Abstract
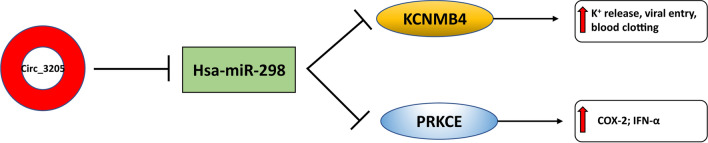
Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) is a new member of the Betacoronaviridae family, responsible for the recent pandemic outbreak of COVID-19. To start exploring the molecular events that follow host cell infection, we queried VirusCircBase and identified a circular RNA (circRNA) predicted to be synthesized by SARS-CoV-2, circ_3205, which we used to probe: (i) a training cohort comprised of two pools of cells from three nasopharyngeal swabs of SARS-CoV-2 infected (positive) or uninfected (negative, UCs) individuals; (ii) a validation cohort made up of 12 positive and 3 negative samples. The expression of circRNAs, miRNAs and miRNA targets was assayed through real-time PCR. CircRNA–miRNA interactions were predicted by TarpMiR, Analysis of Common Targets for circular RNAs (ACT), and STarMir tools. Enrichment of the biological processes and the list of predicted miRNA targets were retrieved from DIANA miRPath v3.0. Our results showed that the predicted SARS-CoV-2 circ_3205 was expressed only in positive samples and its amount positively correlated with that of SARS-CoV-2 Spike (S) mRNA and the viral load (r values = 0.80952 and 0.84867, Spearman’s correlation test, respectively). Human (hsa) miR-298 was predicted to interact with circ_3205 by all three predictive tools. KCNMB4 and PRKCE were predicted as hsa-miR-298 targets. Interestingly, the function of both is correlated with blood coagulation and immune response. KCNMB4 and PRKCE mRNAs were upregulated in positive samples as compared to UCs (6 and 8.1-fold, p values = 0.049 and 0.02, Student’s t test, respectively) and their expression positively correlated with that of circ_3205 (r values = 0.6 and 0.25, Spearman’s correlation test, respectively). We propose that our results convincingly suggest that circ_3205 is a circRNA synthesized by SARS-CoV-2 upon host cell infection and that it may behave as a competitive endogenous RNA (ceRNA), sponging hsa-miR-298 and contributing to the upregulation of KCNMB4 and PRKCE mRNAs.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00018-021-04119-8.
Keywords: Coronavirus disease 2019, circRNA, miRNA sponge, microRNA, Gene expression regulatory network
Introduction
Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) is an enveloped virus classified as a new member of the Coronaviridae family, Betacoronavirus genus, whose genome consists of a single stranded positive ( +) RNA molecule about 30 kilobases long [1, 2]. SARS-CoV-2 is etiologically involved in the life-threatening coronavirus disease 2019 (COVID-19) and is responsible for a pandemic outbreak in March, 2020 [3]. From December 2019 to September 2021, 3,508 SARS-CoV-2 genomes were sampled and 20 clades were identified around the world, according to the GISAID database (https://nextstrain.org/ncov/gisaid/global). Emergence of new SARS-CoV-2 genotypes alerts the scientific community to the possibility that some variants of concern may bypass the immune barrier given by the vaccines currently used and highlights the importance of the identification of therapeutic targets helpful for the management of this clinically very relevant disease [4].
Circular RNAs (circRNAs) are a recently discovered class of RNAs, mainly synthesized through backsplicing and characterized by the covalent bond between their 5’ and 3’ termini [5, 6]. CircRNAs follow tissue- and developmental-specific expression patterns and are mainly localized in the cell cytoplasm [7, 8]. The best characterized functions of circRNAs consist in sponging microRNAs (miRNAs) and RNA-binding proteins (RBPs) [5, 9]: in the first case, circRNAs may be typically involved in competitive endogenous RNA (ceRNA) networks [10–13]; in the second case, circRNAs may regulate biological processes within eukaryotic cells, such as assembly of preinitiation complex (PIC) at the beginning of transcription or also splicing [14, 15]. Moreover, circRNAs may function either as a template for the synthesis of generally short peptides, thanks to the presence of Internal Ribosomes Entry Sites (IRESs) within their sequences, or as a scaffold for the regulation of host gene transcription [16–18]. CircRNAs have been found aberrantly expressed in many cancers and degenerative diseases [19, 20] and are associated with several biological processes, both in physiological and pathological conditions [21–24]. Due to their intrinsic resistance to the activity of exoribonucleases and their presence in several human body fluids as well as within extracellular vesicles, circRNAs have been suggested as good candidate diagnostic and prognostic biomarkers for several diseases [25–28].
Recent evidence has shown the etiological involvement of circRNAs in viral infections. Most specifically, cross-talk between host cell circRNA biogenesis and RBPs linked to immune response (e.g.: immune factors NF90/NF110) has been described [29]. Influenza virus-infected A549 cells showed the induction of a circRNA that acts as a sponge for miRNAs regulating the expression of interferon beta (IFN-β) enhanceosome [30]. At the same time, circRNAs have been demonstrated to be synthesized from the genome of several DNA viruses (e.g.: Herpesviruses), contributing to the infection’s pathogenesis [31–33].
Recently, thanks to RNA-seq data analysis from cells infected with Middle East Respiratory Syndrome Coronavirus (MERS-CoV), Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV) and SARS-CoV-2 RNA (+) betacoronaviruses, several circRNAs of viral origin have been identified and characterized [34, 35]. At the same time, analysis of human circRNAome revealed several differentially expressed (DE) circRNAs in lung epithelial cells as well as in the peripheral blood of individuals infected with SARS-CoV-2 [36, 37]. Based on gene expression datasets, perturbation of ceRNA networks (circRNAs-miRNAs-mRNAs) in host cells following SARS-CoV-2 infection has also been predicted [38]. To improve our knowledge of the molecular dynamics of SARS-CoV-2 infection and prospectively identify new candidate therapeutic targets, in this study we focused on circRNAs synthesized from the viral genome, suggesting their involvement in COVID-19 pathogenesis.
Materials and methods
Sample preparation and diagnosis
Three ml of universal transport medium (UTM™) (COPAN Italia SpA, Brescia, Italy) from nasopharyngeal swabs of individuals suspected to be infected by SARS-CoV-2 were used for diagnostic purposes. A residual 1 ml of UTM™ was centrifuged at 350×g for 5 min at 4 °C to pellet cell debris. Supernatants were discarded and cell pellets stored at − 80 °C until further processing. Nucleic acids were isolated directly from UTM™ through a ThermoFisher Flex apparatus by the MagMAXTM Viral Pathogen Kit (ThermoFisher Scientific, Monza, Italy) for diagnostic purposes. Diagnosis was performed by a multiplex real-time PCR, Allplex™ SARS-CoV-2 Master assay (Seegene Inc., Arrow Diagnostics, Genoa, Italy), following the instructions of the manufacturer. The method amplifies SARS-CoV-2 E, N, RdRp, and S genes, according to World Health Organization (WHO)’s guidelines. Amplification cycles were performed on a Bio-Rad CFX96 real-time PCR instrument (Bio-Rad, Segrate–Milan, Italy). Only samples showing cycle threshold (Ct) values ≤ 35 for all the transcripts assayed were considered positive. A total of 15 positive and 6 and negative samples were assayed in this retrospective study. Data on biological specimens anonymously collected in this study were processed in accordance with the ethical principles reported in the Declaration of Helsinki.
RNA extraction, PCR amplification and Sanger sequencing
RNA was extracted from cells previously collected by nasopharyngeal swab using TRIzol® (ThermoFisher Scientific, ThermoFisher Scientific, Waltham, MA, USA), according to the manufacturer’s instruction [39]. RNA was quantified by a GenQuant pro spectrophotometer (Biochrom, Cambridge, UK). CircRNAs and mRNAs were amplified using a Power SYBR® Green RNA-to-CT™ 1-Step Kit (ThermoFisher Scientific). MiRNAs were reverse transcribed into cDNA by TaqMan™ MicroRNA Reverse Transcription Kit (ThermoFisher Scientific) and amplified through TaqMan™ Universal Master Mix II (ThermoFisher Scientific). PCRs were run on a 7900HT real-time PCR instrument (ThermoFisher Scientific). PCR products of the amplified circ_3205 were then sent to BMR Genomics, Padua, Italy (www.bmr-genomics.it) for purification through ExoSap (Applied Biosystems™, ThermoFischer Scientific). Sanger sequencing was performed with BigDyeTM Terminator v3.1 Cycle Sequencing Kit (ThermoFisher Scientific) on a ABI 3730xl DNA Analyzer (Applied Biosystems™, ThermoFisher Scientific). Sequences and IDs of specific primer pairs and TaqMan probes used in this manuscript are listed in Supplemental Table 1.
Candidate circRNAs selection
SARS-CoV-2 candidate circRNA sequences were retrieved from VirusCircBase (v. 1.1) [40], based on RNA-Seq experiments performed on Calu3 cells infected with the virus for 12 h and 24 h (GSE148729). Candidate circRNAs were filtered through the following criteria: (i) backsplice junction reads counted by at least two circRNA predictive algorithms among CIRI2; circRNA_finder; find_circ [6, 41, 42], for the same circRNA; (ii) abundancy of circRNA (at least two backsplice junction reads for each predictive algorithm).
Prediction of circRNA/miRNA interactions
Interactions between candidate circRNA and miRNAs were predicted by TarpMiR [43], Analysis of Common Targets for circular RNAs (ACT) [44], and STarMir [45]. FASTA sequences of human miRNAs (from miRBase 22 release [46]) and candidate circRNA were given as input to each of the three predictive algorithms. TarpMiR was set choosing human model and a probability cutoff of 0.5. Human V-CLIP data were used to train STarMir predictions [47]. Only miRNAs predicted to interact with SARS-CoV-2 candidate circRNA by all the three tools were considered as potentially implied in the ceRNA network.
Gene Ontology (GO) analysis and miRNA target selection
MiRNAs predicted to interact with candidate circRNA were given as input to DIANA miRPath v3.0 [48] and Biological Process (BP) GO’s subcategory was analyzed. MicroT-CDS (MicroT and False Discovery Rate (FDR)-corrected p value thresholds = 0.8 and 0.05, respectively) was selected as the algorithm for the prediction of miRNA-mRNA interactions. BP GO analysis filtered miRNA targets based on: (i) their potential involvement in blood clotting and immune response pathways, known to be related to COVID-19; (ii) dysregulated expression in lung or nasopharyngeal cells from SARS-CoV-2 infected individuals.
Protein–protein interaction (PPI) network analysis
First and second neighbor interactants of the candidate miRNA targets were retrieved from the HUman Reference protein Interactome (HuRI) database [49]. The list of interactions was given as input to Cytoscape (version 3.8.2) [50] and the network generated was analyzed through g:GOSt, within the g:Profiler web interface [51], and the Cytoscape plugin cytoHubba [52] to study gene functional enrichment and topological features, respectively. Topological analysis focused on the centrality parameters: betweenness; bottleneck; closeness; eccentricity; radiality and stress; for each of them, a corresponding subnetwork has been generated and analyzed for GO BP enrichment.
Statistical analysis
Real-time PCR data were analyzed through the 2−DDCt method [53] and two-sided Student’s t test was applied to identify DE transcripts. Spearman’s correlation test was used to identify positive or negative correlations among transcripts. Modified Fisher’s exact test followed by false discovery rate methodology was used to calculate p values for GO analysis. p values ≤ 0.05 were considered statistically significant.
Results
circRNA_3205 is a candidate viral circRNA expressed in Calu3 infected with SARS-CoV-2
Based on VirusCircBase, a total of 3473 circRNAs were detected by at least one tool among CIRI2; circRNA_finder; find_circ (Supplemental Table 2). SARS-CoV-2 circRNAs 363, 368, 2667, 2670, 2685, 2795, 3058, 3205 were the top eight most expressed circRNAs in Calu-3 infected with SARS-CoV-2 at least in one time point, according at least to two out of three algorithms among CIRI2; circRNA_finder; find_circ (Table 1).
Table 1.
Candidate SARS-CoV-2 circRNAs in Calu-3 infected with SARS-CoV-2 for 12 h and 24 h (data from GSE148729)
| SARS-CoV-2 CircRNA ID (VirusCircBase) | Start | End | Strand | SARS-CoV-2 host gene | Mean JRC 12 h circRNA_finder | Mean JRC 12 h find_circ | Mean JRC 12 h CIRI2 | Mean JRC 24 h circfinder | Mean JRC 24 h find_circ | Mean JRC 24 h CIRI2 |
|---|---|---|---|---|---|---|---|---|---|---|
| circ_363 | 2571 | 3263 | − | ORF1ab | N/A | 2 | 9.5 | N/A | 4.5 | 30 |
| circ_368 | 2571 | 3539 | − | ORF1ab | N/A | 2 | 31 | N/A | 2.5 | 76 |
| circ_2667 | 25,672 | 26,212 | − | ORF3a | N/A | 2.5 | 26.5 | N/A | 3.5 | 33 |
| circ_2670 | 25,672 | 26,241 | − | ORF3a | N/A | 10.5 | 25 | N/A | 10 | 104 |
| circ_2685 | 25,672 | 26,570 | − | ORF3a;M;E | N/A | 3 | 26.5 | N/A | 12.5 | 60 |
| circ_2795 | 26,825 | 27,225 | + | ORF6;M | 12 | 15 | N/A | 12.5 | 17.5 | N/A |
| circ_3058 | 28,404 | 29,080 | + | N | 4.5 | 5.5 | N/A | 19 | 20.5 | N/A |
| circ_3205 | 28,610 | 28,895 | − | N | N/A | 12 | 17 | N/A | 7 | 15.5 |
For each circRNA, the arithmetic mean of the junction read counts (JCR) calculated from each tool for two biological replicates, is reported
N/A JCR not annotated by the tool
Circ_3205 is expressed only in positive samples and its amount positively correlates with that of viral Spike (S) mRNA
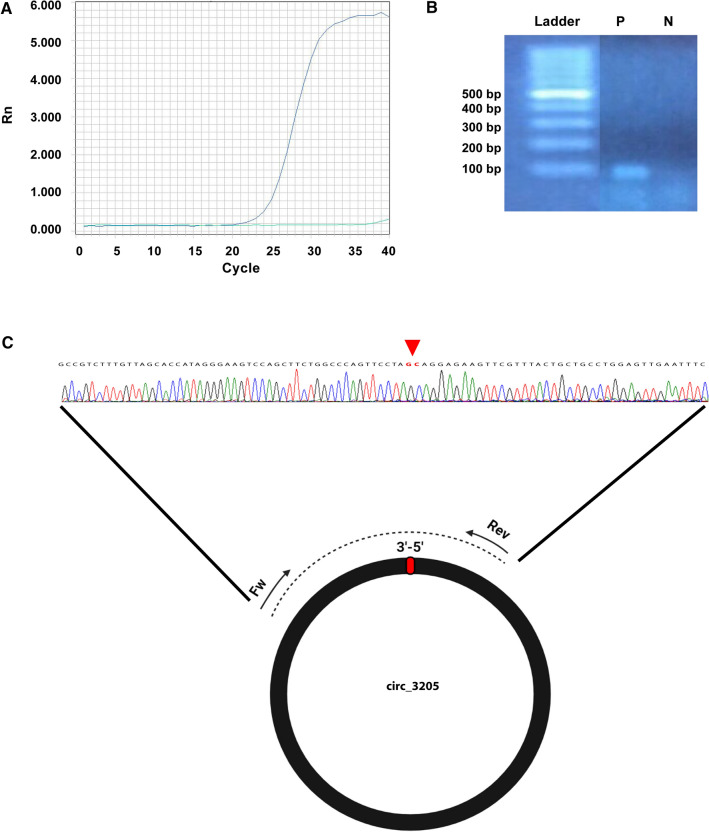
The expression of SARS-CoV-2 circRNAs 363, 368, 2667, 2670, 2685, 2795, 3058, and 3205 was assayed in a discovery cohort made of a pool of three positive samples and three UCs. Based on real-time PCR and gel electrophoresis data, we focused on circ_3205, a 286 nt-long circRNA whose sequence is embedded within the open reading frame (ORF) coding for the nucleocapside (N) protein of SARS-CoV-2 (nucleotide position 28,609–28,898, Wuhan-Hu-1 reference genome, NCBI Reference Sequence: NC_045512.2), clearly present and highly expressed only in positive samples (Fig. 1A and 1B). Sanger sequencing of the PCR amplification product of circ_3205 confirmed the presence of the 3’-5’ junction, specific of this circRNA (Fig. 1C). Gene expression assay in the validation cohort confirmed that circ_3205 was expressed only in positive samples and its amount positively correlated with that of Spike (S) mRNA and SARS-CoV-2 viral load (r values = 0.80952 and 0.84867, p values (two sided) = 0.015 and 0.016, respectively, Spearman’s correlation test) (Fig. 2). The expression of S mRNA positively correlated with SARS-CoV-2 viral load, too (r value = 0.92582, p values (two sided) = 0.003) (Fig. 2).
Fig. 1.
A Real-time PCR amplification plot of circ_3205 for a representative positive (blue curve) and negative (green curve) sample. B Agarose gel (2%) electrophoresis of the real-time PCR products in (A). Ladder = 100 bp DNA ladder (ThermoFisher Scientific); P and N = representative positive and negative sample, respectively. C Graphical representation of the divergent primers used to amplify SARS-CoV-2 circ_3205 and Sanger sequencing of the resulting PCR amplicon (from a representative positive sample). Dotted line represents the PCR amplicon obtained through the use of divergent primers; the red triangle above the electropherogram highlights the 3ʹ–5ʹ junction of circ_3205
Fig. 2.
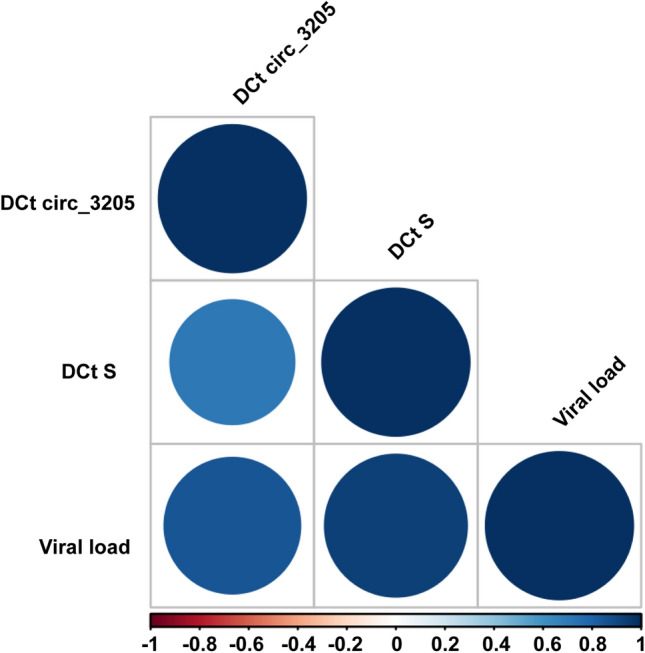
Correlogram showing correlations among the expression of circ_3205, S mRNA and the viral load. Expression of circ_3205 and S mRNA is reported as DCt (Ct of the transcript of interest – Ct of GAPDH, used as endogenous control). Viral load was estimated on the basis of the mean of the Cts of SARS-CoV-2 E, N, RdRp, and S genes. The color of the circle is related to the correlation coefficient (r value), estimated through the Spearman correlation test: the deeper is the blue, the more positive is the correlation, the deeper is the red, the more negative is the correlation, as reported in the legend of the figure. The size of the circle is proportional to the significance of the correlation: the higher is the size of the circle, the lower is the p value
Human (hsa) miRNAs 298, 3940-5p, 4640-5p, 6081, and 6133 are predicted to interact with circ_3205 and their targets are potentially involved in cell response to viral infection
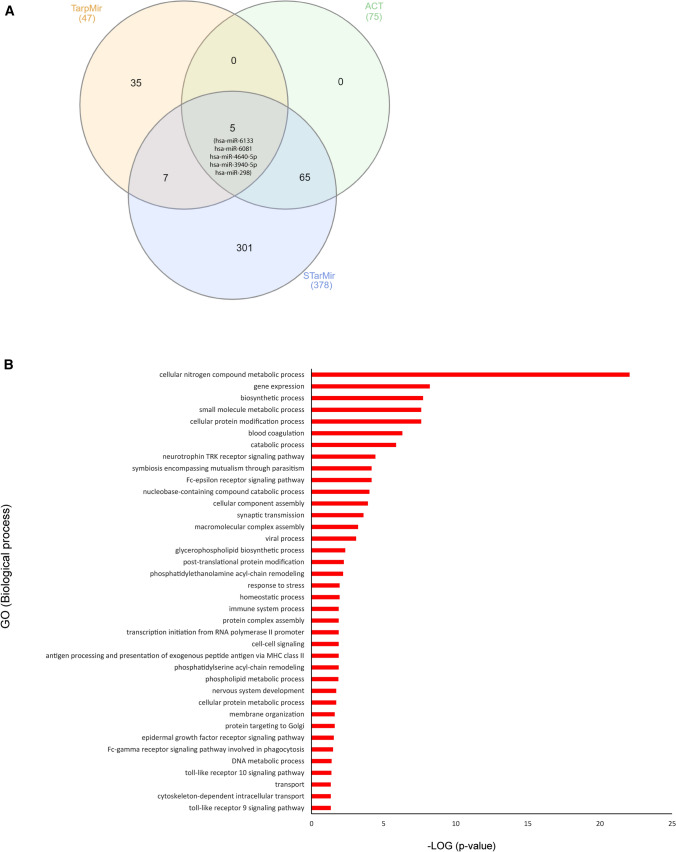
Hsa miRNAs 298, 3940-5p, 4640-5p, 6081 and 6133 were predicted to interact with circ_3205 by all the three predictive algorithms queried (TarpMiR, ACT and STarMir) (Fig. 3A). BP GO analysis highlighted a potential involvement of the mRNA targets regulated by hsa miRNAs 298, 3940-5p, 4640-5p, 6081 and 6133 in SARS-CoV-2 infection-related processes (blood coagulation and immune response) (Fig. 3B).
Fig. 3.
A Venn diagrams showing the number of miRNAs predicted to interact with circ_3205 by TarpMir, ACT and STarMiR predictive tools. The total number of miRNAs predicted by each tool is reported in brackets, while the number of miRNAs, either specifically predicted by only one or by more than one predictive tool, is reported (without brackets) within the Venn diagrams. The name of the five candidate miRNAs predicted to interact with circ_3205 by all the three predictive tools is reported in extenso. B Horizontal bar chart representing the enrichment of biological processes carried out by the predicted targets of hsa miRNAs 298, 3940-5p, 4640-5p, 6081, and 6133. Biological processes are reported in y-axis; statistical significance is reported as -LOG (p value) (x-axis)
Hsa-miR-298 is predicted to target mRNAs coding for proteins involved in blood coagulation and immune response
Based on (i) literature data, (ii) our GO analysis, and (iii) the high probability of interaction with SARS-CoV-2_circ_3205, we focused on hsa-mir-298 (Fig. 4A). Has-miR-298 was predicted to target 18 and 30 mRNAs involved in blood coagulation and immune response, respectively, based on DIANA miRPath analysis (Supplemental Table 3). Ten predicted mRNA targets (CPB2; DCTN1; FN1; KCNMB4; MERTK; MYO1C; PDE1A; PIK3C3; PRKCE; SERPINB2) were selected as candidates because of their known upregulation in biological specimens from SARS-CoV-2-infected individuals, as compared with UCs (Supplemental Table 4).
Fig. 4.
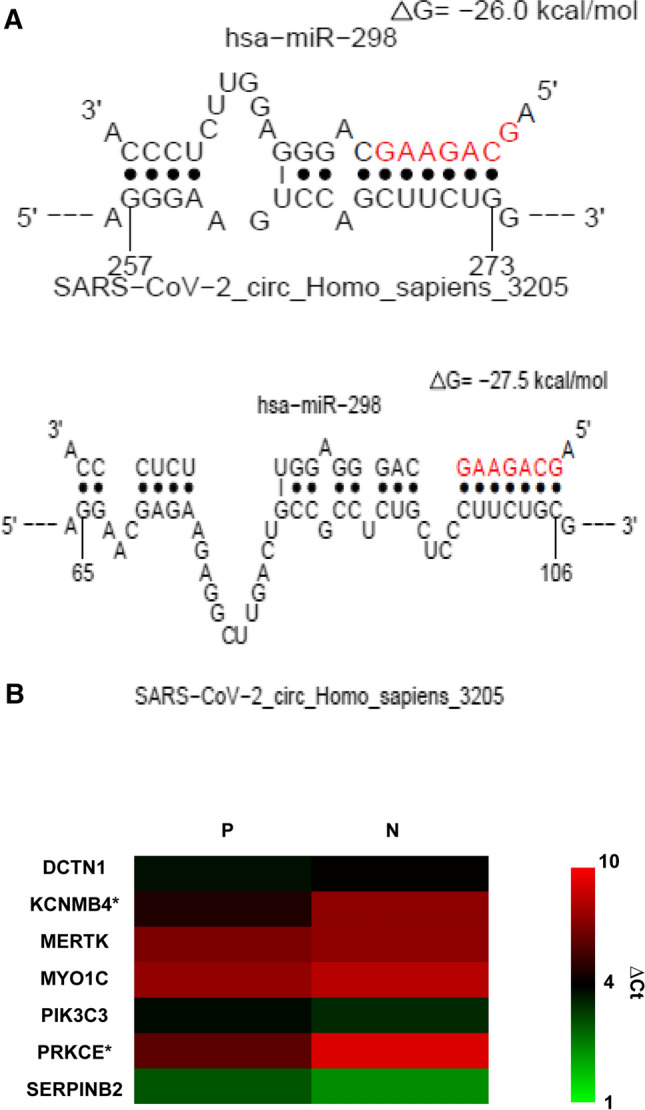
A Graphical representation of the interaction predicted by STarMiR between hsa-miR-298 and circ_3205. MiRNA seed region is showed in red. The numbers below vertical bars indicate the nucleotide position of circ_3205. B Heatmap showing the expression of seven predicted hsa-miR-298 targets whose expression was revealed in positive (P) and negative (N) samples by real-time PCR. Expression is reported as DCt: the lower is this value, the higher is the expression of the target and viceversa. * = statistically significant DE transcript (p value < 0.05, Student’s t test)
Potassium calcium-activated channel subfamily M regulatory beta subunit 4 (KCNMB4) and protein kinase C epsilon (PRKCE) mRNAs are upregulated in positive samples as compared to UCs and their expression positively correlates with that of circ_3205
Quantitative real-time PCR detected the expression of seven (DCTN1; KCNMB4; MERTK; MYO1C; PIK3C3; PRKCE; SERPINB2) out of the ten predicted hsa-miR-298 targets in the analyzed samples. Among them, KCNMB4 and PRKCE were 6 and 8.1-fold more expressed in positive samples as compared to UCs (p values = 0.049 and 0.02, Student’s t test, respectively) and their expression positively correlated with that of circ_3205 (r values = 0.6 and 0.25, Spearman’s correlation test, respectively) (Fig. 4B).
PPI network of KCNMB4 and PRKCE is enriched in biological processes related to immune response and blood coagulation
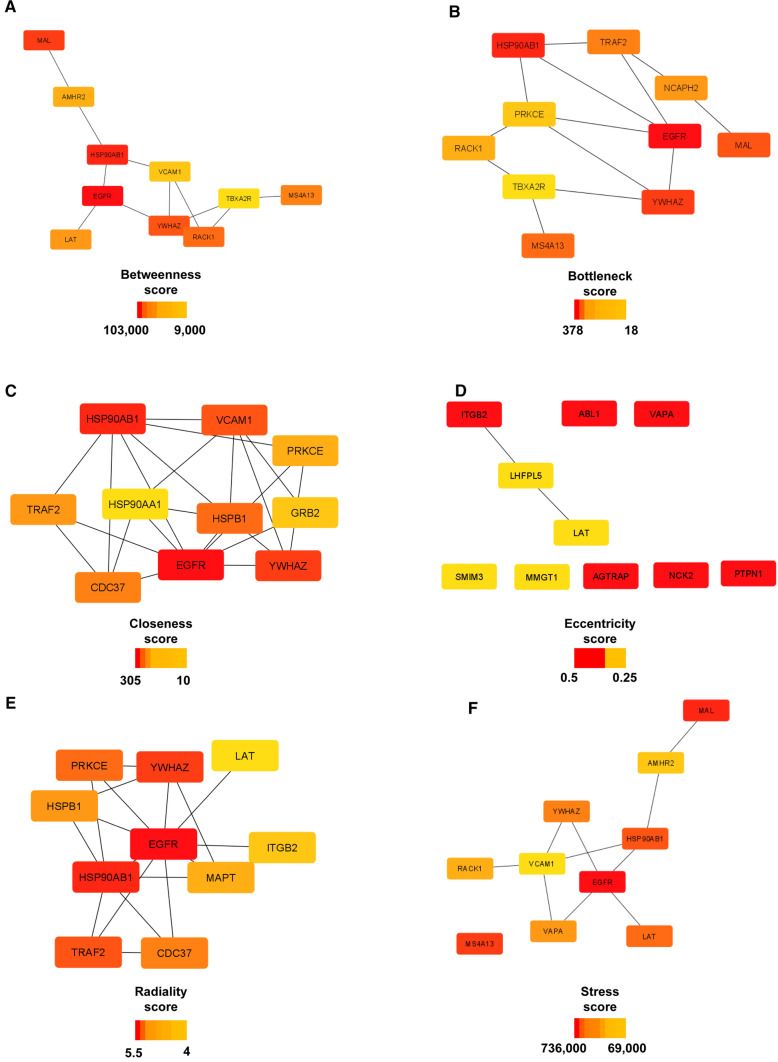
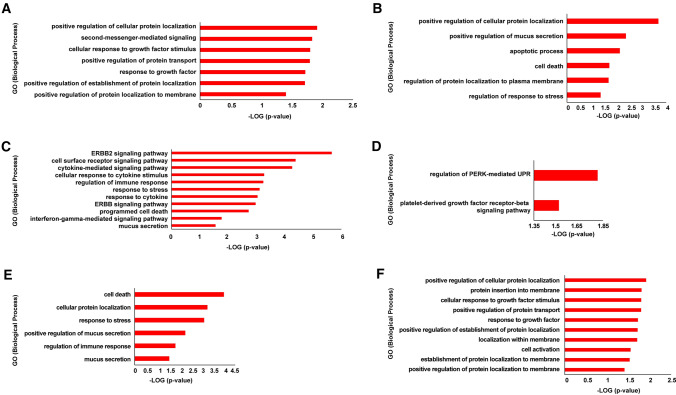
PPI network generated by HuRI consisted of 482 nodes and 1452 edges. GO analysis of the whole network revealed an over-representation of biological functions linked to blood coagulation, immune response, and inflammation (Supplemental Fig.1). The analysis of centralities of the network revealed a total of 27 most central proteins: among them, EGFR, HSP90AB1, YWHAZ occurred in five out of six subnetworks made of the most central nodes (Fig. 5). The generated subnetworks revealed an enrichment in BPs related to SARS-CoV-2 infection program (Fig. 6).
Fig. 5.
Analysis of centralities of KCNMB4 and PRKCE’s PPI network. Subnetworks show the top ten most central nodes calculated by A betweenness, B bottleneck, C closeness, D eccentricity, E radiality, F stress methods. For each subnetwork the scalebar reporting the scores of centrality is shown
Fig. 6.
Functional enrichment of the subnetworks generated by A betweenness, B bottleneck, C closeness, D eccentricity, E radiality, F stress methods. Statistical significance of the functional enrichment within each subnetwork is reported as -LOG (p value) and BPs are arranged in y-axis from the higher to the lower -LOG (p value)
Discussion
The capability of viruses to synthesize circRNAs upon infection has been ascertained, especially in DNA viruses [54–60]. As an example, Epstein Barr virus (EBV), a double stranded DNA virus belonging to the Herpesviridae family, is known to produce about 30 circRNAs during different phases of its infection [61] and some of them (e.g.: circBART2.2) contribute to virus-induced carcinogenesis through the immune escape of nasopharyngeal carcinoma cells [62]. Kaposi’s sarcoma-associated herpesvirus (KSHV) generates circRNAs found to be inserted into virions and implied in several steps of the infection [63].
In this study, we first assayed the expression of eight circRNAs predicted to be synthesized by SARS-CoV-2 upon host cell infection. Based on our experimental results, we then focused on circRNA 3205. Based on data stored in VirusCircBase, circ_3205 was predicted to be synthesized from the negative RNA strand of SARS-CoV-2, specifically from a sequence embedded in the ORF coding for the N protein of the virus. Although the mechanism of circRNA biogenesis from RNA (+) viruses is under investigation, some hypotheses may be proposed: in a recent study, it was found that SARS-CoV-2 genome may be in part reverse transcribed and integrated as DNA into the host genome through a LINE1-mediated mechanism, leading to the production of chimeric viral-host cellular transcripts [64, 65]. Based on this study, it is conceivable that viral circRNAs could be generated through backsplicing from these chimeric viral-host transcripts. Nevertheless, the hypothesis of an integration of SARS-CoV-2 genome (or a part) into the host genome is debated [66]. An alternative path of circRNA biogenesis from RNA (+) viral genomes may consist in splicing-independent mechanisms occurring in the cytoplasm of host cells: this mechanism was previously described for IRE1alpha-mediated XBP1 mRNA splicing in mammalian cells, for IL1b transcripts in platelets and for the recently suggested miR-7-mediated circularization of the CDR1AS transcript [67–69]. Expression of viral circRNAs may perturb the ceRNA networks originally present within host cells or may create new ones [70]. Specifically, our data suggest that once synthesized within the host cell, circ_3205 may function as sponge for hsa-miR-298, allowing for the upregulation of targets involved in the progression through the infection (Fig. 7). MiR-298, together with miR-296, belongs to a genomic locus that is imprinted both in mice and humans [71, 72]. Interestingly, hsa-miR-298 has been predicted to bind the 5’-UTR of the SARS-CoV-2 genome, potentially altering its secondary structure and negatively impacting on its capability to be translated after infection of the host cell [73]. Chopra N. et al. identified hsa-miR-298 as a potential therapeutic agent for Alzheimer’s disease, because of its capability to negatively regulate the expression of human amyloid-β precursor protein (APP), β-site APP-converting enzyme 1 (BACE1) and specific tau protein isoforms [74]. Hsa-miR-298 has also been defined as oncomiRNA in several cancers, thanks to its ability to downregulate proapoptotic proteins such as BAX and PTEN [75, 76]. These literature data support our hypothesis that the circ_3205 sponge effect against hsa-miR-298 may contribute to the progression of the infection, by stabilizing the SARS-CoV-2 genome and triggering biological processes such as inflammation and apoptosis. Our data also shed light on two predicted targets of hsa-miR-298 (KCNMB4 and PRKCE), which we found to be upregulated in positive samples and whose expression positively correlated with that of circ_3205. KCNMB4 encodes a β4 subunit of a voltage-dependent K+ channel, belonging to the Ca2+-activated Slo subfamily (BK) [77, 78]. Upregulation of KCNMB4 correlates with the increased intracellular concentration of Ca2+ observed during SARS-CoV-2 infection [79]. Even though the function exerted by BK channels during the SARS-CoV-2 infection needs further investigation, the role of K+ concentration and K+ channels in facilitating the entry of some viruses into the host cells has been convincingly ascertained [80, 81]. Furthermore, abnormalities in electrolyte serum concentrations (especially sodium, potassium, calcium and chloride) have been found to be related with the prognosis of COVID-19 patients and with the possibility to develop blood clots [82–84]. PRKCE encodes a Ca2+-independent protein kinase, belonging to the subfamily of nonconventional protein kinase C (PKCs); it has been described as involved in SARS-CoV infection, through the calcium-independent PI3K/PKCε/JNK/CREB pathway; this, in turn, induces COX-2 expression upon the interaction between viral S protein and cellular receptors [85]. COX-2 has been found upregulated in several cell types also after SARS-CoV-2 infection [86]. PRKCE expression is further induced by Interferon-α (IFN-α), one of the first cytokines synthesized in infected cells on innate immune response [87]. Due to an abnormal recruitment of proinflammatory cells, IFN-α signaling over a prolonged period of time is known to cause an uncontrolled inflammatory response and potential organ failure in tissues infected by SARS-CoV-2 as well as other respiratory viruses [88, 89]. The study of the PPI network, generated starting from KCNMB4 and PRKCE, revealed Heat Shock Protein 90, Alpha family, class B member 1 (HSP90AB1) as one of the most central nodes, according to five out of six parameters of network centrality. HSP90AB1 is a first neighbor interactant of PRKCE and belongs to the Heat Shock Protein 90 (HSP90) family. Some members of HSP90 family have been recently suggested to foster MERS-CoV, SARS-CoV and SARS-CoV-2 replication and proinflammatory cytokine expression [90, 91]. Subnetworks generated by the study of centralities further revealed an enrichment in BPs strictly related to the local and systemic effects of SARS-CoV-2 infection, such as remodeling of protein trafficking within infected host cell [92], mucus hypersecretion [93], ErbB protein family and growth factor receptor signaling [94, 95], and unfolded protein response [96]. Collectively, these findings corroborate our hypothesis of a functional involvement of KCNMB4 and PRKCE in SARS-CoV-2 infection.
Fig. 7.
Schematic diagram of the ceRNA network mediated by SARS-CoV-2 circ_3205 in human infected cells
Conclusions
Based on the integration of our experimental data and predictive analysis, we propose SARS-Cov-2_circ_3205/hsa-miR-298/KCNMB4 and SARS-Cov-2_circ_3205/hsa-miR-298/PRKCE molecular axes as involved in the progression of SARS-Cov-2 infection and, more in detail, in the related processes of blood clotting and immune response, respectively (Fig. 7).
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
We thank Dr Elisabetta Tendi for her kind support in the first phases of this work. We also thank the Scientific Bureau of the University of Catania for language support. Dr Cristina Barbagallo is actually receiving a protempore contribution (assegno di ricerca) thanks to a funding assigned to Prof. Michele Purrello by Dipartimento di Medicina Clinica e Sperimentale, Università di Catania, Italy, EU. This study was supported by “PIAno di inCEntivi per la RIcerca (PIA.CE.RI.) di Ateneo 2020/2022”—“linea di intervento 3, STARTING GRANT” from the University of Catania and by “Fondi di ateneo 2020/2022, Università di Catania, linea Open Access”.
Author contributions
Conceptualization, D.B., M.P., G.S.; methodology, D.B., C.I.P.; validation, C.B., R.B., A.C.; formal analysis, D.B., C.I.P.; investigation, D.B., C.I.P.; resources, G.S., C.I.P.; data curation, V.S., M.R, C.D.P.; writing—original draft preparation, D.B. and M.P.; writing—review and editing, D.B. and M.P.; visualization, D.B.; supervision, M.P. and G.S.; funding acquisition, M.P. and D.B. All authors have read and agreed to the published version of the manuscript.
Funding
Not aplicable.
Availability of data and material
Data available on request from the authors.
Declarations
Conflict of interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Ethical approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Cinzia Di Pietro, Guido Scalia and Michele Purrello: Senior authors.
References
- 1.Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R, et al. A Novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, Si HR, Zhu Y, Li B, Huang CL, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu F, Zhao S, Yu B, Chen YM, Wang W, Song ZG, Hu Y, Tao ZW, Tian JH, Pei YY, et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Phelan AL, Katz R, Gostin LO. The novel coronavirus originating in Wuhan, China: challenges for global health governance. JAMA. 2020;323:709–710. doi: 10.1001/jama.2020.1097. [DOI] [PubMed] [Google Scholar]
- 5.Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 6.Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 7.Salzman J, Chen RE, Olsen MN, Wang PL, Brown PO. Cell-type specific features of circular RNA expression. PLoS Genet. 2013;9:e1003777. doi: 10.1371/journal.pgen.1003777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS ONE. 2012;7:e30733. doi: 10.1371/journal.pone.0030733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barbagallo D, Caponnetto A, Barbagallo C, Battaglia R, Mirabella F, Brex D, Stella M, Broggi G, Altieri R, Certo F, et al. The GAUGAA Motif Is Responsible for the Binding between circSMARCA5 and SRSF1 and Related Downstream Effects on Glioblastoma Multiforme Cell Migration and Angiogenic Potential. Int J Mol Sci. 2021 doi: 10.3390/ijms22041678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146:353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505:344–352. doi: 10.1038/nature12986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tang CM, Zhang M, Huang L, Hu ZQ, Zhu JN, Xiao Z, Zhang Z, Lin QX, Zheng XL, Yang M, et al. CircRNA_000203 enhances the expression of fibrosis-associated genes by derepressing targets of miR-26b-5p, Col1a2 and CTGF, in cardiac fibroblasts. Sci Rep. 2017;7:40342. doi: 10.1038/srep40342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barbagallo C, Brex D, Caponnetto A, Cirnigliaro M, Scalia M, Magnano A, Caltabiano R, Barbagallo D, Biondi A, Cappellani A, et al. LncRNA UCA1, upregulated in CRC biopsies and downregulated in serum exosomes, controls mRNA expression by RNA-RNA interactions. Mol Therapy Nucleic Acids. 2018;12:229–241. doi: 10.1016/j.omtn.2018.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Conn SJ, Pillman KA, Toubia J, Conn VM, Salmanidis M, Phillips CA, Roslan S, Schreiber AW, Gregory PA, Goodall GJ. The RNA binding protein quaking regulates formation of circRNAs. Cell. 2015;160:1125–1134. doi: 10.1016/j.cell.2015.02.014. [DOI] [PubMed] [Google Scholar]
- 15.Du WW, Yang W, Liu E, Yang Z, Dhaliwal P, Yang BB. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 2016;44:2846–2858. doi: 10.1093/nar/gkw027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang M, Huang N, Yang X, Luo J, Yan S, Xiao F, Chen W, Gao X, Zhao K, Zhou H, et al. A novel protein encoded by the circular form of the SHPRH gene suppresses glioma tumorigenesis. Oncogene. 2018;37:1805–1814. doi: 10.1038/s41388-017-0019-9. [DOI] [PubMed] [Google Scholar]
- 17.Feng Y, Yang Y, Zhao X, Fan Y, Zhou L, Rong J, Yu Y. Circular RNA circ0005276 promotes the proliferation and migration of prostate cancer cells by interacting with FUS to transcriptionally activate XIAP. Cell Death Dis. 2019;10:792. doi: 10.1038/s41419-019-2028-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li H, Yang F, Hu A, Wang X, Fang E, Chen Y, Li D, Song H, Wang J, Guo Y, et al. Therapeutic targeting of circ-CUX1/EWSR1/MAZ axis inhibits glycolysis and neuroblastoma progression. EMBO Mol Med. 2019;11:e10835. doi: 10.15252/emmm.201910835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barbagallo D, Condorelli A, Ragusa M, Salito L, Sammito M, Banelli B, Caltabiano R, Barbagallo G, Zappalà A, Battaglia R, et al. Dysregulated miR-671-5p/CDR1-AS/CDR1/VSNL1 axis is involved in glioblastoma multiforme. Oncotarget. 2016;7:4746–4759. doi: 10.18632/oncotarget.6621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dube U, Del-Aguila JL, Li Z, Budde JP, Jiang S, Hsu S, Ibanez L, Fernandez MV, Farias F, Norton J, et al. An atlas of cortical circular RNA expression in Alzheimer disease brains demonstrates clinical and pathological associations. Nat Neurosci. 2019;22:1903–1912. doi: 10.1038/s41593-019-0501-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang W, Liu Y, Min Z, Liang G, Mo J, Ju Z, Zeng B, Guan W, Zhang Y, Chen J, et al. circMine: a comprehensive database to integrate, analyze and visualize human disease-related circRNA transcriptome. Nucleic Acids Res. 2021 doi: 10.1093/nar/gkab809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ragusa M, Barbagallo D, Chioccarelli T, Manfrevola F, Cobellis G, Di Pietro C, Brex D, Battaglia R, Fasano S, Ferraro B, et al. CircNAPEPLD is expressed in human and murine spermatozoa and physically interacts with oocyte miRNAs. RNA Biol. 2019;16:1237–1248. doi: 10.1080/15476286.2019.1624469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu W, Ji P, Zhao F. CircAtlas: an integrated resource of one million highly accurate circular RNAs from 1070 vertebrate transcriptomes. Genome Biol. 2020;21:101. doi: 10.1186/s13059-020-02018-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu M, Wang Q, Shen J, Yang BB, Ding X. Circbank: a comprehensive database for circRNA with standard nomenclature. RNA Biol. 2019;16:899–905. doi: 10.1080/15476286.2019.1600395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stella M, Falzone L, Caponnetto A, Gattuso G, Barbagallo C, Battaglia R, Mirabella F, Broggi G, Altieri R, Certo F, et al. Serum extracellular vesicle-derived circHIPK3 and circSMARCA5 are two novel diagnostic biomarkers for glioblastoma multiforme. Pharmaceuticals (Basel, Switzerland) 2021 doi: 10.3390/ph14070618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xia D, Gu X. Plasmatic exosome-derived circRNAs panel act as fingerprint for glioblastoma. Aging. 2021;13:19575–19586. doi: 10.18632/aging.203368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen C. Serum hsa_circ_101178 as a Potential Biomarker for Early Prediction of Osteoarthritis. Clin Lab. 2020 doi: 10.7754/Clin.Lab.2020.191251. [DOI] [PubMed] [Google Scholar]
- 28.Wu WP, Pan YH, Cai MY, Cen JM, Chen C, Zheng L, Liu X, Xiong XD. Plasma-derived exosomal circular RNA hsa_circ_0005540 as a novel diagnostic biomarker for coronary artery disease. Dis Markers. 2020;2020:3178642. doi: 10.1155/2020/3178642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li X, Liu CX, Xue W, Zhang Y, Jiang S, Yin QF, Wei J, Yao RW, Yang L, Chen LL. Coordinated circRNA biogenesis and function with NF90/NF110 in viral infection. Mol Cell. 2017;67:214–227.e217. doi: 10.1016/j.molcel.2017.05.023. [DOI] [PubMed] [Google Scholar]
- 30.Qu Z, Meng F, Shi J, Deng G, Zeng X, Ge J, Li Y, Liu L, Chen P, Jiang Y, et al. A novel intronic circular RNA antagonizes influenza virus by absorbing a microRNA that degrades CREBBP and accelerating IFN-β production. MBio. 2021;12:e0101721. doi: 10.1128/mBio.01017-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang JT, Chen JN, Gong LP, Bi YH, Liang J, Zhou L, He D, Shao CK. Identification of virus-encoded circular RNA. Virology. 2019;529:144–151. doi: 10.1016/j.virol.2019.01.014. [DOI] [PubMed] [Google Scholar]
- 32.Tagawa T, Oh D, Santos J, Dremel S, Mahesh G, Uldrick TS, Yarchoan R, Kopardé VN, Ziegelbauer JM. Characterizing expression and regulation of gamma-herpesviral circular RNAs. Front Microbiol. 2021;12:670542. doi: 10.3389/fmicb.2021.670542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Avilala J, Becnel D, Abdelghani R, Nanbo A, Kahn J, Li L, Lin Z. Role of virally encoded circular RNAs in the pathogenicity of human oncogenic viruses. Front Microbiol. 2021;12:657036. doi: 10.3389/fmicb.2021.657036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cai Z, Lu C, He J, Liu L, Zou Y, Zhang Z, Zhu Z, Ge X, Wu A, Jiang T, et al. Identification and characterization of circRNAs encoded by MERS-CoV, SARS-CoV-1 and SARS-CoV-2. Brief Bioinform. 2021;22:1297–1308. doi: 10.1093/bib/bbaa334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang S, Zhou H, Cruz-Cosme R, Liu M, Xu J, Niu X, Li Y, Xiao L, Wang Q, Zhu H, et al. Circular RNA profiling reveals abundant and diverse circRNAs of SARS-CoV-2, SARS-CoV and MERS-CoV origin. bioRxiv. 2020 doi: 10.1101/2020.12.07.415422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang M, Qi M, Xu L, Huang P, Wang X, Sun J, Shi J, Hu Y. Differential host circRNA expression profiles in human lung epithelial cells infected with SARS-CoV-2. Infect Genet Evol. 2021;93:104923. doi: 10.1016/j.meegid.2021.104923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu Y, Zhao T, Deng R, Xia X, Li B, Wang X. A study of differential circRNA and lncRNA expressions in COVID-19-infected peripheral blood. Sci Rep. 2021;11:7991. doi: 10.1038/s41598-021-86134-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Demirci YM, SaçarDemirci MD. Circular RNA-MicroRNA-MRNA interaction predictions in SARS-CoV-2 infection. J Integr Bioinform. 2021;18:45–50. doi: 10.1515/jib-2020-0047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Battaglia R, Vento ME, Ragusa M, Barbagallo D, La Ferlita A, Di Emidio G, Borzí P, Artini PG, Scollo P, Tatone C, et al. MicroRNAs are stored in human MII oocyte and their expression profile changes in reproductive aging. Biol Reprod. 2016;95:131. doi: 10.1095/biolreprod.116.142711. [DOI] [PubMed] [Google Scholar]
- 40.Cai Z, Fan Y, Zhang Z, Lu C, Zhu Z, Jiang T, Shan T, Peng Y. VirusCircBase: a database of virus circular RNAs. Brief Bioinform. 2021;22:2182–2190. doi: 10.1093/bib/bbaa052. [DOI] [PubMed] [Google Scholar]
- 41.Gao Y, Zhang J, Zhao F. Circular RNA identification based on multiple seed matching. Brief Bioinform. 2018;19:803–810. doi: 10.1093/bib/bbx014. [DOI] [PubMed] [Google Scholar]
- 42.Westholm JO, Miura P, Olson S, Shenker S, Joseph B, Sanfilippo P, Celniker SE, Graveley BR, Lai EC. Genome-wide analysis of drosophila circular RNAs reveals their structural and sequence properties and age-dependent neural accumulation. Cell Rep. 2014;9:1966–1980. doi: 10.1016/j.celrep.2014.10.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ding J, Li X, Hu H. TarPmiR: a new approach for microRNA target site prediction. Bioinformatics (Oxford, England) 2016;32:2768–2775. doi: 10.1093/bioinformatics/btw318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lin YC, Lee YC, Chang KL, Hsiao KY. Analysis of common targets for circular RNAs. BMC Bioinform. 2019;20:372. doi: 10.1186/s12859-019-2966-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kanoria S, Rennie W, Liu C, Carmack CS, Lu J, Ding Y. STarMir Tools for Prediction of microRNA Binding Sites. Methods Mol Biol (Clifton NJ) 2016;1490:73–82. doi: 10.1007/978-1-4939-6433-8_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kozomara A, Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014;42:D68–73. doi: 10.1093/nar/gkt1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kishore S, Jaskiewicz L, Burger L, Hausser J, Khorshid M, Zavolan M. A quantitative analysis of CLIP methods for identifying binding sites of RNA-binding proteins. Nat Methods. 2011;8:559–564. doi: 10.1038/nmeth.1608. [DOI] [PubMed] [Google Scholar]
- 48.Vlachos IS, Zagganas K, Paraskevopoulou MD, Georgakilas G, Karagkouni D, Vergoulis T, Dalamagas T, Hatzigeorgiou AG. DIANA-miRPath v3.0: deciphering microRNA function with experimental support. Nucleic acids Res. 2015;43:W460–466. doi: 10.1093/nar/gkv403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Luck K, Kim DK, Lambourne L, Spirohn K, Begg BE, Bian W, Brignall R, Cafarelli T, Campos-Laborie FJ, Charloteaux B, et al. A reference map of the human binary protein interactome. Nature. 2020;580:402–408. doi: 10.1038/s41586-020-2188-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Raudvere U, Kolberg L, Kuzmin I, Arak T, Adler P, Peterson H, Vilo J. g:Profiler: a web server for functional enrichment analysis and conversions of gene lists (2019 update) Nucleic Acids Res. 2019;47:W191–w198. doi: 10.1093/nar/gkz369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chin CH, Chen SH, Wu HH, Ho CW, Ko MT, Lin CY. cytoHubba: identifying hub objects and sub-networks from complex interactome. BMC Syst Biol. 2014;8(4):S11. doi: 10.1186/1752-0509-8-s4-s11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods (San Diego, California) 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 54.Tan KE, Lim YY. Viruses join the circular RNA world. FEBS J. 2021;288:4488–4502. doi: 10.1111/febs.15639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Toptan T, Abere B, Nalesnik MA, Swerdlow SH, Ranganathan S, Lee N, Shair KH, Moore PS, Chang Y. Circular DNA tumor viruses make circular RNAs. Proc Natl Acad Sci USA. 2018;115:E8737–e8745. doi: 10.1073/pnas.1811728115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Abere B, Zhou H, Li J, Cao S, Toptan T, Grundhoff A, Fischer N, Moore PS, Chang Y. Merkel cell polyomavirus encodes circular RNAs (circRNAs) enabling a dynamic circRNA/microRNA/mRNA regulatory network. MBio. 2020 doi: 10.1128/mBio.03059-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tagawa T, Gao S, Koparde VN, Gonzalez M, Spouge JL, Serquiña AP, Lurain K, Ramaswami R, Uldrick TS, Yarchoan R, et al. Discovery of Kaposi's sarcoma herpesvirus-encoded circular RNAs and a human antiviral circular RNA. Proc Natl Acad Sci USA. 2018;115:12805–12810. doi: 10.1073/pnas.1816183115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ungerleider NA, Jain V, Wang Y, Maness NJ, Blair RV, Alvarez X, Midkiff C, Kolson D, Bai S, Roberts C, et al. Comparative analysis of gammaherpesvirus circular RNA repertoires: conserved and unique viral circular RNAs. J Virol. 2019 doi: 10.1128/jvi.01952-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lu S, Zhu N, Guo W, Wang X, Li K, Yan J, Jiang C, Han S, Xiang H, Wu X, et al. RNA-Seq Revealed a circular RNA-microRNA-mRNA regulatory network in hantaan virus infection. Front Cell Infect Microbiol. 2020;10:97. doi: 10.3389/fcimb.2020.00097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Niu M, Ju Y, Lin C, Zou Q. Characterizing viral circRNAs and their application in identifying circRNAs in viruses. Brief Bioinform. 2021 doi: 10.1093/bib/bbab404. [DOI] [PubMed] [Google Scholar]
- 61.Ungerleider N, Concha M, Lin Z, Roberts C, Wang X, Cao S, Baddoo M, Moss WN, Yu Y, Seddon M, et al. The Epstein Barr virus circRNAome. PLoS Pathog. 2018;14:e1007206. doi: 10.1371/journal.ppat.1007206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ge J, Wang J, Xiong F, Jiang X, Zhu K, Wang Y, Mo Y, Gong Z, Zhang S, He Y, et al. Epstein-barr virus-encoded circular RNA CircBART2.2 promotes immune escape of nasopharyngeal carcinoma by regulating PD-L1. Cancer Res. 2021;81:5074–5088. doi: 10.1158/0008-5472.can-20-4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Abere B, Li J, Zhou H, Toptan T, Moore PS, Chang Y. Kaposi's sarcoma-associated herpesvirus-encoded circRNAs are expressed in infected tumor tissues and are incorporated into virions. MBio. 2020 doi: 10.1128/mBio.03027-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang L, Richards A, Barrasa MI, Hughes SH, Young RA, Jaenisch R. Reverse-transcribed SARS-CoV-2 RNA can integrate into the genome of cultured human cells and can be expressed in patient-derived tissues. Proc Natl Acad Sci USA. 2021 doi: 10.1073/pnas.2105968118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yin Y, Liu XZ, He X, Zhou LQ. Exogenous coronavirus interacts with endogenous retrotransposon in human cells. Front Cell Infect Microbiol. 2021;11:609160. doi: 10.3389/fcimb.2021.609160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Parry R, Gifford RJ, Lytras S, Ray SC, Coin LJM. No evidence of SARS-CoV-2 reverse transcription and integration as the origin of chimeric transcripts in patient tissues. Proc Natl Acad Sci USA. 2021 doi: 10.1073/pnas.2109066118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Back SH, Lee K, Vink E, Kaufman RJ. Cytoplasmic IRE1alpha-mediated XBP1 mRNA splicing in the absence of nuclear processing and endoplasmic reticulum stress. J Biol Chem. 2006;281:18691–18706. doi: 10.1074/jbc.M602030200. [DOI] [PubMed] [Google Scholar]
- 68.Denis MM, Tolley ND, Bunting M, Schwertz H, Jiang H, Lindemann S, Yost CC, Rubner FJ, Albertine KH, Swoboda KJ, et al. Escaping the nuclear confines: signal-dependent pre-mRNA splicing in anucleate platelets. Cell. 2005;122:379–391. doi: 10.1016/j.cell.2005.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Belter A, Popenda M, Sajek M, Woźniak T, Naskręt-Barciszewska MZ, Szachniuk M, Jurga S, Barciszewski J. A new molecular mechanism of RNA circularization and the microRNA sponge formation. J Biomol Struct Dyn. 2020 doi: 10.1080/07391102.2020.1844802,1-8. [DOI] [PubMed] [Google Scholar]
- 70.Ghosal S, Das S, Sen R, Chakrabarti J. HumanViCe: host ceRNA network in virus infected cells in human. Front Genet. 2014;5:249. doi: 10.3389/fgene.2014.00249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Robson JE, Eaton SA, Underhill P, Williams D, Peters J. MicroRNAs 296 and 298 are imprinted and part of the GNAS/Gnas cluster and miR-296 targets IKBKE and Tmed9. RNA (New York, NY) 2012;18:135–144. doi: 10.1261/rna.029561.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Barbagallo D, Piro S, Condorelli AG, Mascali LG, Urbano F, Parrinello N, Monello A, Statello L, Ragusa M, Rabuazzo AM, et al. miR-296-3p, miR-298-5p and their downstream networks are causally involved in the higher resistance of mammalian pancreatic α cells to cytokine-induced apoptosis as compared to β cells. BMC Genom. 2013;14:62. doi: 10.1186/1471-2164-14-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pierce JB, Simion V, Icli B, Pérez-Cremades D, Cheng HS, Feinberg MW. Computational analysis of targeting SARS-CoV-2, viral entry proteins ACE2 and TMPRSS2, and interferon genes by host microRNAs. Genes. 2020 doi: 10.3390/genes11111354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chopra N, Wang R, Maloney B, Nho K, Beck JS, Pourshafie N, Niculescu A, Saykin AJ, Rinaldi C, Counts SE, et al. MicroRNA-298 reduces levels of human amyloid-β precursor protein (APP), β-site APP-converting enzyme 1 (BACE1) and specific tau protein moieties. Mol Psychiatry. 2020 doi: 10.1038/s41380-019-0610-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhao H, Zhao D, Tan G, Liu Y, Zhuang L, Liu T. Bufalin promotes apoptosis of gastric cancer by down-regulation of miR-298 targeting bax. Int J Clin Exp Med. 2015;8:3420–3428. [PMC free article] [PubMed] [Google Scholar]
- 76.Arabsorkhi Z, Gharib E, YaghmoorianKhojini J, Farhadieh ME, Nazemalhosseini-Mojarad E, Zali MR. miR-298 plays a pivotal role in colon cancer invasiveness by targeting PTEN. J Cell Physiol. 2020;235:4335–4350. doi: 10.1002/jcp.29310. [DOI] [PubMed] [Google Scholar]
- 77.Brenner R, Jegla TJ, Wickenden A, Liu Y, Aldrich RW. Cloning and functional characterization of novel large conductance calcium-activated potassium channel beta subunits, hKCNMB3 and hKCNMB4. J Biol Chem. 2000;275:6453–6461. doi: 10.1074/jbc.275.9.6453. [DOI] [PubMed] [Google Scholar]
- 78.Wang B, Rothberg BS, Brenner R. Mechanism of beta4 subunit modulation of BK channels. J Gen Physiol. 2006;127:449–465. doi: 10.1085/jgp.200509436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Danta CC. SARS-CoV-2, hypoxia, and calcium signaling: the consequences and therapeutic options. ACS pharmacol Transl Sci. 2021;4:400–402. doi: 10.1021/acsptsci.0c00219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hover S, King B, Hall B, Loundras EA, Taqi H, Daly J, Dallas M, Peers C, Schnettler E, McKimmie C, et al. Modulation of potassium channels inhibits bunyavirus infection. J Biol Chem. 2016;291:3411–3422. doi: 10.1074/jbc.M115.692673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Punch EK, Hover S, Blest HTW, Fuller J, Hewson R, Fontana J, Mankouri J, Barr JN. Potassium is a trigger for conformational change in the fusion spike of an enveloped RNA virus. J Biol Chem. 2018;293:9937–9944. doi: 10.1074/jbc.RA118.002494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tezcan ME, DoganGokce G, Sen N, ZorlutunaKaymak N, Ozer RSB. Baseline electrolyte abnormalities would be related to poor prognosis in hospitalized coronavirus disease 2019 patients. New Microbes New Infect. 2019;2020(37):100753. doi: 10.1016/j.nmni.2020.100753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Su H, Yang M, Wan C, Yi LX, Tang F, Zhu HY, Yi F, Yang HC, Fogo AB, Nie X, et al. Renal histopathological analysis of 26 postmortem findings of patients with COVID-19 in China. Kidney Int. 2020;98:219–227. doi: 10.1016/j.kint.2020.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Noori M, Nejadghaderi SA, Sullman MJM, Carson-Chahhoud K, Kolahi AA, Safiri S. Epidemiology, prognosis and management of potassium disorders in Covid-19. Rev Med Virol. 2021 doi: 10.1002/rmv.2262,e2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Liu M, Yang Y, Gu C, Yue Y, Wu KK, Wu J, Zhu Y. Spike protein of SARS-CoV stimulates cyclooxygenase-2 expression via both calcium-dependent and calcium-independent protein kinase C pathways. FASEB J. 2007;21:1586–1596. doi: 10.1096/fj.06-6589com. [DOI] [PubMed] [Google Scholar]
- 86.Chen JS, Alfajaro MM, Wei J, Chow RD, Filler RB, Eisenbarth SC, Wilen CB. Cyclooxgenase-2 is induced by SARS-CoV-2 infection but does not affect viral entry or replication. bioRxiv. 2020 doi: 10.1101/2020.09.24.312769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wang C, Constantinescu SN, MacEwan DJ, Strulovici B, Dekker LV, Parker PJ, Pfeffer LM. Interferon alpha induces protein kinase C-epsilon (PKC-epsilon) gene expression and a 4.7-kb PKC-epsilon-related transcript. Proc Natl Acad Sci USA. 1993;90:6944–6948. doi: 10.1073/pnas.90.15.6944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Israelow B, Song E, Mao T, Lu P, Meir A, Liu F, Alfajaro MM, Wei J, Dong H, Homer RJ, et al. Mouse model of SARS-CoV-2 reveals inflammatory role of type I interferon signaling. J Exp Med. 2020 doi: 10.1084/jem.20201241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Makris S, Paulsen M, Johansson C. Type I Interferons as Regulators of Lung Inflammation. Front Immunol. 2017;8:259. doi: 10.3389/fimmu.2017.00259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Li C, Chu H, Liu X, Chiu MC, Zhao X, Wang D, Wei Y, Hou Y, Shuai H, Cai J, et al. Human coronavirus dependency on host heat shock protein 90 reveals an antiviral target. Emerg Microbes Infect. 2020;9:2663–2672. doi: 10.1080/22221751.2020.1850183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wyler E, Mösbauer K, Franke V, Diag A, Gottula LT, Arsiè R, Klironomos F, Koppstein D, Hönzke K, Ayoub S, et al. Transcriptomic profiling of SARS-CoV-2 infected human cell lines identifies HSP90 as target for COVID-19 therapy. Science. 2021;24:102151. doi: 10.1016/j.isci.2021.102151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Banerjee AK, Blanco MR, Bruce EA, Honson DD, Chen LM, Chow A, Bhat P, Ollikainen N, Quinodoz SA, Loney C, et al. SARS-CoV-2 disrupts splicing, translation, and protein trafficking to suppress host defenses. Cell. 2020;183:1325–1339.e1321. doi: 10.1016/j.cell.2020.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Khan MA, Khan ZA, Charles M, Pratap P, Naeem A, Siddiqui Z, Naqvi N, Srivastava S. Cytokine storm and mucus hypersecretion in COVID-19: review of mechanisms. J Inflamm Res. 2021;14:175–189. doi: 10.2147/jir.s271292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Saul S, Karim M, Huang PT, Ghita L, Chiu W, Kumar S, Bhalla N, Leyssen P, Cohen CA, Huie K, et al. Discovery of pan-ErbB inhibitors protecting from SARS-CoV-2 replication, inflammation, and lung injury by a drug repurposing screen. bioRxiv. 2021 doi: 10.1101/2021.05.15.444128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Klann K, Bojkova D, Tascher G, Ciesek S, Münch C, Cinatl J. Growth factor receptor signaling inhibition prevents SARS-CoV-2 replication. Mol Cell. 2020;80:164–174.e164. doi: 10.1016/j.molcel.2020.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chan CP, Siu KL, Chin KT, Yuen KY, Zheng B, Jin DY. Modulation of the unfolded protein response by the severe acute respiratory syndrome coronavirus spike protein. J Virol. 2006;80:9279–9287. doi: 10.1128/jvi.00659-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data available on request from the authors.