Abstract
Trichophyton rubrum is the commonest cause of dermatophytosis of skin and nail tissue. Molecular characterization of the T. rubrum ribosomal DNA nontranscribed-spacer region revealed two novel tandemly repetitive subelements (TRSs): TRS-1, containing a 27-bp palindromic sequence, and TRS-2. Specific amplification of TRS-1 produced strain-characteristic banding patterns (PCR types), with 21 TRS-1 PCR types recognized from 101 clinical isolates. Four simple patterns representing 1 to 4 copies of TRS-1 accounted for 75 (75%) of all 101 strains, whereas more complex patterns were observed for 21 (20%) of the 101 isolates. The copy number of TRS-2 was 0 to 3 repeats per cistron, with a majority of isolates having two copies of this element. Eleven isolates were polymorphic for TRS-2, and in combination, 23 separate PCR types were recognized by amplification of both TRS-1 and TRS-2. The PCR patterns from both elements were stable and reproducible. Elements with homology to TRS-1 were present in three phylogenetically related species, Trichophyton violaceum, Trichophyton gourvilii, and Trichophyton soudanense, but these elements were not identified in other dermatophyte taxa. There was no clear correlation of PCR type with specimen (skin or nail tissue), but certain PCR types appeared to show a bias in geographic distribution. This new method of typing T. rubrum will enable important questions about pathogenesis and epidemiology of this fungus to be addressed.
The dermatophytes are keratinophilic fungi belonging to three genera, Trichophyton, Microsporum, and Epidermophyton. The anthropophilic species Trichophyton rubrum is an important etiologic agent of tinea (ringworm) infections, including onychomycosis (nail infection), tinea cruris (groin infection), and tinea pedis (“athlete's foot”). These conditions are relatively benign and easy to treat, except in immunocompromised patients (6), but therapy is costly and can fail in a significant proportion of cases (14, 19).
A lack of adequate methods for strain identification in T. rubrum has impeded efforts to identify strain-related differences in infective potential or transmissibility. A typing system would be useful to determine, for example, whether certain strains are more likely to cause onychomycosis than skin infection. In cases of recurrent posttreatment onychomycosis (19), strain fingerprinting could determine whether the original isolate is responsible for reinfection (i.e., whether treatment failed) or a new strain has been acquired. There are several potential epidemiological applications for typing, including identifying strains endemic to a particular area and determining common sources of infection.
Attempts to differentiate isolates of T. rubrum using molecular methods (7, 10, 12, 21) have been largely unsuccessful, and the extent of interstrain genomic variation within this apparently clonal species appears limited (7). However, we have recently identified substantial polymorphism in the ribosomal DNA (rDNA) repeat region of T. rubrum (9). In this report we describe two novel tandemly repetitive subelements (TRSs), TRS-1 and TRS-2, located in the T. rubrum rDNA nontranscribed spacer (NTS). Specific amplification of the NTS region identified strain-specific variations in the copy number of these different subrepeat elements. The characteristic fingerprints generated by this PCR assay provided a rapid, stable, and reproducible molecular typing system to study the epidemiology and pathogenicity of T. rubrum infection.
MATERIALS AND METHODS
Dermatophyte isolates.
The strains of T. rubrum and their origins are listed in Table 1. Isolates were recovered from a range of clinical lesions. There were no evident epidemiological links between any of the strains in the study, with the exception that one pair of isolates (WCH 01 and WCH 02) was derived from two different sites on a single patient. All isolates were identified to species level using standard mycological procedures (15), and species identity was confirmed by restriction analysis of the amplified internal transcribed spacer (ITS) regions I and II (9).
TABLE 1.
Strain details, rDNA type, and PCR type assignations.
| Isolate no. | Sourcea | Clinical site | rDNA type | TRS-1 PCR type | TRS-2 PCR type |
|---|---|---|---|---|---|
| 97-12329 | UKI | Skin | A | 1 | I |
| 97-12444 | UKI | Skin, toe | A | 1 | I |
| 97-12912 | UKI | Toenail | A | 1 | I |
| 97-12959 | UKI | Toenail | A | 1 | II |
| 97-13550 | UKI | Toenail | A | 1 | I |
| 97-13826 | UKI | Toenail | B | 1 | II |
| 97-14279 | UKI | Skin | B | 1 | II |
| 97-14358 | UKI | Skin, toe web | A | 1 | I |
| 97-14468 | UKI | NKb | A | 1 | II |
| 97-14482 | UKI | Toenail | B | 1 | II |
| 97-15521 | UKI | Toenail | A | 1 | II |
| 98-8070 | UKI | Skin, body | A | 1 | I |
| 99-3854 | UKI | Skin, ankle | A | 1 | II |
| 99-4116 | UKI | Skin, groin | A | 1 | NDc |
| 99-4143 | UKI | Skin, groin | A | 1 | NDc |
| FMG 1063 | TUN | NKb | A | 1 | I |
| FMG 3443 | FRA | NKb | A | 1 | NDc |
| 96-T2/1852 | GER | Toenail | A | 1 | NDc |
| 96-T2/1895 | GER | Toenail | A | 1 | NDc |
| 96-T2/2404 | GER | Toenail | A | 1 | II |
| 96-T2/3891 | HOL | Toenail | A | 1 | II |
| 96-T2/3962 | FIN | Toenail | A | 1 | II |
| WCH 01 | AUS | Skin, neck | A | 1 | II |
| WCH 02 | AUS | Skin, foot | A | 1 | II |
| WCH 03 | AUS | Toenail | A | 1 | II |
| WCH 06 | AUS | Skin, foot | A | 1 | NDc |
| Si 60 | JAP | Skin, foot | A | 1 | II |
| 95-T2/7 | ICE | Toenail | A | 1 | II |
| 95-T2/14 | ICE | Toenail | A | 1 | II |
| 95-T2/17 | ICE | Toenail | A | 1 | II |
| 95-T2/24 | ICE | Toenail | B | 1 | II |
| 95-T2/27 | ICE | Toenail | B | 1 | II |
| 96-T2/2244 | ICE | Toenail | A | 1 | II |
| 96-T2/2248 | ICE | Toenail | A | 1 | II |
| 97-N221 | ICE | Toenail | A | 1 | NDc |
| 97-N250 | ICE | Toenail | A | 1 | II |
| 97-N251 | ICE | Toenail | A | 1 | II |
| 97-N366 | ICE | Toenail | A | 1 | II |
| 97-N393 | ICE | Toenail | A | 1 | III |
| 99-N741 | ICE | Toenail | A | 1 | II |
| CBS 303.38 | VIEb | Skin, bodyb | A+ | 1+ | I |
| LM 101 | UKI | NKb | B | 2 | II |
| 94-6597 | UKI | Skin, leg | B | 2 | NDc |
| 95-1295 | UKI | Skin, leg | B | 2 | II |
| 97-12335 | UKI | Toenail | C | 2 | NDc |
| 97-12698 | UKI | Toenail | B | 2 | II |
| 97-14166 | UKI | Skin, groin | C | 2 | II + III |
| 97-14462 | UKI | Skin, foot | C | 2 | II |
| 99-7537 | UKI | Skin | B | 2 | II |
| 99-8204 | UKI | Skin, hand | B | 2 | II |
| 99-9133 | UKI | Skin, flank | B | 2 | II |
| 96-T2/1894 | GER | Toenail | B | 2 | II |
| WCH 07 | AUS | Skin, groin | B | 2 | II |
| S-08 | IND | NKb | B | 2 | II |
| Menon 60 | IND | Skin, body | B | 2 | II |
| S-05 | IND | NKb | B | 2 | II |
| Si 40 | JAP | Skin, foot | B | 2 | II |
| Si 44 | JAP | Skin, body | B | 2 | II |
| NCPF 295 | UKI | Skin, foot | C | 3 | II |
| 96-T2/2240 | ICE | Toenail | C | 3 | II |
| WCH 10403 | AUS | Skin, arm | C | 3 | II |
| WCH 20064 | AUS | Skin, groin | C | 3 | II |
| WCH 05 | AUS | Skin, foot | C | 3 | II |
| Menon 168 | IND | Skin, groin | C | 3 | II |
| 95-T2/11 | ICE | Toenail | D | 3 | II |
| 95-T2/253 | FIN | Toenail | D | 3 | II |
| Si 10 | JAP | Skin, body | C | 3 | II |
| 97-12063 | UKI | Skin, foot | D | 3 | II |
| 98-551 | UKI | Skin, foot | D | 3 | NDc |
| 98-259 | UKI | Skin, breast | C | 3 | II |
| 99-8147 | UKI | Skin, hand | C | 3 | II |
| 93-DS/TrP | UKI | Skin, finger | D | 4 | II |
| 98-7838 | UKI | Skin, back | D | 4 | II |
| WCH 08 | AUS | Toenail | D | 4 | II |
| Nakamura | JAP | Nail | C | 4 | II |
| FMG 10008 | TUN | NKb | D | 4 | II |
| 96-T2/3877 | ICE | Toenail | E | 5 | II |
| 99-4647 | UKI | NKb | E | 5 | II |
| Si 42 | JAP | Skin, foot | E | 5 | II |
| 97-12332 | UKI | Toenail | F | 6 | II |
| 96-T2/3964 | FIN | Toenail | G | 7 | II |
| S-9-01 | IND | NKb | O | 8 | II |
| S-9-02 | IND | NKb | O | 8 | II |
| 97-15477 | UKI | Toenail | M | 9 | II |
| 96-8533 | UKI | Toenail | H | 10 | II |
| 96-T2/2833 | GER | Toenail | P | 10 | NDc |
| 99-7824 | UKI | Skin | M | 10 | NDc |
| 95-T2/20 | ICE | Toenail | L | 11 | NDc |
| 97-12496 | UKI | Toenail | N | 11 | NDc |
| 95-764 | UKI | Skin | I | 12 | II |
| 98-5693 | UKI | Toenail | J | 12 | 0 |
| 97-12790 | UKI | Toenail | M | 13 | II |
| Si 19 | JAP | Skin, foot | M | 13 | II |
| S-06 | IND | NKb | K | 14 | II |
| LM 102 | UKI | NKb | I | 15 | II |
| 97-14581 | UKI | Skin, foot | K | 15 | II |
| Si 62 | JAP | Skin, body | Q | 16 | II |
| 98-1973 | UKI | Toenail | Q | 17 | II |
| Si 8 | JAP | Skin, foot | R | 18 | II |
| 98-9514 | UKI | Toenail | S | 19 | II |
| S-900008 | IND | NK | T | 20 | III |
Strain origins: AUS, Australia; FIN, Finland; FRA, France; GER, Germany; HOL, Holland; ICE, Iceland; IND, India; JAP, Japan; TUN, Tunisia; UKI, United Kingdom; VIE, Vietnam; NCPF 295, National Collection of Pathogenic Fungi, PHLS Mycology Reference Laboratory, Bristol, United Kingdom; CBS 303.38, Centraalbureau voor Schimmelcultures, Baarn, The Netherlands.
NK, not known.
ND, not determined.
Additional dermatophyte species were kindly provided by Gillian Midgley, Institute of Dermatology, St. Thomas' Hospital, London, United Kingdom.
Extraction of T. rubrum genomic DNA from fungal plate cultures.
The nucleic acid extraction procedure described previously (9) was modified to allow DNA extraction directly from primary plate cultures. Approximately 2 mg of hyphal tissue was placed in a 0.6-ml microcentrifuge tube containing 500 mg of 0.45- to 0.5-mm-diameter sterile glass beads (B. Braun Biotech International GmbH, Melsungen, Germany). Three hundred microliters of lysis buffer (400 mM Tris-HCl [pH 8.0], 60 mM EDTA, 150 mM NaCl, 1% sodium dodecyl sulfate, 50 μg of proteinase K per ml) was added, and two freeze-thaw cycles at −80°C were carried out. The sample was vortexed for 3 min and incubated for 1 h at 60°C. The base of the microcentrifuge tube was pierced with a sterile needle and placed in a 1.5-ml microcentrifuge tube, and the liquid contents were transferred to the larger tube by centrifugation (13,000 × g, 30 s). Fifty microliters of 5 M sodium perchlorate was added, and incubation continued for a further 15 min at 60°C. Nucleic acids were phenol-chloroform extracted, ethanol precipitated, and washed as described previously (9), and the purified DNA was resuspended in 30 μl of sterile water.
PCR amplification of the rDNA NTS region of T. rubrum NCPF 295.
Two primers were designed from conserved regions of fungal 25S and 18S genes to amplify the NTS region of T. rubrum. The 25S consensus primer 25SCON2 (5′-TAGACCGTCGTGAGACAG-3′) was designed from an alignment of DNA sequences from the 3′ ends of five fungal 25S genes obtained from the GenBank (1) database. This primer corresponds to bases 2,969 through 2,986 of the Candida albicans 25S sequence (GenBank accession no. X70659). The second primer, NS1-R (5′-GAGACAAGCATATGACTAC-3′) was constructed using the reverse complement sequence of a universal fungal 18S rDNA primer, NS-1 (21).
Genomic DNA of T. rubrum NCPF 295 was prepared as described previously (9) and treated with RNase ONE ribonuclease (Promega Corp, Madison, Wis.) according to the manufacturer's instructions to remove RNA. The NTS region of T. rubrum NCPF 295 was amplified using the Expand Long Template PCR system (Boehringer Mannheim UK Ltd., Lewes, East Sussex, United Kingdom). Master Mix 1 contained 500 μM deoxynucleoside triphosphates (dNTPs; dATP, dCTP, dGTP, and dTTP) and 300 nM upstream (25SCON2) and downstream (NS1-R) primers, made up in a total volume of 90 μl with sterile water. Three 30-μl aliquots of Mix 1 were distributed into thin-walled PCR tubes. Three microliters of diluted T. rubrum NCPF 295 genomic DNA was added to give approximately 10 and 20 ng of template in two of the tubes, and 3 μl of water was placed in the third tube.
Master Mix 2 contained 1× Expand Long PCR buffer 3 (Boehringer Mannheim; patent formulation) with magnesium chloride at 2.25 mM and 3 μl of PCR enzyme mix (Boehringer Mannheim; patent formulation) in a total volume of 100 μl. Immediately prior to thermal cycling, 33 μl of Mix 2 was added to the three 33-μl aliquots of Mix 1. The amplification reaction was carried out on a Primus 96 thermal cycler (MWG-BIOTECH GmbH, Ebersberg, Germany) with an initial denaturation for 2 min at 93°C, followed by 30 cycles of primer annealing at 51°C for 0.5 min, extension at 68°C for 12 min, and denaturation at 93°C for 0.5 min. A terminal extension step of 68°C for 7 min completed the PCR.
Subcloning and sequencing of the NTS region of T. rubrum NCPF 295.
The PCR produced a single 3-kb product. This fragment was gel fractionated and purified using a StrataPrep DNA gel extraction kit (Stratagene Cloning Systems, La Jolla, Calif.). It was then subcloned into the pGEM-T vector system (Promega) and transformed into the Escherichia coli host strain XL1-Blue (Stratagene). The recombinant plasmid pGEM-tTrNTS was purified by sodium dodecyl sulfate lysis and matrix binding using a Qiagen plasmid miniprep kit (Qiagen GmbH, Hilden, Germany). The cloned insert was sequenced in both orientations using a primer walking strategy. Sequencing was carried out using dye terminator chemistry on an Applied Biosystems 377 automated sequencer. Repetitive regions of DNA were identified by using a nucleic acid dot matrix self-comparison program found at http://arbl.cvmbs.colostate.edu/molKit/dnadot/.
PCR assay for strain characterization.
Two primer pairs were designed from sequences flanking the two repetitive elements, TRS-1 and TRS-2. Primers TrNTSF-2 (5′-ACCGTATTAAGCTAGCGCTGC-3′) and TrNTSR-4 (5′-TGCCACTTCGATTAGGAGGC-3′) were used to amplify TRS-1, and primers TrNTSR-1 (5′-CTCAGTCGAACCGTGAGGC-3′) and TrNTSC-1 (5′-CGAGACCACGTGATACATGCG-3′) amplified TRS-2.
For amplification of TRS-1, a reaction mixture was made containing reaction buffer (50 mM KCl, 10 mM Tris-HCl [pH 9.0], 0.1% Triton X-100), 1.5 mM magnesium chloride, a 0.25 mM concentration of each dNTP (dATP, dCTP, dGTP, and dTTP), 34 μg of bovine serum albumin (BSA), 30 pmol each of primers TrNTSF-2 and TrNTSR-4, 5 U of Taq polymerase (Promega), and approximately 10 ng of diluted template DNA. The final reaction volume was made up to 100 μl with pure water. The amplification reaction was carried out on a Primus 96 thermal cycler (MWG-BIOTECH GmbH) with an initial denaturation for 2 min at 94°C, followed by 30 cycles of primer annealing at 58°C for 0.5 min, extension at 72°C for 3 min, and denaturation at 94°C for 0.5 min. A terminal extension step of 72°C for 10 min completed the PCR. Amplification of TRS-2 was carried out using the same reaction mixture as TRS-1 but without BSA. Cycling conditions for this reaction included denaturation for 1 min at 94°C, followed by 30 cycles of primer annealing at 55°C for 0.5 min, extension at 72°C for 2 min, denaturation at 94°C for 0.5 min, and a terminal extension step of 72°C for 10 min. Amplification products were separated by electrophoresis in 2% agarose gels, visualized by staining with ethidium bromide, and photographed.
PCR types 1 (strain 96-T2/2248) and 3 (strain NCPF 295) were run as size markers to assist in the scoring of simple PCR types 1 through 4. The designation of complex PCR types was made by visual comparison of the separate pattern types run contemporaneously on agarose gels. Multiple comparisons were made to differentiate similar patterns with minor band variations. Samples were amplified de novo for each comparative gel, and all bands, including minor ones, were reproducible.
Confirmation of assay specificity.
A single copy each of the TRS-1 repeat and flanking regions was amplified from strain 96-T2/2248 using primers TrNTSF-2 and TrNTSR-4. The product was labeled by incorporation of digoxigenin-labeled dNTPs using a random primed DNA labeling kit (Boehringer Mannheim UK Ltd.). Genomic DNA was prepared from 22 dermatophyte species and two isolates each of C. albicans and Aspergillus fumigatus, as described previously (9). EcoRI restriction digests of the genomic DNAs were electrophoresed, transferred to a nylon membrane, and hybridized with the single TRS-1 repeat probe. Conditions were as previously described (9), except that the two 15-min stringent washes were carried out using increased salt (2× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate] instead of 0.1× SSC) and a lower wash temperature (20°C instead of 68°C) to reduce the stringent conditions. Specificity was also confirmed by preparing a 1/25 dilution of genomic DNA from each species and using it as template in the PCR strain-typing assay above.
rDNA typing.
An rDNA type was determined for all T. rubrum isolates by Southern hybridization using the methods described previously (9).
The species and GenBank accession numbers of the five fungal 25S sequences used to design the 25SCON2 consensus primer were C. albicans (X70659), Saccharomycopsis fibuligera (U09238), Cryptococcus neoformans (L14067), Pneumocystis carinii (M86760), and Arxula adeninivorans (Z50840).
Nucleotide sequence accession numbers.
The nucleic acid sequence data for the NTS regions and/or subrepeat elements of T. rubrum strains NCPF 295, 96-T2/2248, 97-12332, and CBS 303.38 have been deposited in the GenBank/EMBL/DDBJ databases with the accession numbers AF222887, AF222888, AF222889, and AF222890.
RESULTS
Sequence characterization of the NTS region of T. rubrum NCPF 295 and identification of subrepeat elements.
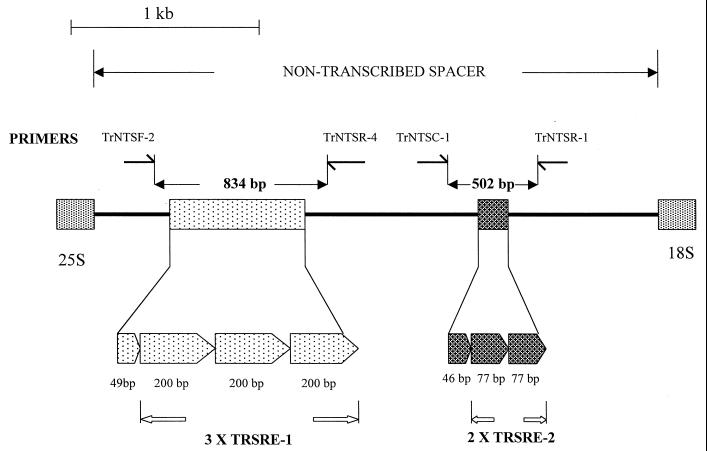
The putative NTS region was 2,486 bp in length and contained two sets of TRSs (Fig. 1). The first repeat locus was located 480 bp downstream from the putative 3′ terminus of the large-subunit (25S) RNA gene. Three tandemly repetitive copies of a 200-bp element (T. rubrum TRS-1) were identified, with an adjacent 49-bp partial repeat. A truncated, highly degenerate copy of a single repeat unit was located on the immediate downstream (18S) side of this repeat region. A second set of shorter subrepeat elements (T. rubrum TRS-2) was located 590 bp upstream from the predicted 5′ end of the 18S gene. This repeat unit was present as two tandem copies, 77 bp in length, followed by a third 46-bp partial repeat (Fig. 1).
FIG. 1.
Schematic representation of the NTS region of T. rubrum NCPF 295, showing the location of two sets of repetitive subelements, TRS-1 and TRS-2. The locations of the primer pairs used to amplify these repetitive regions for strain typing are indicated. The expanded section indicates the length in base pairs (bp) and the positions of the five complete and two partial repetitive elements.
Nucleotide homology searches (using Basic Local Alignment Search Tool Nucleotide [BLASTN] software) found no significant similarities between the T. rubrum NTS region and other sequences in the GenBank database. A number of possible open reading frames were identified from the NTS sequence, encoding predicted proteins up to 148 amino acids in length. None of these showed significant homology to any sequences deposited in SWISS-PROT or other protein databases, nor was a putative 5S gene identified. A region of secondary structure, with multiple homopolymer tracts up to 19 residues in length, was located immediately downstream of the predicted 3′ terminus of the 25S gene.
Sequence analysis of TRS-1 repeat units from T. rubrum isolates of different rDNA types.
Length variations in the NTS region of T. rubrum were identified during a previous study (9). Sequence analysis was carried out to confirm that these polymorphisms were primarily due to variations in the copy number of TRS-1. The first strain analyzed, T. rubrum NCPF 295, represented an rDNA type C strain (9) and contained three complete copies of TRS-1. The sequence of an rDNA type A strain, 96-T2/2248, contained a single complete copy of TRS-1. Incomplete sequence data from an rDNA type F strain (strain 97-12332) suggested the presence of up to seven copies of this element. Strain CBS 303.38 was a variant rDNA type A strain, and analysis of the single TRS-1 element present showed it had undergone major rearrangements, including loss of 19 bp from the central region (Fig. 2). In addition, one copy of the smaller TRS-2 subrepeat elements had been lost in this isolate.
FIG. 2.
Sequence variation in the TRS-1 elements of T. rubrum strains 96-T2/2248, NCPF 295, 97-12332, and CBS 303.38. Dots indicate residues which are identical to the 2248rl sequence, and dashes represent bases which are absent from 30338rl, the truncated TRS-1 element of strain CBS 303.38. The sequences of five TRS-1 elements, rl from NCPF 295 and rl, r2, r5, and r6 from 97-12332, are identical to the 2248 rl sequence (data not shown). The 49-bp partial TRS-1 element represented by 2248pr is identical in strains 96-T2/2248, NCPF 259, and 97-12332. The partial element 30338pr from strain CBS 303.38 has the CGA→ATG change also present in 295r3. The 27-bp palindromic sequence is shown in bold and overlined. Dots indicate residues which are identical to the 2248rl sequence, and dashes represent bases which are absent from 30338rl, the truncated TRS-1 element of strain CBS 303.38
Sequence alignments showed that most copies of TRS-1, both within and between strains, were identical (Fig. 2) or nearly so, with two repeats showing minor G→A or C→A transitions or transversions. However, the third TRS-1 element from T. rubrum NCPF 295 plus two of the partial 49-bp elements had a specific alteration in bases 28 through 30 (CGA→ATG) followed by a G→C transversion at position 34. These changes are also present in the incomplete sequence obtained from a fourth copy of TRS-1 in strain 97-12332 (data not shown). Finally, an almost perfect palindromic sequence of 27 bp, highlighted in Fig. 2, is present in each full copy of TRS-1, as well as in the partial and degenerate TRS-1 sequences described above.
Strain typing of T. rubrum by specific amplification of the TRS-1 and TRS-2 repeat regions.
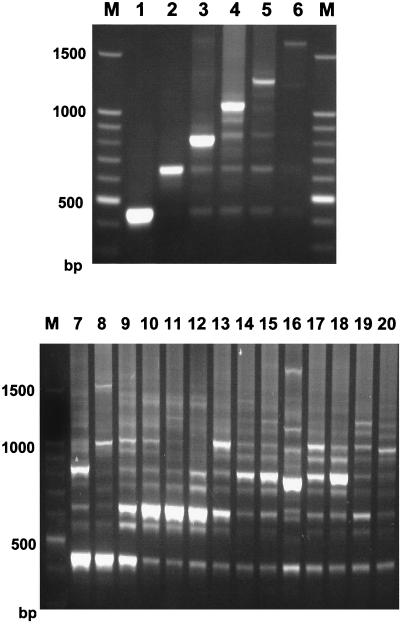
Primers TrNTSF-2 and TrNTSR-4 were used to amplify the TRS-1 repeat regions from six T. rubrum strains of rDNA types A through F. The major fragment amplified from each rDNA type showed an incremental length variation equivalent to one TRS-1 repeat unit, except the variation in length between types E and F, which may have represented seven rather than six copies of the subrepeat. A ladder of minor bands ranging in size from 634 to 1,434 bp and stepping up in increments of 200 bp was also present. These simple patterns were designated PCR types 1 through 6 (Fig. 3, lanes 1 through 6).
FIG. 3.
Amplification of the TRS-1 subrepeat element produced 21 PCR types from 101 clinical isolates of T. rubrum. (PCR type 1+ from strain CBS 303.38 is not shown). PCR types 1 through 6 represent strains with 1 to 6 copies of TRS-1 in the NTS. The copy number and chromosomal distribution of TRS-1 in strains with complex pattern types 7 through 20 are undetermined. M, molecular weight marker.
The assay was then used to examine 101 random clinical isolates of T. rubrum (Table 1). The majority of these isolates produced band patterns characteristic of PCR types 1 through 6, with the PCR type usually but not always related to the corresponding rDNA type (Table 1). Forty strains (40%) were PCR type 1, 17 (17%) were PCR type 2, and 13 (13%) were PCR type 3. Altogether, 80 strains (80%) were PCR types 1 through 6 (Fig. 3, lanes 1 through 6). The remaining 21 strains (20%) had complex PCR patterns, consisting of multiple bands in several different registers. The assignment of individual PCR types to these strains was complicated by difficulties in accurately scoring the presence of minor bands and in estimating band intensities. These complex patterns were derived from isolates that also had multiple polymorphic bands on rDNA typing.
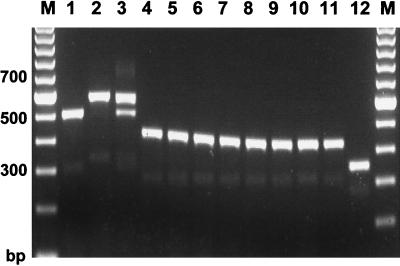
The NTS region containing TRS-2 was amplified using primers TrNTSR-1 and TrNTSC-1. A single 502-bp product (Fig. 1), representing two complete copies of TRS-2, was obtained from the majority of strains tested (75 of 87 [86%]). In seven isolates, the fragment appeared smaller by a length equivalent to one 77-bp TRS-2 repeat unit (Fig. 4). All of these strains had a single copy of TRS-1 (PCR type 1). Strain CBS 303.38, which had a degenerate form of TRS-1, also had a single TRS-2 element. In strains 97-N393 and S-900008, the fragment length (577 bp) suggested the presence of three copies of TRS-2, while the product amplified from isolate 98-5693 (an rDNA type J/PCR type 12 strain) had lost a fragment equivalent in size (144 bp) to both copies of TRS-2 (Fig. 4). Strain 97-14166 (PCR type 2) contained two forms of the TRS-2 locus, representing three and two copies of TRS-2, respectively.
FIG. 4.
Amplification of the TRS-2 subrepeat element, using primers TrNTSR-1 and TrNTSC-1. A 502-bp product (lane 1) representing two complete copies of TRS-2 was amplified from 86% of all isolates. Variation in the TRS-2 fragment occurred mainly in strains with a single copy of TRS-1 (PCR type 1). Lanes: 1, NCPF 295; 2, 97-12329; 3, 97-12444; 4, 97-12912; 5, 97-13550; 6, 97-14358; 7, 98-8070; 8, CBS 303.38; 9, FMG 1063; 10, 97-N393; 11, 97-14166; 12, 98-5693. M, molecular weight marker.
Individual PCR types did not show statistically significant associations with specific geographic regions, but no strains of the commonest PCR type (type 1) were present in 7 samples originating from the Indian subcontinent (Table 1), and only one type 1 strain was present in 11 strains from Japan. There are no clear relationships between PCR type and tissue type (e.g., skin or nail) or site of a clinical lesion.
Reproducibility and specificity of PCR strain typing.
The PCR types remained constant throughout different batches of primers and PCR reagents and using different thermal-cycling machines. The addition of BSA to the reaction mixture increased the efficiency of amplification but did not affect the size or number of bands detected. DNA extracted from T. rubrum NCPF 295 subcultured on Sabouraud agar on four separate occasions over a 2-year period gave identical PCR types. The use of a high-quality DNA template was important for consistent results, and the rapid DNA extraction procedure gave identical results to those for a template purified by the original method (9). The rapid DNA extraction procedure gave best results for fresh mycelial growth, although successful strain typing was achieved using template extracted from 4-month-old plate cultures.
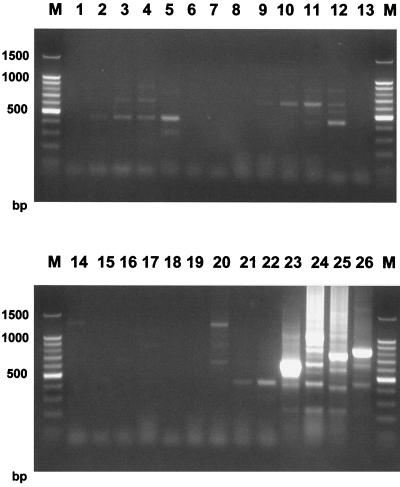
The specificity of PCR was determined by carrying out a standard PCR typing reaction on 22 different dermatophyte species, plus 2 clinical isolates each of C. albicans and A. fumigatus. Specific amplification products were obtained from T. rubrum, Trichophyton gourvilii, Trichophyton violaceum, and Trichophyton soudanense. All other species either produced weak bands which were interpreted as nonspecific or there were no visible products (Fig. 5). The specificity was further confirmed by probing Southern blots of these species using a single, labeled TRS-I element. The probe hybridized to the same four dermatophyte taxa that produced specific amplification products, although some weak hybridization to other species was detected at low stringency (data not shown).
FIG. 5.
Specificity of the PCR. Amplification was performed using primers TrNTSF-2 and TrNTSR-4 of 22 different dermatophyte species, plus two laboratory isolates of A. fumigatus and C. albicans. Lanes: 1, A. fumigatus strain MRL 2-1; 2, A. fumigatus strain MRL 2-5; 3, C. albicans strain MRL 10-1; 4, C. albicans strain MRL 12-2; 5, Epidermophyton floccosum strain DS.EF; 6, Microsporum audouinii strain SJ.EM 4875; 7, Microsporum canis strain 97/12400; 8, Microsporum gypseum strain SJ.EM 3928; 9, Microsporum persicolor strain NCPF 356; 10, Trichophyton concentricum strain NCPF 600; 11, Trichophyton erinacei strain NCPF 375; 12, Trichophyton equinum strain SJ.A1; 13, T. mentagrophytes var. interdigitale strain LM 43; 14, T. mentagrophytes var. mentagrophytes strain DS.TMvM; 15, Arthroderma benhamiae strain PV3000; 16, Arthroderma simii strain CBS448.65; 17, Arthroderma vanbreuseghemii strain A27961; 18, Trichophyton quinckeanum strain LM 54; 19, Trichophyton schoenleinii strain SJ.EV 327; 20, Trichophyton terrestre strain LM 39; 21, Trichophyton tonsurans strain SJ.EM 6717; 22, Trichophyton verrucosum strain 98/4545; 23, T. gourvilii NCPF 5019; 24, T. rubrum strain NCPF 295; 25, T. soudanense strain 98/7676; 26, T. violaceum strain SJ.EM 7115; M, molecular weight marker.
DISCUSSION
Strain identification in the dermatophyte fungus T. rubrum has been attempted using a range of molecular-typing procedures. These investigations have mainly involved the use of random amplified polymorphic-DNA methods and typically identified just a single strain (7, 10, 12) or a small number of strain lineages (21). The lack of differentiation and problems of reproducibility associated with random amplified polymorphic-DNA procedures have prompted us to evaluate alternative genotyping approaches. We previously described a method for differentiating strains of this species by analysis of restriction fragment length polymorphisms in the nontranscribed region of the rDNA spacer (9). In this study, we confirmed these polymorphisms and demonstrated the presence of two novel subrepeat elements, TRS-1 and TRS-2, in the NTS region. We further showed that variations in the copy number of these elements are responsible for the NTS length polymorphisms detected previously. Specific amplification of the NTS regions containing these elements provides a simple, rapid, and discriminatory method for strain typing of T. rubrum.
A similar PCR assay has been described for typing strains of the pathogenic yeast Candida krusei (2, 3). This method is based on the 164-bp NTS subrepeat element CKRS-1, which was present as six full-length copies in the strain analyzed (2). These authors reported a unique CKRS-1 PCR pattern for each of 95 independent C. krusei strains or patients they examined, which is in marked contrast to the situation with T. rubrum, in which 40% of strains belong to a single pattern type (type 1) and 75% of all isolates show one of four simple patterns (types 1 through 4). The reason for this limited range of common PCR types may relate to the presumed asexual nature of T. rubrum (the anamorph of this species has not been described), as genomic diversity at the rDNA locus may be restricted by the absence of sexual recombination. An alternative hypothesis involves the contemporary geographic radiation of this species from foci of endemicity in Southeast Asia and West Africa, which may have occurred as recently as the mid-nineteenth century (5, 13). This global spread of T. rubrum may have resulted from the rapid proliferation of a limited number of comparatively undifferentiated clones, reflecting the restricted number of prevalent PCR types reported here. Strains of these common PCR types may demonstrate specific characteristics of pathogenicity or infectivity that have enhanced their spread.
The extensive diversity present in the rDNA NTS contrasts with a recent account of the strictly clonal nature of T. rubrum (7). The variations in copy number of the NTS subrepeats presumably result from the process of unequal sister chromatid exchange at mitosis (17). Polymorphisms in tracts of repetitive DNA generated by unequal crossing-over are localized and are not linked to variation due to recombination and mutation elsewhere in the genome. The localized nature of micro- and minisatellite polymorphisms means that they may not be detected by random PCR-based methods that index whole-genome variation or by isoenzyme analysis (multilocus enzyme electrophoresis). Unusually, therefore, we have demonstrated the presence of at least one variable minisatellite locus in the T. rubrum genome, which otherwise appears to be clonal (7). The short evolutionary history and the apparent clonality of this species suggest that such markers may represent the primary or indeed only variable loci. Our results have encouraged us to seek other cryptic, polymorphic repetitive sequences for strain differentiation in T. rubrum. The identification of such additional unlinked variable markers would also permit allelic analysis using current population genetic methods (18).
Amongst isolates in the United Kingdom, the largest number of strains were TRS-1 type 1, the second largest type 2, etc. One explanation for this pattern assumes that multiplication in TRS-1 copy number by unequal crossover is a rare event. Thus, if all strains with multiple copies of TRS-1 are derived initially from type 1 isolates, then there will be progressively smaller numbers of strains with increased copy numbers of this repeat. There may also be coincidental selection for strains with a single copy or low copy numbers of TRS-1 among clinical isolates. Further work on other genetic markers and strains from patients with “carrier status” may enable these questions to be answered. We were surprised to discover that variation in copy number of the shorter TRS-2 element occurs predominantly in strains which have a single copy of TRS-1. Although two copies of TRS-2 are found in most isolates, the coincidental presence of two or more copies of TRS-1 seems to result in unequal crossover occurring preferentially between the larger elements.
While the structure of these repeat elements was stable in a strain cultured in vitro over a period of 1 year, it remains possible that in vivo recombination and changes in copy number occur more frequently, particularly in strains with large copy numbers of TRS-1. Finally, it should be noted that the process of crossover fixation could produce identical events in strains that do not share a common ancestry. Consequently, epidemiological interpretations placed on strains which share the same PCR type have to be made with care and with due regard to additional factors such as the temporal and geographical relationships between isolates.
Among the simple types (PCR types 1 through 6), there are often ladders of minor bands which appear to be PCR artifacts caused by the loss of entire copies of the TRS-1 repeat during amplification. We identified a 27-bp palindromic sequence in TRS-1 (Fig. 2) which has the potential to initiate hairpin formation during PCR, resulting in “looping out” of the interpalindromic region. Hence, in a strain with three copies of TRS-1, hairpin formation between different repeats would produce minor fragments representing one or two copies of TRS-1 in addition to the main amplification product representing three copies of the element. The use of hot starting and inclusion of dimethyl sulfoxide and detergents such as Triton X-100 in the PCR mixture were ineffective in eliminating or reducing these amplification artifacts.
The molecular basis and identification of the common PCR types (PCR types 1 through 6) are straightforward, but they are more difficult to pinpoint in strains with more complex banding patterns (PCR types 7 through 20). These multiple bands may represent the presence of rRNA cistrons with different TRS-1 and -2 copy numbers, rearrangements within NTS regions to duplicate primer sites, or even heterokaryons.
We also noted several discrepancies (Table 1) between the PCR type and rDNA type for some isolates. In certain strains, the rDNA type determined by Southern hybridization was one size register greater than the corresponding PCR type; for example, a PCR type 1 appeared as an rDNA type B. These anomalies may be due to misinterpretation of the rDNA type, as the 200-bp increments representing each rDNA type occur in EcoRI fragments of 4 to 5 kb in overall length. In addition, the 77-bp size difference caused by loss of one copy of TRS-2 introduced minor incremental differences in the fragment size of some rDNA type A strains.
Several isolates with multiple band patterns had identical PCR types but different rDNA types (Table 1). These anomalies probably arise from the influence of various technical differences between PCR and probe hybridization methodologies. For instance, in strains with complex rDNA and PCR patterns, the distribution and copy number of TRS-1 and/or TRS-2 between different cistrons of the rDNA repeat may not be the same. These heterogeneous repeats may be evident by probe hybridization, but they may not be detected by the PCR assay, particularly if the amplified fragments are relatively long and the heterogeneous repeat copy numbers low. These explanations do not account for all of the discrepancies in the PCR and rDNA results of the complex types. Technical differences between the two methods and the extent to which either typing system is able to render variation visible are potential sources of inconsistency. Resolution of these issues requires the sequencing of the NTS regions of complex types in order to determine the most reliable method of assay. While this work is ongoing, use of both PCR and rRNA analysis by Southern blotting to type complex types is recommended.
The TRS-1 sequence variations found in strain CBS 303.38 (Fig. 2) are also difficult to understand. This strain was first isolated from a case of tinea corporis in Vietnam in 1938, and it is therefore one of the oldest isolates of T. rubrum studied (4). This isolate may represent a progenitor of “modern” T. rubrum strains, which subsequently evolved to infect nail and skin tissue in shod feet. Alternatively, the strain may be one of an original group of isolates that did not spread to other parts of the world due to characteristics of low transmissibility or pathogenicity (5, 13). Further characterization of strain CBS 303.38 and of isolates with multiple banding patterns may provide answers to some of these questions.
The primers used in the T. rubrum assay allow specific amplification from three closely related species, T. gourvilii, T. soudanense, and T. violaceum (8, 11, 16), but not from other dermatophyte species. We have recently found that isolates of the dermatophytes Trichophyton mentagrophytes var. mentagrophytes and T. mentagrophytes var. interdigitale also show extensive polymorphism in the NTS region (T. Mochizuki, C. J. Jackson, R. C. Barton, M. Moore, and E. G. V. Evans, unpublished data). The specificity of the T. rubrum PCR assay and the lack of hybridization of the TRS-1 probe with members of the T. mentagrophytes group indicate that the variable NTS region of this dermatophyte complex bears little genetic homology to the TRS-1 region of T. rubrum.
There was no clear correlation between strain PCR type and the clinical source of the isolate, i.e., skin or nail specimens. However, the distribution of PCR types was not the same across all geographic regions. For example, there was only a single type 1 strain among 11 Japanese isolates examined, whereas in the United Kingdom and most other countries, type 1 was by far the most common PCR type seen. Further isolates from this and other regions are required to confirm these divergences, and they may enable us to map the spread of T. rubrum that has occurred over the last 150 years.
The analysis of subrepeat elements in the rDNA of T. rubrum affords a valuable technique for strain identification in this genetically invariant fungus. Variable repetitive sequences in this and other Trichophyton species will continue to provide important tools for dermatophyte strain typing and epidemiology. This new approach will enable us to reassess our current knowledge of the epidemiology of dermatophyte infections and also provide new insights into pathogenicity and the patterns of disease caused by these organisms.
ACKNOWLEDGMENTS
Strains were generously provided by D. Ellis, Mycology Unit, Womans and Childrens Hospital, North Adelaide, Australia; A. Chakrabarti, Department of Medical Microbiology, Postgraduate Institute for Medical Education and Research, Chandigarh, India; T. Mochizuki, Department of Dermatology, Kanazawa Medical University, Ishikawa, Japan; and R. Grillot, Department of Medical and Molecular Mycology, Faculté de Médecine de Grenoble, France.
We thank the Janssen Research Foundation, Beerse, Belgium, for their financial support for this project.
REFERENCES
- 1.Benson D A, Boguski M S, Lipman D J, Ostell J, Ouellette B F F, Rapp B A, Wheeler D L. GenBank. Nucleic Acids Res. 1999;27:12–17. doi: 10.1093/nar/27.1.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carlotti A, Srikantha T, Schröppel K, Kvaal C, Villard J, Soll D R. A novel repeat sequence (CKRS-1) containing a tandemly repeated sub-element (kre) accounts for differences between Candida krusei strains fingerprinted with the probe CkF1,2. Curr Genet. 1997;31:255–263. doi: 10.1007/s002940050203. [DOI] [PubMed] [Google Scholar]
- 3.Carlotti A, Chaib F, Couble A, Bourgeois N, Blanchard V, Villard J. Rapid identification and fingerprinting of Candida krusei by PCR-based amplification of the species-specific repetitive polymorphic sequence CKRS-1. J Clin Microbiol. 1997;35:1337–1343. doi: 10.1128/jcm.35.6.1337-1343.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Catanei A. On the classification of Trichophyton rubrum: observation and experimental investigation of a new strain of this fungus. Arch Inst Pasteur. 1938;16:227–231. [Google Scholar]
- 5.Charif M A, Elewski B. A historical perspective on onychomycosis. Dermatol Ther. 1997;3:43–45. [Google Scholar]
- 6.Cribier B, Mena M L, Rey D, Partisani M, Fabien V, Lang J M, Grosshans E. Nail changes in patients infected with human immunodeficiency virus: a prospective controlled study. Arch Dermatol. 1998;134:1216–1220. doi: 10.1001/archderm.134.10.1216. [DOI] [PubMed] [Google Scholar]
- 7.Gräser Y, Kühnisch J, Presber W. Molecular markers reveal exclusively clonal reproduction in Trichophyton rubrum. J Clin Microbiol. 1999;37:3713–3717. doi: 10.1128/jcm.37.11.3713-3717.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harmsen D, Schwinn A, Brocker E B, Frosch M. Molecular differentiation of dermatophyte fungi. Mycoses. 1999;42:67–70. doi: 10.1046/j.1439-0507.1999.00261.x. [DOI] [PubMed] [Google Scholar]
- 9.Jackson C J, Barton R C, Evans E G V. Species identification and strain differentiation of dermatophyte fungi by analysis of ribosomal-DNA spacer regions. J Clin Microbiol. 1999;37:931–936. doi: 10.1128/jcm.37.4.931-936.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu D, Coloe S, Pedersen J, Baird R. Use of arbitrarily primed polymerase chain reaction to differentiate Trichophyton dermatophytes. FEMS Microbiol Lett. 1996;136:147–150. doi: 10.1111/j.1574-6968.1996.tb08040.x. [DOI] [PubMed] [Google Scholar]
- 11.Liu D, Coloe S, Baird R, Pedersen J. Molecular determination of dermatophyte fungi using the arbitrarily primed polymerase chain reaction. Br J Dermatol. 1997;137:351–355. doi: 10.1046/j.1365-2133.1997.18481941.x. [DOI] [PubMed] [Google Scholar]
- 12.Mochizuki T, Sugie N, Uehara M. Random amplification of polymorphic DNA is useful for the differentiation of several anthropophilic dermatophytes. Mycoses. 1997;40:405–409. doi: 10.1111/j.1439-0507.1997.tb00175.x. [DOI] [PubMed] [Google Scholar]
- 13.Rippon J W, editor. Medical mycology: the pathogenic fungi and pathogenic actinomycetes. Philadelphia, Pa: The W. B. Saunders Co.; 1988. p. 178. [Google Scholar]
- 14.Suhonen R E, Dawber P R, Ellis D H. Fungal infections of the skin, hair and nails. London, United Kingdom: Martin Dunitz; 1999. [Google Scholar]
- 15.Summerbell R C, Kane J. Physiological and other special tests for identifying dermatophytes. In: Kane J, Summerbell R, editors. Laboratory handbook of dermatophytes. Belmont, Calif: Star Publishing Co.; 1997. pp. 45–79. [Google Scholar]
- 16.Summerbell R C, Haughland R A, Li A, Gupta A K. rRNA gene internal transcribed spacer 1 and 2 sequences of asexual, anthropophilic dermatophytes related to Trichophyton rubrum. J Clin Microbiol. 1999;37:4005–4011. doi: 10.1128/jcm.37.12.4005-4011.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Szostak J, Wu R. Unequal crossing over in the ribosomal DNA of Saccharomyces cerevisiae. Nature. 1980;284:426–430. doi: 10.1038/284426a0. [DOI] [PubMed] [Google Scholar]
- 18.Taylor J W, Geiser D M, Burt A, Koufopanou V. The evolutionary biology and population genetics underlying fungal strain typing. Clin Microbiol Rev. 1999;12:126–146. doi: 10.1128/cmr.12.1.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tosti A, Piraccini B M, Stinchi C, Colombo M D. Relapses of onychomycosis after successful treatment with systemic antifungals: a three-year follow-up. Dermatology. 1998;197:162–166. doi: 10.1159/000017990. [DOI] [PubMed] [Google Scholar]
- 20.White T J, Bruns T, Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis M A, editor. PCR protocols: a guide to methods and applications. London, United Kingdom: Academic Press Ltd.; 1990. pp. 315–322. [Google Scholar]
- 21.Zhong Z, Li R, Li D, Wang D. Typing of common dermatophytes by random amplification of polymorphic DNA. Jpn J Med Mycol. 1997;38:239–246. [Google Scholar]