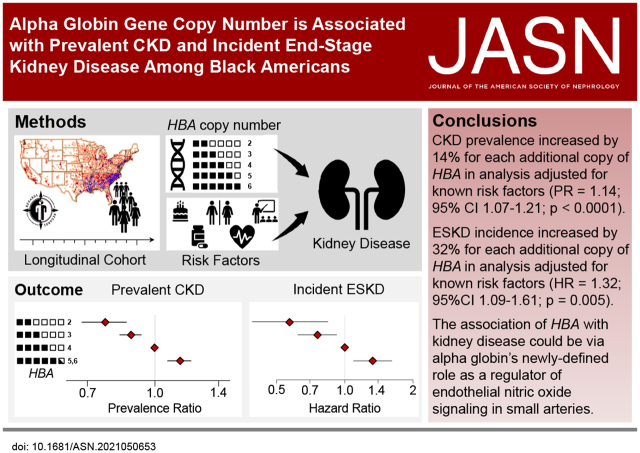
Significance Statement
Resistance artery endothelial cells express α-globin, which limits nitric oxide signaling and enhances α-adrenergic–mediated vasoconstriction—two signaling pathways involved in renal blood flow regulation and kidney disease pathogenesis. A common HBA gene deletion might therefore confer protection against kidney disease by increasing endothelial nitric oxide signaling and decreasing vasoconstriction in response to α-1–adrenergic stimuli. Among Black Americans, HBA copy number varied from 2 to 6; an increase by one HBA gene copy was associated with 14% greater risk of CKD and 32% greater hazard of incident ESKD. This study identifies HBA deletions as protective against CKD and ESKD and highlights the importance of understanding the role of α-globin in renovascular pathophysiology.
Keywords: human genetics, nitric oxide, hemodynamics and vascular regulation, alpha globin, alpha thalassemia, kidney disease, African Americans, prevalence, gene dosage
Visual Abstract
Abstract
Background
α-Globin is expressed in endothelial cells of resistance arteries, where it limits endothelial nitric oxide signaling and enhances α-adrenergic–mediated vasoconstriction. α-Globin gene (HBA) copy number is variable in people of African descent and other populations worldwide. Given the protective effect of nitric oxide in the kidney, we hypothesized that HBA copy number would be associated with kidney disease risk.
Methods
Community-dwelling Black Americans aged ≥45 years old were enrolled in a national longitudinal cohort from 2003 through 2007. HBA copy number was measured using droplet digital PCR. The prevalence ratio (PR) of CKD and the relative risk (RR) of incident reduced eGFR were calculated using modified Poisson multivariable regression. The hazard ratio (HR) of incident ESKD was calculated using Cox proportional hazards multivariable regression.
Results
Among 9908 participants, HBA copy number varied from 2 to 6. In analyses adjusted for demographic, clinical, and genetic risk factors, a one-copy increase in HBA was associated with 14% greater prevalence of CKD (PR, 1.14; 95% CI, 1.07 to 1.21; P<0.0001). While HBA copy number was not associated with incident reduced eGFR (RR, 1.06; 95% CI, 0.94 to 1.19; P=0.38), the hazard of incident ESKD was 32% higher for each additional copy of HBA (HR, 1.32; 95% CI, 1.09 to 1.61; P=0.005).
Conclusions
Increasing HBA copy number was associated with a greater prevalence of CKD and incidence of ESKD in a national longitudinal cohort of Black Americans.
Black Americans develop kidney disease at a younger age than other Americans and are three times more likely to develop end-stage kidney disease (ESKD), even after accounting for socioeconomic factors and comorbid medical conditions.1–6 DNA sequence variants that increase the risk of kidney disease (including one in the hemoglobin β locus [HBB]), sickle cell trait,7,8 and apolipoprotein L-1 (APOL1)9–11 are more common among Black Americans, yet only partly explain the racial disparity in kidney disease. The evaluation of genetic risk factors for diseases common in minority populations in the United States remains a high priority. However, Black Americans and other minority populations are underrepresented in many large-scale longitudinal US population studies, hindering genetic investigations into kidney disease and other common chronic diseases.12,13
Basic science14–20 and clinical studies21–23 have identified nitric oxide signaling as important in protection against and recovery from ischemic or oxidative injury to the kidney; however, to date there have been few clear associations between genetic variants in nitric oxide signaling pathways in the kidney and kidney function or kidney disease.24–27 α-Globin has been proposed to function as a regulator of nitric oxide signaling in the endothelial cells of small resistance arteries.28 Genetic or pharmacologic disruption of α-globin or its stabilizing protein leads to enhanced nitric oxide signaling and decreased vasoconstriction in response to α-adrenergic stimuli,29–31 potentially conferring protection against kidney injury.
α-Globin is expressed by the HBA1 and HBA2 genes that are organized in tandem on chromosome 16. The sequence similarity of the HBA1 and HBA2 genes makes them susceptible to deletions, which are common among individuals from Africa, Southeast Asia, the Pacific Islands, and the Mediterranean.32 The 3.7-kilobase insertion/deletion variant is common among Black Americans. Because of the unique challenges of genotyping this structural variant, it has been omitted from genome-wide association studies. This deletion spans parts of the HBA2 and HBA1 genes and reduces the functional gene copy number by one, leading to a decrease in total α-globin gene (HBA) expression in red cell precursors.33 The quantitative relationship between HBA copy number and α-globin expression in red blood cell (RBC) precursors is the molecular basis for the spectrum of α-thalassemia syndromes.33–35 In light of the newly recognized function of α-globin as a restrictor of endothelial nitric oxide signaling,28 we hypothesized that α-globin gene deletions would be associated with protection against prevalent CKD, incident reduced eGFR, and incident ESKD among Black Americans.
Methods
Study Design
The REasons for Geographic and Racial Differences in Stroke (REGARDS) study is a longitudinal cohort study designed to determine the reasons for racial disparities in stroke and cognitive decline in Black and White Americans aged ≥45 years.36 REGARDS enrolled 30,239 community-dwelling participants from the 48 contiguous United States from 2003 to 2007. A total of 12,514 (41%) participants were Black, and 56% were from states considered to be in the stroke belt, where stroke incidence in the United States is highest. Exclusion criteria included self-reported race other than Black or White, residence in or on the waiting list for a nursing home, active cancer within the past year, or inability to communicate in English.
Baseline variables were collected via standardized computer-assisted telephone interview, self-administered questionnaire, and in-home physical examination.36,37 Trained personnel administered computer-assisted telephone interviews to collect study participant age, sex, region of residence, insurance status, education level, income, and reports of physician-diagnosed comorbid conditions. Trained personnel conducted in-home examinations to measure height, weight, and BP; collect blood and urine specimens; and review medication bottles. Baseline measurements were repeated in some participants during a second in-home visit conducted a median (25th, 75th percentile) of 9.5 (8.7, 9.9) years after baseline.
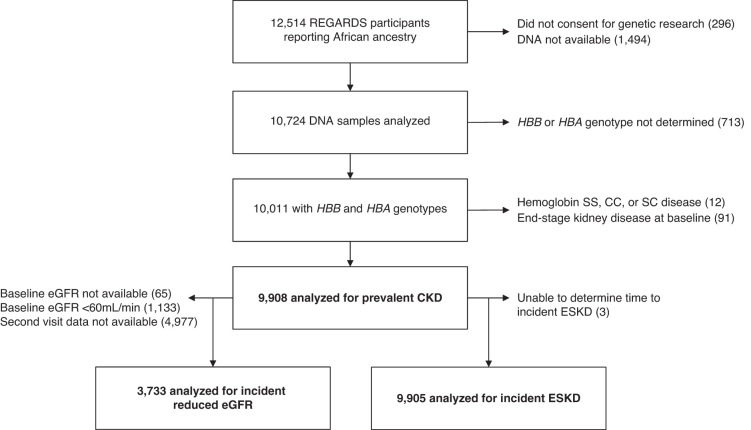
All participants provided oral and written informed consent. The REGARDS study was approved by participating center Institutional Review Boards. All self-reported Black participants consenting to genetic research were eligible to participate in this study. Participants were excluded if they had hemoglobin SS (n=5), SC (n=1), or CC (n=6); or had ESKD at enrollment (n=91) (Figure 1). This study followed the Strengthening the Reporting of Observational Studies in Epidemiology reporting guideline.
Figure 1.
Cohort flow diagram.
Main Exposure Variable
HBA copy number was evaluated as a numeric variable with values of 2, 3, 4, 5, or 6 as determined by droplet digital PCR (ddPCR) analysis of genomic DNA. The ddPCR copy number assay targeted a unique sequence within the 3.7-kb insertion/deletion polymorphism. We determined copy number of this target relative to a reference gene, EIF2C1 (human eukaryotic translation initiation factor 2C, 1). Two-dimensional clusters of droplet counts for target and reference genes were manually gated using Quantasoft (Bio-Rad) per the manufacturer's protocols. Droplet counts, copy number variant values, and 95% CIs for copy number variant were extracted and visualized, and genotype was assigned using custom scripts in the R computing environment without user intervention (see the Supplemental Material).
Outcome Measures
CKD prevalence was defined by an eGFR<60 ml/min per 1.73 m2 or a urine albumin-creatinine ratio ≥30 mg/g or at baseline. The Chronic Kidney Disease Epidemiology Collaboration equation was used to estimate GFR from serum creatinine concentrations. In addition, prevalence of eGFR<60 ml/min per 1.73 m2 and a urine albumin-creatinine ratio ≥30 mg/g at baseline were evaluated as separate outcome measures. Incident reduced eGFR was defined by an eGFR<60 ml/min per 1.73 m2 at the follow-up in-home visit and >40% decline in eGFR from baseline, among those who had eGFR≥60 ml/min per 1.73 m2 at baseline.38 This definition of incident reduced eGFR emphasized the clinically relevant transition across the 60-ml/min per 1.73 m2 threshold and did not examine changes among those who were below that threshold at baseline. The incident reduced eGFR analysis was limited to those participants with available follow-up visit data (Figure 1). Incident ESKD was identified by linkage to United States Renal Data System (USRDS) data through December 31, 2018.
Covariates
Hypertension was defined as one or more of the following: SBP≥140 mm Hg or DBP≥90 mm Hg (recorded as the average of two measurements performed according to a standardized protocol39); self-reported current use of medication to control BP; or two or more antihypertensive medications found on medication bottle review during the baseline in-home study visit. A clinical definition of hypertension was employed as a covariate instead of measured BP because measured BP would be variably affected by antihypertensive use. Hemoglobin, RBC count, and mean cell volume (MCV) values were measured with an automated hematology analyzer and mean cell hemoglobin (MCH), mean corpuscular hemoglobin concentration (MCHC), and red cell distribution width–coefficient of variation were calculated using standard formulae.40 Diabetes mellitus was defined by a fasting glucose level ≥126 mg/dl, a random glucose ≥200 mg/dl, or use of antidiabetic medication. Body mass index was calculated from measured height and weight. Smoking status was self-reported and categorized as never, past, or current smoker. Region was defined as three geographic areas: stroke belt buckle (coastal plains of North Carolina, South Carolina, and Georgia), stroke belt (the rest of North Carolina, South Carolina, and Georgia and the entire states of Tennessee, Alabama, Mississippi, Arkansas, and Louisiana), and stroke non-belt (the remaining contiguous United States).36,41 Age, sex, race, health insurance (yes or no), highest education level obtained (less than high school, high school, some college, college or more), and annual income (≤$20K, $20–34K, $35–74K, ≥$75K) were self-reported. Sickle cell trait and hemoglobin C genotypes were measured by a TaqMan single nucleotide polymorphism (SNP) genotyping assay (Applied Biosystems/Thermo Fisher Scientific).8 APOL1 high-risk genotype was defined as the presence of two renal risk alleles compared with a reference of less than two high-risk alleles (Supplemental Material).42,43
Statistical Analysis
The analytic plan and outcome measure definitions were prespecified and approved by the REGARDS Cohort Study Executive Committee in December of 2018.
Unadjusted Analyses of Participant Characteristics by HBA Copy Number
For continuous measures of participant characteristics, medians and 25th and 75th percentiles were calculated by HBA copy number. Differences between copy number groups were assessed by Kruskal–Wallis nonparametric ANOVA. Categoric variables were calculated as percentages within each category and differences were assessed by chi-squared tests of association.
Adjusted Analyses of HBA Copy Number and Kidney Disease Outcome Measures
The primary outcome measure was the presence or absence of CKD at baseline (CKD prevalence) and the main exposure variable was HBA copy number. In addition, secondary outcome measures for incident reduced eGFR and incident ESKD were evaluated. Thirteen known risk factors for kidney disease were included in multivariable regression models for CKD prevalence, incident reduced eGFR, and incident ESKD (see ‘Covariates’ section above). A modified Poisson regression model with robust variance estimation was used to estimate the prevalence ratio (PR) and 95% confidence interval (95% CI) for CKD, the separate outcomes of eGFR<60 ml/min per 1.73 m2 or urine albumin-creatinine ratio ≥30 mg/g at baseline, and the relative risk (RR) and 95% CI for incident reduced eGFR. These models estimated the difference in log-transformed PR or RR for each additional HBA allele. The use of Poisson regression with robust variance estimation allows for estimating the RR directly (reported here as the PR for prevalent CKD and RR for incident reduced eGFR), rather than the odds ratio that arises from logistic regression. The RR is regarded as a more clinically meaningful and transparent assessment of risk than the odds ratio in cohort studies.44 Incident ESKD hazard ratio (HR) and 95% CI were estimated with a multivariable Cox proportional hazards model for the time between baseline and date of ESKD onset from the USRDS. Baseline CKD and eGFR were not included as covariates in the incident ESKD model because they are intermediate variables (not independent variables) on the causal pathway to ESKD. We performed a time-to-event analysis including the time from baseline until (1) a participant dies; (2) is registered in USRDS for ESKD; or (3) follow-up stops. The timing of events for each genotype, unadjusted for covariates, is provided in Supplemental Figure 1.
Missing Data Considerations
Missing data for the outcome measures and explanatory variables were rare (<0.5%) with some exceptions, e.g., complete blood count results (Table 1). A multiple imputation approach was employed in all multivariable regression models (Supplemental Material).45
Table 1.
Clinical and demographic characteristics according to HBA copy number
| Characteristic | All Participants | HBA Copy Number | P Valueb | |||
|---|---|---|---|---|---|---|
| 2 | 3 | 4 | ≥5a | |||
| Participants, N (%) | 9908 (100) | 393 (4) | 2744 (28) | 6668 (67) | 103 (1) | NA |
| Age, years | 63.0 (57.0, 70.0) |
64.0 (58.0, 70.0) |
64.0 (57.0, 71.0) |
63.0 (57.0, 70.0) |
66.5 (61.0, 72.0) |
0.01 |
| Female sex, N (%) | 6098 (62) | 246 (63) | 1681 (61) | 4114 (62) | 57 (55) | 0.57 |
| Body mass index, kg/m2 | 29.7 (26.2, 34.4) |
30.3 (26.8, 34.7) |
29.9 (26.1, 34.4) |
29.7 (26.1, 34.3) |
30.6 (27.6, 34.3) |
0.22 |
| Smoking status, N (%) | 0.54 | |||||
| Never | 4473 (45) | 169 (43) | 1255 (46) | 3004 (45) | 45 (44) | |
| Past | 3672 (37) | 160 (41) | 1005 (37) | 2463 (37) | 44 (43) | |
| Current | 1718 (17) | 61 (16) | 470 (17) | 1173 (18) | 14 (14) | |
| Region, N (%) | 0.06 | |||||
| Non-belt | 4928 (50) | 170 (43) | 1394 (51) | 3304 (50) | 60 (58) | |
| Belt | 3286 (33) | 142 (36) | 881 (32) | 2235 (33) | 28 (27) | |
| Buckle | 1694 (17) | 81 (21) | 469 (17) | 1129 (17) | 16 (16) | |
| Medically insured, N (%) | 8919 (90) | 355 (91) | 2487 (91) | 5981 (90) | 96 (93) | 0.29 |
| Education level, N (%) | 0.29 | |||||
| Less than high school | 1935 (20) | 71 (18) | 547 (20) | 1296 (19) | 21 (20) | |
| High school graduate | 2728 (28) | 111 (28) | 787 (29) | 1801 (27) | 29 (28) | |
| Some college | 2657 (27) | 105 (27) | 754 (27) | 1769 (27) | 29 (28) | |
| College graduate or more | 2577 (26) | 105 (27) | 655 (24) | 1793 (27) | 24 (23) | |
| Income, N (%) | 0.12 | |||||
| ≤$20K | 2653 (30) | 104 (30) | 746 (31) | 1773 (30) | 30 (32) | |
| $20K–34K | 2607 (30) | 109 (31) | 722 (30) | 1743 (30) | 33 (35) | |
| $35K–74K | 2536 (29) | 106 (30) | 718 (30) | 1688 (29) | 26 (25) | |
| ≥$75K | 914 (10) | 32 (9) | 214 (9) | 662 (11) | n<11 | |
| Hemoglobin, g/dl | 13.1 (12.2, 14.0) |
12.3 (11.6, 13.2) |
12.9 (12.0, 13.8) |
13.2 (12.4, 14.1) |
13.0 (12.0, 14.1) |
<0.0001 |
| RBC count, million cells/mm3 | 4.5 (4.2, 4.8) |
5.2 (4.7, 5.6) |
4.7 (4.3, 5.0) |
4.4 (4.1, 4.7) |
4.4 (4.0, 4.7) |
<0.0001 |
| MCV, fl | 88.0 (84.0, 92.0) |
74.0 (72.0, 77.0) |
84.0 (82.0, 87.0) |
90.0 (87.0, 93.0) |
88.0 (86.0, 92.0) |
<0.0001 |
| MCH, pg | 29.6 (27.9, 30.9) |
23.8 (22.8, 24.8) |
27.9 (26.9, 29.0 |
30.3 (29.2, 31.4) |
29.9 (29.1, 30.9) |
<0.0001 |
| MCHC, g/dl | 33.4 (32.9, 33.9) |
32.1 (31.6, 32.5) |
33.0 (32.6, 33.5) |
33.7 (33.2, 34.1) |
33.7 (33.2, 34.0) |
<0.0001 |
| RDW-CV, % | 14.0 (13.3, 14.8) |
15.0 (14.4, 15.9) |
14.2 (13.5, 15.0) |
13.8 (13.2, 14.6) |
13.6 (13.2, 14.2) |
<0.0001 |
| eGFR<60 ml/min per 1.73 m2, N (%) | 1133 (11) | 37 (9) | 300 (11) | 779 (12) | 17 (17) | 0.15 |
| Urine albumin-creatinine ratio >30 mg/g, N (%) | 1794 (19) | 59 (16) | 498 (19) | 1214 (19) | 23 (24) | 0.22 |
| CKD prevalence,c N (%) | 2476 (25) | 80 (20) | 672 (25) | 1688 (25) | 36 (35) | 0.008 |
| Hypertension, N (%) | 7305 (77) | 298 (78) | 2029 (77) | 4894 (76) | 84 (84) | 0.24 |
| Diabetes mellitus, N (%) | 2846 (29) | 103 (26) | 817 (30) | 1897 (29) | 29 (28) | 0.42 |
| Sickle cell trait, N (%) | 743 (8) | 25 (6) | 211 (8) | 501 (8) | n<11 | 0.73 |
| APOL1 high-risk, N (%) | 1250 (13) | 60 (15) | 349 (13) | 830 (13) | 11 (11) | 0.39 |
Values are median (25th, 75th percentile) except where otherwise indicated. Missing data are as follows: smoking status (n=45, 0.5%); insured (n=11, 0.1%); education (n=12, 0.1%); income (n=1198 refused, 12.1%); eGFR (n=65, 0.7%); urine albumin-creatinine ratio (n=398, 4.0%); hemoglobin (n=3189, 32.2%); RBC count (n=3189, 32.2%); MCV (n=3195, 32.3%); MCH (n=3189, 32.2%); MCHC (n=3189, 32.2%); RDW-CV (n=3202, 32.3%); prevalent CKD (n=367, 3.7%); hypertension (n=389, 3.9%); diabetes mellitus (n=51, 0.5%); body mass index (n=73, 0.7%); APOL1 (n=64, 0.6%). Denominators for percentages in the table are on the basis of the number of nonmissing responses. Because of rounding, percentages may not sum to 100%. NA, not applicable; K, thousand; MCV, mean cell volume; MCH, mean cell hemoglobin; MCHC, mean corpuscular hemoglobin concentration; RDW-CV, red cell distribution width-coefficient of variation; APOL1, apolipoprotein-L1.
102 participants were found to have five HBA gene copies and two were found to have six HBA gene copies.
P values for tests of differences by HBA copy number generated from the chi-squared test for categoric variables and the Kruskal–Wallis nonparametric ANOVA test for continuous variables.
CKD prevalence was defined by a urine albumin-creatinine ratio ≥30 mg/g or an eGFR<60 ml/min per 1.73 m2 at baseline.
Sensitivity Analyses and Tests for Interaction
To address the possibility of association by population stratification, we performed a sensitivity analysis including the first ten principal components of ancestry among 7641 (77%) participants for whom Infinium Expanded Multi-Ethnic Genotyping Array data were available.
Prespecified tests for interaction between HBA copy number and sickle cell trait on the outcomes of CKD prevalence, incident reduced eGFR, and incident ESKD were performed for the fully adjusted multivariable models. Additionally, prespecified tests for interaction between HBA copy number and each of the following variables, APOL1, age, sex, and hypertension, were performed on the outcome of CKD prevalence for the fully adjusted multivariable model. A post hoc sensitivity analysis was performed in which HBA copy number was analyzed as a categoric variable instead of as a quantitative variable in the analysis of prevalent CKD and incident ESKD outcomes.
Relationships between the Structural Variant and Flanking SNPs
To exclude the possibility that the associations with kidney disease risk measured at the structural variant in the HBA1 and HBA2 loci were due to linkage disequilibrium with flanking SNPs, we analyzed SNP genotypes available on 8841 participants from the Infinium Expanded Multi-Ethnic Genotyping Array. Pairwise linkage disequilibrium was calculated between the −3.7-kb structural variant and each of 13,549 SNPs in the first 1 Mb of chromosome 16 using the r2 statistic in the R (v.3.6.2) package genetics. Associations between individual SNPs and CKD prevalence were calculated using PLINK (v.1.9) in a regression model that included age, sex, and the first four principal components of ancestry. The threshold for significance for a 1-Mb segment of the genome was estimated to be P<1.5 × 10−4 on the basis of the threshold for genome-wide significance (P<5 × 10−8) divided by the fraction of the genome (1 Mb of 3097 Mb) analyzed.46
Population Preventable Fraction
To estimate the fraction of kidney disease prevented by the protective genotypes of HBA copy numbers 2 or 3 among Black Americans, we calculated the population preventable fraction and the upper and lower bounds of the 95% CI for the fully adjusted models (further methods in the Supplemental Material).
Results
HBA Copy Number Variation
HBA copy number was variable among the 9918 participants who met inclusion and exclusion criteria (Figure 1): 393 participants (4%) had two HBA copies, 2744 (28%) had three HBA copies, 6668 (67%) had four HBA copies, 101 (1%) had five HBA copies, and two (<1%) participants had six HBA copies (Table 1). CKD prevalence increased with HBA copy number. CKD prevalence was 20% among those with two HBA copies; 25% among those with three copies; 25% among those with four copies; and 35% among those with ≥5 copies (P=0.008; Table 1). RBC parameters differed according to HBA copy number (P<0.0001): a hypochromic, microcytic anemia was observed in those with two copies of HBA.
HBA Copy Number and Kidney Disease Outcome Measures
Prevalent CKD
A total of 2476 (25%) participants had CKD at baseline. In an unadjusted analysis, each additional copy of HBA was associated with an 8% greater prevalence of CKD (PR, 1.08; 95% CI, 1.02 to 1.15; P=0.01). After adjusting for 13 CKD risk factors selected a priori, each additional copy of HBA was associated with a 14% greater prevalence of CKD (PR, 1.14; 95% CI, 1.07 to 1.21; P<0.0001; Table 2 ). In separate fully adjusted models analyzing the outcomes of prevalent eGFR<60 ml/min per 1.73 m2 or urine albumin-creatinine ratio ≥30 mg/g, each additional HBA copy increased the prevalence of reduced eGFR (PR, 1.22; 95% CI, 1.11 to 1.34; P<0.0001) and increased the prevalence of elevated urine albumin-creatinine ratio (PR, 1.09; 95% CI, 1.01 to 1.17; P=0.02; Table 2).
Table 2.
Association of HBA gene copy number with CKD prevalence—analysis adjusted for all listed covariates
| Characteristic | CKDa (n=9908) Modified Poisson |
eGFR<60 ml/min per 1.73 m2 (n=1133) Modified Poisson |
Urine albumin-creatinine ratio >30 mg/g (n=1794) Modified Poisson |
||||||
|---|---|---|---|---|---|---|---|---|---|
| PR | 95% CI | P valueb | PR | 95% CI | P valueb | PR | 95% CI | P valueb | |
| HBA copy number, per gene copy | 1.14 | (1.07 to 1.21) | <0.0001 | 1.22 | (1.11 to 1.34) | <0.0001 | 1.09 | (1.02 to 1.18) | 0.01 |
| Sickle cell trait | 1.44 | (1.30 to 1.59) | <0.0001 | 1.29 | (1.09 to 1.51) | 0.006 | 1.56 | (1.38 to 1.77) | <0.0001 |
| APOL1 high-risk | 1.17 | (1.07 to 1.29) | 0.0007 | 1.16 | (1.00 to 1.35) | 0.06 | 1.26 | (1.12 to 1.40) | <0.0001 |
| Hemoglobin, per 1 g/dl | 0.90 | (0.87 to 0.92) | <0.0001 | 0.76 | (0.72 to 0.79) | <0.0001 | 0.93 | (0.89 to 0.97) | 0.00005 |
| Age, per year | 1.03 | (1.03 to 1.04) | <0.0001 | 1.07 | (1.06 to 1.07) | <0.0001 | 1.02 | (1.01 to 1.02) | <0.0001 |
| Female sex | 0.78 | (0.72 to 0.84) | <0.0001 | 0.74 | (0.65 to 0.84) | <0.0001 | 0.73 | (0.66 to 0.81) | <0.0001 |
| Body mass indexc | 1.08 | (1.05 to 1.12) | <0.0001 | 1.08 | (1.03 to 1.15) | 0.004 | 1.08 | (1.04 to 1.13) | 0.0003 |
| Hypertension | 1.86 | (1.66 to 2.07) | <0.0001 | 2.97 | (2.32 to 3.80) | <0.0001 | 1.82 | (1.57 to 2.11) | <0.0001 |
| Diabetes mellitus | 1.65 | (1.54 to 1.76) | <0.0001 | 1.36 | (1.22 to 1.52) | <0.0001 | 1.94 | (1.77 to 2.11) | <0.0001 |
| Smoking status | |||||||||
| Never (ref) | — | — | |||||||
| Past | 0.98 | (0.91 to 1.05) | 0.52 | 0.92 | (0.82 to 1.03) | 0.16 | 1.01 | (0.92 to 1.11) | 0.81 |
| Present | 1.29 | (1.17 to 1.43) | <0.0001 | 1.04 | (0.87 to 1.24) | 0.67 | 1.43 | (1.28 to 1.60) | <0.0001 |
| Medically insured | 0.91 | (0.81 to 1.04) | 0.16 | 1.10 | (0.86 to 1.43) | 0.44 | 0.85 | (0.74 to 0.98) | 0.03 |
| Region | |||||||||
| Non-belt (ref) | — | — | |||||||
| Belt | 0.97 | (0.90 to 1.05) | 0.45 | 0.99 | (0.88 to 1.11) | 0.81 | 0.93 | (0.85 to 1.02) | 0.13 |
| Buckle | 1.00 | (0.91 to 1.09) | 0.99 | 0.94 | (0.81 to 1.10) | 0.45 | 1.03 | (0.93 to 1.16) | 0.55 |
| Education level | |||||||||
| <HS grad (ref) | — | — | |||||||
| HS grad | 0.94 | (0.86 to 1.03) | 0.16 | 0.96 | (0.83 to 1.11) | 0.61 | 0.95 | (0.85 to 1.02) | 0.13 |
| Some college | 0.93 | (0.90 to 1.08) | 0.75 | 0.99 | (0.85 to 1.15) | 0.87 | 1.01 | (0.90 to 1.14) | 0.84 |
| ≥College grad | 0.92 | (0.82 to 1.02) | 0.29 | 1.07 | (0.90 to 1.27) | 0.42 | 0.90 | (0.79 to 1.04) | 0.15 |
| Income | |||||||||
| <$20K (ref) | — | — | |||||||
| $20K - $34K | 0.98 | (0.90 to 1.07) | 0.64 | 1.04 | (0.91 to 1.19) | 0.54 | 0.95 | (0.85 to 1.05) | 0.30 |
| $35K - $74K | 0.93 | (0.84 to 1.02) | 0.13 | 0.91 | (0.76 to 1.09) | 0.31 | 0.87 | (0.77 to 1.00) | 0.05 |
| ≥$75K | 0.75 | (0.63 to 0.90) | 0.0001 | 0.78 | (0.57 to 1.07) | 0.13 | 0.64 | (0.52 to 0.79) | <0.0001 |
All variables shown in the table were included in the multivariable model. Multiple imputations were performed for missing data. The number of participants available for this analysis was 9908. APOL1, apolipoprotein-L1; ref, reference category; –, reference category; HS, high school; grad, graduate; K, thousand.
CKD prevalence was defined by a urine albumin-creatinine ratio ≥30 mg/g or an eGFR<60 ml/min per 1.73 m2 at baseline.
Analysis was performed using a modified Poisson multivariable regression model employing a linear effect of HBA allele count on the log of the PR.
Body mass index was scaled by SD.
Incident Reduced eGFR
Data were available for 3733 participants who were evaluated at a second in-home visit (Figure 1). A total of 655 (17.5%) developed incident reduced eGFR over a median (25th, 75th percentile) of 9.5 (8.7, 9.9) years. In unadjusted analysis there was no association between HBA copy number and incident reduced eGFR (RR, 1.01; 95% CI, 0.90 to 1.15; P=0.83).
After adjusting for relevant risk factors, there was no association between HBA copy number and incident reduced eGFR (RR, 1.06; 95% CI, 0.94 to 1.19; P=0.38; Table 3).
Table 3.
Association of HBA gene copy number with incident reduced eGFR—analysis adjusted for all listed covariates
| Characteristic | Incident Reduced eGFRa (n=3733) Modified Poisson |
||
|---|---|---|---|
| RR | 95% CI | P Valueb | |
| HBA copy number, per gene copy | 1.06 | (0.94 to 1.19) | 0.38 |
| Sickle cell trait | 1.08 | (0.84 to 1.39) | 0.53 |
| APOL1 high-risk | 1.02 | (0.83 to 1.26) | 0.84 |
| Hemoglobin, per 1 g/dl | 0.91 | (0.85 to 0.97) | 0.007 |
| Age, per year | 1.04 | (1.03 to 1.05) | <0.0001 |
| Female sex | 1.18 | (0.98 to 1.41) | 0.08 |
| Body mass indexc | 1.08 | (1.00 to 1.15) | 0.04 |
| Hypertension | 1.15 | (0.98 to 1.36) | 0.09 |
| Diabetes mellitus | 1.47 | (1.27 to 1.70) | <0.0001 |
| Smoking status | |||
| Never (ref) | — | — | |
| Past | 0.99 | (0.85 to 1.15) | 0.89 |
| Present | 1.25 | (1.01 to 1.54) | 0.04 |
| Medically insured | 0.99 | (0.76 to 1.29) | 0.95 |
| Region | |||
| Non-belt (ref) | — | — | |
| Belt | 0.96 | (0.82 to 1.13) | 0.65 |
| Buckle | 1.13 | (0.94 to 1.35) | 0.18 |
| Education level | |||
| <HS grad (ref) | — | — | |
| HS grad | 0.99 | (0.79 to 1.23) | 0.90 |
| Some college | 1.03 | (0.83 to 1.29) | 0.78 |
| ≥College grad | 0.93 | (0.73 to 1.19) | 0.57 |
| Income | |||
| <$20K (ref) | — | — | |
| $20K–34K | 1.00 | (0.83 to 1.20) | 0.96 |
| $35K–74K | 0.93 | (0.76 to 1.16) | 0.53 |
| ≥$75K | 1.05 | (0.79 to 1.41) | 0.72 |
All variables shown in the table were included in multivariable model. Multiple imputations were performed for missing data. The number of participants available for this analysis was 3733. n=238 participants were missing eGFR at the second visit. APOL1, apolipoprotein-L1; ref, reference category; –, reference category; HS, high school; grad, graduate; K, thousand.
Incident reduced eGFR was defined by an eGFR<60 ml/min at the follow-up in-home visit and >40% decline in eGFR from baseline, among those who had eGFR≥60 ml/min at baseline.
Analysis was performed using a modified Poisson multivariable regression model employing a linear effect of HBA allele count on the log of the RR.
Body mass index was scaled by SD.
Incident ESKD
Out of 9905 eligible participants for the ESKD analysis, 342 participants developed ESKD over a median (25th, 75th percentile) of 10.7 (5.5, 14.0) years of follow-up. In an unadjusted analysis there was no significant association between HBA copy number and the hazard of incident ESKD (HR, 1.07; 95% CI, 0.88 to 1.29; P=0.50; unadjusted survival curves are reported in Supplemental Figure 1). In contrast, an analysis adjusted for 13 risk factors selected a priori found that each additional copy of HBA was associated with a 32% increase in the hazard of incident ESKD (HR, 1.32; 95% CI, 1.09 to 1.61; P=0.005; Table 4).
Table 4.
Association of HBA gene copy number with incident ESKD– analysis adjusted for all listed covariates
| Characteristic | Incident ESKDa (n=9905) Cox Proportional Hazards |
||
|---|---|---|---|
| HR | 95% CI | P Valueb | |
| HBA copy number, per gene copy | 1.32 | (1.09 to 1.61) | 0.005 |
| Sickle cell trait | 1.85 | (1.34 to 2.56) | 0.0002 |
| APOL1 high-risk | 1.72 | (1.31 to 2.27) | 0.0001 |
| Hemoglobin, per 1 g/dl | 0.61 | (0.55 to 0.67) | <0.0001 |
| Age, per year | 1.00 | (0.99 to 1.02) | 0.66 |
| Female sex | 0.41 | (0.32 to 0.54) | <0.0001 |
| Body mass indexc | 1.05 | (0.93 to 1.18) | 0.46 |
| Hypertension | 4.01 | (2.49 to 6.44) | <0.0001 |
| Diabetes mellitus | 3.42 | (2.70 to 4.32) | <0.0001 |
| Smoking status | |||
| Never (ref) | — | — | |
| Past | 1.19 | (0.93 to 1.18) | 0.16 |
| Present | 1.97 | (1.43 to 2.72) | <0.0001 |
| Medically insured | 0.91 | (0.62 to 1.36) | 0.65 |
| Region | |||
| Non-belt (ref) | — | — | |
| Belt | 0.85 | (0.66 to 1.09) | 0.20 |
| Buckle | 0.97 | (0.72 to 1.30) | 0.85 |
| Education level | |||
| <HS grad (ref) | — | — | |
| HS grad | 1.24 | (0.90 to 1.70) | 0.19 |
| Some college | 1.04 | (0.73 to 1.46) | 0.84 |
| ≥College grad | 1.30 | (0.89 to 1.89) | 0.17 |
| Income | |||
| <$20K (ref) | — | — | |
| $20K–34K | 1.03 | (0.77 to 1.38) | 0.83 |
| $35K–74K | 0.70 | (0.49 to 1.00) | 0.05 |
| ≥$75K | 0.42 | (0.22 to 0.80) | 0.008 |
All variables in the table were included in the multivariable model. Multiple imputations were performed for missing data. The number of participants available for this analysis was n=9905. APOL1, apolipoprotein-L1; ref, reference category; –, reference category; HS, high school; grad, graduate; K, thousand.
Incident ESKD was identified by linkage to the USRDS through December 31, 2018.
Analysis was performed using Cox proportional hazards multivariable regression employing a linear effect of HBA allele count on the log of the HR.
Body mass index was scaled by SD.
Sensitivity Analyses and Tests of Interaction
The point estimates of the associations between HBA copy number and the outcomes of CKD prevalence, incident reduced eGFR, and incident ESKD were not substantially changed and remained significant for both CKD prevalence and incident ESKD after adjustment for the first ten principal components of ancestry (Supplemental Table 1). There was no interaction between HBA copy number and sickle cell trait for the outcomes of CKD prevalence, incident reduced eGFR, or incident ESKD (Supplemental Table 2), nor was there an interaction between HBA copy number and each of age, sex, hypertension, or APOL1 on the outcome of CKD prevalence (Supplemental Table 3). The P value of each crossproduct interaction term was ≥0.20 in the fully adjusted models. When HBA copy number was analyzed as a categoric variable instead of as a quantitative variable, the risks associated with each copy number genotype for prevalent CKD and incident ESKD were similar to risks determined from prespecified models that analyzed copy number as a quantitative variable; however, the risk estimates for the categoric analysis of the 5–6 copy category lacked precision due to the small number of observations for these genotypes (Supplemental Tables 4 and 5).
Relationships between the HBA Structural Variant and Nearby Sequence Variants
To assess whether associations with HBA copy number could be attributed to sequence variants flanking the HBA1 and HBA2 genes, we examined pairwise linkage disequilibrium between the −3.7-kb structural variant and 13,549 SNPs within the first 1 Mb of the chromosome. Pairwise linkage disequilibrium was weak between the structural variant and each SNP; the maximum r2 value was 0.25 and most SNPs had r2 values close to zero (Supplemental Figure 2, A and B). Each SNP was tested for association with CKD prevalence. None of the SNPs reached the threshold for significance for a 1-Mb genome region (Supplemental Figure 2, C and D), suggesting that the association between the structural variant and kidney disease risk was not due to linkage disequilibrium with SNPs in flanking gene regions.
Population Preventable Fraction
To assess the fraction of kidney disease prevented by the protective two- and three-copy HBA genotypes, we estimated the population preventable fraction. The two- and three-copy HBA genotypes prevent 4.3% (95% CI, 2.0 to 6.5) of CKD prevalence and 11.4% (95% CI, 2.2 to 20.6) of incident ESKD among Black Americans (Supplemental Material).
Discussion
In this national longitudinal cohort study of Black Americans, each additional HBA copy was associated with a 14% greater prevalence of CKD and a 32% greater risk of incident ESKD after adjustment for established genetic, biomedical, demographic, and social risk factors. In addition, there was a significant association between HBA copy number and each component of the prevalent CKD outcome, low eGFR and elevated urine albumin-creatinine ratio. Kidney disease risk increased with higher HBA copy number, suggesting that the level of HBA gene expression, rather than a specific allele, was responsible for this genetic association. These findings support the hypothesis that higher HBA copy number is associated with greater risk of kidney disease among Black Americans; in other words, the common 3.7-kb deletion is protective. Moreover, the protective two- and three-copy HBA genotypes (i.e., homozygotes and heterozygotes for the 3.7-kb gene deletion) were estimated to prevent approximately 4% of prevalent CKD and 11% of incident ESKD among Black Americans at the population level.
HBA copy number was associated with both a greater prevalence of CKD at baseline and greater incidence of ESKD over time; HBA copy number was not associated with incident reduced eGFR. To be included in the incident reduced eGFR analysis, participants had to undergo a second in-home visit. Participants in the group providing second in-home visit data were healthier (less smoking, hypertension, and diabetes mellitus at baseline) and had a higher socioeconomic status (higher education and income) at baseline than those who did not take part in the second in-home visit (see Supplemental Table 6). Together, the reduced disease severity, smaller sample size available for analysis (n=3733), and the stringent prespecified definition of reduced eGFR (>40% decrease among those with eGFR>60 ml/min per 1.73 m2 at baseline) may have reduced the power to detect an effect of HBA copy number on this secondary end point.
We performed ddPCR to quantify HBA copy number, which allowed a variety of underlying genotypes (−a/−a, −a/aa, aa/aa, aa/aaa, and aaa/aaa) to be summarized as an integer copy number (2, 3, 4, 5, 6) that reflects the number of functional HBA gene copies per genome. We evaluated a prespecified additive relationship between HBA copy number and kidney disease risk. Although the specific relationship between HBA copy number, HBA transcript abundance, and α-globin protein function in the kidney has yet to be defined, hematologic observations demonstrate a quantitative relationship between HBA copy number and α-globin protein abundance.35,47,48 Alternative models, in which HBA copy number was analyzed as a categoric variable instead of as a quantitative variable, further supported the conclusion that CKD and ESKD risk increased with HBA copy number (Supplemental Tables 4 and 5). Genetic associations can arise from population structure, i.e., underlying differences in genetic ancestry between those who develop kidney disease and those who do not. To address this, we performed a prespecified sensitivity analysis that included principal components of ancestry and confirmed that the association between HBA copy number and CKD prevalence was not due to population structure (Supplemental Table 1). The association with HBA copy number was unlikely due to linkage disequilibrium with sequence variants in nearby genes, given the established relationship between HBA copy number and functional gene expression; the rapid decline of linkage disequilibrium away from the HBA loci49; and the additive mode of inheritance (i.e., a dose-response relationship between HBA copy number and CKD risk) which transcends any single HBA allele. Nevertheless, we measured pairwise linkage disequilibrium between the structural variant and SNPs directly and measured association between each SNP and CKD prevalence. We found no SNPs that could explain the observed association between HBA copy number and kidney disease risk (Supplemental Figure 2). Changes in the level of HBA gene expression determined by functional gene copy number remains the most plausible genetic mechanism responsible for the associations with kidney disease risk.
α-Thalassemia is known to modify the risk of sickle cell disease nephropathy50,51; therefore, we considered whether the reduction in HBA copy number mitigated the effect of sickle cell trait on kidney disease risk. We found the association between HBA copy number and kidney disease risk to be independent of sickle cell trait, and there was no interaction between HBA copy number and sickle cell trait on kidney disease outcomes in the prespecified analysis. A previous study reported that HBA genotype modified CKD risk among those with sickle cell trait, but found no association between HBA and CKD in the general population.52 This study differs substantially from the prior report in the following ways: (1) ddPCR was used to quantify HBA copy number from 2 to 6; (2) a prespecified log-linear effect of HBA copy number was employed; (3) this study population was more than three times larger, with older individuals and a higher prevalence of CKD; and (4) additional established social, demographic, and clinical factors were included in the current analysis. Together, the precise estimation of copy number, increased sample size, and additional covariates may have increased the power to detect an association between HBA copy number and kidney disease risk in a general population sample of Black Americans.
There are several potential mechanisms through which HBA copy number variation could modify kidney disease risk among Black Americans. One such mechanism is through decreased erythroid expression of HBA leading to anemia, a risk factor for kidney disease. However, HBA alleles associated with protection against kidney disease tended to be associated with lower, not higher, hemoglobin levels. Moreover, the associations between HBA copy number and kidney disease risk remained significant when adjusted for hemoglobin level. We considered whether anemia associated with CKD could have contributed to overestimation of the effect of HBA copy number in the CKD prevalence model. In a post hoc sensitivity analysis omitting hemoglobin, the association between HBA copy number and CKD prevalence remained (Supplemental Table 7). Red cell microcytosis associated with lower HBA copy numbers could potentially confer protection against kidney disease through improved blood rheology53; however, this mechanism does not explain the increased risk associated with higher HBA copy numbers (5, 6) that have normal MCV. Thus, a clear hematologic mechanism explaining the association between HBA copy number and kidney disease risk is not apparent.
α-Globin is expressed not only in red cell precursors, but also in the endothelium of resistance arteries where it regulates nitric oxide signaling.28 In human omental arteries, α-globin binds with eNOS at the myoendothelial junction where it regulates vasoconstriction to phenylephrine in a NOS-dependent fashion.54(preprint) In mice, pharmacologic disruption of the α-globin/eNOS complex in endothelial cells increased nitric oxide release and led to a fall in BP in both normal and hypertensive animals, but had no effect in endothelial eNOS–knockout mice.29,30 Genetic deletion of Hba or α-hemoglobin stabilizing protein (Ahsp) in mice altered small artery vasoreactivity by reducing vasoconstriction to α-adrenergic stimuli in a NOS-dependent manner.28,31 Individuals with HBA deletions had enhanced tissue perfusion after transient arterial occlusion or in response to temperature changes, consistent with increased endothelial nitric oxide release.55,56 Together, these human and mouse studies identify variation in α-globin expression to be associated with changes in vascular nitric oxide signaling and vasoconstriction to α-1–adrenergic stimuli. We found HBA copy number to be independently associated with each component of CKD, low eGFR or elevated urine albumin-creatine ratio. We speculate that decreased endothelial HBA expression in individuals with decreased HBA copy number could lead to increases in endothelial nitric oxide signaling in human arteries and decreased vasoconstriction in response to sympathetic nervous system activity. This could confer protection against kidney disease in a manner consistent with the protective role of increased nitric oxide signaling in experimental and human clinical studies of kidney injury.15,16,21,22,57 Further investigation is necessary to understand how reduced HBA expression affects renal function, response to injury, and progression of kidney disease.
This study has strengths and limitations. The REGARDS study is one of the largest available cohorts of Black Americans and has clearly defined kidney disease outcome measures and data on social, biomedical, and genetic factors. This allows adjustment for established kidney disease risk factors when estimating risk. We prespecified the analysis plan to reduce the chance of spurious associations. We employed a robust and quantitative ddPCR method to genotype the insertion/deletion polymorphism and the gene triplication in the HBA loci. We evaluated genetic risk factors for CKD specific to Black individuals, including sickle cell trait and high-risk APOL1 status, and determined that HBA copy number was associated with kidney disease independently of these factors. This analysis focused on a Black American population, in which the 3.7-kb gene deletion is common. A limitation of this study is the lack of an independent replication cohort. Although the REGARDS study provides one of the largest longitudinal cohorts with which to study kidney disease in Black Americans, the novel associations reported here would be further strengthened by replication in similarly powered cohorts of Black Americans and in other populations in which HBA deletions are common.
The major genetic variants that determine kidney disease risk among Black Americans arose in Africa as mutations conferring resistance to infectious diseases: HBA and HBB variants (like those that cause α-thalassemia and sickle cell trait) confer protection against malaria whereas APOL1 variants protect against trypanosomiasis, also known as African sleeping sickness.58–60 In contrast to the variant alleles of HBB (such as sickle cell trait) and APOL1, which confer protection against infectious diseases at the cost of increased kidney disease risk, the 3.7-kb deletion in HBA is associated with protection against both malaria and kidney disease (Supplemental Figure 3).
In conclusion, we report that higher HBA copy number was independently associated with greater CKD prevalence and ESKD incidence after accounting for known clinical, demographic, and genetic risk factors in this national longitudinal study of Black Americans. The high frequency of the HBA gene deletion found in Black Americans may act to reduce the overall burden of kidney disease in this population.
Disclosures
M. Cushman reports Scientific Advisor or Membership: International Society on Thrombosis and Haemostasis—journal editor, Research and Practice in Thrombosis and Haemostasis. O.M. Gutierrez discloses receiving grant funding and consulting fees from Akebia Therapeutics; grant funding and consulting fees from Amgen; grant funding from GlaxoSmithKline; consulting fees from QED Therapeutics; and Honoraria from Akebia, Amgen, Ardelyx, AstraZeneca, and Reata. J.B. Kopp reports a Patent (Vpr antibodies) with the National Institutes of Health; Scientific Advisor or Membership as a Board Member of the National Kidney Foundation serving the National Capitol Area, NIH Foundation for the Advancement of Education in the Sciences; Editorial Boards of the American Journal of Nephrology and American Journal of Physiology—Renal; and Other Interests/Relationships with the National Institute of Diabetes and Digestive and Kidney Diseases via Confidentiality disclosure agreement with Intermune, Ionis, and Third Rock; Collaboration research and development agreement with Sanofi; and Research collaboration agreement with Astra Zeneca, Genentech, Merck, and Third Rock. B.T. Mott reports Consultancy Agreements with Geminus Therapeutics; Ownership Interest in Geminus Therapeutics; and Patents and Inventions with Chinook Therapeutics and Geminus Therapeutics. R.P. Naik reports Consultancy Agreements with Elsevier; and Research Funding from Rigel. L.H. Pecker reports Scientific Advisor or Membership as Sickle Cell Reproductive Health Education Directive. N.A. Zakai reports Honoraria from the American Society of Hematology and the International Society on Thrombosis and Haemostasis; and Scientific Advisor or Membership via Editorial Board for Research and Practice of Thrombosis and Haemostasis, Various committees for the Hemophilia and Thrombosis Research Society, and the American Society of Hematology. All remaining authors have nothing to disclose.
Funding
This is an ancillary study supported by cooperative agreement U01 NS041588 cofunded by the National Institute of Neurological Disorders and Stroke (NINDS) and the National Institute on Aging (NIA), National Institutes of Health, Department of Health and Human Service. This research was supported in part by the Divisions of Intramural Research, National Institute of Allergy and Infectious Diseases (NIAID) (project AI001150) (A.P. Ruhl, H.C. Ackerman) and the National Heart, Lung, and Blood Institute (NHLBI) (project HL006196) (A.P. Ruhl, Y. Yang, H.C. Ackerman). This work was also funded in part by the National Cancer Institute (NCI) Intramural Research Program under contract HHSN26120080001E (C.A. Winkler), the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) Z01 DK04312 (J.B. Kopp), and the NHLBI grants K08HL12510 (R.P. Naik) and K08HL096841 (N.A. Zakai).
Supplementary Material
Acknowledgments
The authors thank the investigators, staff, and participants of the Reasons for Geographic and Racial Differences in Stroke (REGARDS) study for their valuable contributions. A full list of participating REGARDS investigators and institutions can be found at http://www.regardsstudy.org. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NINDS,NIA,NIAID,NCI,NIDDK, or NHLBI. The content of this publication does not necessarily reflect the view or policy of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the government. Some of the data reported here have been supplied by the United States Renal Data System (USRDS). The interpretation and reporting of these data are the responsibility of the authors and in no way should be seen as an official policy or interpretation of the US government.
Dr. A. Parker Ruhl and Dr. Hans C. Ackerman conceived and designed the study and drafted the first version of the manuscript. Dr. Yu Yang, Dr. Bryan T. Mott, Dr. Cheryl A. Winkler, and Dr. Jeffrey B. Kopp performed sample analyses. Dr. Leslie A. Lange performed the principal components of ancestry analysis. Dr. Amit Patki performed the linkage disequilibrium analysis. Dr. Neal Jeffries designed and implemented the statistical analyses. All authors contributed to data interpretation and revised and approved the manuscript.
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
Supplemental Material
This article contains the following supplemental material online at http://jasn.asnjournals.org/lookup/suppl/doi:10.1681/ASN.2021050653/-/DCSupplemental.
I. Supplemental Figure and Tables
Supplemental Table 1. Association of HBA copy number with CKD prevalence, incident reduced eGFR, and incident ESKD—fully adjusted models including ten principal components of ancestry.
Supplemental Table 2. Prespecified test for interaction between HBA copy number and SCT on the outcomes of CKD prevalence, incident reduced eGFR, and incident ESKD in fully adjusted models.
Supplemental Table 3. Prespecified tests for interaction between each of age, sex, hypertension, or APOL1 and HBA on the outcome of CKD prevalence in fully adjusted models.
Supplemental Table 4. Post-hoc sensitivity analysis of the association of categorical HBA copy number with CKD prevalence.
Supplemental Table 5. Post-hoc sensitivity analysis of the association of categorical HBA copy number with ESKD incidence.
Supplemental Table 6. Comparison of participants with and without second visit data.
Supplemental Table 7. Post-hoc sensitivity analysis of the association of HBA copy number with CKD prevalence when hemoglobin is omitted from the model.
Supplemental Figure 1. Unadjusted survival curves of ESKD onset by genotype.
Supplemental Figure 2. Linkage disequilibrium and association analysis of sequence variants in the first 1 Mb of chromosome 16 flanking HBA1 and HBA2.
Supplemental Figure 3. Comparison of prevalence ratios for HBA, APOL1, and HBB genetic risk factors for CKD.
II. Estimation of population preventable fraction of HBA copy number on kidney disease.
III. Additional Methods.
a. HBA Genotyping Methods
b. APOL1 Genotyping Methods
c. ESKD Time-to-event Analysis Methods
d. Multiple Imputation Procedure
e. Diagnostic Modeling Description
f. Assessment of the Missing at Random Assumption
IV. Supplemental References.
References
- 1.Hsu CY, Lin F, Vittinghoff E, Shlipak MG: Racial differences in the progression from chronic renal insufficiency to end-stage renal disease in the United States. J Am Soc Nephrol 14: 2902–2907, 2003 [DOI] [PubMed] [Google Scholar]
- 2.Tarver-Carr ME, Powe NR, Eberhardt MS, LaVeist TA, Kington RS, Coresh J, et al. : Excess risk of chronic kidney disease among African-American versus white subjects in the United States: a population-based study of potential explanatory factors. J Am Soc Nephrol 13: 2363–2370, 2002 [DOI] [PubMed] [Google Scholar]
- 3.United States Renal Data System: 2018 USRDS Annual Data Report: Epidemiology of kidney disease in the United States. National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases, Bethesda, MD, 2018. Available at: https://www.usrds.org/annual-data-report/previous-adrs/. Accessed: July 1, 2021
- 4.Burrows NR, Li Y, Williams DE: Racial and ethnic differences in trends of end-stage renal disease: United States, 1995 to 2005. Adv Chronic Kidney Dis 15: 147–152, 2008 [DOI] [PubMed] [Google Scholar]
- 5.Nally JV Jr: Chronic kidney disease in African Americans: puzzle pieces are falling into place. Cleve Clin J Med 84: 855–862, 2017 [DOI] [PubMed] [Google Scholar]
- 6.McClellan W, Warnock DG, McClure L, Campbell RC, Newsome BB, Howard V, et al. : Racial differences in the prevalence of chronic kidney disease among participants in the Reasons for Geographic and Racial Differences in Stroke (REGARDS) Cohort Study. J Am Soc Nephrol 17: 1710–1715, 2006 [DOI] [PubMed] [Google Scholar]
- 7.Naik RP, Derebail VK, Grams ME, Franceschini N, Auer PL, Peloso GM, et al. : Association of sickle cell trait with chronic kidney disease and albuminuria in African Americans. JAMA 312: 2115–2125, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Naik RP, Irvin MR, Judd S, Gutiérrez OM, Zakai NA, Derebail VK, et al. : Sickle cell trait and the risk of ESRD in blacks. J Am Soc Nephrol 28: 2180–2187, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Genovese G, Friedman DJ, Ross MD, Lecordier L, Uzureau P, Freedman BI, et al. : Association of trypanolytic ApoL1 variants with kidney disease in African Americans. Science 329: 841–845, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Parsa A, Kao WHL, Xie D, Astor BC, Li M, Hsu CY, et al. ; AASK Study Investigators; CRIC Study Investigators : APOL1 risk variants, race, and progression of chronic kidney disease. N Engl J Med 369: 2183–2196, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kopp JB, Winkler CA: Genetics, genomics, and precision medicine in end-stage kidney disease. Semin Nephrol 38: 317–324, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kannel WB: The Framingham Study: its 50-year legacy and future promise. J Atheroscler Thromb 6: 60–66, 2000 [DOI] [PubMed] [Google Scholar]
- 13.Rocca WA, Yawn BP, St Sauver JL, Grossardt BR, Melton LJ 3rd: History of the Rochester Epidemiology Project: half a century of medical records linkage in a US population. Mayo Clin Proc 87: 1202–1213, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Amador-Martínez I, Pérez-Villalva R, Uribe N, Cortés-González C, Bobadilla NA, Barrera-Chimal J: Reduced endothelial nitric oxide synthase activation contributes to cardiovascular injury during chronic kidney disease progression. Am J Physiol Renal Physiol 317: F275–F285, 2019 [DOI] [PubMed] [Google Scholar]
- 15.Emans TW, Janssen BJ, Joles JA, Krediet CTP: Nitric oxide synthase inhibition induces renal medullary hypoxia in conscious rats. J Am Heart Assoc 7: e009501, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krishnan SM, Kraehling JR, Eitner F, Bénardeau A, Sandner P: The impact of the nitric oxide (NO)/soluble guanylyl cyclase (sGC) signaling cascade on kidney health and disease: a preclinical perspective. Int J Mol Sci 19: E1712, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ortiz PA, Garvin JL: Cardiovascular and renal control in NOS-deficient mouse models. Am J Physiol Regul Integr Comp Physiol 284: R628–R638, 2003 [DOI] [PubMed] [Google Scholar]
- 18.Forbes MS, Thornhill BA, Park MH, Chevalier RL: Lack of endothelial nitric-oxide synthase leads to progressive focal renal injury. Am J Pathol 170: 87–99, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang W, Mitra A, Poole B, Falk S, Lucia MS, Tayal S, et al. : Endothelial nitric oxide synthase-deficient mice exhibit increased susceptibility to endotoxin-induced acute renal failure. Am J Physiol Renal Physiol 287: F1044–F1048, 2004 [DOI] [PubMed] [Google Scholar]
- 20.Zhou H-L, Zhang R, Anand P, Stomberski CT, Qian Z, Hausladen A, et al. : Metabolic reprogramming by the S-nitroso-CoA reductase system protects against kidney injury. Nature 565: 96–100, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lei C, Berra L, Rezoagli E, Yu B, Dong H, Yu S, et al. : Nitric oxide decreases acute kidney injury and stage 3 chronic kidney disease after cardiac surgery. Am J Respir Crit Care Med 198: 1279–1287, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baylis C: Nitric oxide synthase derangements and hypertension in kidney disease. Curr Opin Nephrol Hypertens 21: 1–6, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gu X, Herrera GA: Expression of eNOS in kidneys from hypertensive patients. Int J Nephrol Renovasc Dis 3: 11–19, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Casas JP, Cavalleri GL, Bautista LE, Smeeth L, Humphries SE, Hingorani AD: Endothelial nitric oxide synthase gene polymorphisms and cardiovascular disease: a HuGE review. Am J Epidemiol 164: 921–935, 2006 [DOI] [PubMed] [Google Scholar]
- 25.Zhou T-B, Yin S-S: Association of endothelial nitric oxide synthase Glu298Asp gene polymorphism with the risk of end-stage renal disease. Ren Fail 35: 573–578, 2013 [DOI] [PubMed] [Google Scholar]
- 26.Yun Z, Yu-Ping Y, Zong-Wu T, Yang S, Fang Y, Fang S: Association of endothelial nitric oxide synthase gene polymorphisms with end-stage renal disease: a systematic review and meta-analysis. Ren Fail 36: 987–993, 2014 [DOI] [PubMed] [Google Scholar]
- 27.Chand S, Chue CD, Edwards NC, Hodson J, Simmonds MJ, Hamilton A, et al. : Endothelial nitric oxide synthase single nucleotide polymorphism and left ventricular function in early chronic kidney disease. PloS One 10: e0116160, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Straub AC, Lohman AW, Billaud M, Johnstone SR, Dwyer ST, Lee MY, et al. : Endothelial cell expression of haemoglobin α regulates nitric oxide signalling. Nature 491: 473–477, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Straub AC, Butcher JT, Billaud M, Mutchler SM, Artamonov MV, Nguyen AT, et al. : Hemoglobin α/eNOS coupling at myoendothelial junctions is required for nitric oxide scavenging during vasoconstriction. Arterioscler Thromb Vasc Biol 34: 2594–2600, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Keller TC 4th, Butcher JT, Broseghini-Filho GB, Marziano C, DeLalio LJ, Rogers S, et al.: Modulating vascular hemodynamics with an alpha globin mimetic peptide (HbαX). Hypertension 68:1494<en dash>1503, 2016 [DOI] [PMC free article] [PubMed]
- 31.Lechauve C, Butcher JT, Freiwan A, Biwer LA, Keith JM, Good ME, et al. : Endothelial cell α-globin and its molecular chaperone α-hemoglobin-stabilizing protein regulate arteriolar contractility. J Clin Invest 128: 5073–5082, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Piel FB, Weatherall DJ: The α-thalassemias. N Engl J Med 371: 1908–1916, 2014 [DOI] [PubMed] [Google Scholar]
- 33.Dozy AM, Kan YW, Embury SH, Mentzer WC, Wang WC, Lubin B, et al. : α-Globin gene organisation in blacks precludes the severe form of -thalassaemia. Nature 280: 605–607, 1979 [DOI] [PubMed] [Google Scholar]
- 34.King AJ, Higgs DR: Potential new approaches to the management of the Hb Bart’s hydrops fetalis syndrome: the most severe form of α-thalassemia. Hematology (Am Soc Hematol Educ Program) 2018: 353–360, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ma SK, Au WY, Chan AY, Chan LC: Clinical phenotype of triplicated -globin genes and heterozygosity for β0-thalassemia in Chinese subjects. Int J Mol Med 8: 171–175, 2001 [PubMed] [Google Scholar]
- 36.Howard VJ, Cushman M, Pulley L, Gomez CR, Go RC, Prineas RJ, et al. : The reasons for geographic and racial differences in stroke study: objectives and design. Neuroepidemiology 25: 135–143, 2005 [DOI] [PubMed] [Google Scholar]
- 37.Gillett SR, Boyle RH, Zakai NA, McClure LA, Jenny NS, Cushman M: Validating laboratory results in a national observational cohort study without field centers: the Reasons for Geographic and Racial Differences in Stroke cohort. Clin Biochem 47: 243–246, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schrauben SJ, Shou H, Zhang X, Anderson AH, Bonventre JV, Chen J, et al. ; CKD Biomarkers Consortium and the Chronic Renal Insufficiency Cohort (CRIC) Study Investigators : Association of multiple plasma biomarker concentrations with progression of prevalent diabetic kidney disease: findings from the Chronic Renal Insufficiency Cohort (CRIC) study. J Am Soc Nephrol 32: 115–126, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chobanian AV, Bakris GL, Black HR, Cushman WC, Green LA, Izzo JL Jr, et al. ; National Heart, Lung, and Blood Institute Joint National Committee on Prevention, Detection, Evaluation, and Treatment of High Blood Pressure; National High Blood Pressure Education Program Coordinating Committee : The Seventh Report of the Joint National Committee on Prevention, Detection, Evaluation, and Treatment of High Blood Pressure: the JNC 7 report. JAMA 289: 2560–2572, 2003 [DOI] [PubMed] [Google Scholar]
- 40.Zakai NA, McClure LA, Prineas R, Howard G, McClellan W, Holmes CE, et al. : Correlates of anemia in American blacks and whites: the REGARDS Renal Ancillary Study. Am J Epidemiol 169: 355–364, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Howard G, Howard VJ: Twenty years of progress toward understanding the Stroke Belt. Stroke 51: 742–750, 2020 [DOI] [PubMed] [Google Scholar]
- 42.Kopp JB, Nelson GW, Sampath K, Johnson RC, Genovese G, An P, et al. : APOL1 genetic variants in focal segmental glomerulosclerosis and HIV-associated nephropathy. J Am Soc Nephrol 22: 2129–2137, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.David VA, Binns-Roemer EA, Winkler CA: Taqman assay for genotyping CKD-associated APOL1 SNP rs60910145: a cautionary note. Kidney Int Rep 4: 184–185, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zou G: A modified Poisson regression approach to prospective studies with binary data. Am J Epidemiol 159: 702–706, 2004 [DOI] [PubMed] [Google Scholar]
- 45.Van Buuren S, Goorthius-Oudshoom K: mice: Multivariate Imputation by Chained Equations in R. J Stat Softw 45: 1–67, 2011 [Google Scholar]
- 46.Pe’er I, Yelensky R, Altshuler D, Daly MJ: Estimation of the multiple testing burden for genomewide association studies of nearly all common variants. Genet Epidemiol 32: 381–385, 2008 [DOI] [PubMed] [Google Scholar]
- 47.Liebhaber SA, Kan YW: Differentiation of the mRNA transcripts originating from the alpha 1- and alpha 2-globin loci in normals and alpha-thalassemics. J Clin Invest 68: 439–446, 1981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Clark B, Shooter C, Smith F, Brawand D, Steedman L, Oakley M, et al. : Beta thalassaemia intermedia due to co-inheritance of three unique alpha globin cluster duplications characterised by next generation sequencing analysis. Br J Haematol 180: 160–164, 2018 [DOI] [PubMed] [Google Scholar]
- 49.Danjou F, Zoledziewska M, Sidore C, Steri M, Busonero F, Maschio A, et al. : Genome-wide association analyses based on whole-genome sequencing in Sardinia provide insights into regulation of hemoglobin levels. Nat Genet 47: 1264–1271, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Guasch A, Zayas CF, Eckman JR, Muralidharan K, Zhang W, Elsas LJ: Evidence that microdeletions in the α globin gene protect against the development of sickle cell glomerulopathy in humans. J Am Soc Nephrol 10: 1014–1019, 1999 [DOI] [PubMed] [Google Scholar]
- 51.Geard A, Pule GD, Chetcha Chemegni B, Ngo Bitoungui VJ, Kengne AP, Chimusa ER, et al. : Clinical and genetic predictors of renal dysfunctions in sickle cell anaemia in Cameroon. Br J Haematol 178: 629–639, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Raffield LM, Ulirsch JC, Naik RP, Lessard S, Handsaker RE, Jain D, et al. ; NHLBI Trans-Omics for Precision Medicine (TOPMed) Consortium, Hematology & Hemostasis, Diabetes, and Structural Variation TOPMed Working Groups : Common α-globin variants modify hematologic and other clinical phenotypes in sickle cell trait and disease. PloS Genet 14: e1007293, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lamarre Y, Romana M, Lemonne N, Hardy-Dessources MD, Tarer V, Mougenel D, et al. : Alpha thalassemia protects sickle cell anemia patients from macro-albuminuria through its effects on red blood cell rheological properties. Clin Hemorheol Microcirc 57: 63–72, 2014 [DOI] [PubMed] [Google Scholar]
- 54.Brooks SD, Kamenyeva O, Ganesan S, et al. : Hemoglobin interacts with endothelial nitric oxide synthase to regulate vasodilation in human resistance arteries. medRxiv. 2021.04.06.21255004 (Preprint posted April 9, 2021) [Google Scholar]
- 55.Denton CC, Shah P, Suriany S, et al. : Loss of alpha-globin genes in human subjects is associated with improved nitric oxide-mediated vascular perfusion. Am J Hematol 96: 277–281, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Romana M, Reminy K, Moeckesch B, Charlot K, Hardy-Dessources MD, Doumdo L, et al. : Loss of alpha globin genes is associated with improved microvascular function in patients with sickle cell anemia. Am J Hematol 96: E165–E168, 2021 [DOI] [PubMed] [Google Scholar]
- 57.Singh RR, Easton LK, Booth LC, Schlaich MP, Head GA, Moritz KM, et al. : Renal nitric oxide deficiency and chronic kidney disease in young sheep born with a solitary functioning kidney. Sci Rep 6: 26777, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Allison AC: Protection afforded by sickle-cell trait against subtertian malareal infection. BMJ 1: 290–294, 1954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wambua S, Mwangi TW, Kortok M, Uyoga SM, Macharia AW, Mwacharo JK, et al. : The effect of α+-thalassaemia on the incidence of malaria and other diseases in children living on the coast of Kenya. PloS Med 3: e158, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vanhamme L, Paturiaux-Hanocq F, Poelvoorde P, Nolan DP, Lins L, Van Den Abbeele J, et al. : Apolipoprotein L-I is the trypanosome lytic factor of human serum. Nature 422: 83–87, 2003 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.