd-1 or CNOT9, is a subunit of the highly conserved CCR4–NOT deadenylase complex, a multiprotein assembly involved in posttranscriptional and translational regulation of gene expression (Miller and Reese 2012; Wahle and Winkler 2013; Buschauer et al. 2020). The complex catalyzes the removal of poly(A) tails in mRNAs, thus leading to their translational repression and degradation. It is also involved in the deadenylation and degradation of mRNA targets for proper spermatogenesis (Legrand and Hobbs 2018). By integrating several regulatory processes, the complex is considered a key regulator of eukaryotic gene expression (Collart 2016). Among the subunits of the CCR4–NOT complex, CAF40 acts as an important hub for the recruitment of the complex by mRNA-binding proteins (Sgromo et al. 2017, 2018; Keskeny et al. 2019). Moreover, it was shown to act independently of the complex, by interacting with transcription factors and altering their activation potential (Garapaty et al. 2008).
Zeus, retroposed 5 Ma, was already shown to play an important role in male fertility in Drosophila, by binding and regulating the expression of a large set of target genes, many not shared with the parental CAF40 (Chen et al. 2012). In order to uncover how newly duplicated genes are integrated into ancestral networks and reshape the conserved pathways of important functions, we first describe the divergence between CAF40 and its two retroduplicated genes, Poseidon and Zeus, in gene sequence and expression patterns. Second, using RNAi-knockdown and CRISPR-Cas9-deletions, we further explore their phenotypic importance for viability and male fertility. Third, we demonstrate that both duplicates are able to repress a tethered mRNA reporter, and that Poseidon protein, but not Zeus, retained the ability to interact with the CCR4–NOT complex. Finally, our RNA-seq data demonstrate that the independent silencing of each paralog impacts the regulation of a distinct set of genes, likely due to diverse functions between regulatory processes in which the paralogs are acting in. Together, our data show that both young duplicates are integrated into Drosophila male germline regulatory pathways, interact with highly conserved regulatory mechanisms, and impact the gene expression network in different ways.
Results
Rapid Evolution of Poseidon and Zeus Out of the Extremely Conserved CAF40
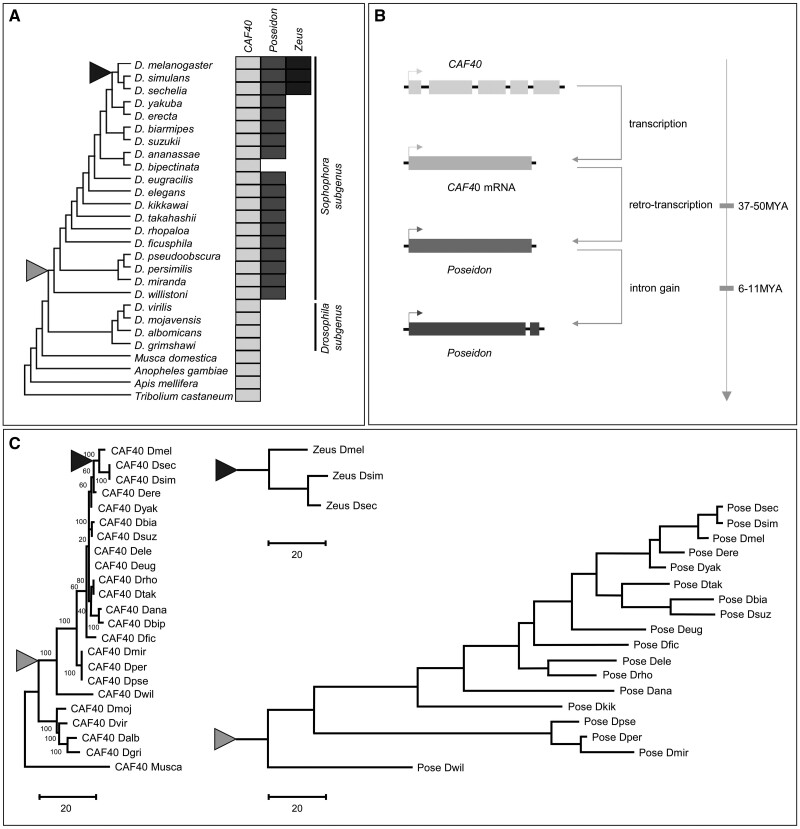
Comparative genomic analyses from public databases had previously identified Zeus as a diverged copy of CAF40 (Zhang et al. 2010), which prompted its functional description as a duplicated gene (Quezada-Diaz et al. 2010; Chen et al. 2012). Curiously, our analyses using the CAF40 sequence as query in sequence search against Drosophila genomes had also revealed the presence of another annotated paralogous gene, although it had not been studied until now (CG2053 in D. melanogaster). We named this gene Poseidon, as a reference to Zeus’ brother in the Greek mythology (PSI-BLAST e-value = 7×10−56, coverage = 81% between CAF40 and Poseidon in D. melanogaster).
Our search revealed that the intact Open Reading Frames of Poseidon are present in the third chromosome of 18 Drosophila species (fig. 1A), all from the subgenus Sophophora. Reciprocal BLAST searches using the Poseidon sequence as query did not find any other significant match in eukaryotes except for CAF40 and Zeus orthologs. The phylogenetic distribution suggests that Poseidon is a relatively young gene that appeared 36 Ma, before D. willistoni diverged from other Sophophora species(Russo et al. 1995; Clark et al. 2007; Markow and O’Grady 2007). Zeus originated after the split of the most recent common ancestor of D. melanogaster and D. yakuba 3–6 Ma (Russo et al. 1995; Clark et al. 2007; Markow and O’Grady 2007; Quezada-Diaz et al. 2010; Chen et al. 2012). Despite the recent origination of these two new genes from different CAF40 ancestries, these genes show a high level of divergence in their protein sequences (supplementary fig. S1A, Supplementary Material online).
Fig. 1.
Origination of Poseidon and Zeus from CAF40 in Drosophila. (A) Distribution of the three paralogs among Drosophila species, with other insects as outgroups. Filled boxes represent the presence of each gene. (B) Scheme depicting Poseidon origination process through retroduplication, and insertion into an autosome. (C) Phylogenetic relationship in CAF40, Poseidon, and Zeus reconstructed respectively through the Maximum Parsimonious method using protein sequences. Dmel, Drosophila melanogaster; Dsec, Drosophila sechellia; Dsim, Drosophila simulans; Dere, Drosophila erecta; Dyak, Drosophila yakuba; Dbia, Drosophila biarmipes; Dsuz, Drosophila suzukii; Dele, Drosophila elegans; Deug, Drosophila eugracilis; Drho, Drosophila rhopaloa; Dtak, Drosophila takahashii; Dana, Drosophila ananassae; Dbip, Drosophila bipectinate; Dfic, Drosophila ficusphila; Dmir, Drosophila miranda; Dkik, Drosophila kikkawai; Dper, Drosophila persimilis; Dpse, Drosophila pseudoobscura; Dwil, Drosophila willistoni; Dmoj, Drosophila mojavensis; Dvir, Drosophila virilus; Dalb, Drosophila albomicans; Dgri, Drosophila grimshawi; Musca, Musca domestica (Housefly). The black and gray triangles represent the origination branch of Zeus and Poseidon, respectively. The scale for branch lengths is number of substitutions per site.
The presence of introns is useful for determining the mechanisms of origination of new genes (Long et al. 2013). CAF40 orthologs have between four and six introns in Drosophila species. Poseidon, however, has no introns in all detected species except one small intron in its 3′-end in the D. melanogaster subspecies group, which is unrelated in sequence or position to any intron found in CAF40. The lack of ancestral introns in the duplicated gene suggests that Poseidon initially originated through an X-to-autosome retroposition event, with the insertion of the duplicate in the third chromosome. Subsequently in the most recent common ancestor of the D. melanogaster subgroup species that diverged 6–11 Ma, Poseidon gained a new intron (fig. 1B).
Phylogenetic analyses of the three genes using the Maximum Parsimonious method in the MEGA platform (Kumar et al. 2018) suggest that Poseidon and Zeus originated through two independent RNA-based duplications from CAF40 in different branches of the Drosophila phylogeny (fig. 1C). The phylogenetic position of Zeus is consistent with its retroduplication after the split of the most recent common ancestor of D. melanogaster and D. yakuba (3–6 Ma), as previously described (Quezada-Diaz et al. 2010; Chen et al. 2012). The branch lengths in figure 1C also reveal that Poseidon and Zeus sequences rapidly diverged, in contrast to the extremely slow evolution of the parental CAF40. As an illustration, the CAF40 protein sequences from D. melanogaster and D. willistoni, which split ∼36 Ma (Russo et al. 1995; Clark et al. 2007; Markow and O’Grady 2007), diverged in only 7.7% of the sites in accordance with the extreme conservation in the protein sequences encoded by the CAF40 homologs across all multicellular organisms (supplementary fig. S1B, Supplementary Material online), whereas Poseidon and Zeus diverged 48.6% and 22.5%, respectively, from CAF40 in D. melanogaster (supplementary table S1, Supplementary Material online). An orthologous comparison of CAF40 and Poseidon protein sequences between D. melanogaster and D. willistoni, revealed an amino acid substitution rate of 0.12% per million years and 0.68% per million years, respectively. The comparison of Zeus protein sequence between D. melanogaster and D. simulans, which diverged by 3 Ma (Russo et al. 1995; Clark et al. 2007; Markow and O’Grady 2007), obtains an unusually high substitution rate of 4.92% per million years.
The Duplicates Diverged at Highly Conserved Sites
In order to understand whether the duplicated proteins accumulated replacements at conserved residues in the ancestral protein, or merely at the highly variable termini of the protein, we estimated the Shannon entropy (H) for each residue in an alignment of CAF40 homologs from 42 eukaryotes, and contrasted it with the replaced residues in the duplicates (supplementary fig. S1, Supplementary Material online). We found that amino acid replacements in Poseidon and Zeus occurred even at extremely conserved sites of CAF40. In both duplicates, replacements are distributed throughout the protein structure, including the charged groove formed by the conserved armadillo-repeat domain (Garces et al. 2007), which was shown to be important for CAF40 interactions (Chen, Boland, et al. 2014; Sgromo et al. 2017, 2018; Keskeny et al. 2019).
For the sake of comparison, out of the 49 completely conserved residues in CAF40 among eukaryotes (there are 49 conserved residues with which their H = 0; supplementary fig. S1C and D, Supplementary Material online), Poseidon diverged in 25, and Zeus, in 11 of these completely conserved residues. Furthermore, several diverged residues that were experimentally shown to be functionally relevant for CAF40 interaction with its protein partners in previous studies were replaced in the duplicates (supplementary table S2, Supplementary Material online; Chen, Boland, et al. 2014; Mathys et al. 2014; Sgromo et al. 2017). The extensive replacement of amino acids that are highly constrained in the parental protein suggests that Poseidon and Zeus functional properties may have diverged substantially from CAF40.
The Duplicates Acquired a Restricted Expression Pattern
Part of the phenotypic divergence between duplicated genes and their parents may result from their differential expression patterns. We used extensive transcriptome data from publicly available databases to investigate to which extent Poseidon and Zeus diverged from CAF40 at the expression level. First, a comparison of expression of these genes in several tissues from D. melanogaster evidences that both Poseidon and Zeus have acquired a narrower expression pattern when compared with CAF40 (fig. 2A). The duplicates are only expressed at larval imaginal discs, adult male reproductive tissues, and pupae at low or intermediate levels, in sharp contrast with CAF40, which is expressed at all assayed tissues and development stages, from intermediate to high levels. We experimentally confirmed the distinct expression of the three paralogs in D. melanogaster larvae, head, and testis through RNA extraction followed by RT–PCR (supplementary fig. S2, Supplementary Material online).
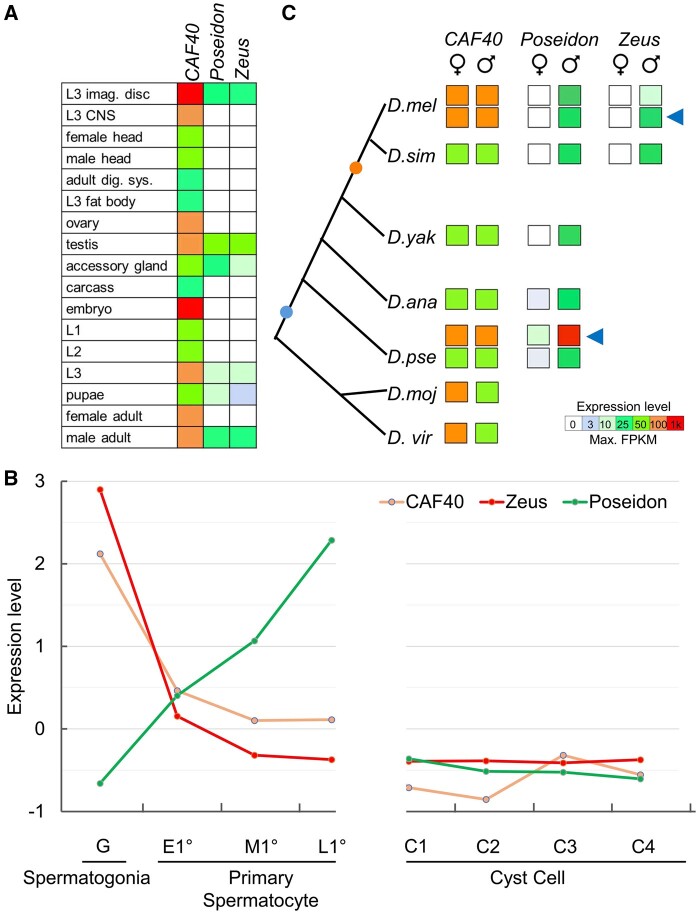
Fig. 2.
CAF40, Poseidon, and Zeus have distinct expression patterns. (A) Summary of the expression intensity of each paralog in D. melanogaster tissues and development times. Expression summarized as average FPKM. CAF40 broad expression contrasts with the duplicates restricted expression pattern. (B) Expression level, expressed as Z scores of TPM values of each gene to normalize by experimental variance (The Z score of TPM normalized reads for each gene were directly retrieved from “Gene Level Data” Excel sheet in Supplementary Data Set 2 from Mahadevaraju et al. 2021), at eight different cell types in D. melanogaster testes (spermatogonia [Spmt, mitosis, G], primary spermatocyte [meiosis, early (E1°), middle (M1°), and late(L1°)], cyst cell differentiation [four stages from C1 to C4]) (Mahadevaraju et al. 2021). These data provide evidence for the compensation of the autosomal Poseidon for the significantly reduced expression of the X-linked CAF40 in the spermatocyte stages. Z scores mean the number of SDs from the mean. Z=(x−μ)/σ, x is the value to be standardized, μ is the mean, and σ is the SD. (C) Summary of expression intensity in female/male adults for Drosophila species with available data for each paralog. Circles in the phylogeny represent the duplication event for Poseidon (blue) and Zeus (orange). Blue arrows indicate the expression intensity in reproductive organs (testes and ovaries) of D. melanogaster (D. mel) and D. pseudoobscura (D. pse). Male-biased expression for Poseidon and Zeus are conserved across the phylogeny. Data extracted from Brown et al. (2014), VanKuren and Vibranovski (2014), and Vibranovski, Zhang, et al. (2009).
We also compared the expression of each paralog at different testis cell types in D. melanogaster. We directly retrieved the Z score of transcriptions per kilobase million (TPM) normalized reads for each gene from a recently published data set of genome expression during spermatogenesis using sing cell RNA-seq (scRNA-seq) from eight spermatogenesis phases in mitosis, meiosis, and cyst cell differentiation (see “Gene Level Data” Excel sheet in Supplementary Data Set 2 from Mahadevaraju et al. 2021). We show that the three paralogs have distinct expression dynamics (fig. 2B). The scRNA-seq detected expression patterns of the three genes are similar to those detected previously from dissected testis tissues (Vibranovski, Zhang, et al. 2009), revealing the expression dynamics of these X-linked (CAF40) and autosomal genes (Poseidon and Zeus) in spermatogenesis with a much higher resolution. Although CAF40 expression level is high in spermatogonia, it reduces expression in meiotic and cyst cells, suggesting CAF40 is negatively impacted by the inactivation of male X chromosome in spermatocytes. Zeus shows a strong peak of expression, higher than CAF40, in the early mitotic phase, and subsequent drop of expression in the meiotic and postmeiotic stages. Starting in a low level of expression in spermatogonia, Poseidon increases expression in the middle primary spermatocyte (M1°), eventually showing a peak of expression in the late primary spermatocyte (L1°), a common pattern for autosomal retrogenes expressed in the testis that compensates for the lowly expressed X-linked paralogs (Vibranovski, Zhang, et al. 2009). Such a difference in expression is expected for retrogenes, which are inserted in genomic contexts diverse from the parental copy and may promptly acquire and/or evolve new cis-regulatory elements (Bai et al. 2008). The retroposed genes in autosomes may also help avoid the meiotic X chromosome inactivation (MXCI) when functioning in the meiosis stages of spermatogenesis (Betran et al. 2002; Vibranovski et al. 2012; Mahadevaraju et al. 2021).
Finally, we analyzed transcriptome data from other six Drosophila species in order to understand whether the duplicates male-biased expression pattern is conserved across the phylogeny. In these six species, we found that CAF40 is expressed at intermediate or high levels in adults, consistent with its role as a housekeeping gene. In contrast, these species exhibit significant male-biased expression of the duplicated genes (four additional species for Poseidon, and one for Zeus), similar to the pattern observed for D. melanogaster (fig. 2C).
Poseidon and Zeus Impact Viability and Fertility
Restricted expression pattern of the duplicates, along with their conserved sex-specific expression across fly species, suggest that the duplicates may have been integrated into developmental and/or reproductive processes. First, we tested this hypothesis by using RNAi-mediated knockdown and CRISPR-based knockout to assay the functional effects of the three paralogs on D. melanogaster viability and fertility.
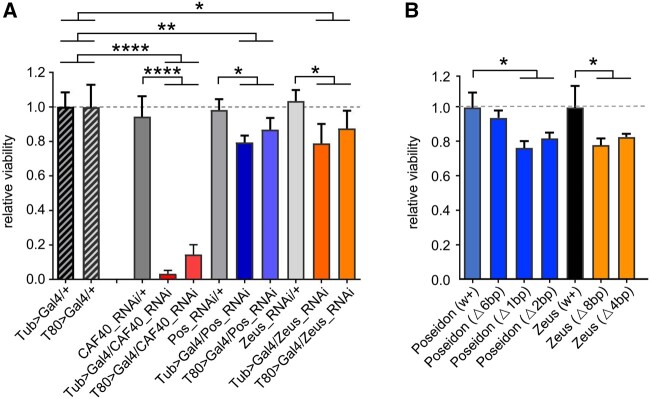
RNAi-knockdown using both a ubiquitous (Tub84B>GAL4) and an imaginal disc-specific driver (T80>GAL4) confirmed that CAF40 expression is essential for survival. Less than 3% of the flies developed into adults when the gene was silenced with the ubiquitous driver (fig. 3A and supplementary fig. S3, Supplementary Material online). An essential role of CAF40 in cellular processes is also observed in distant eukaryotes, as evidenced by knockdown experiments in C. elegans (Kamath et al. 2003) and humans (Wang et al. 2015). Our knockdown assays detected a significant phenotypic impact on viability for Poseidon and Zeus. RNAi-knockdown of these genes reduced the relative fly viability by ∼20%, with a slightly stronger effect of the ubiquitous driver over the imaginal disc one (fig. 3A). A more negative impact on viability was detected with ∼25% reduction when the genes were knocked out using CRISPR/Cas9 methods that created frameshift mutations by deleting one and two nucleotides in Poseidon and four and eight nucleotides in Zeus (fig. 3B). In comparison, an in-frame mutant by a deletion of six nucleotides in Poseidon did not have significant effect (fig. 3B).
Fig. 3.
CAF40, Poseidon, and Zeus impact on viability. (A) Viability of RNAi-expressing flies, relative to the control genotype from each individual cross. The two hatched bars in the left show the relative viability of the two control GAL4-expressing drivers, Tub (ubiquitous) and T80 (imaginal-discs). CAF40-RNAi-expressing flies in red; Poseidon in blue and Zeus in orange. (B) Viability of homozygous flies for different CRISPR-Cas9 deletions for Poseidon (blue) and Zeus (orange) relative to the control from the same background strain (black). Notice the nonsignificant effect of the only nonframeshift deletion for Poseidon (first blue bar on the left); t-test: *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. Error bars indicate SD.
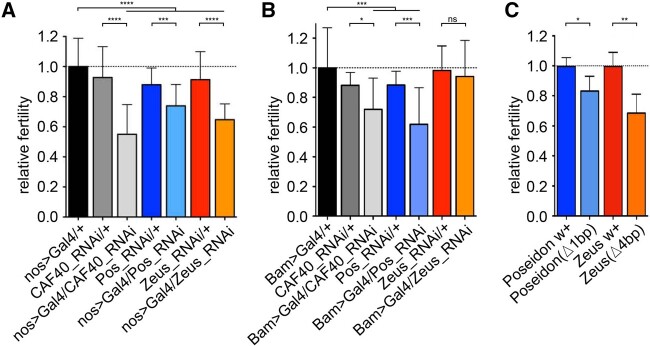
Given the expression of the duplicates in testes, we further investigated the effect of these on male fertility. We used RNAi-knockdown with two different testis-specific drivers: nanos>GAL4 that silenced the gene expression at spermatogonia and male germline stem cells, and Bam>GAL4 that silenced gene expression at late spermatogonia and early spermatocytes stages (White-Cooper 2012). These drivers allowed us to assay the fertility effects at different spermatogenesis phases. Independent silencing by the two drivers detected significant fertility effects in all three genes (fig. 4). The CAF40 knockdown showed the strongest effect (∼40% fertility reduction with nanos), in accordance with its function as a fundamental housekeeping gene. It is worth noting that Chen et al. (2012) used a KK RNAi line of CAF40 (KK101462) with same driver and showed no significant fertility effect (see Materials and Methods). The nanos driver also caused a 20% and 30% fertility decrease when knocking down Poseidon and Zeus, respectively. In contrast, the knockdown using the Bam driver caused the strongest fertility defect for Poseidon (30% fertility reduction) and a lower but significant effect for CAF40 (20% fertility reduction), suggesting CAF40 may play a more important role in an earlier stage (spermatogonia) of spermatogenesis than Poseidon with a more critical role in spermatocytes and/or later stages of spermatogenesis. Zeus appeared not to play any significant role in the later stages of spermatogenesis that was silenced by the Bam-driver (t-test, P = 0.59). Instead, Zeus was knocked down with a significant effect at the earlier stage by the nanos driver (Chen et al. 2012). These observations are in accordance with the expression pattern of the paralogs during spermatogenesis (fig. 2).
Fig. 4.
CAF40, Poseidon, and Zeus impact on male fertility. Relative fertility of males expressing RNAi using testis-specific drivers: (A) nanos>GAL4 (early spermatogenesis); and (B) Bam>GAL4 (late spermatogenesis). Black bars represent the controls, CAF40-RNAi-expressing flies in gray, Poseidon in blue, Zeus in orange. (C) Fertility of males homozygous for frameshift CRISPR-Cas9 deletions for Poseidon (blue) and Zeus (orange) compared with the control of the same background strain (darker bars). t-test: *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. Error bars indicate SD.
Finally, Poseidon and Zeus impact on male fertility was also confirmed by the knockout analyses using CRISPR/Cas9-generated frameshift deletions, significantly decreasing male fertility by 17% and 32%, respectively (fig. 4C). In summary, a combination of knockdown and knockout analyses reveals that Poseidon and Zeus carry out important functions in support of viability and male fertility in the Sophophora subgenus (Poseidon) or the D. melanogaster subgroup (Zeus).
Poseidon and Zeus Interaction with the CCR4–NOT Complex
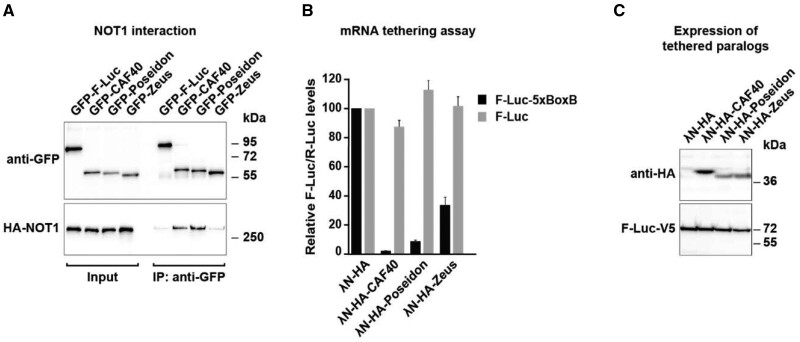
CAF40 is a highly conserved subunit of the CCR4–NOT complex (Miller and Reese 2012) and evolved slowly as a highly conserved gene in aforementioned analyses (fig. 1). However, both Poseidon and Zeus have intense protein sequence divergence with CAF40. We then try to test whether Poseidon and Zeus, the two duplicates retroposed from CAF40, maintained its ancestral functions or evolved novel gene functions. First, we independently expressed a GFP-tagged version of each paralog in Dm S2 cells and assayed their interaction with HA-tagged NOT1 through co-immunoprecipitation followed by Western-blotting analysis. NOT1 was selected because it is the central scaffold subunit of the CCR4–NOT complex and it was shown to directly interact with CAF40 via a CAF40/CNOT9-binding domain (CN9BD) (Chen, Boland, et al. 2014; Mathys et al. 2014; Collart and Panasenko 2017). Interestingly, we found that Poseidon maintained the ability to interact with NOT1, whereas Zeus either lost it, or binds NOT1 only weakly (fig. 5A).
Fig. 5.
CAF40, Poseidon, and Zeus protein interaction with the CCR4–NOT complex. (A) Co-immunoprecipitation assay showing the interaction of CAF40 paralogs with NOT1 in Dm S2 cells. Co-immunoprecipitation was conducted in the presence of RNase A to exclude RNA-mediated interactions. Cell lysates expressing GFP-tagged CAF40, Poseidon and Zeus, and HA-tagged NOT1. GFP-F-Luc served as a negative control. Input samples consist of 3% for the GFP-tagged proteins and 1% for the HA-tagged proteins, and immunoprecipitated samples of 10% for the GFP-tagged proteins and 30% for the HA-tagged proteins. Protein size markers are shown on the right in each panel. (B) Tethering assay using λN-HA-tagged CAF40, Poseidon and Zeus and the F-Luc-5BoxB reporter in Dm S2 cells (black bars). A plasmid expressing R-Luc served as a transfection control, and an F-Luc reporter lacking the 5BoxB binding sites for λN- was used as control (gray bars). F-Luc activity levels were normalized to those of the R-Luc control and set to 100% in cells expressing the λN-HA peptide alone. Error bars indicate SD of five replicates. CAF40 and Poseidon exhibit similar abilities of repressing the luciferase reporter (black bar) compared with the control (gray bar). Zeus exhibits lower, though still significant, repression ability. (C) Western-blot analysis showing the equivalent expression of the λN-HA-tagged proteins used in the tethering assays shown in (B). Protein size markers (kDa) are shown on the right of the panel.
The conservation of the interaction with NOT1 observed for Poseidon suggests that it could be incorporated into the CCR4–NOT complex, in contrast to Zeus. We further infer that, if this is true, Poseidon should have conserved the repressive effect on targeted mRNAs observed for CAF40 (Bawankar et al. 2013; Sgromo et al. 2017). We tested this hypothesis by measuring each paralog’s ability to repress a luciferase reporter mRNA in a λN/BoxB tethering assay. In the tethering assay the λN-tagged CAF40 paralogs are efficiently recruited to the reporter RNA, carrying five BoxB elements due to the strong binding of the λN peptide to the BoxB elements. In agreement with previous reports, tethering of λN-tagged CAF40 to a luciferase reporter mRNA carrying five BoxB elements in the 3′-UTR (F-Luc-5xBoxB; black bar) strongly represses the protein synthesis of the reporter. In contrast, CAF40 did not affect the expression of the control F-Luc mRNA lacking BoxB elements (gray bar) (fig. 5B). Intriguingly, Poseidon is also able to reduce the reporter expression to similar levels (∼10% of the control level, fig. 5B), indicating that Poseidon can act together with posttranscriptional regulators such as the CCR4–NOT complex to modulate the luciferase mRNA. In contrast, Zeus exhibits a weaker repressive ability compared with the other paralogs, although it is clearly significant (∼40% of the control level). All paralogs were expressed at comparable levels (fig. 5C).
Taken together, these results suggest that Poseidon conserved CAF40’s ability to interact with the CCR4–NOT complex through interactions with NOT1 most likely leading to the degradation of targeted transcripts. Zeus, however, lost or weakened its CCR4-NOT recruitment ability. Nevertheless, the fact that Zeus is still able to decrease reporter protein levels when tethered to an mRNA, suggests that it either evolved new protein interactions involved in mRNA regulation or that the weaker CCR4-NOT recruitment ability is sufficient to mediate repression in the tethering assay.
CAF40, Poseidon, and Zeus Impact Gene Regulation
Given the central role of CAF40 in several cellular regulatory processes, we investigated the impact of the three paralogs on global gene expression. Moreover, since the two duplicates, Poseidon and Zeus, are highly diverged at their protein sequence and expression profile from the parental CAF40, they provide a suitable system to assay global expression regulation among the paralogs.
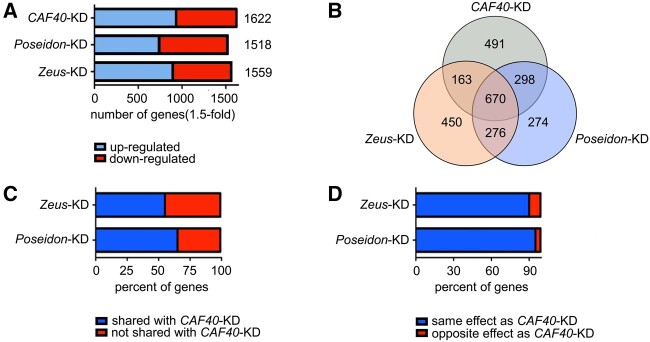
We conducted a genome-wide transcriptome analysis of adult testes to assay the impact of CAF40, Poseidon, and Zeus on global gene expression using germline-specific knockdown with nanos-GAL4 drivers (supplementary table S3 and figs. S4–S6, Supplementary Material online). Our transcriptome data showed that RNAi-silencing was effective and specific for each paralog, reducing mRNA levels of each gene in at least 60% compared with the control, while not impacting the other paralogs (supplementary table S4, Supplementary Material online). The knockdown of each of the three genes detected over 1,000 differentially expressed genes (DEGs) (fig. 6A and B; supplementary tables S5–S7, Supplementary Material online). Putting together, the knockdowns of the three genes affected the expression of totally 2,622 genes (union set of different expressed genes from the three KD lines) (fig. 6B), corresponding to more than a fifth of the genes mapped in our transcriptome (11,491) (supplementary figs. S7 and S8 and table S3, Supplementary Material online). Such a widespread effect on gene expression suggests that Poseidon also plays an important role on gene regulation in spermatogenesis, as previously shown for CAF40 and Zeus (Chen et al. 2012).
Fig. 6.
Impact of CAF40, Poseidon, and Zeus knockdowns on global gene expression. (A) Number of genes with >1.5-fold change in expression compared with the control in the knockdown of each paralog measured through RNA-seq. Upregulated genes shown in blue, downregulated shown in red. (B) Venn diagram representing the number of genes differentially expressed upon the knockdown of each paralog (CAF40 in gray, Poseidon in blue, Zeus in orange) and their intersections. Notice the large number of genes (670) commonly affected by the three knockdowns, as well as genes exclusively perturbed by one paralog (1,215). (C) Proportion of genes differentially expressed upon Poseidon and Zeus knockdown that are also affected by CAF40 knockdown. Notice that Poseidon affects a higher proportion of genes shared with CAF40 compared with Zeus (63.7% vs. 53.3%; χ2=50.7, P < 10−10). (D) Proportion of genes differentially expressed upon the knockdown of Poseidon and Zeus that are affected in the same direction (i.e., up- or downregulated) in the CAF40-KD. Notice that, despite the fact that the large majority of genes are affected in the same direction by the duplicates and the parental knockdown, Zeus knockdown affects a larger proportion of genes in the opposite direction of that of CAF40 when compared with Poseidon (9.1% of perturbed genes in Zeus-KD vs. 3.8% in Poseidon-KD were in the opposite direction of CAF40-KD, χ2=21.3, P < 10−5).
We also investigated the differential impact of these paralogs. We classified the genes with perturbed expression compared with the controls in each knockdown sample as male or female-biased, based on two independent Drosophila databases (Zhang et al. 2010; Assis et al. 2012). KD of both Zeus and Poseidon shows bias towards both downregulating male-biased genes and upregulating female-biased genes (supplementary table S8, Supplementary Material online, χ2 test, P < 0.05). These results indicate that both Poseidon and Zeus evolved function of activating male-biased genes expression and repressing female-biased genes expression (a consistent result with Chen et al. 2012). However, we could not conclude here that the upregulated female-biased genes of Zeus-KD were significantly enriched on the X chromosome (supplementary table S8, Supplementary Material online, χ2 test, P = 0.4431). This may be caused by the very low number of the intersection genes between X-linked female-biased genes and those down/upregulated genes of the three KDs (supplementary table S8, Supplementary Material online). However, the downregulated male-biased genes of both Poseidon-KD and Zeus-KD were overrepresented on autosomes (supplementary table S8, Supplementary Material online, χ2 test, both P < 0.01), consistent with previously reported chromosomal distribution patterns of sex-biased genes(Chen et al. 2012).
A large set of 670 genes (25.5% of total 2,622 impacted genes) was perturbed when any of the three paralogs was individually silenced (fig. 6B). Interestingly, the cellular processes with the most significant enrichment in these genes were proteolysis (GO:0006508, adj. P < 10−7, both of the results by the tools Gorilla and g: Profiler confirm this cellular processes) and reproduction (GO:0032504, adj. P < 10−7) (supplementary table S9, Supplementary Material online). Poseidon shares the most genes with CAF40, in comparison to other two two-gene comparisons (298 vs. 163, 276, respectively) whereas Zeus shared least number (163) of genes with CAF40 (fig. 6B). Furthermore, in the shared genes with CAF40, Poseidon has a significantly higher proportion of genes in the same direction towards up- or downregulation whereas Zeus has opposite changes, for example, a gene with upregulation for CAF40 and downregulation for Zeus (fig. 6C and D; supplementary table S10, Supplementary Material online) (Chi-squared test, P = 0.000699, <0.01). These observations are in accordance to the finding that Poseidon and CAF40 behave more similarly in regard to protein–protein interactions and repressive activity in comparison to Zeus (fig. 5).
Nevertheless, a substantial set of genes (1,215 genes, 46.3%) was perturbed by only one of the knockdowns, but not shared with the other two, which reveals the distinct impact that each paralog has in the global regulatory network (fig. 6B). This suggests that three paralogs have evolved peculiar interactions with large numbers of nonoverlapping genes. Interestingly, Poseidon has the least number of such peculiar gene interactions (274 vs. 491 [CAF40], 450 [Zeus], respectively). Additionally, we calculated the DEGs intersection numbers of those subgroups between our study and Chen et al. (2012) (Microarray) and found those DEGs intersection numbers between two studies are all relatively small. This may be caused by different experiments conditions (see Materials and Methods).
Discussion
We showed that a functionally important and conserved member of the CCR4–NOT complex, CAF40, gave rise to two gene duplicates through retroposition, Poseidon and Zeus, in recent evolution of Drosophila species. We demonstrated that Poseidon and Zeus are functionally important genes that have quickly diverged from CAF40 in protein sequence and expression shortly after duplication, whereas the parental CAF40 remained highly conserved (fig. 1C). Remarkably, even residues that have been conserved in CAF40 for a long evolutionary time (e.g., amino acids identical in all eukaryotic homologs) were extensively substituted in the duplicates, which may impact conserved functions of the protein (supplementary fig. S1, Supplementary Material online).
However, our molecular functional analyses show that both Poseidon and Zeus are important spermatogenesis genes. Poseidon retained mRNA suppression functions of CAF40, whereas Zeus evolved divergent functions as a suppressor of female genes in males (Chen et al. 2012).
Our co-immunoprecipitation assay showed that Poseidon protein conserved CAF40 ability to interact with NOT1. Moreover, such interaction is consistent with CAF40 and Poseidon, both showing a strong repressive effect on a tethered reporter transcript (fig. 5). Zeus retains a lower repressive activity, which probably reflects its divergent ability on acting in protein stability. These data suggest that, whereas Poseidon likely has inherited the CAF40 role in the CCR4–NOT complex (although with a distinct impact on gene regulation, as discussed below), Zeus likely functions independently of this complex as a suppressor of femininized genes (Wu and Xu 2003), as the genomic DNA binding has shown, revealed by ChIP-chip analysis (Chen et al. 2012).
Given the high expression and the conserved functions of CAF40 in posttranscriptional gene regulation, an important question is raised: why did the Sophophora subgenus evolve an additional copy, Poseidon, to encode a similar function in mRNA regulation?
The duplications of the X-linked CAF40 in form of the autosomal Poseidon and Zeus are likely a consequence of natural selection acting to comply with MXCI in evolution of sex chromosomes in Drosophila (Betran et al. 2002; Vibranovski, Zhang, et al. 2009; Mahadevaraju et al. 2021). An autosomal location can help these new genes to play their functional roles by escaping expression suppression by MXCI as its X-linked CAF40 experiences during inactivation. The fitness effects that these new gene duplicates brought under natural selection were detected to be critically important in viability during development and male specific fertility. Given the divergence of Zeus from Poseidon, these data suggest likely different roles of the two paralogs. Poseidon may compensate for CAF40 whose expression is suppressed by MXCI, similar to the function of the autosomal retroposed RPL10L in humans (Jiang et al. 2017; Long and Emerson 2017). However, Zeus is not completely redundant to Poseidon. The divergence in DNA sequence and expression make them functionally distinct in spermatogenesis. Additionally, a more complicated model was the SAXI model of the sexual antagonistic selection on the sex chromosome (Wu and Xu 2003). The model argued that the selection leading to demasculinization of X chromosomes before the establishment of the silencing of X chromosome (or regions) in evolution can also pressure a X->A gene traffic through duplication including retroposition (Emerson et al. 2004). Related to this, female germlines, which are not subject to the X inactivation in Drosophila, may also express in a lower level the X-linked male genes that are antagonistically selected against. These genes in females can serve as substrate for retroposition as well.
Our genome-wide transcriptome analyses demonstrated that the independent perturbation of the three paralogs impacts the regulation of thousands of genes in the testes (fig. 6). This is in agreement with the important role of CAF40 in transcriptional and posttranscriptional regulation (Collart 2016), as well as with the significant role of Zeus as a suppressor of female genes in males (Chen et al. 2012). The analysis of the genes that are commonly perturbed by the paralogs’ knockdown compared with the control (fig. 6B), revealed a strong enrichment for genes related to catabolic function (supplementary table S9, Supplementary Material online), such as serine-type endopeptidase activity (GO:0004252, P < 10−28), serine hydrolase activity (GO:0017171, P < 10−27), and catalytic activity (GO:0003824, P < 10−6). Those enrichment suggests that the knockdown of the paralogs affects the regulatory balance between transcription, translation, and degradation of numerous downstream genes, given the importance of the parental gene in coordinating and integrating different regulatory pathways (Miller and Reese 2012).
Taken together, the analyses presented here suggest that CCR4-NOT, a multifunctional complex that controls gene expression at multiple levels within the cell, evolved a new member Poseidon, through retroposition from CAF40, in Drosophila. Poseidon and Zeus, despite their relatively recent origination in Drosophila, were integrated into fundamental cellular and molecular processes with profound impacts in the regulatory network and phenotype. They were selected for compensation for the inactivated X-linked CAF40 during male meiosis or unrelated new functions, respectively. They reveal that a fundamentally important and conserved gene function also evolved with quick gene evolution, driven by evolution of sex chromosomes with its ancestral generated MXCI.
Materials and Methods
Molecular Evolutionary Analyses
Poseidon (CG2053) had been previously computationally identified as a putative young gene (Zhang et al. 2010). Gene and protein sequences were retrieved from Flybase and NCBI, aligned with MUSCLE (Edgar 2004) and manually curated. Reciprocal PSI-BLAST (NCBI) searches were employed to survey for CAF40, Poseidon, and Zeus orthologs in eukaryotes. The proper substitution model for the alignment (GTR+G) was selected through a likelihood ratio test using jModeltTest (Posada 2008). The phylogenetic relationship among the paralogs was firstly inferred through Bayesian analysis in MrBayes (Ronquist et al. 2012) by putting the three genes together. MCMC analysis was run with four chains for 2 million generations, with trees begin sampled every 500 generations, and the first 25% of samples were discarded as burn-in (supplementary fig. S9, Supplementary Material online). However, we found that there is no way to avoid long branch attraction (LBA) when invoking both rapidly evolving genes, Zeus and Poseidon, or branch length attraction (BLA) when invoking extremely slowly evolving CAF40 and the two rapidly evolving genes to generate congruent trees between genes tree and species three. We have also tried the three classical ways (ML, NJ, and MP) to construct three phylogenetic trees by putting the all the orthologs of the three paralogs together, and find they have same problem as supplementary figure S9, Supplementary Material online. However, all those three phylogenetic trees indicated that the Zeus orthologs’ cluster are always closer to CAF40 orthologs’ cluster than Poseidon. This indicates that Zeus originated from CAF40 (Bai et al. 2007; Quezada-Diaz et al. 2010). Finally, to avoid LBA and BLA effect, we respectively constructed the congruent phylogenetic trees using the classical maximum parsimonious method by separating the three paralogs (fig. 1C).
For the divergence of expression analysis, we retrieved the summary of expression data (FPKM [fragments per kilobase per million mapped reads]) from modENCODE and public RNA sequencing data of diverse fly species (Brown et al. 2014; Chen, Sturgill, et al. 2014; VanKuren and Vibranovski 2014). Expression values of each paralog at different spermatogenesis stages in D. melanogaster was compared using data from the SpermPress database (Vibranovski, Zhang, et al. 2009).
Shannon Entropy Analyses
Shannon’s entropy was calculated for an alignment of CAF40 orthologous protein sequences from 56 eukaryotes (supplementary fig. S1, Supplementary Material online), and the entropy value (H) for each residue was plotted onto CAF40 protein structure from D. melanogaster (Sgromo et al. 2017) using PyMOL. We used Shannon entropy (H) (Shannon 1948), with calculation of H score which represents standard entropy for a 22-letter alphabet. The calculation by bio3d follows: http://thegrantlab.org/bio3d/, or https://bitbucket.org/Grantlab/bio3d/downloads/, https://github.com/Grantlab/bio3d.git, bio3d_2.2-2.tar.gz, last accessed date: May 2019 (Mirny and Shakhnovich 1999; Grant 2006).
Knockdown and Knockout Phenotypes
In order to assay the knockdown effect of each paralog on egg to adult viability, homozygous UAS-TRiP RNAi lines (Perkins et al. 2015) were crossed to a balanced constitutive driver line (Tub84B>GAL4/TM3) and an imaginal disc-specific driver line (T80>GAL4/CyO) (supplementary table S11, Supplementary Material online, shows the list of lines used). At least ten independent replicates of three couples were allowed to cross and lay eggs for 7 days at 23 °C. All F1 adults in the progeny were scored, and the proportion of wild/balancer phenotypic markers for all replicates was compared with control crosses (TRiP background line BDSC 36303 crossed to the driver lines). Male fertility effects were assayed for the three paralogs by driving GAL4 expression using male germline-specific nanos-GAL4 and Bam-GAL4 drivers, which are expressed in early and late spermatogenesis, respectively (supplementary table S11, Supplementary Material online).
Of particular note is that Chen et al. (2012) used a RNAi line (KK101462) of CAF40 from KK (phiC31) RNAi line library in VDRC (https://stockcenter.vdrc.at/control/library_rnai, last accessed May 2019). Based on our lab empirical data, the KK library may have a lower knockdown efficiency than GD library (Xia et al. 2021), we tried to seek other mutant to reanalysis the effect of this gene. Finally, we used the UAS-TRiP RNAi lines in this study. TRiP uses a short but specific dsRNA (∼21 bp), whereas KK uses a longer dsRNA (81–799 bp, average: 357 bp) and they used different vector with varying KD efficiency (KK: pkC26; TRiP: pVALIUM20). Ni et al. (2011) found that the VALIUM20 (TRiP line construction vector) gives a stronger knockdown than the long-hairpin–based vector VALIUM10 in the soma and works well in the germline. This may cause the different phenotype effect of CAF40 between our study and Chen et al. (2012). The second potential reason may be the different target site of the two lines: KK101462 targets the last exon and the 3′-UTR region of CAF40, whereas TRIP.HMS05850 targets the third exon of CAF40. Therefore, we believe that dsRNA vector and target position may cause the different phenotypes of one same gene.
At least 15 replicates with 3- to 5-day-old knockdown males were individually crossed to two virgin females from background line BDSC 36303 for one day. Females were allowed to lay eggs for 7 days, and all the F1 adults were counted. Knockdown efficiency for each paralog was well confirmed (the expression was reduced to 15–35% of the control) through RT–PCR (supplementary fig. S3, Supplementary Material online). RNA samples were extracted in triplicate using RNeasy kit (Qiagen, Cat. No. 74104), digested with DNase I (Invitrogen, Cat. No. 18047019) to remove genomic DNA contamination, and reverse transcribed with SuperScript III Reverse Transcriptase (Invitrogen, Cat. No. 18080093) using oligo(dT) primers. RT–PCR was performed using iTaq Universal SYBR Green Supermix (Bio-Rad, Cat. No. 1725121), with three technical replicates for each biological replicate. Quantitative PCR values were normalized using the ΔΔCT method to the Rp49 control product.
CRISPR-Cas9 frameshift deletions were induced for Poseidon and Zeus. Guide RNAs were designed using CRISPR Optimal Target Finder (supplementary table S12, Supplementary Material online, http://targetfinder.flycrispr.neuro.brown.edu/, last accessed May 2020) to target early portions of the exon, and injected (300 ng/μl) along with Cas9 protein (PNA Bio Lab: CP01, 500 ng/μl) into embryos from the BDSC 25710 line (Bassett and Liu 2014). F1 mutant individuals were screened and crossed to balancer lines (w+; Sb/TM3; and w+; Sco/CyO, respectively). Small frameshift deletions were confirmed through Sanger sequencing and created early stop codons in the transcribed genes (supplementary fig. S10, Supplementary Material online). Viability and male fertility assays were performed with knockout flies as described above, using the injected line BDSC 25710 as control.
Co-immunoprecipitation Assay
DNA constructs with the coding region of D. melanogaster genes CAF40, NOT1, used were described before (Sgromo et al. 2017, 2018). Plasmids encoding Poseidon and Zeus were generated by inserting the corresponding cDNA (Thermo Scientific) into the pAc5.1-λN-HA and pAc5.1-GFP vectors (Rehwinkel et al. 2005; Tritschler et al. 2007) using HindIII and XhoI restriction sites. All constructs were confirmed by Sanger sequencing. For the co-immunoprecipitation assay in D. melanogaster S2 cells (ATCC), 2.5 × 106 cells were seeded per well in 6-well plates and transfected using Effectene transfection reagent (Qiagen, Cat. No. 301425). The transfection mixtures in figure 5A contained 1, 1.8, and 1.8 μg of plasmids expressing GFP-tagged CAF40, Poseidon, and Zeus, respectively. About 1 μg of HA-tagged NOT1 was used.
Cells were harvested 3 days after transfection, and co-immunoprecipitation assays were performed using RIPA buffer (20 mM HEPES [pH 7.6], 150 mM NaCl, 2.5 mM MgCl2, 1% NP-40, 1% sodium deoxycholate supplemented with protease inhibitors as previously described; Tritschler et al. 2008; Sgromo et al. 2018). All co-immunoprecipitation assays in S2 cell lysates were performed in the presence of RNase A as previously described (Sgromo et al. 2017). All Western blots were developed using an ECL western-blotting detection system (GE Healthcare, RPN2232). The antibodies used in this study are listed in supplementary table S13, Supplementary Material online.
Luciferase Assay
For the λN-tethering assays in D. melanogaster S2 cells, 2.5 × 106 cells per well were seeded in 6-well plates and transfected using Effectene transfection reagent (Qiagen, Cat. No. 301425). The transfection mixtures contained the following plasmids: 0.1 μg of Firefly luciferase reporters (F-Luc-5BoxB or F-Luc-V5), 0.4 μg of the Renilla Luciferase (R-luc) transfection control, and various amounts of plasmids expressing the λN-HA-tagged paralogs (0.01 μg for CAF40, 0.1 μg for Poseidon, 0.02 μg for Zeus). The plasmids for tethering assays in S2 cells (F-Luc-5BoxB, F-Luc-V5, and R-Luc) were previously described (Behm-Ansmant et al. 2006; Zekri et al. 2013). Cells were harvested 3 days after transfection and Firefly and Renilla luciferase activities were measured by using a Dual-Luciferase Reporter Assay System (Promega, Cat. No. E1910). The mean values ±SD from five independent experiments are shown.
RNA-Seq Analysis
Total RNA was extracted with Arcturus PicoPure RNA Isolation kit (Applied Biosystems, LOT 00665884) from testes of 3- to 5-day-old knockdown males and controls, with three biological replicates. A total amount of 1 μg RNA per sample was used to construct the cDNA library, using NEBNext Ultra RNA Library Prep Kit for Illumina (NEB, No. E7770) following manufacturer’s recommendations. Briefly, poly(A) mRNA was purified from total RNA using oligo(dT)-attached magnetic beads, reverse-transcribed to double-stranded cDNA with random primers, end-repaired and ligated with NEB adaptors for Illumina, before sequencing (HiSeq 4000, University of Chicago Genomics Core Facility).
Raw reads were processed and mapped to D. melanogaster reference genome (dm6) using STAR with default parameters (Dobin et al. 2013), and evaluation of transcriptional expression was carried out using featuresCounts (Liao et al. 2014). For the differential expression analysis, methods DESeq2 (Love et al. 2014, v1.21.22), edgeR (Robinson et al. 2010, v3.23.5), and limma (Ritchie et al. 2015, v3.37.7) were independently employed. Genes were considered as differentially expressed if they were consensually called by the three methods (supplementary fig. S11, Supplementary Material online), with an expression fold change of at least 1.5 compared with the control at false discovery rate <0.1. For DEGs, enriched biological processes and molecular functions were identified using both GOrilla (Eden et al. 2009) and g: Profiler (Raudvere et al. 2019), with P < 10−4, and a false discovery rate of 0.1. Both of the two tools showed same result. The interaction network of proteins with enrichment for catabolic processes was visualized using STRING (von Mering et al. 2003), selecting only experimentally validated interactions with high confidence. The analyses of DEGs with male/female-biased expression followed two independent Drosophila databases (Zhang et al. 2010; Assis et al. 2012).
We calculated the DEGs intersection numbers of those subgroups between our study and Chen et al. (2012) (Microarray). For example, only 241/1,784 = 13.5% genes out of CAF40_DEGs_Chen was overlapped with CAF40_DEGs_Xia. The intersection between those 833 genes (intersection between CAF40_DEGs_Xia and Zeus_DEGs_Xia) and 664 genes (intersection between CAF40_DEGs_Chen and Zeus_DEGs_Chen) is only 50 genes (supplementary table S10, Supplementary Material online). In addition, only 36 genes were overlapped between CAF40_downregulated DEGs in Chen et al. (2012) and CAF40_downregulated DEGs in our study. Those small intersection numbers between our study and Chen et al. (2012) were mainly caused by different approaches, RNAi lines and developmental stages: firstly, Chen et al. (2012) used GD49820 and KK101462 as Zeus and CAF40 RNAi lines, respectively. However, our study used TRiP RNAi lines for both Zeus and CAF40. Secondly, Chen et al. (2012) applied microarray expression profiling to obtain the DEGs of Zeus and CAF40 KD, whereas our study used RNA-seq. Thirdly, Chen et al. (2012) used testis from 1- to 7-day-old males, whereas our study used testes of 3- to 5-day-old knockdown males. Therefore, even those small intersection numbers between our study and Chen et al. (2012), among the DEGs in Zeus and CAF40 knockdown genotypes, our study also showed <50% DEGs show changes in the same direction (46.6% and 48.5%, supplementary table S10, Supplementary Material online). Additionally, Poseidon knockdown genotypes showed a slightly higher proportion of DEGs (57.3% and 61.2%, supplementary table S10, Supplementary Material online) which showed changes in the same direction with CAF40.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
We dedicated this article to Elisa Izaurralde, to show our deep respect for her devotedness to science and warm collegiality, besides her monumental contribution to the investigation of posttranscriptional and translational regulation. We remember that, in her last months, she enthusiastically arranged a fruitful collaboration with the Chicago team. She gave insightful discussion and designed with A.S. the experiments related to understanding the molecular function and evolution of Poseidon. We also appreciate Nicholas VanKuren’s constructive discussion with several details in our study. I.M.V. identified existence of Poseidon in the Sophophora subgenus of Drosophila. S.X. created the CRISPR mutants in this study. I.M.V., S.X., and S.H. conducted analyses of fitness effects, expression, and evolution of Poseidon and Zeus. S.X. conducted phylogeny analyses, sequence, and RNA-seq analyses. A.S., A.B. with E.I., investigated the molecular functions of Poseidon and Zeus. Wen-Qing Chan at the University of Chicago Center for Research Informatics provided valuable help in the RNA-seq analyses. M.L. and I.M.V. conceived the study of the new gene systems, M.L., I.M.V., S.X., and A.S. composed the manuscript. I.M.V. was supported by the Science without Borders Scholarship (BEX18816/12-6). M.L. was supported by NSF1026200 and NIH R01GM116113.
Data Availability
The data underlying this article are available in the article and in its Supplementary Material online.
References
- Abrusan G. 2013. Integration of new genes into cellular networks, and their structural maturation. Genetics 195:1407–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assis R, Zhou Q, Bachtrog D.. 2012. Sex-biased transcriptome evolution in Drosophila. Genome Biol Evol. 4(11):1189–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai YS, Casola C, Betran E.. 2008. Evolutionary origin of regulatory regions of retrogenes in Drosophila. BMC Genomics 9:241–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai YS, Casola C, Feschotte C, Betran E.. 2007. Comparative genomics reveals a constant rate of origination and convergent acquisition of functional retrogenes in Drosophila. Genome Biol. 8(1):R11–R19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker CC, Fuller MT.. 2007. Translational control of meiotic cell cycle progression and spermatid differentiation in male germ cells by a novel eIF4G homolog. Development 134(15):2863–2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett A, Liu JL.. 2014. CRISPR/Cas9 mediated genome engineering in Drosophila. Methods 69(2):128–136. [DOI] [PubMed] [Google Scholar]
- Bawankar P, Loh B, Wohlbold L, Schmidt S, Izaurralde E.. 2013. NOT10 and C2orf29/NOT11 form a conserved module of the CCR4-NOTcomplex that docks onto the NOT1 N-terminal domain. RNA Biol. 10(2):228–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behm-Ansmant I, Rehwinkel J, Doerks T, Stark A, Bork P, Izaurralde E.. 2006. MRNA degradation by miRNAs and GW182 requires both CCR4: NOT deadenylase and DCP1: DCP2 decapping complexes. Genes Dev. 20(14):1885–1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belote JM, Zhong L.. 2009. Duplicated proteasome subunit genes in Drosophila and their roles in spermatogenesis. Heredity (Edinb). 103(1):23–31. [DOI] [PubMed] [Google Scholar]
- Betran E, Thornton K, Long M.. 2002. Retroposed new genes out of the X in Drosophila. Genome Res. 12(12):1854–1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JB, Boley N, Eisman R, May GE, Stoiber MH, Duff MO, Booth BW, Wen JY, Park S, Suzuki AM, et al. 2014. Diversity and dynamics of the Drosophila transcriptome. Nature 512(7515):393–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buschauer R, Matsuo Y, Sugiyama T, Chen YH, Alhusaini N, Sweet T, Ikeuchi K, Cheng JD, Matsuki Y, Nobuta R, et al. 2020. The Ccr4-Not complex monitors the translating ribosome for codon optimality. Science 368(6488):281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carelli FN, Hayakawa T, Go Y, Imai H, Warnefors M, Kaessmann H.. 2016. The life history of retrocopies illuminates the evolution of new mammalian genes. Genome Res. 26(3):301–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casola C, Betran E.. 2017. The genomic impact of gene retrocopies: what have we learned from comparative genomics, population genomics, and transcriptomic analyses? Genome Biol Evol. 9(6):1351–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Ni X, Krinsky BH, Zhang YE, Vibranovski MD, White KP, Long M.. 2012. Reshaping of global gene expression networks and sex-biased gene expression by integration of a young gene. EMBO J. 31(12):2798–2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Zhang YE, Long M.. 2010. New genes in Drosophila quickly become essential. Science 330(6011):1682–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Boland A, Kuzuoğlu-Öztürk D, Bawankar P, Loh B, Chang C-T, Weichenrieder O, Izaurralde E.. 2014. A DDX6-CNOT1 complex and W-binding pockets in CNOT9 reveal direct links between miRNA target recognition and silencing. Mol Cell. 54(5):737–750. [DOI] [PubMed] [Google Scholar]
- Chen Z-X, Sturgill D, Qu J, Jiang H, Park S, Boley N, Suzuki AM, Fletcher AR, Plachetzki DC, FitzGerald PC, et al. 2014. Comparative validation of the D. melanogaster modENCODE transcriptome annotation. Genome Res. 24(7):1209–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark AG, Eisen MB, Smith DR, Bergman CM, Oliver B, Markow TA, Kaufman TC, Kellis M, Gelbart W, Iyer VN, et al. 2007. Evolution of genes and genomes on the Drosophila phylogeny. Nature 450:203–218. [DOI] [PubMed] [Google Scholar]
- Collart MA. 2016. The Ccr4-Not complex is a key regulator of eukaryotic gene expression. Wiley Interdiscip Rev RNA. 7(4):438–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collart MA, Panasenko OO.. 2017. The Ccr4-not complex: architecture and structural insights. Subcell Biochem. 83:349–379. [DOI] [PubMed] [Google Scholar]
- Dai HZ, Yoshimatsu TF, Long MY.. 2006. Retrogene movement within- and between-chromosomes in the evolution of Drosophila genomes. Gene 385:96–102. [DOI] [PubMed] [Google Scholar]
- Ding Y, Zhao L, Yang S, Jiang Y, Chen Y, Zhao R, Zhang Y, Zhang G, Dong Y, Yu H, et al. 2010. A young Drosophila duplicate gene plays essential roles in spermatogenesis by regulating several Y-linked male fertility genes. PLoS Genet. 6(12):e1001255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR.. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29(1):15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eden E, Navon R, Steinfeld I, Lipson D, Yakhini Z.. 2009. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics 10(1):48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. 2004. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5(1):113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson JJ, Kaessmann H, Betran E, Long MY.. 2004. Extensive gene traffic on the mammalian X chromosome. Science 303(5657):537–540. [DOI] [PubMed] [Google Scholar]
- Erwin DH, Davidson EH.. 2009. The evolution of hierarchical gene regulatory networks. Nat Rev Genet. 10(2):141–148. [DOI] [PubMed] [Google Scholar]
- Garapaty S, Mahajan MA, Samuels HH.. 2008. Components of the CCR4-NOT complex function as nuclear hormone receptor coactivators via association with the NRC-interacting factor NIF-1. J Biol Chem. 283(11):6806–6816. [DOI] [PubMed] [Google Scholar]
- Garces RG, Gillon W, Pai EF.. 2007. Atomic model of human Rcd-1 reveals an armadillo-like-repeat protein with in vitro nucleic acid binding properties. Protein Sci. 16(2):176–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant BJ, Rodrigues AP, ElSawy KM, McCammon JA, Caves LS.. 2006. Bio3d: an R package for the comparative analysis of protein structures. Bioinformatics 22(21):2695–2696. [DOI] [PubMed] [Google Scholar]
- Halfon MS. 2017. Perspectives on gene regulatory network evolution. Trends Genet. 33(7):436–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison PW, Wright AE, Zimmer F, Dean R, Montgomery SH, Pointer MA, Mank JE.. 2015. Sexual selection drives evolution and rapid turnover of male gene expression. Proc Natl Acad Sci U S A. 112(14):4393–4398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiller M, Chen X, Pringle MJ, Suchorolski M, Sancak Y, Viswanathan S, Bolival B, Lin TY, Marino S, Fuller MT.. 2004. Testis-specific TAF homologs collaborate to control a tissue-specific transcription program. Development 131(21):5297–5308. [DOI] [PubMed] [Google Scholar]
- Jiang L, Li T, Zhang XX, Zhang BB, Yu CP, Li Y, Fan SX, Jiang XH, Khan T, Hao QM, et al. 2017. RPL10L is required for male meiotic division by compensating for RPL10 during meiotic sex chromosome inactivation in mice. Curr Biol. 27(10):1498–1505. [DOI] [PubMed] [Google Scholar]
- Kaessmann H, Vinckenbosch N, Long M.. 2009. RNA-based gene duplication: mechanistic and evolutionary insights. Nat Rev Genet. 10(1):19–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaessmann H. 2010. Origins, evolution, and phenotypic impact of new genes. Genome Res. 20(10):1313–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath RS, Fraser AG, Dong Y, Poulin G, Durbin R, Gotta M, Kanapin A, Le Bot N, Moreno S, Sohrmann M, et al. 2003. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421(6920):231–237. [DOI] [PubMed] [Google Scholar]
- Kasinathan B, Colmenares SU 3rd, McConnell H, Young JM, Karpen GH, Malik HS.. 2020. Innovation of heterochromatin functions drives rapid evolution of essential ZAD-ZNF genes in Drosophila. eLife 9:e63368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keskeny C, Raisch T, Sgromo A, Igreja C, Bhandari D, Weichenrieder O, Izaurralde E.. 2019. A conserved CAF40-binding motif in metazoan NOT4 mediates association with the CCR4–NOT complex. Genes Dev. 33(3-4):236–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondrashov FA. 2012. Gene duplication as a mechanism of genomic adaptation to a changing environment. Proc Biol Sci. 279(1749):5048–5057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K.. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YCG, Ventura IM, Rice GR, Chen DY, Colmenares SU, Long M.. 2019. Rapid evolution of gained essential developmental functions of a young gene via interactions with other essential genes. Mol Biol Evol. 36(10):2212–2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legrand JMD, Hobbs RM.. 2018. RNA processing in the male germline: mechanisms and implications for fertility. Semin Cell Dev Biol. 79:80–91. [DOI] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, Shi W.. 2014. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30(7):923–930. [DOI] [PubMed] [Google Scholar]
- Long MY, Emerson JJ.. 2017. Meiotic sex chromosome inactivation: compensation by gene traffic. Curr Biol. 27(13):R659–R661. [DOI] [PubMed] [Google Scholar]
- Long MY, VanKuren NW, Chen SD, Vibranovski MD.. 2013. New gene evolution: little did we know. Annu Rev Genet. 47:307–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S.. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15(12):550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahadevaraju S, Fear JM, Akeju M, Galletta BJ, Pinheiro MM, Avelino CC, Cabral-de-Mello DC, Conlon K, Dell’Orso S, Demere Z, et al. 2021. Dynamic sex chromosome expression in Drosophila male germ cells. Nat Commun. 12:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markow TA, O’Grady PM.. 2007. Drosophila biology in the genomic age. Genetics 177(3):1269–1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathys H, Basquin J, Ozgur S, Czarnocki-Cieciura M, Bonneau F, Aartse A, Dziembowski A, Nowotny M, Conti E, Filipowicz W.. 2014. Structural and biochemical insights to the role of the CCR4-NOT complex and DDX6 ATPase in microRNA repression. Mol Cell. 54(5):751–765. [DOI] [PubMed] [Google Scholar]
- Matsuno M, Compagnon V, Schoch GA, Schmitt M, Debayle D, Bassard J-E, Pollet B, Hehn A, Heintz D, Ullmann P, et al. 2009. Evolution of a novel phenolic pathway for pollen development. Science 325(5948):1688–1692. [DOI] [PubMed] [Google Scholar]
- Mering CV, Huynen M, Jaeggi D, Schmidt S, Bork P, Snel B.. 2003. STRING: a database of predicted functional associations between proteins. Nucleic Acids Res. 31(1):258–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miklos GL, Rubin GM.. 1996. The role of the genome project in determining gene function: insights from model organisms. Cell 86(4):521–529. [DOI] [PubMed] [Google Scholar]
- Miller JE, Reese JC.. 2012. Ccr4-Not complex: the control freak of eukaryotic cells. Crit Rev Biochem Mol Biol. 47(4):315–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirny LA, Shakhnovich EI.. 1999. Universally conserved positions in protein folds: reading evolutionary signals about stability, folding kinetics and function. J Mol Biol. 291(1):177–196. [DOI] [PubMed] [Google Scholar]
- Ni J-Q, Zhou R, Czech B, Liu L-P, Holderbaum L, Yang-Zhou D, Shim H-S, Tao R, Handler D, Karpowicz P, et al. 2011. A genome-scale shRNA resource for transgenic RNAi in Drosophila. Nat Methods. 8(5):405–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins LA, Holderbaum L, Tao R, Hu Y, Sopko R, McCall K, Yang-Zhou D, Flockhart I, Binari R, Shim H-S, et al. 2015. The transgenic RNAi project at Harvard Medical School: resources and validation. Genetics 201(3):843–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posada D. 2008. jModelTest: phylogenetic model averaging. Mol Biol Evol. 25(7):1253–1256. [DOI] [PubMed] [Google Scholar]
- Quezada-Diaz JE, Muliyil T, Rio J, Betran E.. 2010. Drcd-1 related: a positively selected spermatogenesis retrogene in Drosophila. Genetica 138(9-10):925–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raudvere U, Kolberg L, Kuzmin I, Arak T, Adler P, Peterson H, Vilo J.. 2019. g: profiler: a web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res. 47(W1):W191–W198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehwinkel J, Behm-Ansmant I, Gatfield D, Izaurralde E.. 2005. A crucial role for GW182 and the DCP1:DCP2 decapping complex in miRNA-mediated gene silencing. RNA 11(11):1640–1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richler C, Soreq H, Wahrman J.. 1992. X-inactivation in mammalian testis is correlated with inactive X-specific transcription. Nat Genet. 2(3):192–195. [DOI] [PubMed] [Google Scholar]
- Ritchie ME, Phipson B, Wu D, Hu YF, Law CW, Shi W, Smyth GK.. 2015. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43(7):e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK.. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26(1):139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Hohna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP.. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross BD, Rosin L, Thomae AW, Hiatt MA, Vermaak D, de la Cruz AF, Imhof A, Mellone BG, Malik HS.. 2013. Stepwise evolution of essential centromere function in a Drosophila neogene. Science 340(6137):1211–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo CA, Takezaki N, Nei M.. 1995. Molecular phylogeny and divergence times of drosophilid species. Mol Biol Evol. 12(3):391–404. [DOI] [PubMed] [Google Scholar]
- Saleem S, Schwedes CC, Ellis LL, Grady ST, Adams RL, Johnson N, Whittington JR, Carney GE.. 2012. Drosophila melanogaster p24 trafficking proteins have vital roles in development and reproduction. Mech Dev. 129(5-8):177–191. [DOI] [PubMed] [Google Scholar]
- Sgromo A, Raisch T, Backhaus C, Keskeny C, Alva V, Weichenrieder O, Izaurralde E.. 2018. Drosophila Bag-of-marbles directly interacts with the CAF40 subunit of the CCR4-NOT complex to elicit repression of mRNA targets. RNA 24(3):381–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sgromo A, Raisch T, Bawankar P, Bhandari D, Chen Y, Kuzuoğlu-Öztürk D, Weichenrieder O, Izaurralde E.. 2017. A CAF40-binding motif facilitates recruitment of the CCR4-NOT complex to mRNAs targeted by Drosophila Roquin. Nat Commun. 8:14307–14316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon CE. 1948. A mathematical theory of communication. Syst Tech J. 27(3):379–422. [Google Scholar]
- Toups MA, Hahn MW.. 2010. Retrogenes reveal the direction of sex-chromosome evolution in mosquitoes. Genetics 186(2):763–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tritschler F, Eulalio A, Helms S, Schmidt S, Coles M, Weichenrieder O, Izaurralde E, Truffault V.. 2008. Similar modes of interaction enable Trailer Hitch and EDC3 to associate with DCP1 and Me31B in distinct protein complexes. Mol Cell Biol. 28(21):6695–6708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tritschler F, Eulalio A, Truffault V, Hartmann MD, Helms S, Schmidt S, Coles M, Izaurralde E, Weichenrieder O.. 2007. A divergent Sm fold in EDC3 proteins mediates DCP1 binding and P-body targeting. Mol Cell Biol. 27(24):8600–8611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanKuren NW, Long MY.. 2018. Gene duplicates resolving sexual conflict rapidly evolved essential gametogenesis functions. Nat Ecol Evol. 2(4):705–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanKuren NW, Vibranovski MD.. 2014. A novel dataset for identifying sex-biased genes in Drosophila. J Genomics. 2:64–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vibranovski MD, Lopes HF, Karr TL, Long M.. 2009. Stage-specific expression profiling of Drosophila spermatogenesis suggests that meiotic sex chromosome inactivation drives genomic relocation of testis-expressed genes. PLoS Genet. 5(11):e1000731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vibranovski MD, Zhang Y, Long M.. 2009. General gene movement off the X chromosome in the Drosophila genus. Genome Res. 19(5):897–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vibranovski MD, Zhang YE, Kemkemer C, Lopes HF, Karr TL, Long M.. 2012. Re-analysis of the larval testis data on meiotic sex chromosome inactivation revealed evidence for tissue-specific gene expression related to the drosophila X chromosome. BMC Biol. 10(1):49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahle E, Winkler GS.. 2013. RNA decay machines: deadenylation by the CCR4–NOT and Pan2-Pan3 complexes. Biochim Biophys Acta. 1829(6-7):561–570. [DOI] [PubMed] [Google Scholar]
- Wang T, Birsoy K, Hughes NW, Krupczak KM, Post Y, Wei JJ, Lander ES, Sabatini DM.. 2015. Identification and characterization of essential genes in the human genome. Science 350(6264):1096–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White-Cooper H. 2010. Molecular mechanisms of gene regulation during Drosophila spermatogenesis. Reproduction 139(1):11–21. [DOI] [PubMed] [Google Scholar]
- White-Cooper H. 2012. Tissue, cell type and stage-specific ectopic gene expression and RNAi induction in the Drosophila testis. Spermatogenesis 2(1):11–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witt E, Benjamin S, Svetec N, Zhao L.. 2019. Testis single-cell RNA-seq reveals the dynamics of de novo gene transcription and germline mutational bias in Drosophila. eLife 8:e47138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CI, Xu EY.. 2003. Sexual antagonism and X inactivation–the SAXI hypothesis. Trends Genet. 19(5):243–247. [DOI] [PubMed] [Google Scholar]
- Xia S, VanKuren NW, Chen C, Zhang L, Kemkemer C, Shao Y, Jia H, Lee U, Advani AS, Gschwend A, et al. 2021. Genomic analyses of new genes and their phenotypic effects reveal rapid evolution of essential functions in Drosophila development. PLoS Genet. 17(7):e1009654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zekri L, Kuzuoğlu-Öztürk D, Izaurralde E.. 2013. GW182 proteins cause PABP dissociation from silenced miRNA targets in the absence of deadenylation. EMBO J. 32(7):1052–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang WY, Landback P, Gschwend AR, Shen BR, Long MY.. 2015. New genes drive the evolution of gene interaction networks in the human and mouse genomes. Genome Biol. 16:202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YE, Vibranovski MD, Krinsky BH, Long MY.. 2010. Age-dependent chromosomal distribution of male-biased genes in Drosophila. Genome Res. 20(11):1526–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong L, Belote JM.. 2007. The testis-specific proteasome subunit Pros alpha 6T of D-melanogaster is required for individualization and nuclear maturation during spermatogenesis. Development 134(19):3517–3525. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article are available in the article and in its Supplementary Material online.