Abstract
Insect Trichoplusia ni High Five™ (Hi5) cells have been widely explored for production of heterologous proteins, traditionally mostly using the lytic baculovirus expression vector system (BEVS), and more recently using virus-free transient gene expression systems. Stable expression in such host cells would circumvent the drawbacks associated with both systems when it comes to scale-up and implementation of more efficient high-cell density process modes for the manufacturing of biologics. In this study, we combined Flipase (Flp) recombinase-mediated cassette exchange (RMCE) with fluorescence-activated cell sorting (FACS) for generating a stable master clonal Hi5 cell line with the flexibility to express single or multiple proteins of interest from a tagged genomic locus. The 3-step protocol herein implemented consisted of (i) introducing the RMCE docking cassette into the cell genome by random integration followed by selection in Hygromycin B and FACS (Hi5-tagging population), (ii) eliminating cells tagged in loci with low recombination efficiency by transfecting the tagging population with an eGFP-containing target cassette followed by selection in G418 and FACS (Hi5-RMCE population), and (iii) isolation of pure eGFP-expressing cells by FACS and expansion to suspension cultures (Hi5-RMCE master clone). Exchangeability of the locus in the master clone was demonstrated in small-scale suspension cultures by replacing the target cassette by one containing a single protein (i.e., iCherry, as an intracellular protein model) or two proteins (i.e., influenza HA and M1 for virus-like particles production, as an extracellular protein model). Overall, the stable insect Hi5 cell platform herein assembled has the potential to assist and accelerate biologics development.
Keywords: stable expression, insect Hi5 cells, RMCE, FACS, influenza VLPs
Introduction
The superiority of insect cells over bacteria or yeast for expressionof complex proteins [e.g., glycoproteins, G-protein-coupled receptors (GPCRs), virus-like particles (VLPs), and “difficult-to-express” mammalian proteins] is well-demonstrated (Stolt-Bergner et al. 2018). When considering insect cells for the production of recombinant proteins to meet pharmaceutical needs (Cox 2012; Krammer and Palese 2015), or to feed structural, functional and drug screening studies, the leading choice is the Baculovirus Expression Vector System (BEVS) (Assenberg et al. 2013; Fernández and Vega 2013). The BEVS is flexible, rapid and often generates high titers of complex and multi-subunit gene products during the late phase of viral infection, even from the most challenging cellular locations (such as cell membranes, among others) (Drugmand et al. 2012). It is used to produce the approved commercial vaccine FluBlok for human use (Krammer and Palese 2015). Nevertheless, generating and preserving high-quality recombinant baculovirus is rather time-consuming and labor-intensive. Moreover, reports indicate that membrane and secreted proteins are frequently expressed poorly and heterogeneously (McCarroll and King 1997), suggesting that the insect cell secretory pathway may be compromised due to the virus infection process (Ailor and Betenbaugh 1999). Degradation of produced recombinant proteins is also likely to occur due to the release of intracellular proteases from the virus-lysed host cells, thus reducing recovery of the overexpressed protein (Summers 1991; Farrell et al. 1998).
Virus-free expression in insect cells has been increasingly explored to circumvent BEVS-related drawbacks. Transient transfection is a promising approach to produce a wide variety of recombinant proteins in short periods, suitable for target screening and at early stages of the development of biopharmaceutical products (Puente-Massaguer et al. 2018). However, it has several limitations when it comes to scale-up (e.g., need for high DNA amounts, mixing of DNA with the transfection reagent, partial-to-complete medium replacement prior to transfection), commonly leading to increased manufacturing costs and batch-to-batch variability (Ansorge et al. 2009; Gutiérrez-Granados et al., 2016 ; Bleckmann et al. 2019; Fuenmayor et al. 2019). Stable gene expression from a recombinant cell line has several advantages over transient transfection, including lower demand of plasmid DNA and transfection reagent, higher batch-to-batch consistency and easier scale-up.
One of the critical steps when developing a stable cell line is the rapid selection of cell clones which stably express high quantities of the gene of interest (GOI). This can be a time-consuming and labor-intensive task if classical cell line development methods are used (Bleckmann et al. 2016). Recombinase-mediated cassette exchange (RMCE) techniques can foster cell line generation by eliminating the need for single cell cloning and cell line screening, the most time-consuming steps (Qiao et al. 2009). In a classical RMCE approach, the sequence of steps involve (i) random integration of a reporter cassette (tagging cassette) in the parental cells’ genome, (ii) limiting dilution followed by screening of isolated clones with a desired trait (e.g., growth performance and high expression levels of the transgene from a single locus to meet the long term stability), and (iii) evaluation of clones’ ability to support RMCE, identifying those that can be used to express the GOI by means of targeted integration. The latter task is labor-, time-, and cost-intensive, mainly because many of the clones selected at this stage turn out to be incapable of supporting cassette replacement (Vidigal et al. 2018). To lessen the probability of selecting such “useless” clones, analyzing a higher number of cells at the population level and rapidly selecting those showing amenability for cassette replacement with high-throughput screening methods (e.g., fluorescence-activated cell sorting—FACS) may be an option. This would allow for the identification of loci supporting both high transgene expression and high Flp-mediated recombination efficiency (the so-called “hot-spots”). In addition, the RMCE reaction at the population level (before moving to subcloning) would allow further enrichment by FACS of a population of cells that are both high expressing and amenable to cassette exchange.
In this study, we applied the aforementioned concept (i.e., RMCE combined with FACS) and implemented for the first time Flp-RMCE in insect Hi5 cells, generating a stable master clonal cell line with flexibility to express (single and multiple) proteins of interest from a tagged genomic locus. As proof of concept, iCherry (as an intracellular protein product) and Influenza-VLPs displaying M1 core protein and HA antigen (as an extracellular protein product) were produced in this platform.
Materials and methods
Plasmid design and construction
The primers and templates used in plasmid construction are described in Supplementary information (see Supplementary Table S1). All restriction enzymes were purchased from New England Biolabs (NEB) and used according to manufacturer’s instructions. The polymerase chain reactions (PCRs) were carried out using the Phusion DNA Polymerase (Thermo Fisher Scientific), and cloning reactions were conducted using the In-Fusion HD Cloning Plus kit (Clontech). All inserts were verified by Sanger sequencing.
Generation of pTagging-iCherry-hyg:
pTagging-iCherry-hyg vector (GenBank MW246024) was generated by cloning the hygromycin resistance gene and SV40 polyA fragment [PCR amplified from pTag-OpIE2dsRed (Fernandes et al. 2012)], into NheI digested pTagging-iCherry backbone. The pTagging-iCherry vector (GenScript, GenBank MW246023) was in silico designed using Vector NTI and synthesized by GenScript (USA). This cassette is composed by the following elements: the OpIE2 promoter from the Orgyia pseudotsugata multicapsid nucleopolyhedrosis virus (OpMNPV) immediate-early 2 (ie-2) gene, FRT wt (Fwt) sequence, icherry gene (a Spodoptera frugiperda codon-optimized version of mCherry designed using the GenScript Rare Codon Analysis Tool) with a polyA from the OpMNPV ie-2 gene, FRT F5 (F5) sequence, and the OpIE1 promoter, all cloned in pUC57.
Generation of phr5-AcIE1-iFlp:
Constitutive expression of iflp (a S. frugiperda codon-optimized Flp) from the AcIE1 promoter was obtained by excising iflp from OpIE2-iFlp-IEterm plasmid (GenBank MG051710) and cloning it into BamHI/HindIII digested pIEx™-10 (Novagen, EMD Millipore) vector to generate the phr5-AcIE1-iFlp vector (GenBank MW246025). The OpIE2-iFlp-IEterm plasmid was kindly provided by the Protein Sample Production Facility (PSPF) at the Helmholtz Centre for Infection Research (Braunschweig, Germany).
Generation of pTarget-eGFP-neo:
Enhanced green fluorescent protein (egfp) gene with a polyA from the OpMNPV ie-2 gene was amplified from pTarget-OpIE2-eGFP and the neomycin gene was amplified from pTag-OpIE2-dsRed (Fernandes et al. 2012). The vector backbone fragment containing F5 and Fwt was amplified from the pTarget-OpIE2-eGFP cassette. The three fragments were cloned simultaneously using the In-Fusion HD Cloning Plus kit (Clontech) to generate the pTarget-eGFP-neo vector (GenBank MW246027).
Generation of pTarget-eGFP-iFlp-neo:
The hr5-AcIE1 promoter together with the iflp gene and ie1 terminator were cloned between the egfp and neomycin genes of pTarget-eGFP-neo upon SfiI/PacI digestion to generate pTarget-eGFP-iFlp-neo vector (Genbank MW246026).
Generation of pTarget-iCherry-zeo:
pTarget-eGFP-neo vector was digested with NdeI/NotI and the egfp gene was replaced by icherry gene amplified from pTagging-iCherry-hyg. The neomycin gene in the resulting vector was excised by inverse PCR and reconstituted ATG zeocin gene from pIZT/V5-His (Invitrogen) was cloned in the excised site to generate pTarget-iCherry-zeo vector (GenBank MW246028).
Generation of pFluTarget:
The HA and M1 genes from a subtype H3 Influenza virus strain were PCR amplified from a vector kindly provided by RedBiotech AG (Switzerland). The HA gene was cloned into a NotI pIZT/V5-His plasmid, resulting in pIZT-HA. M1 gene was cloned into a SacI digested pIZT/V5-His plasmid, resulting in pIZT-HA and pIZT-M1, respectively. The amplified fragment containing the OpIE2 promoter, HA gene, and polyA from pIZT-HA was cloned into pIZT-M1 linearized by inverted PCR, generating the pIZT-M1-HA vector. Finally, the iCherry gene was excised from the pTarget-iCherry-zeo vector using NdeI and PacI, and replaced by the fragment containing OpIE2 promoter, M1 gene, polyA, OpIE2 promoter, HA gene, and polyA, which was amplified from pIZT-M1-HA.
Cell lines and culture media
Trichoplusia ni BTI-TN5B1-4 (High Five™, Hi5) cells (Cat. no. B855-02, Invitrogen Corporation, Paisley, UK) were cultured in Insect-XPRESS™ (Lonza, Basel, Switzerland) media. The routine culture was performed in 125 or 500 mL shake flasks (10% working volume) at 100 rpm in an Innova 44 R incubator (Eppendorf) at 27°C. Cells were sub-cultured to 0.2–0.3×106 cell/mL every 3–4 days.
Cell transfection with the pTagging-iCherry and pTagging-iCherry-hyg
To randomly integrate the pTagging-iCherry or pTagging-iCherry-hyg cassettes in the genome of Hi5 cells, parental cells were transfected at 0.3×106 cell/mL with 0.3μg of pTagging-iCherry or pTagging-iCherry-hyg, using 8μL of Cellfectin® II Reagent (Invitrogen) per 1×106 cells, defined as 1 unit of transfection (u.t.). Cells transfected with pTagging-iCherry-hyg were submitted to antibiotic selection after 48 hours post-transfection (hpt) in medium containing Hygromycin B (200µg/mL; InvivoGen).
Flp-mediated cassette exchange
Flp-mediated cassette exchange was performed in shake flask cultures, using a protocol described elsewhere (Fernandes et al. 2012). The antibiotic selection marker (G418 300µg/mL or Zeocin 200µg/mL, InvivoGen) was added 48 hpt. When the cell viability dropped below 60% (about 120 hpt), the cells were transferred to six-well plates (BD Falcon™) and kept in selection. Cell expansion in adherent was performed until cells achieved enough cell number and high cell viability to be able to grow in suspension.
Fluorescence-activated cell sorting
Cells were sorted in a MoFlo high-speed cell sorter (Beckman Coulter, Fort Collins, USA) using a previously developed protocol (Vidigal et al. 2013). Cell populations expressing iCherry or eGFP were collected into ∼1 ml of PBS, also supplemented with PF68 maintained at 4°C. After sorting, cells were pelleted (200×g for 10 minutes), seeded in 6 well plates (BD Falcon™), and kept for one week in culture medium with Antibiotic–Antimycotic (Invitrogen). Single cells resulting from the sorting of the Hi5-eGFP population were collected into 96 well plates (BD Falcon™) containing conditioned culture medium supplemented with 10% FBS (Gibco) and Antibiotic–Antimycotic (Invitrogen) and monitored every 2–3 days. Wells containing multiple colonies were discarded. After adaptation to these culture conditions, robust clones were obtained and expanded until they could support to growth in suspension culture.
Analytics
Cell concentration and viability:
Cell concentration and viability were analyzed by trypan blue exclusion method daily. Cell counting was performed in a Fuchs-Rosenthal hemocytometer chamber (Brand, Wertheim, Germany) or in the Cedex HiRes Analyzer (Roche, Germany).
Fluorescence microscopy:
Engineered Hi5 cells expressing the reporter proteins iCherry or eGFP were visually inspected by fluorescence microscopy (DMI6000, Leica). The ImageJ software (available at http://rsb.info.nih.gov/ij/, last accessed on 25/05/2021) was used to merge channels and adjust the linear brightness and contrast of the images.
Flow cytometry:
CyFlow® space (Partec GmbH, Münster, Germany) was used to evaluate transfection and recombination efficiencies, to characterize tagging populations, target populations, and clones in terms of eGFP or iCherry fluorescence intensity and percentage. Exponentially growing cells were harvested and diluted in PBS. eGFP was detected using the FL1 channel (emission filter: 520±5 nm) and iCherry by using the FL5 channel (emission filter 630 ±22 nm). Fluorescence intensity from 10 000 events per sample was collected and analyzed using the FlowMax (2009, Quantum Analysis GmbH) and FlowJo™ (2019, Becton, Dickinson, and Company), respectively.
Western blot:
Culture samples were centrifuged at 200×g for 10 minutes and supernatants were collected and stored at –20°C until further analysis. eGFP and iCherry were analyzed in cellular extracts after cell lysis using I-PER™, according to manufacturer’s instructions (Thermo Fisher Scientific). M1 and HA were analyzed in culture supernatants. For M1, 250µL sample was precipitated in cold ethanol (1:4 parts) and resuspended in loading buffer (42µL); after denaturation, 20µL/well was applied in the gel. For HA, 15µL/well of the supernatant was loaded. Proteins were denatured at 95°C for 5 minutes, separated under reducing conditions in a 4–12% SDS gel (Thermo Fisher Scientific), and transferred to a nitrocellulose (eGFP) or a PVDF membrane (iCherry, M1, and HA) using iBlot® Transfer Stack (Thermo Fisher Scientific). For eGFP identification, membrane was incubated with an anti-GFP monoclonal antibody (dil. 1:2000) (Sigma) during 2 hours at room temperature. For iCherry identification, membrane was incubated with an anti-dsRed polyclonal antibody (di. 1:1000, Clontech) during 2 hours at room temperature. For M1 protein identification, membrane was incubated with a goat polyclonal antibody (dil. 1:2000) (Abcam, Cat# ab20910) during 1 hour at room temperature. For HA identification, membrane was incubated with a combination of two polyclonal antibodies (dil. 1:2000) from National Institute for Biological Standards and Control (NIBSC) for 1 hour at room temperature. As secondary antibodies, an anti-mouse (dil. 1:5000) (Sigma, Cat# A5420), an anti-rabbit (dil. 1:5000) (GE Healthcare, catalog #NA931-1ML), an anti-goat IgG (dil. 1:2000) (Life Technologies, Cat# 81-1620), and an anti-sheep (dil. 1:5000) (Santa Cruz Biotechnology, Cat# sc-2473) antibody conjugated with horseradish peroxidase labeling were used for 1 hour incubation at room temperature. Protein band detection was performed with the enhanced chemiluminescence (ECL) detection system (Amersham Biosciences) and captured digitally with no illumination for chemiluminescence using the ChemiDoc™ XR (Bio-Rad). Molecular weight marker SeeBlue™ Plus 2 prestained standard 1x (Invitrogen) was used and stain-free imaging was performed using ChemiDoc™ XR, Bio-Rad. The equal-sized transects images obtained by the ChemiDoc™ XR system were superimposed to display the molecular weight marker and protein bands in the same image. The GFP standard (Sigma) was used as a positive control.
Genomic DNA extraction:
To confirm the presence of the tagging or target cassettes in clones, genomic DNA was extracted using an in-house protocol. Briefly, 8×106 cells were pelleted and then added to a solution of 1 mL of Bradley’s solution, 5µL of proteinase K and 2 µL of RNase, followed by overnight incubation at 55°C. Bradley’s solution is composed by 10 mM Tris/HCl (pH 7.5), 2 mM EDTA, 10 mM NaCl and 0.5% SDS. 250 µL of 5 mM NaCl was added for 5 minutes on ice, and the solution centrifuged for 15 minutes at 10,000 rpm. Isopropanol was added to the resulting supernatant to precipitate DNA. After centrifugation at 10,000 rpm for 10 minutes at 4°C, the supernatant was discarded, the pellet was washed with 1.2 mL of 70% cold ethanol and allowed to air-dry. Pellet was dissolved in nuclease-free water.
Genomic PCR:
PCR was performed using 100 ng of DNA and amplification was done as described above with primer annealing at 69°C and extension step at 72°C for 2 minutes and 51 seconds. Forward and reverse primer sequences to cover the entire tagging cassette, target cassette from the OpIE2 promoter to the OpIE1 promoter (GCCGCGCGTTATCTCATGCGC and GCCGTTGGTGGCGTGAGGCATGTAA) or both, were designed. Agarose gel electrophoresis was performed in a 0.7% agarose concentration for 1 hour at 90 V, and the NZY DNA Ladder III (NZYTech; ref. MB04402) was used.
Hemagglutination assay:
The assay used is a plate-based assay in which the concentration of HA in bulk samples can be determined by comparing the hemagglutination profile of these samples with that of a standard of known HA concentration. Briefly, samples were twofold serially diluted in PBS and incubated at 4°C for 30 minutes with 25 µL of 1% chicken red blood cells (RBC) (Lohmann Tierzucht GMBH, Germany). Hemagglutination of RBC was identified by the formation of a network (lattice structure) of interconnected RBC and HA (positive results); if there is not enough HA to bind to RBC, they settle to the bottom of the well (negative result). As standard, an influenza vaccine with a known HA concentration was added to each assay experiment. The HA titer of a sample was determined by calculating the maximum dilution that gave a positive outcome and comparing it to the one obtained for the standard.
Data availability
Plasmid sequences and annotations have been deposited in the GenBank database (accession numbers MW246023, MW246024, MW246025, MW246026, MW246027, and MW246028). The vector pFluTarget is proprietary rights and its sequence can be specified upon request. Supplementary Table S1 contains the primers used to amplify the fragments for plasmid constructions and restriction enzymes used for vector digestion. Cell populations and cell lines generated in this study are the property of Instituto de Biologia Experimental e Tecnológica (IBET) and will be provided upon request addressed to aroldao@ibet.pt. Supplementary material is available at figshare: https://doi.org/10.25387/g3.14544066.
Results
Establishment of a master Hi5-RMCE clonal cell line
Generation of a high expression tagging cell population:
A two-step process was used to generate a tagging cell population with high iCherry expression (named “Hi5-tagging population” from now on): (i) the RMCE docking cassette (pTagging-icherry) was introduced into the genome of insect Hi5 cells by random integration, and (ii) the resulting tagging cell population was sorted by FACS to select the top 30% iCherry-expressing cells (Figure 1, step 1). The RMCE tagging cassette comprises the iCherry (mCherry codon-optimized for insect cells) gene flanked by the heterologous flipase recognition target (FRT) sites Fwt and F5, with the promoter driving its expression (POpIE2) placed upstream of the Fwt site. Flow cytometric analysis of transfected Hi5 cells (so-called unsorted tagging pool) over 4 consecutive passages showed that the intensity of iCherry fluorescence signal and the number of iCherry positive cells rapidly decreased along passages (Figure 2A), suggesting that stringent selection methods should be applied at a tagging population level to limit such loss of expression. Therefore, the pTagging-iCherry vector was modified to include the hygromycin selection marker gene inside the FRT flanking region, to be expressed from the OpIE1 promoter placed downstream of the F5 site. Parental Hi5 cells transfected with this vector and Hygromycin B (hygro) selected during one week were sorted by FACS gating for the top 30% iCherry expressing cells (Figure 2B). The stability of the resulting sorted tagging population was evaluated along time by comparing (i) the levels of iCherry expression given by the geometric mean fluorescence intensity (Geo MFI), and (ii) the percentage of iCherry positive cells given by flow cytometry analysis (Figure 2C). The percentage of iCherry positive cells in the sorted population remained above 96%, and the obtained Geo MFI values are relatively constant, around 2 AU (arbitrary units).
Figure 1.
Establishment of Hi5-RMCE master clone and producer cells. Scheme describing the generation of Hi5-RMCE master clones (steps 1–3) and Hi5-RMCE producer cells (step 4). In the 1st RMCE, the tagging cassette vector, encoding the iCherry reporter and the hygromycin selection marker flanked by FRT sites FRT wt (Fwt) and FRT F5 (F5) and OpIE2 and OpIE1 promoters, is randomly integrated into the parental cell genome. Flipase RMCE of the FRT-flanked eGFP target cassette sequences results in expression of the eGFP reporter gene along with the expression of the G418 selection marker gene. In the 2nd RMCE, the target cassette including the GOI and the zeocin selection marker gene is integrated into the locus through Flp-RMCE.
Figure 2.
Generation of a tagging cell population with iCherry expression. (A) Flow cytometric analysis of unsorted tagging pool along passages under no selective pressure. (B) Flow cytometric analysis of unsorted and sorted (30% highest iCherry fluorescent cells) tagging pool under Hygromycin B selective pressure. (C) Stability of sorted tagging population over time. Geo MFI—Geometric Mean Fluorescence Intensity. Dashed line and shaded region represent the average and standard deviation of Geo MFI value, respectively.
Generation of a cell population amenable to Flp-RMCE:
A high eGFP-expressing cell population amenable to Flp-RMCE (named “Hi5-RMCE population” from now on) was generated using a two-step process: (i) Flp-RMCE to introduce the eGFP-encoding “exchange cassette” into the docking sites of the Hi5-tagging population, and (ii) FACS for selection of high eGFP-expressing cells (Figure 1, step 2).
Two different Flp-RMCE strategies were evaluated: (i) single-transfection in which besides eGFP and the G418 selection marker, also iFlp is delivered in the “exchange cassette”; and, (ii) co-transfection in which iFlp is delivered in a second plasmid. The “exchange cassette” is flanked by the same set of FRTs as in the “tagging cassette,” and importantly does not encode promoters for eGFP and G418 selection marker. Therefore, the eGFP and G418 genes are only expressed when cassette exchange successfully occurs. This design allows for monitoring the efficiency of the recombination process. Different plasmid DNA concentrations and Target: iFlp plasmid ratios were also evaluated. Single-transfection using a DNA concentration of 0.3µg/u.t. proved to be the most efficient strategy, with the highest percentage of eGFP positive cells amongst all conditions screened (Figure 3A). This population of cells was then placed under selective pressure (G418) for 3 weeks (Figure 3B, unsorted pool), sorted for eGFP positive and iCherry negative cells, and enriched for the top 1% eGFP expressing cells (Figure 3B, sorted pool). The sorted population showed stability along passages, with a constant percentage of eGFP positive cells (>93%) and expression of eGFP (average Geo MFI of 7 AU) (Figure 3C).
Figure 3.
Generation of a high eGFP-expressing cell population amenable to Flp-RMCE. (A) Flow cytometric analysis of Hi5-tagging population upon Flp-RMCE using different transfection strategies (single- vs co-transfection), Target DNA quantities (0.1 vs 0.3 µg/u.t.), and constitutive promoters driving iFlp expression (POpIE2 vs Phr5-IE1). Values are normalized to the control (co-transfection with 0.1 µg Target DNA/u.t. and iFlp expression driven by Phr5-IE1). (B) Flow cytometric analysis of Hi5-RMCE population resulting from single transfection upon 3 weeks in selection with G418, before (unsorted pool) and after sorting (sorted pool). (C) Stability of sorted Hi5-RMCE population over multiple passages. Geo MFI—Geometric Mean Fluorescence Intensity. Dashed line and shaded region represent the average and standard deviation of Geo MFI value, respectively.
Generation of a high-producer, amenable to Flp-RMCE cell clone:
To generate a high-producer, amenable to Flp-RMCE cell clone (named “Hi5-RMCE master clone” from now on), the Hi5-RMCE population was subjected to a second round of FACS for single cell seeding into 96 well plates (Figure 1, step 3). Monoclonal cells were monitored by fluorescence microscopy and nonstable positive cells, cells with low eGFP expression and cells with slow growth rate were discarded. The remaining colonies were expanded, and four of them were characterized in suspension cultures. Site-specific integration of eGFP target cassette into the locus was confirmed by genomic PCR analysis (Figure 4A). Clones were screened for eGFP fluorescence intensity by flow cytometry, with clone #44 reporting the highest value (up to fourfold more than in the sorted pool) (Figure 4B) and no iCherry fluorescence signal (Figure 4C). Notably, all isolated clones showed stable eGFP expression over several weeks and were single-positive as determined by flow cytometry (data not shown).
Figure 4.
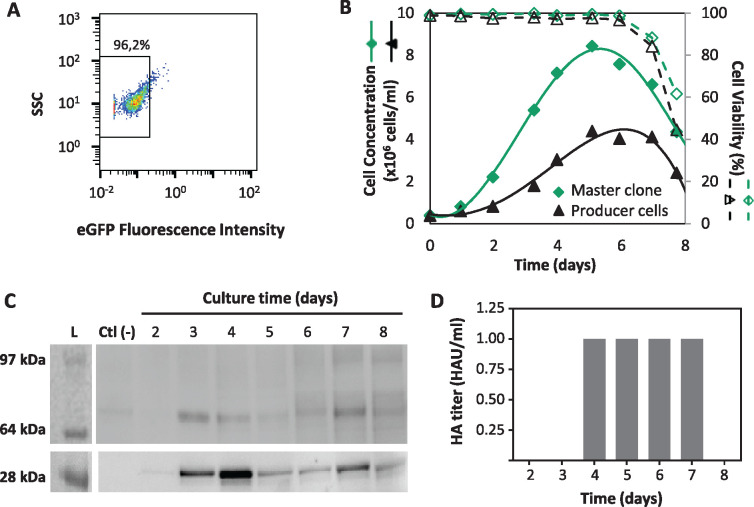
Generation of a high-producer, amenable to Flp-RMCE cell clone. (A) Genomic PCR analysis to confirm site-specific integration of the eGFP target cassette (5691 bp) in the tagged locus (2951 bp) of Hi5-RMCE clones #11, #18, #38, and #44. (B) Flow cytometric analysis of Hi5-RMCE clones #11, #18, #38, and #44. Sorted pool (control) is the Hi5-RMCE population. (C) iCherry and eGFP fluorescence of Hi5-RMCE master clone. (D) Flow cytometric analysis of Hi5-RMCE master clone upon Flp-RMCE using two transfection strategies (single- vs co-transfection) and two Target DNA quantities (0.1 vs 0.3 µg/u.t.). (E) iCherry and eGFP fluorescence of Hi5-RMCE master clone upon 3 weeks in zeo selection after cassette exchange. (F) Cell growth kinetics of Hi5-RMCE master clone. (G) eGFP fluorescence intensity of Hi5-RMCE master clone.
The exchangeability of the locus in Hi5-RMCE clone #44 was assessed by replacing the existing eGFP target cassette by another containing iCherry and a zeocin (zeo) resistance marker—Hi5-RMCE iCherry producer cells (Figure 1, step 4). Flow cytometry data shows that constitutive iFlp expression levels are insufficient to promote cassette exchange as no iCherry positive cells were obtained after 72 hpt (Figure 4D). To overcome this bottleneck, co-transfection of the iCherry target cassette together with iFlp plasmid was attempted and it proved to be successful. The highest percentage of iCherry positive cells was observed with a Target DNA concentration of 0.3µg/u.t. and Target: iFlp plasmids ratio of 1:3. Transfected cells were placed under selective pressure for 3 weeks, after which an isogenic population of iCherry-positive and eGFP-negative cells was obtained (Figure 4E). Clone #44 displayed promising results in terms of RMCE locus accessibility and ease of generating an isogenic population after zeo selection, and therefore was further characterized regarding cell growth (Figure 4F) and eGFP expression (Figure 4G). The growth kinetics of the Hi5-RMCE master clone is typical of a parental, nonengineered insect (Hi5) cell line, but with a growth rate significantly lower (µ=0.029 h−1 vs µ=0.041 h−1). The expression of eGFP was confirmed by flow cytometry, and a typical Gaussian-like distribution was observed, consistent with the clonal origin of Hi5-RMCE master clone. Importantly, the levels achieved are similar to those obtained for iCherry in Hi5-RMCE producer cells (Supplementary Figure S1).
Production of influenza HA-VLPs
The feasibility of the Hi5-RMCE master clone herein developed for expression of heterologous proteins was demonstrated with the production of influenza HA-VLPs. To do this, a target cassette containing the genes coding for HA (the antigen) and M1 (the scaffold) influenza proteins was constructed and used for RMCE in the master cell line (named “Hi5-RMCE producer cells” from now on) (Figure 1, step 4). Upon 3 weeks in zeo selection, Hi5-RMCE producer cells lost their eGFP fluorescence, thus confirming successful cassette exchange (Figure 5A). The growth kinetics of these cells was assessed and compared to that of the Hi5-RMCE master clone (Figure 5B). The differences were (i) reduced cell growth rate (µ=0.025 h−1 vs µ=0.029 h−1), and (ii) lower peak cell concentration (4x106 cell/mL vs 8x106 cell/mL). The expression of M1 and HA proteins was assessed by Western blot, and both proteins, with the expected molecular weight (28 and 68 kDa, respectively), could be identified in supernatant samples collected from shake flask cultures (Figure 5C). Expression of HA was also assessed by hemagglutination assay, and a HA titer of 1 HAU/mL was reached after day 4 (Figure 5D). The results obtained confirm the feasibility of the Hi5-RMCE master clone herein developed for the production of influenza HA-VLPs.
Figure 5.
Production of recombinant Influenza HA-VLPs. (A) Flow cytometry analysis of Hi5-RMCE master clone after 3 weeks in zeo selection upon cassette exchange. (B) Cell growth kinetics of Hi5-RMCE-FluVLPs producer cells and Hi5-RMCE master clone. (C) Western blot analysis of culture supernatant samples; expected MW of influenza HA and M1 proteins are 68 and 28 kDa, respectively. (D) Hemagglutination assay of culture supernatant samples.
Discussion
In this study, we have combined RMCE technology with FACS for the generation of a stable insect Hi5-RMCE master clone with flexibility to express single or multiple proteins of interest from a tagged genomic locus. To achieve this goal, a protocol for selection of Hi5 clones with high recombinant protein expression and amenable to RMCE was devised. The first round of cassette exchange was performed at the tagging population level, with subcloning of high expressing cells prone to RMCE done afterward by FACS to discard cells tagged in loci that do not support RMCE. The co-selection of high eGFP-expressing and RMCE competent clones reduced significantly the number of clones to screen and subsequently the time needed from gene transfer to protein production (Hacker and Balasubramanian 2016). Similar strategies have been applied before to other cell hosts to achieve comparable outcomes (Qiao et al. 2009; Baser et al. 2016; Vidigal et al. 2018).
Gene expression stability is one of the key properties of an engineered cell line. For example, stable expression in suspension cultures reduces batch-to-batch variability and makes process scale-up easier. More importantly, process intensification becomes possible by changing from batch to perfusion or continuous cultures (Zitzmann et al. 2017). In this study, the tagging cassette was randomly integrated into parental Hi5 cell’s genome (guaranteed by the selection with Hygromycin B), and not as an episomal element, thus limiting the loss of GOI expression over time (Fernandes et al. 2012). Indeed, the percentage of positive cells and fluorescence intensities of reporter proteins expressed by the Hi5-tagging population, Hi5-RMCE population, and the Hi5-RMCE master clone remained constant along culture passages, hence demonstrating the potential of insect Hi5 cells for stable protein production and their scale-up.
Another desired attribute in cell line development is the integration of a single copy of the transgene into cell’s genome. This is because when multiple copies are integrated homologous recombination can occur potentially leading to loss of expression of the GOI over time and/or deletion of large regions of the genome host that compromise cell line integrity (Lakso et al. 1996). In this study, the strategy devised encloses a two-step selection process (i.e., FACS and antibiotic) following RMCE with the objective of minimizing the generation of clones with multiple copies. Although the results presented confirm the inexistence of copies of tagging and target cassettes simultaneously in Hi5-RMCE master clone genome (which otherwise would per se indicate multicopy integration), the possibility of a second (or more) copy of the target cassette in its genome cannot be ruled out. To ascertain this, karyotyping by Fluorescence In Situ Hybridization (FISH), digital PCR or Southern blot could have been used. However, these methods rely on the availability of detailed genomic information of the host cell line, which for insect Hi5 cells is currently lacking (Fu et al. 2018; Talsania et al. 2019). PCR-based sequencing assays [e.g., inverse PCR, splinkerette-PCR (Potter and Luo 2010; Mabashi-Asazuma and Jarvis 2017), linear amplification mediated PCR, targeted locus amplification] or whole-genome sequencing are alternatives to karyotyping, being routinely used to identify integration sites of the transgene and determine the number of copies present in the genome. This holds true for cell lines whose genome sequence is very complete and well noted, allowing the design of efficient primers and probes as well as the selection of internal references, which is as mentioned above not yet the case for insect Hi5 cells. Therefore, and with the genomic information available at the moment, none of these techniques would have provided a conclusive answer (at maximum a hint or hypothesis) to whether there is one or multiple insertions. New developments in omics technologies [e.g., single-cell RNA sequencing (Zhang et al. 2015; Feng et al. 2020)] will be critical to assist on the complete genome assembly and annotation of the insect Hi5 cell line (incl. impact of cis and trans acting regulatory elements on transgene expression) and thus allow the use of these techniques for estimation of the copy number of inserted target cassette in the Hi5-RMCE master clone.
Clonality is another key property of an engineered cell line. Having a producer cell line with high assurance of monoclonality is essential for the approval of a given product by regulatory agencies (Frye et al. 2016). In this study, subcloning after the first RMCE resulted in clones with defined integration sites which are prone to iFlp-RMCE. These clones have RMCE-integration loci that facilitate generation of isogenic cell lines (Oumard et al. 2006; Balasubramanian et al. 2016). Moreover, due to its re-usable locus, clones can be rapidly converted into producer cells by RMCE, thus avoiding the risk of stochastic gene expression resulting from random gene integration (Fernandes et al. 2012). The final RMCE generates an isogenic cell pool with stable expression of the protein(s) of interest, avoiding additional subcloning steps.
The design of the exchange cassette was crucial for success of the RMCE/FACS strategy herein implemented. A “promoter trap” based approach was followed, where strong promoters are placed outside the FRT sites and ultimately drive the expression of the GOI and/or resistance gene, so that stringent selection procedures could be enforced (i.e., combining antibiotic pressure and FACS). This promoter-less exchange cassette prevents simultaneous expression of the GOI and the antibiotic resistance gene when the plasmid randomly integrates into other genomic sites besides the pre-selected site. Also, this design provides flexibility for protein expression as not only the expression locus can be re-used but also the target protein(s) can be expressed in combination with the accessory proteins or scaffolds of interest.
The number of transient transfection protocols for Hi5 cells has been steadily increasing. However, most still require high DNA amounts, usually in the range of 1–28.6µg of plasmid DNA per 106 cells (Ogay et al. 2006; Shen et al. 2015; Mori et al. 2017; Bleckmann et al. 2019). The stable cell line herein developed required a reduced amount of plasmid DNA (0.3µg Target DNA per 106 cells) to be delivered to the cell in a single transfection reaction. The ratio and absolute quantities of the exchange and recombinase plasmids influence cassette exchange efficiency (Figures 3A and 4C) and an excess of Flp plasmid in relation to the exchange cassette was used as recommended in other RMCE studies (Lauth et al. 2002; Schebelle et al. 2010; Sorrell et al. 2010; Vidigal et al. 2013). Moreover, greater efficiency in the Flp-RMCE was obtained when the iFlp was delivered as part of the exchange cassette. Although a direct comparison between single transfection and co-transfection has not been reported so far, delivery of Flp in the same plasmid as the exchange cassette has been reported in other systems (Oumard et al. 2006; Vidigal et al. 2018).
The potential of the Hi5-RMCE master clone herein established was demonstrated by expressing Influenza HA-VLPs from its tagged locus. Although the target proteins (i.e., HA and M1) could be identified in the supernatant of cell cultures by Western blot and HA assay, the overall productivity was relatively low suggesting that the GOI may be expressed at residual levels in genomic DNA. One can speculate about the promoter strength and/or silencing of expression locus as plausible reasons for such low expression. However, two pieces of evidence point in the opposite direction: (i) the expression locus was chosen based on the strong constitutive promoter (POpIE2) that controls the expression of the transgenes (Nehlsen et al. 2011), and (ii) silencing of the expression locus by methylation of the promoters will hardly occur in the cell line herein established as low CpG ratios across the plasmids were introduced. It is then clear that molecular engineering strategies are needed to overcome this bottleneck. For example, the use of alternate start codons may promote higher transcription rates, as it did for CHO (Baumann et al. 2017). To improve activity and stability of the target protein, other insect cell promoters, regulatory elements and/or insulators may be used (Sarkar et al. 2006; Bleckmann et al. 2015, 2019; Baser et al. 2016; Shirk and Furlong 2016). For the generation of the tagging population, other DNA delivery methods ( Yáñez and Porter, 1999 ) and systems for site-specific integration in transcriptionally active genome regions (e.g., CRISPR/Cas) may be tested. At the process level, tailor-made refeed strategies and bioprocess operations such as perfusion can be explored to increase cell density and subsequently production yields (Fernandes et al. 2014; Sequeira et al. 2018).
Conclusions
This study demonstrates the suitability of RMCE technology and FACS high-throughput screening as a powerful combination for the establishment of a stable Hi5-RMCE master cell clone. The final RMCE results in an isogenic cell pool with stable expression of the protein(s) of interest in a two-step protocol, avoiding additional subcloning steps. It constitutes a valuable expression tool to overcome specific bottlenecks associated with stable cell line development (relying on random integration), baculovirus-based or other transient expression systems, and a fast strategy to produce recombinant proteins to cope with emergent needs of biologics.
Acknowledgments
The authors wish to thank the technical support from C. Bispo, C. Andrade, and R. Gardner [Flow Cytometry Facility, Instituto Gulbenkian de Ciência (IGC)].
Funding
This work was supported by EU-funded projects “ComplexINC” (grant agreement no. 270089) and “EDUFLUVAC” (FP7-HEALTH-2013-INNOVATION-1, GA n. 602640), and by the Portuguese “Fundação para a Ciência e a Tecnologia” (FCT) through the following initiatives: R&D Projects (PTDC/EBB-EBI/102266/2008, EXPL/BBB-BIO/1541/2013, and IF/01704/2014/CP1229/CT0001), PhD fellowships (SFRH/BD/90564/2012 and SFRH/BD/86744/2012), and “Investigador FCT” Program (IF/01704/2014).
Conflicts of interest
None declared.
Literature cited
- Ailor E, Betenbaugh MJ.. 1999. Modifying secretion and post-translational processing in insect cells. Curr Opin Biotechnol. 10:142–145. [DOI] [PubMed] [Google Scholar]
- Ansorge S, Lanthier S, Transfiguracion J, Durocher Y, Henry O, et al. 2009. Development of a scalable process for high-yield lentiviral vector production by transient transfection of HEK293 suspension cultures. J Gene Med. 11:868–876. [DOI] [PubMed] [Google Scholar]
- Assenberg R, Wan PT, Geisse S, Mayr LM.. 2013. Advances in recombinant protein expression for use in pharmaceutical research. Curr Opin Struct Biol. 23:393–402. [DOI] [PubMed] [Google Scholar]
- Balasubramanian S, Wurm FM, Hacker DL.. 2016. Multigene expression in stable CHO cell pools generated with the piggyBac transposon system. Biotechnol Prog. 32:1308–1317. [DOI] [PubMed] [Google Scholar]
- Baser B, Spehr J, Büssow K, van den Heuvel J.. 2016. A method for specifically targeting two independent genomic integration sites for co-expression of genes in CHO cells. Methods. 95:3–12. [DOI] [PubMed] [Google Scholar]
- Baumann M, Gludovacz E, Sealover N, Bahr S, George H, et al. 2017. Preselection of recombinant gene integration sites enabling high transcription rates in CHO cells using alternate start codons and recombinase mediated cassette exchange. Biotechnol Bioeng. 114:2616–2627. [DOI] [PubMed] [Google Scholar]
- Bleckmann M, Fritz MH-Y, Bhuju S, Jarek M, Schürig M, et al. 2015. Genomic analysis and isolation of RNA polymerase II dependent promoters from Spodoptera frugiperda. PLoS One. 10:e0132898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleckmann M, Schürig M, Chen FF, Yen ZZ, Lindemann N, et al. 2016. Identification of essential genetic baculoviral elements for recombinant protein expression by transactivation in Sf21 insect cells. PLoS One. 11:e0149424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleckmann M, Schürig M, Endres M, Samuels A, Gebauer D, et al. 2019. Identifying parameters to improve the reproducibility of transient gene expression in High Five cells. PLoS One. 14:e0217878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox MMJ. 2012. Recombinant protein vaccines produced in insect cells. Vaccine. 30:1759–1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drugmand JC, Schneider YJ, Agathos SN.. 2012. Insect cells as factories for biomanufacturing. Biotechnol Adv. 30:1140–1157. [DOI] [PubMed] [Google Scholar]
- Farrell PJ, Lu M, Prevost J, Brown C, Behie L, et al. 1998. High-level expression of secreted glycoproteins in transformed lepidopteran insect cells using a novel expression vector. Biotechnol Bioeng. 60:656–663. [DOI] [PubMed] [Google Scholar]
- Feng M, Xia J, Fei S, Wang X, Zhou Y, et al. 2020. 2020 Identification of silkworm hemocyte subsets and analysis of their response to BmNPV infection based on single-cell RNA sequencing 2. bioRxiv. 10:344127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes F, Dias MM, Vidigal J, Sousa MFQ, Patrone M, et al. 2014. Production of rotavirus core-like particles in Sf9 cells using recombinase-mediated cassette exchange. J Biotechnol. 171:34–38. [DOI] [PubMed] [Google Scholar]
- Fernandes F, Vidigal J, Dias MM, Prather KLJ, Coroadinha AS, et al. 2012. Flipase-mediated cassette exchange in Sf9 insect cells for stable gene expression. Biotechnol Bioeng. 109:2836–2844. [DOI] [PubMed] [Google Scholar]
- Fernández FJ, Vega MC.. 2013. Technologies to keep an eye on: alternative hosts for protein production in structural biology. Curr Opin Struct Biol. 23:365–373. [DOI] [PubMed] [Google Scholar]
- Frye C, Deshpande R, Estes S, Francissen K, Joly J, et al. 2016. Industry view on the relative importance of “clonality” of biopharmaceutical-producing cell lines. Biologicals. 44:117–122. [DOI] [PubMed] [Google Scholar]
- Fu Y, Yang Y, Zhang H, Farley G, Wang J, et al. 2018. The genome of the Hi5 germ cell line from Trichoplusia ni, an agricultural pest and novel model for small RNA biology. eLife. 7:e31628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuenmayor J, Cervera L, Gòdia F, Kamen A.. 2019. Extended gene expression for Gag VLP production achieved at bioreactor scale. J Chem Technol Biotechnol. 94:302–308. [Google Scholar]
- Gutiérrez-Granados S, Cervera L, de las Segura MM, Wölfel J, Gòdia F.. 2016. Optimized production of HIV-1 virus-like particles by transient transfection in CAP-T cells. Appl Microbiol Biotechnol. 100:3935–3947. [DOI] [PubMed] [Google Scholar]
- Hacker DL, Balasubramanian S.. 2016. Recombinant protein production from stable mammalian cell lines and pools. Curr Opin Struct Biol. 38:129–136. [DOI] [PubMed] [Google Scholar]
- Krammer F, Palese P.. 2015. Advances in the development of influenza virus vaccines. Nat Rev Drug Discov. 14:167–182. [DOI] [PubMed] [Google Scholar]
- Lakso M, Pichel JG, Gorman JR, Sauer B, Okamoto Y, et al. 1996. Efficient in vivo manipulation of mouse genomic sequences at the zygote stage. Proc Natl Acad Sci USA. 93:5860–5865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauth M, Spreafico F, Dethleffsen K, Meyer M.. 2002. Stable and efficient cassette exchange under non-selectable conditions by combined use of two site-specific recombinases. Nucleic Acids Res. 30:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mabashi-Asazuma H, Jarvis DL.. 2017. CRISPR-Cas9 vectors for genome editing and host engineering in the baculovirus–insect cell system. Proc Natl Acad Sci USA. 114:9068–9073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarroll L, King LA.. 1997. Stable insect cell cultures for recombinant protein production. Curr Opin Biotechnol. 8:590–594. [DOI] [PubMed] [Google Scholar]
- Mori K, Hamada H, Ogawa T, Ohmuro-Matsuyama Y, Katsuda T, et al. 2017. Efficient Production of Antibody Fab Fragment by Transient Gene Expression in Insect Cells. J Biosci Bioeng. 124:221–226. [DOI] [PubMed] [Google Scholar]
- Nehlsen K, da Gama-Norton L, Schucht R, Hauser H, Wirth D.. 2011. Towards rational engineering of cells: recombinant gene expression in defined chromosomal loci. BMC Proc. 5:O6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogay ID, Lihoradova OA, Azimova SS, Abdukarimov AA, Slack JM, et al. 2006. Transfection of insect cell lines using polyethylenimine. Cytotechnology. 51:89–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oumard A, Qiao J, Jostock T, Li J, Bode J.. 2006. Recommended method for chromosome exploitation: RMCE-based cassette-exchange systems in animal cell biotechnology. Cytotechnology. 50:93–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter CJ, Luo L.. 2010. Splinkerette PCR for mapping transposable elements in Drosophila. PLoS One. 5:e10168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puente-Massaguer E, Lecina M, Gòdia F.. 2018. Nanoscale characterization coupled to multi-parametric optimization of Hi5 cell transient gene expression. Appl Microbiol Biotechnol. 102:10495–10510. [DOI] [PubMed] [Google Scholar]
- Qiao J, Oumard A, Wegloehner W, Bode J.. 2009. Novel tag-and-exchange (RMCE) strategies generate master cell clones with predictable and stable transgene expression properties. J Mol Biol. 390:579–594. [DOI] [PubMed] [Google Scholar]
- Sarkar A, Atapattu A, Belikoff EJ, Heinrich JC, Li X, et al. 2006. Insulated piggyBac vectors for insect transgenesis. BMC Biotechnol. 6:27–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schebelle L, Wolf C, Stribl C, Javaheri T, Schnütgen F, et al. 2010. Efficient conditional and promoter-specific in vivo expression of cNAs of choice by taking advantage of recombinase-mediated cassette exchange using FlEx gene traps. Nucleic Acids Res. 38:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sequeira DP, Correia R, Carrondo MJT, Roldão A, Teixeira AP, et al. 2018. Combining stable insect cell lines with baculovirus-mediated expression for multi-HA influenza VLP production. Vaccine. 36:3112–3123. [DOI] [PubMed] [Google Scholar]
- Shen X, Pitol AK, Bachmann V, Hacker DL, Baldi L, et al. 2015. A simple plasmid-based transient gene expression method using High Five cells. J Biotechnol. 216:67–75. [DOI] [PubMed] [Google Scholar]
- Shirk PD, Furlong RB.. 2016. Insect cell transformation vectors that support high level expression and promoter assessment in insect cell culture. Plasmid. 83:12–19. [DOI] [PubMed] [Google Scholar]
- Sorrell DA, Robinson CJ, Smith JA, Kolb AF.. 2010. Recombinase mediated cassette exchange into genomic targets using an adenovirus vector. Nucleic Acids Res. 38:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolt-Bergner P, Benda C, Bergbrede T, Besir H, Celie PHN, et al. 2018. Baculovirus-driven protein expression in insect cells: a benchmarking study. J Struct Biol. 203:71–80. [DOI] [PubMed] [Google Scholar]
- Summers M. 1991. Catalytic subunit of human DNA polymerase a overproduced from baculovirus-infected insect cells. J Biol Chem. 266:22739–22748. [PubMed] [Google Scholar]
- Talsania K, Mehta M, Raley C, Kriga Y, Gowda S, et al. 2019. Genome assembly and annotation of the Trichoplusia ni Tni-FNL insect cell line enabled by long-read technologies. Genes (Basel). 10:79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidigal J, Dias MM, Fernandes F, Patrone M, Bispo C, et al. 2013. A cell sorting protocol for selecting high-producing sub-populations of Sf9 and High Five™ cells. J Biotechnol. 168:436–439. [DOI] [PubMed] [Google Scholar]
- Vidigal J, Fernandes B, Dias MM, Patrone M, Roldão A, et al. 2018. RMCE-based insect cell platform to produce membrane proteins captured on HIV-1 Gag virus-like particles. Appl Microbiol Biotechnol. 102:655–666. [DOI] [PubMed] [Google Scholar]
- Yáñez RJ, Porter ACG.. 1999. Influence of DNA delivery method on gene targeting frequencies in human cells. Somat Cell Mol Genet. 25:27–31. [DOI] [PubMed] [Google Scholar]
- Zhang L, Inniss MC, Han S, Moffat M, Jones H, et al. 2015. Recombinase-mediated cassette exchange (RMCE) for monoclonal antibody expression in the commercially relevant CHOK1SV cell line. Biotechnol Prog. 31:1645–1656. [DOI] [PubMed] [Google Scholar]
- Zitzmann J, Sprick G, Weidner T, Schreiber C, Czermak P.. 2017. Process Optimization for Recombinant Protein Expression in Insect Cells. S.J.T. Gowder (Ed.), New Insights Cell Cult. Technol. (1st ed.), InTech Open, Rijeka (2017), p. 43–98. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Plasmid sequences and annotations have been deposited in the GenBank database (accession numbers MW246023, MW246024, MW246025, MW246026, MW246027, and MW246028). The vector pFluTarget is proprietary rights and its sequence can be specified upon request. Supplementary Table S1 contains the primers used to amplify the fragments for plasmid constructions and restriction enzymes used for vector digestion. Cell populations and cell lines generated in this study are the property of Instituto de Biologia Experimental e Tecnológica (IBET) and will be provided upon request addressed to aroldao@ibet.pt. Supplementary material is available at figshare: https://doi.org/10.25387/g3.14544066.