Abstract
Globally, coronavirus disease (COVID-19) has badly affected the medical system and economy. Sometimes, the deadly COVID-19 has the same symptoms as other chest diseases such as pneumonia and lungs cancer and can mislead the doctors in diagnosing coronavirus. Frontline doctors and researchers are working assiduously in finding the rapid and automatic process for the detection of COVID-19 at the initial stage, to save human lives. However, the clinical diagnosis of COVID-19 is highly subjective and variable. The objective of this study is to implement a multi-classification algorithm based on deep learning (DL) model for identifying the COVID-19, pneumonia, and lung cancer diseases from chest radiographs. In the present study, we have proposed a model with the combination of Vgg-19 and convolutional neural networks (CNN) named BDCNet and applied it on different publically available benchmark databases to diagnose the COVID-19 and other chest tract diseases. To the best of our knowledge, this is the first study to diagnose the three chest diseases in a single deep learning model. We also computed and compared the classification accuracy of our proposed model with four well-known pre-trained models such as ResNet-50, Vgg-16, Vgg-19, and inception v3. Our proposed model achieved an AUC of 0.9833 (with an accuracy of 99.10%, a recall of 98.31%, a precision of 99.9%, and an f1-score of 99.09%) in classifying the different chest diseases. Moreover, CNN-based pre-trained models VGG-16, VGG-19, ResNet-50, and Inception-v3 achieved an accuracy of classifying multi-diseases are 97.35%, 97.14%, 97.15%, and 95.10%, respectively. The results revealed that our proposed model produced a remarkable performance as compared to its competitor approaches, thus providing significant assistance to diagnostic radiographers and health experts.
Keywords: COVID-19, Deep learning, Chest radiographs, Coronavirus
Introduction
In December 2019, several patients with pneumonia with unknown causes were reported in Wuhan, China [1]. Most of the reported patients worked at or lived in the region of the local seafood wholesale market of Huanan [2], where live animals were also available for sale. The initial stages of this pneumonia of unidentified aetiology had severe acute respiratory syndrome (SARS), while some patients had rapidly increasing acute respiratory distress disorder, and other serious complications were also observed [3]. On the day of 7th January, a novel coronavirus (nCoV) was diagnosed from the throat swab sample of a patient by the “Chinese Center for Disease Control and Prevention” (CDC) in Wuhan and was subsequently named 2019nCoV by “World Health Organization” (WHO) [4]. Coronaviruses (CoVs) can cause multiple infections primarily respiratory problems in humans, such as the Middle East respiratory syndrome (MERS) and SARS [5], 6. CoVs have been diagnosed in various mammals, including bats, dogs, mice, and camels, as well as in different avian hosts [7], 8. Among the several CoVs, most of the patients were associated with minor clinical symptoms and a good prognosis [9]. In November 2002, Guangdong, southern China, was affected by a novel beta coronavirus and resulted in more than 8000 human infections [10]. Furthermore, approximately 800 deaths in 37 countries were reported between 2002 and 2003 [11]. In 2012, MERS-CoV was diagnosed in Saudi Arabia, and was responsible for more than 2400 laboratory-confirmed cases of CoV [12]. Moreover, 858 fatalities were reported since September 2012 [13]. A total of 156,784,309 coronavirus -infected cases, 3,272,172 death cases, and 134,181,315 recovered cases have been reported since May 7, 2021, around the globe [14]. Initially, a real-time reverse transcription-polymerase chain reaction (RT-PCR) test was used for the detection of COVID-19 [15]. The execution process of this test is to obtain the DNA of the infected person by applying the reverse transcription method and then assigned to PCR for strengthening DNA for analysis. Hence, it can diagnose coronavirus, because this virus only carries RNA sequences [16]. Nevertheless, the PCR test contains a few limitations such as it takes from few hours to more than 2 days in diagnosing the COVID-19 virus. The results obtained from these PCR kits are not very reliable, because it also produces false-negative outputs [17]. Therefore, our health experts are facing severe issues in diagnosing COVID-19 at an early stage. The researchers are trying to design an efficient and effective solution in classifying COVID-19-infected patients. They choose the X-ray imaging analysis for the identification of coronavirus [18]. Chest X-ray imaging can play a significant role in diagnosing the coronavirus infection by observing some distinct features such as lungs opacity or vague patterns. The manual process of classifying COVID-19 infected chest X-rays may be time-consuming and inappropriate. Therefore, a more advanced and automated system is required for diagnosing coronavirus, which reduces the stress and pressure of diagnostician`s amenities. The detection of COVID-19-infected images makes it easy for the health experts to start the treatment process at the initial stage to minimize the spread of the disease. Significant research has been done in the diagnosis of COVID-19 which saves time and reduces the effort of health experts. Automatic detection of coronavirus at an early stage has been a demanding and effective area of recent research studies. Some other appropriate works for the diagnosis of COVID-19 using different artificial intelligence have also been studied. Oh et al. [29] designed CNN models by adopting a patch-based system applied on scarce amounts of data sets. The final result was taken by the system from voting the majority of patching classifiers results. This experiment was performed on a total of 15,043 images including 8851 normal, 6012 Pneumonia, and 180 COVID-19 positive cases. The remarkable results were achieved by CNN with an accuracy of 88.9%, precision of 83.4%, recall of 85.9%, f1-score of 84.4%, and specificity of 96.4%. Zhang et al. [30] proposed an 18-layers of residual CNN on X-ray images and their work was based on three major contributions. First, they considered the CNN module to get features then work on the classification, and in the last step, they applied the anomaly module for the detection score. This experiment was done on a total of 1531 radiographs in which 100 images were COVID positive and 1431 were pneumonia-infected images. They attained a sensitivity of 96% and specificity of 70.65% for COVID-19 detection. Apostolopoulos et al. [31] proposed the CNN model and applied it to a relatively small amount of datasets. This experiment was performed on two different databases. The first database contained a total of 1427 images including 224 COVID-19 positives, 700 bacterial infection images, and the rest of them belonged to normal patients. The second dataset also contained the same amount of COVID-19 images, pneumonia-infected, and normal cases. MobileNetV2 was applied on the second dataset which generated better accuracy of 96.78%, specificity of 96.46%, and sensitivity of 98.66% among all CNN models. Tsiknakis et al. [32] introduced a new automatic COVID-19 detection model based on a deep learning model named Inception-V3. This experiment was applied on 572 cases that have 150 bacterial and viral infections, 122 images contained COVID positive, and 150 normal cases. They achieved a classification accuracy of 76%. Sethy et al. [33] applied nine different transfer learning approaches on the dataset having 381 chest X-ray images to extract the features from them. They used SVM to diagnose the COVID-19 by these extracted features. The resnet-50 proves the best one for getting features from datasets. A resnet-50 and SVM model shows the accuracy and f1-score of 95.33% and 95.34%, respectively. Saha et al. [34] designed new methods of corona detection based on X-ray images. They named their proposed system EMCNet which was based on simple CNN structure used to extract the features from the images. After extracting the features, they applied an ensemble of machine learning [35] classifiers to classify the COVID-19 infected cases. The EMCNet achieved a significant accuracy of 98.91. Mahmud et al. [36] introduced new automatic COVID detection models using a deep learning model named CovXNet which automatically extracted features from X-ray images. Their deep learning model used depth-wise convolution phenomena for automatic detection. They performed their experiment on pneumonia and normal X-ray images. After this, they applied their proposed model on the classification of COVID-19 and pneumonia images of X-rays. The model consists of gradient-based discriminative localization and a slack algorithm for automatic detection. The CovXNet model showed good accuracy of 97.4% among normal and COVID-19 cases, while generated 90.2% accuracy among all other cases like viral and bacterial infections and COVID-19 cases. Horry et al. [37] applied four renowned transfer learning classifiers for COVID-19 detection on the total 60,798 images of datasets including 322 pneumonia patients, 115 COVID-19 positive cases, and 60,361 normal cases. In all of these four models, VGG16 and VGG19 showed the best results for classification dilemmas. The Vgg-19 achieved the 81% of accuracy in diagnosing the COVID-19-positive cases.
In the disease diagnosis process, deep learning (DL) classifiers have opened a new door [41]. With the collaboration of convolutional neural networks (CNNs), the healthcare system has achieved significant and effective diagnostics advancement in disease detection such as chest disease detection [42], identification of cancer cells [43], brain and breast tumor classification [44], and analysis of genomic sequences [45]. In this study, we have proposed a novel multi-classification model named Best Diagnostic Classifier Network (BDCNet) based on CNN and VGG-19 for diagnosing COVID-19, pneumonia, and lung cancer patients from chest X-ray images. The objective of this paper is to accurately and efficiently classify the COVID-19 and other chest diseases, i.e., pneumonia and lungs cancer-infected patients using chest radiography and to assist our frontline health workers in finding the abnormal patterns that occurred due to these diseases. To the best of our knowledge, there is no other study that introduces the single CNN model for the classification of a collection of chest tract diseases. It also reduces the burden of the health experts to apply different kinds of applications for the diagnosis of each chest disease separately. Furthermore, BDCNet is also compared with four well-renowned medical classifiers such as VGG-16 & vgg-19 [46], ResNet-50 [47], and inception v3 [49].
In summary, the main contribution of this study is stated as follows:
The authors have proposed BDCNet (combination of VGG-19 & CNN) which can extract dominant features from chest radiography to classify the COVID-19, pneumonia, and lung cancer patients. In addition, depth comparison has been done with state-of-the-art classifiers as well as four baseline classifiers such as Vgg-16, Vgg-19, ResNet-50, and Inception v3 in terms of accuracy, specificity, sensitivity, f1-score, and AUC.
2. The BDCNet has been trained and tested on chest radiology images of COVID-19, pneumonia, and lungs cancer X-ray images. The images of COVID-19 gathered from eight different publically available databases such as SIRM database [19], radiopaedia.org [20], GitHub source [21], TCIA [22], Kaggle repository [23], NIH chest X-ray images [24], and Mendeley [25], 26. The dataset of pneumonia images was collected from the Radiological Society of North America (RSNA) [27]. This dataset contains a total of 5216 X-rays including 1349 normal and 3867 pneumonia images. The lungs cancer dataset of chest X-rays and CT scan images was collected from [28], with about 20,000 chest X-rays and CT scans. Furthermore, the CT scan images of lung cancer are not considered in this study. The collected radiographs are applied to the proposed model with the ratio of 70:20:10 for training, validation, and testing, respectively.
The BDCNet showed outstanding performance in terms of accuracy (99.10%), sensitivity (98.31%), precision (99.9%), and F1-score (99.09%).
Additionally, we also designed a novel framework that can diagnose COVID-19 infected patients using chest radiography.
The structure of this research study is summarized as follows: Sect. 2 consists of data preprocessing, datasets description, proposed BDCNet method, and their structure. Section 3 provides the experimental results and discussions. In Sect. 4, this research has been concluded.
Materials and methods
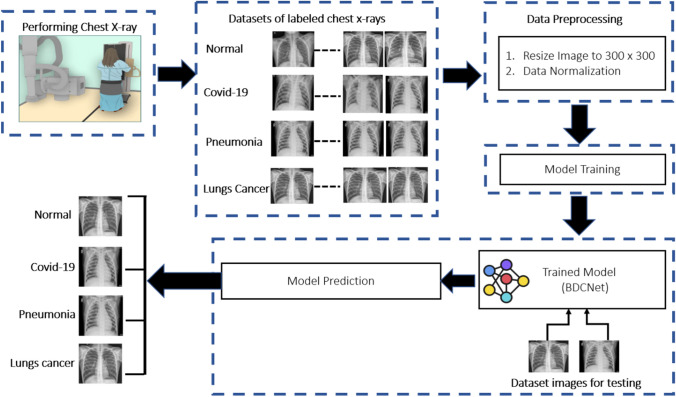
This section consists of an experimental procedure that was conducted to measure the classification accuracy of the proposed model and four baselines pre-trained models such as VGG-16, VGG-19, ResNet-50, and Inception-v3. For this work, we have designed an automated system named BDCNet for the diagnosis of multi-chest diseases, which was trained and tested on ten different datasets including COVID-19, pneumonia, and lung cancer databases. The collected radiograph images have been fixed to the size of 300 × 300 resolution. The data normalization process was also applied on all of the datasets to prevent the model from overfitting. The dataset was divided into three portions such as training, validation, and testing. The proposed model was trained using the training and validation sets of normal and multi-chest disease radiographs. The experimental process was executed for up to 30 epochs. After completing epochs, the proposed model achieved the expected and significant training and validation accuracy. The performance of the proposed system (BDCNet) was evaluated and compared with four pre-trained classifiers in terms of confusion matrix, i.e., accuracy, sensitivity, precision, and f1-score. The model block diagram is illustrated in Fig. 1.
Fig.1.
Datasets image classification approach
Dataset descriptions
For training and testing of the proposed BDCNet, eight COVID-19 datasets were obtained from different resources. Initially, we collected 660 coronaviruses infected chest radiographs from a GitHub repository created by Cohen et al. [38]. This repository contains images collected from different hospitals and open sources. The complete metadata information is not discussed, but the average age of the COVID-19-infected patients was around 55 years. A total of 2371 COVID-19 positive chest radiographs were collected from SIRM database [19], TCIA [39], radiopaedia.org [20], Mendeley [25], 26 including GitHub source [21]. The negative or healthy person chest radiographs were gathered from two repositories such as NIH [24] and Kaggle [40] chest radiology images. The dataset of pneumonia images was collected from the RSNA [27]. This dataset contains a total of 5216 X-rays including 1349 normal and 3867 pneumonia images. The lungs cancer dataset of chest X-rays and CT scan images was collected from [28], with about 20,000 chest X-rays and CT scans. A total of 5000 images of lungs cancer was obtained from the dataset and the rest of the CT scans are not considered in this study. The sample images of COVID-19, pneumonia, lung cancer, and healthy persons are illustrated in Fig. 2.
Fig. 2.
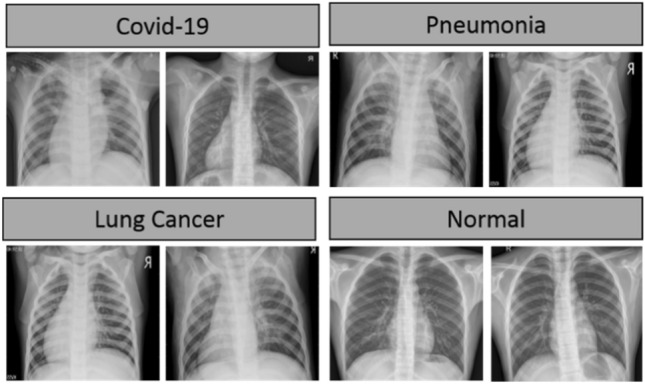
Chest radiograph images
Therefore, the proposed BDCNet was trained and tested on these databases with the split of 70%:10%:20% for training, testing, and validation, respectively. The statistics for the partitioning of the dataset is discussed in Table 1.
Table 1.
Splitting of the dataset for training, testing, and validation
| Dataset splitting | COVID-19 | Pneumonia | Lung cancer | Normal | Total |
|---|---|---|---|---|---|
| Training | 1670 | 2707 | 3500 | 2065 | 9942 |
| Validation | 467 | 387 | 500 | 295 | 1649 |
| Testing | 234 | 773 | 1000 | 589 | 2596 |
| Total | 2371 | 3867 | 5000 | 2949 | 14,187 |
Data preprocessing and normalization
The collected radiology images from publically available databases were of different sizes of resolution. Therefore, we resized the images to the fixed size of resolution which is 300 × 300 pixels. The process of data normalization was also applied for the appropriate training of the proposed method. Hence, our datasets were prepared to be fed into the BDCNet for the training process.
Transfer learning classifiers
This section consists of existing state-of-the-art transfer learning classifiers such as Vgg-16, Vgg-19, AlexNet, ResNet-50, and inception v3, which were applied to achieve the clinical purpose of classification of healthy and COVID-19-infected radiographs. All of these classifiers were trained on the ImageNet (ILSVR) database, which contains thousands of variant kinds of objects used to train and measure the classification performance of the models [48]. Vgg-16 [50] is an open-source framework and is mainly used in a different variety of research contexts [51]. The architecture of the pre-trained Vgg-16 model is based on six steps. The first two steps consist of two convolutional layers with one max-pooling layer having a stride of 2. In further three steps, three convolutional layers are present with one max-pooling layer of stride 2. The final step contains the three fully connected layers (FCLs). In addition, FCL was replaced which allows the model to classify the COVID-19 radiographs using sigmoid functions. Another pre-trained architecture, Vgg-19 [52] is deeper than Vgg-16, and it is more expensive to train the model. ResNet-50 [53] is the 50 layers residual network, and its architecture has four steps. It contains three deep residual networks, in which 1, 64, 64, and 256 kernels were applied. A CNN-based inception model is commonly used for disease classification. Inception is a variation of Inception v3 [54] which was trained on more than millions of images collected from the ImageNet database. The reason behind selecting these four pre-trained classifiers was that they had been widely selected by the researchers [55], evaluated, and recognized as the state-of-the-art classifiers in medical domain image classification.
Proposed CNN architecture
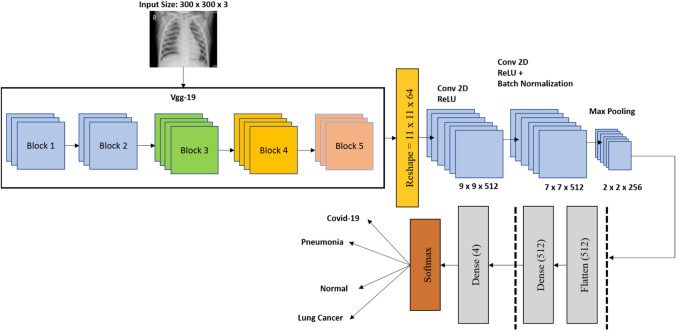
The proposed BDCNet has been designed on pre-trained model VGG19 followed by CNN's network to classify the multi-chest disease using chest radiographs. In general, CNN is based on three variant kinds of layers such as convolutional (Conv) layer, pooling layer (PL), and fully connected layer (FCL). The feature extraction (FE) contains VGG19, followed by two CNN blocks that work as FE portions. The BDCNet model details are represented in Fig. 3.
Fig. 3.
Proposed BDCNet
The input of chest radiographs having a size of 300 × 300 resolutions of radiographs is fed to the proposed model for training purposes. There are RGB channels in the input image. The first layer of our model is the conv layer. This layer starts the process using filters, which are also known as the kernel. The size of the kernel is based on two values, as shown in Eq. 1
| 1 |
where fw represents the filter width, and fh denotes the filter height. In our work, we fixed the size of the filter to the value of 3, and Eq. (1) becomes FS = 3 × 3. These filters help us in obtaining the low-level features of the images such as edges and curves [51], and also called feature identifiers. The three more conv layers have been added to the model to extract the profound features from the radiographs, and thus, provide comprehensive patterns of the images to the model. The filters also start the conv operations with the sub-area of the radiographs. The reason for applying conv operation was to be multiplied and added the filters and the pixel values of the radiographs. The sub-area of the radiographs is also called the receptive field. Then, more conv layers were inserted in the model step by step to increasing the ability of the model for extracting the feature elements. The values of the filter are termed weights, and these weights must be learned by the model at the time of training. The filter executes the conv process from the starting of the radiographs and uninterruptedly moved across the complete radiograph. The method of performing filter conv ends when the whole image was processed. Feature maps during the radiographs yield an array of values. Equation (2) was used to calculate the values of the feature map
| 2 |
where G, I, and FS represent the feature map, input image, and kernel, respectively. i and j were used to obtain the size of the filters. In addition, the indexes of the resultant array were denoted with m and n. The value of moving the filter was confined by one more parameter known as a stride. In our study, we set the value of stride to 1 for all conv layers. By selecting the higher values of stride reduces the spatial dimension and produces some issues; for instance, the receptive field can go outside of the input size. Hence, to overcome these problems, a zero-padding technique was applied which pads the zero around the border of the input radiographs to maintain the output size dimension as same as the input. The value of the stride was obtained using the zero-padding technique as expressed in Eq. (3)
| 3 |
where P represents the zero paddings and h contains the filter height or width, because both of the filter height and width are the same in our study. Conv layers have used different numbers of filters for extracting the multiple and dominant features. The first conv layer of our model contains 16 filters. The number of filters of other conv layers was changed from 16 to 512. The output volume which is also known as the activation map was calculated using Eqs. (4), (5), and (6), respectively
| 4 |
| 5 |
| 6 |
where Iw and Ih represent the height and width of the input image, and fw and fh are the filter size in terms of width and height. P shows the zero-padding and D is for stride, while Fn contains the number of filters applied to the conv layers. The first conv layer of our proposed model (see Table 2) consisted of Iw = 300, Ih = 300, fw = 3, fh = 3, D = 1, P = 0, and Fn = 16. Using Eqs. (4–6), the following values are calculated:
Table 2.
BDCNet model summary
| Layer (type) | Shape | Parameters |
|---|---|---|
| Vgg19 (layers) | (30,30,32) | 9248 |
| reshape (Reshape) | (28,28,32) | 9248 |
| Conv2d (Conv2D) | (26,26,32) | 9248 |
| Max_pooling2d_3 (MaxPooling2D) | (13,13,32) | 0 |
| dropout_3 (Dropout) | (13,13,32) | 0 |
| Conv2d (Conv2D) | (11,11,64) | 18,496 |
| Conv2d (Conv2D) | (9,9,32) | 18,464 |
| Conv2d(Conv2D) | (7,7,32) | 9248 |
| Max_pooling2d (MaxPooling2D) | (3,3,32) | 0 |
| dropout(Dropout) | (3,3,32) | 0 |
| Conv2d (Conv2D) | (1,1,64) | 18,496 |
| flatten (Flatten) | 64 | 0 |
| dense (Dense) | 512 | 33,280 |
| dropout (Dropout) | 512 | 0 |
| Dense (Dense) | 2 | 1026 |
The activation function rectified linear unit (ReLU) was applied on the conv outputs. The process of ReLU activation is to change all the negative results by zero. The purpose to use the ReLU function in our proposed CNN model is that it improves the nonlinearity and helps to increase the computational time without influencing the model accuracy [56], 57. The max-pooling (MP) layer was inserted after the conv layers which helps to reduce the spatial dimension of the input radiographs. In BDCNet, we fixed the filter of the size of 2 × 2 and also set the value of stride to 2 for MP layers. This filter convolves all over the input volume and produced the highest value of the sub-area of the radiographs. It has been observed that MP layers consist of a particular feature`s position about other features. It also reduces the computational cost by decreasing the number of weights and prevents the model from overfitting. Then, the dropout layer was added. We have applied different values of the dropout to our model such as 0.05, 0.20, and 0.25, but only the appropriate and significant threshold of 0.20 was reported in this study. The reason behind applying the different dropout values was to prevent our proposed model from overfitting [51]. The working of this layer was to drop out the activation randomly and also confirm the model to predict the actual label. The flattened layer of our model was applied to convert the two-dimensional feature map into a one-dimensional feature vector. The data generated by the flatten layer were fed to the FCL. The FCL performed the classification process from the obtained one-dimensional feature vector. For this study, FCL contains 512 neurons. The initial dense layer of FCL passes the result of activation to the other dense layer. The last output of the model is developed from a dense layer with four neurons and a softmax activation function, which classifies the output image into one of the chest diseases classes: COVID-19, pneumonia, lung cancer, or normal. The architecture of the proposed BDCNet model is presented in Fig. 3. The overall number of parameters is 15,733,060, which are divided into two portions; the total number of trainable parameters (TP) is 15,733,000, and the non-trainable parameters (NTP) are 60. The difference between TP and NTP is that the TPs are the parameters that change in time of training and are required by training to get the optimal value of these parameters, while NTPs are the parameters that are not changed at the time of the training process. Thus, the NTPs will not contribute during the classification phase. The layer used in our proposed BDCNet and their detailed description are presented in Table 2.
Performance evaluation
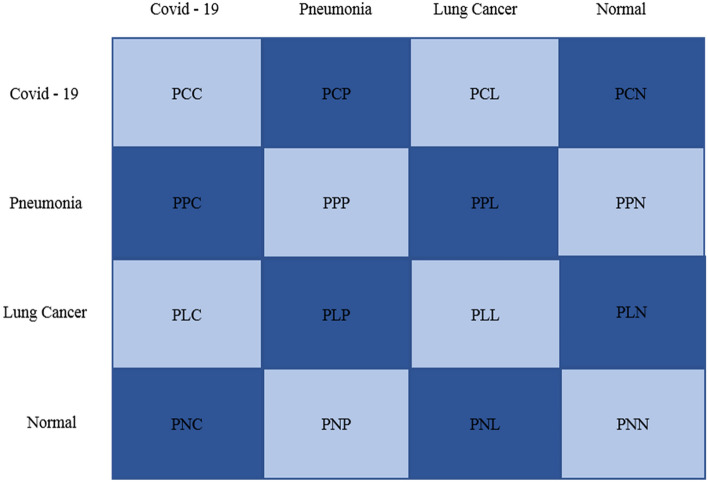
In this study, we solved the multi-classification problem, i.e., COVID-19, pneumonia, lung cancer, and healthy cases, so a confusion matrix was used to evaluate the model’s performance. Figure 4 represents the structure of the confusion matrix.
Fig.4.
Confusion matrix
The parameters of the confusion matrix defined in Fig. 4 are discussed in Table 3.
Table 3.
Parameters used in confusion matrix along with their descriptions
| Parameters | Descriptions |
|---|---|
| PCC | COVID-19 were correctly classified as covid-19 |
| PCP | COVID-19 were incorrectly classified as pneumonia |
| PCL | COVID-19 were correctly classified as lung cancer |
| PCN | COVID-19 were correctly classified as normal |
| PPC | Pneumonia was incorrectly classified as COVID-19 |
| PPP | Pneumonia was correctly classified as pneumonia |
| PPL | Pneumonia was incorrectly classified as lung cancer |
| PPN | Pneumonia was incorrectly classified as normal |
| PLC | Lung cancer was incorrectly classified as COVID-19 |
| PLP | Lung cancer was incorrectly classified as pneumonia |
| PLL | Lung cancer was correctly classified as lung cancer |
| PLN | Lung cancer was incorrectly classified as normal |
| PNC | Normal was incorrectly classified as COVID-19 |
| PNP | Normal was incorrectly classified as pneumonia |
| PNL | Normal was incorrectly classified as lung cancer |
| PNN | Normal was correctly classified as normal |
The performance of our proposed BDCNet was calculated in terms of four metrics including accuracy (Acc), sensitivity (SN) is also called recall (R), precision (P), and F1-score (F). These metrics were measured by the following formulas which are given below in Eqs. (7–10).
| 7 |
| 8 |
| 9 |
| 10 |
We also describe the variables of Eqs. (7–10) for True Positives (TP), True Negatives (TN), False Positives (FP), and False Negatives (TN) as in Table 4.
Table 4.
Confusion matrix equations
| Labels | TP | TN | FP | FN |
|---|---|---|---|---|
| Normal | PNN | PPC + PPL + PCL + PLC + PLP + PCP + PCC + PLL + PPP | PNL + PNC + PNP | PLN + PCN + PPN |
| Pneumonia | PPP | PNL + PNC + PLN + PCN + PNN + PCL + PLC + PCC + PLL | PPL + PPC + PPN | PLP + PCP + PNP |
| COVID-19 | PCC | PLL + PPL + PNL + PNP + PPP + PLP + PLN + PPN + PNN | PCN + PCP + PCL | PNC + PPC + PLC |
| Lung cancer | PLL | PNN + PPN + PCN + PNP + PPP + PCP + PNC + PPC + PCC | PLC + PLP + PLN | PCL + PPL + PNL |
Results and discussion
The VGG19 + CNN-based BDCNet has been designed to classify different chest diseases using chest X-rays. Different hyperparameters such as learning rate, epochs, and batch size of the proposed model were fine-tuned using grid search methods. The BDCNet was trained up to 30 epochs. The layers of all four pre-trained models and BDCNet were set to 0.05 based on the initial learning rate using a “stochastic gradient descent” (SGD) optimizer with the momentum of 0.8. After 10 epochs, the value of the learning rate was reduced by a factor of 0.1. The reason behind doing this was to prevent the model from overfitting. The batch size was set to 20 for our proposed BDCNet. The performance of the proposed BDCNet, Vgg-16, Vgg-19, ResNet-50, and inceptionv3 for each class label was calculated in terms of accuracy, sensitivity, F1-score, precision, confusion matrix, and ROC curve.
Experimental process
The BDCNet and all four pre-trained models were implemented with the help of the Keras library. The methods which were not directly linked with the conv network were programmed in the python programming language. The experiment was run on a Windows-based operating system with 16 GB GPU NVIDIA GeForce GTX and 32 GB RAM.
BDCNet results
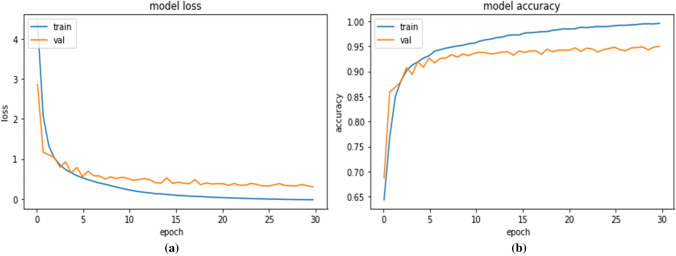
The training accuracy and validation accuracy of our proposed BDCNet relating to 30 epochs are shown in Fig. 5. It has been noticed that the highest obtained training accuracy was 99.92% and validation accuracy was 95.01%. The training and validation loss of our proposed model was 0.0011 and 0.069, respectively. These outputs showed that our proposed BDCNet was trained well and can correctly identify the multi-chest diseases such as COVID-19, pneumonia, lung cancer, and normal radiographs.
Fig.5.
Performance analysis of BDCNet: (a) training and validation loss; (b) training and validation accuracy
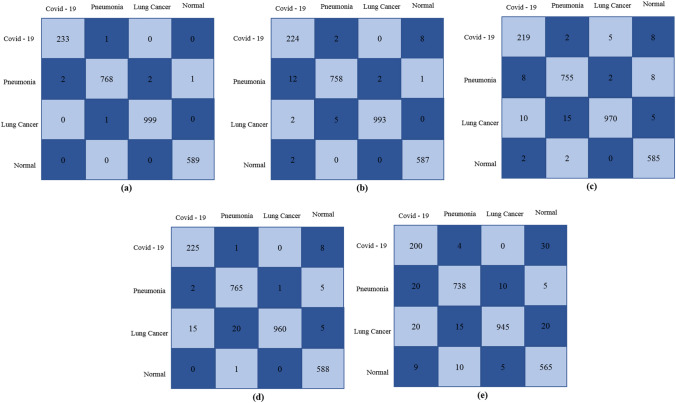
To evaluate the performance of the proposed BDCNet and four pre-trained models, several performance metrics were applied for measuring the classification of COVID-19, pneumonia, lung cancer, and normal X-rays. The dataset was divided with the ratio of 70:20:10 for training, validation, and testing, respectively. Therefore, a total of 2596 X-rays including 234 COVID-19, 773 pneumonia, 1000 lung cancer, and 589 normal images were used to measure the classification accuracy of BDCNet and other transfer learning classifiers. The confusion matrix of all these classifiers is presented in Fig. 6. In the confusion matrix (see Fig. 4), the rows represent the actual cases, while columns show the predicted cases output.
Fig.6.
Confusion matrix: (a) proposed model; (b) Vgg-16; (c) Vgg-19; (d) Resnet-50; (e) Inception v3
Figure 6 illustrates that, in a total of 234 coronavirus infected radiographs, our proposed CNN model accurately identified 233 COVID-19 cases and misclassified 1 case as pneumonia. In classifying pneumonia diagnosed radiographs, our model accurately identified the 768 pneumonia cases and misclassified 2 cases as COVID-19, 2 cases as lung cancer, and 1 case as normal. For lung cancer, among 1000 chest rays, BDCNet accurately classifies 999 cancer-infected X-rays and misclassifies 1 case as pneumonia. Our proposed model also predicted the exact class label for all non-infected cases. The vgg-16 classifier correctly detected the 224 COVID-19, 758 pneumonia, 993 lung cancer, and 587 normal cases. Vgg-19 predicted the 219 coronavirus radiographs and misclassified 2 infected cases as pneumonia, 5 cases of lung cancer, and 8 cases as normal. In addition, Resnet-50 and inception-v3 also diagnosed the 225 and 200 COVID-19 cases respectively, among 234 cases. The results measured from evaluation parameters such as accuracy (Acc), sensitivity (SN) or recall (R), precision (P), and F1-score (F) for proposed BDCNet and other four transfer learning classifiers are shown in Table 5.
Table 5.
Performance comparison of BDCNet with pre-trained classifiers
| Models | Accuracy (%) | Recall (%) | Precision (%) | F1-Score (%) |
|---|---|---|---|---|
| Vgg-16 | 97.35 | 97.14 | 98.99 | 97.46 |
| Vgg-19 | 96.14 | 95.76 | 98.20 | 96.29 |
| ResNet-50 | 97.15 | 98.30 | 98.50 | 98.83 |
| Inception-v3 | 95.10 | 95.80 | 96.66 | 98.83 |
| BDCNet | 99.10 | 98.31 | 99.9 | 99.09 |
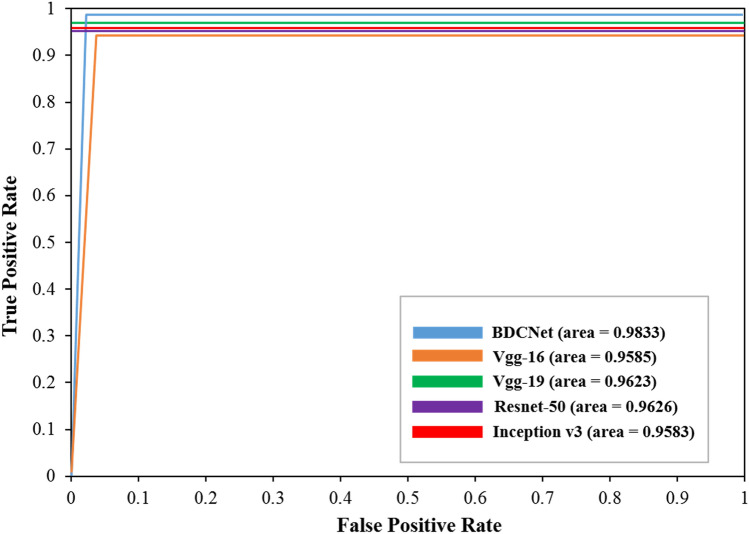
From Table 5, our proposed CNN model achieved remarkable results, i.e., 99.10% accuracy, 98.31% recall or sensitivity, 99.9% precision, and 99.09% f1-score for classifying normal and COVID-19 classes. The vgg-16 model achieved the accuracy of 97.35%, 97.14% precision, 98.99% recall, and 97.46% f1-score. The outputs attained by the resnet-50 were 97.15%, 98.30%, 98.50%, and 98.83% for accuracy, recall, precision, and f1-score, respectively. It has been also noticed that inception-v3 produced poor results as compared to its other competitor approaches. Finally, the classification accuracy of our proposed CNN is greater as compared to the other four pre-trained baseline classifiers. Because all these pre-trained classifiers contain deep networks, and their spatial resolution of the feature map for their last conv layer results has been significantly moderated, which affects their classification accuracy. Moreover, the filter size of these pre-trained classifiers is not appropriate for this kind of dilemma, ignored key features, and developed the great input related to the neuron’s receptive fields. Hence, our proposed CNN model overcomes the issues of low resolution and overlapping in the inflammatory portion of COVID-19-infected radiology images. Our model also contains the significant size of the filters and accelerates the convergence while significantly reducing the negative impact of the structured noise and further improving the classification performance. The model is considered to be appropriate and efficient if it achieved the highest value of area under the curve (AUC) of ROC. The ROC curve is measured using true-positive rate (TPR) and false-positive rates (FPR). Figure 7 represents the AUC (ROC) of our proposed CNN and other four baseline pre-trained classifiers. The proposed CNN classifier achieved an AUC of 0.9833. For Vgg-16, Vgg-19, Resnet-50, and inception-v3, the AUC were 0.9585, 0.9423, 0.9626, and 0.9183, respectively. The results of AUC (ROC) divulge that our proposed classifier outperforms the other four classifiers. In addition, our proposed model would be helpful for human diagnosticians in diagnosing the COVID-19 pandemic infected cases from chest radiography.
Fig. 7.
ROC of BDCNet and pre-trained classifiers
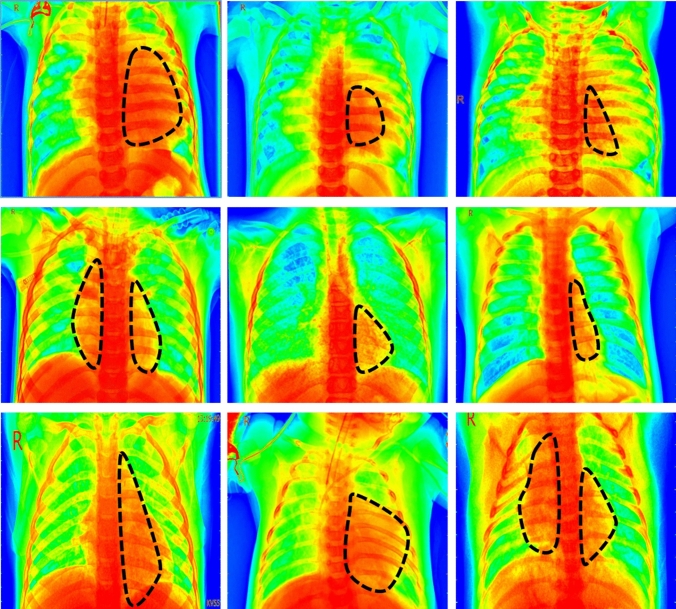
Furthermore, we also applied the Grad-CAM [1] heatmap method to visually illustrate the output produced by our proposed model. The function of the heatmap is to highlight the significant portion that the model emphasizes on the chest radiographs. Figure 8. depicts the heatmap of BDCNet model. In addition, the black dotted line represents the infected portion of the lungs.
Fig. 8.
The first row represents the heatmap of COVID-19, the second shows pneumonia, and the last one is of lung cancer
Comparison with the state-of-the-art
Table 6 determines the comparison of our proposed classifier BDCNet with other state-of-the-art methods in terms of accuracy, precision, recall, and f1-score.
Table 6.
Comparison of BDCNet with state-of-the-art classifiers
| Ref. | Classifiers | Accuracy (%) | Precision (%) | Recall (%) | F1-Score (%) |
|---|---|---|---|---|---|
| [30] | CNN | 72.77 | 73.83 | 71.70 | – |
| [32] | Inception-v3 | 76.0 | 87 | 93 | – |
| [55] | GoogleNet | 80.56 | 84.17 | 80.56 | 82.32 |
| [29] | CNN | 88.9 | 83.4 | 85.9 | 96.4 |
| [56] | Xception + ResNet50 V2 | 91.4 | 21.95 | 87.09 | – |
| [33] | ResNet50 + SVM | 95.33 | – | 95.33 | 95.34 |
| [31] | MobileNet v2 | 96.78 | 96.46 | 98.66 | – |
| [34] | CNN + Ensemble of ML classifiers | 98.91 | 100 | 97.82 | 98.89 |
| [60] | CNN | 93.2 | 98.7 | 96.06 | 97.90 |
| [61] | Xception | 97.97 | 97.0 | 78.0 | 86.0 |
| [62] | PAM-DenseNet | 94.29 | 93.75 | 95.74 | – |
| [63] | ECONET | 96.07 | 96.28 | 96.26 | 96.26 |
| [64] | EfficientNet | 96.70 | 96.69 | 97.54 | 97.11 |
| [65] | C + EffxNet | 99.0 | 98.0 | 98.0 | 98.0 |
| Proposed model | BDCNet | 99.10 | 98.31 | 99.9 | 99.09 |
Discussion
Chest radiographs can be used for the screening of multi-chest disease classification. It provides a comprehensive image of a certain region, using which we can diagnose the disease and internal infected parts. Chest radiography is a more reliable and efficient method especially for the detection of COVID-19, lung cancer, and pneumonia patients as compared to the current process of RT-PCR. However, the COVID-19 cases are increasing day by day; an automatic diagnosis process was needed to initially diagnose this deadly virus. With the use of deep learning (DL) techniques, we can automatically distinguish the COVID-19-positive cases and other chest tract diseases from chest radiography. Therefore, we designed a deep learning-based BDCNet model which significantly diagnoses COVID-19, pneumonia, and lung cancer, which helps the clinical experts to start the treatment process of such patients at the initial stage. The above-performed experimental work shows that our proposed BDCNet is efficiently and significantly trained on COVID-19, lung cancer, and pneumonia infection occurred on the lungs, and our model accurately classifies these infected cases. In comparison to the classification performance of the other four pre-trained classifiers, our BDCNet model has outstanding classification in the domain of healthy, COVID-19, lung cancer, and pneumonia-infected chest radiographs, having an accuracy of 99.10%. Our BDCNet model and transfer learning classifiers such as vgg-16, vgg-19, Resnet-50, and inception-v3 were trained on the datasets with the fixed image resolution of 300 × 300 × 3. In addition, the cross-entropy loss function was used for training the proposed model. The comparative classification performance in terms of different metrics for the proposed BDCNet and all four pre-trained classifiers tested in this study are presented in Table 5. It is observed that our proposed model has achieved notable performance, with the highest AUC of 0.9833, recall of 99.9%, precision of 98.31%, f1-score of 99.09%, and accuracy of 99.10%. The diagnostic performances of other competitors’ methods of transfer learning techniques with pre-trained weights were slightly degraded. The Resnet-50 model also produced a significant AUC score of 0.9626 as compared to other Vgg-16, vgg-19, and inception classifiers. Among all, inception-v3 generates the less score of AUC (0.9583), recall of 95.80%, precision of 96.66%, f1-score of 98.83%, and accuracy of 95.10%. Generally, the selection of CNN-based pre-trained architecture did not influence the overall binary classification dilemma. These pre-trained classifiers performed better at diagnosing disease from multiple categories [58], or for erudite problems such as segmentation. In addition, some researchers [51, 58, 59] believe that these networks did not work appropriately when the number of CNN layers increases for the specific binary classification issues. In Table 6, we also compare the classification accuracy of our proposed model with state-of-the-art classifiers. The analysis of experimental results with state-of-the-art methods demonstrates that our proposed method of classifying COVID-19, lung cancer, and pneumonia-infected radiographs has added significant output in assisting the doctors. Zhang et al., [30], Apostolopoulos et al. [31], Tsiknakis et al.’s [32] and work presented by Saha et al. [34] gained the overall classification performance of the deep learning methods on multi-chest diseases using X-rays were 72.77% (CNN), 96.78% (MobileNet v2), 76% (inception v3), and 98.91% (CNN and ensemble of ML classifiers), respectively. Xiao et al. [62] designed a CNN model called dense connectivity network with parallel attention module (PAM-DenseNet) for the classification of COVID-19 using lungs CT scans. Their proposed model achieves promising results with an accuracy of 94.29%, a precision of 93.75%, a sensitivity of 95.74%, and a specificity of 96.77%. Chowdhury et al. [65] developed an ensemble of Convolutional Neural Network (CNN) based on EfficientNet, named ECOVNet, to detect COVID-19, normal, and pneumonia from chest X-rays. The results show that the ECOVNet significantly improves detection performance with an overall accuracy of 96.07%. Marques et al. [64] proposed an automated system to support the diagnosis of COVID-19 patients using EfficientNet, and they achieved the average accuracy value for multi-classification was 96.70%. Canayaz [65] designed a novel hybrid approach for COVID-19 diagnosis on CT images based on CBAM and EfficientNet. Their approach accurately predicts COVID-19 with a 99% accuracy rate. The results of Table 5 also show that our method is more capable of finding the patterns of abnormalities and extracting the discriminative sequences in classifying different chest diseases from radiographs samples, with the highest accuracy result of 99.10%. The results produced by the different pre-trained classifiers are shown in Table 5 and in conjunction with the examination of the nature of COVID-19, pneumonia, and lung cancer-infected chest radiographs classification task, we provide the detailed elucidation why prior arts show less diagnostics performance. Firstly, the pre-trained image classifier consists of deep networks, and their last conv layers have reduced the feature map of spatial resolution, which confines the classification performance of the models. Also, the filter size of these networks is not appropriate due to which the neurons linked with the input are large which directly ignores the significant features. Our BDCNet model can overcome these issues. We proposed a Vgg-19-based CNN model with the amalgamation of dilated convolution values for classifying the chest diseases using radiographs from different databases. Our BDCNet model has also overcome the problem of low resolution and overlapping in the inflammatory portion of chest radiography. In addition, our model accelerates the convergence while significantly reducing the negative impact of the structured noise and further enhancing the diagnostic performance. Furthermore, we also applied a suitable filter size of 3 × 3 to the proposed model. The analysis of experimental results demonstrates that our proposed method for multi-classification of chest diseases from radiographs based on lungs opacity has added significant and appropriate output in assisting human diagnosticians.
Conclusion
In the present study, a multi-classification BDCNet model was developed and evaluated for detecting multi-chest problems such as COVID-19, pneumonia, and lung cancer from chest radiographs. Nowadays worldwide, the COVID-19 and other chest infection diseases are growing rapidly and affecting the community. With an immense number of cases, a quick and efficient testing process is required. Many people have been died and admitted into ICUs due to inappropriate and slow testing processes, inadequate amenities, and unable to diagnose multiple chest ailments at an early stage. We have proposed a framework for multi-chest diseases detection named as BDCNet which can contribute to automatically detect the COVID-19, lung cancer, and pneumonia-infected patients from chest radiographs. The CNN-based BDCNet can extract foremost patterns from chest radiographs. An extensive experiment shows that our proposed BDCNet has the best diagnostic performance as compared to renowned pre-trained and state-of-the-art classifiers. Considering all these outputs, it is concluded that BDCNet can play a vital role as a helping hand of medical experts. It is also highly capable of relieving stress and pressure on paramedical staff and helps them in controlling the epidemic. In our future work, we will consider more datasets for chest radiographs and extract their features using pre-trained models. Integration of deep learning into radiology systems could be very helpful to suggest at the point of care, and it can enhance the quality, ease of use, and cost of chest diseases diagnosing from chest X-rays worldwide.
Funding
No funding was received.
Availability of data and materials
Not applicable.
Code availability
Not applicable.
Declarations
Conflict of interest
The authors declare no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Lu H, Stratton CW, Tang YW. Outbreak of pneumonia of unknown etiology in Wuhan China: the mystery and the miracle. J Med Virol. 2020 doi: 10.1002/jmv.25678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hui DS, Azhar EI, Madani TA, et al. The continuing 2019nCoV epidemic threat of novel coronaviruses to global health—the latest 2019 novel coronavirus outbreak in Wuhan, China. Int J Infect Dis. 2020;91:264–266. doi: 10.1016/j.ijid.2020.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020 doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.WHO (2020) Clinical management of severe acute respiratory infection when Novel coronavirus (nCoV) infection is suspected: interim guidance. Jan 11, 2020. https://www.who.int/internalpublicationsdetail/clinicalmanagementofsevereacuterespiratoryinfectionwhennovelcoronavirus(ncov)infectionissuspected. Accessed 20 Jan 2020
- 5.Yin Y, Wunderink RG. MERS, SARS and other coronaviruses as causes of pneumonia. Respirology. 2018;23:130–137. doi: 10.1111/resp.13196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Drosten C, Günther S, Preiser W, et al. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N Engl J Med. 2003;348:1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- 7.Cavanagh D. Coronavirus avian infectious bronchitis virus. Vet Res. 2007;38:281–297. doi: 10.1051/vetres:2006055. [DOI] [PubMed] [Google Scholar]
- 8.Ismail MM, Tang AY, Saif YM. Pathogenicity of turkey coronavirus in turkeys and chickens. Avian Dis. 2003;47:515–522. doi: 10.1637/5917. [DOI] [PubMed] [Google Scholar]
- 9.Su S, Wong G, Shi W, et al. Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol. 2016;24:490–502. doi: 10.1016/j.tim.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peiris JS, Guan Y, Yuen KY. Severe acute respiratory syndrome. Nat Med. 2004;10(suppl 12):S88–S97. doi: 10.1038/nm1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chan-Yeung M, Xu RH. SARS: epidemiology. Respirology. 2003;8(suppl):S9–14. doi: 10.1046/j.1440-1843.2003.00518.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zaki AM, van Boheemen S, Bestebroer TM, Osterhaus AD, Fouchier RA. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med. 2012;367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 13.Lee J, Chowell G, Jung E. A dynamic compartmental model for the Middle East respiratory syndrome outbreak in the Republic of Korea: a retrospective analysis on control interventions and superspreading events. J Theor Biol. 2016;408:118–126. doi: 10.1016/j.jtbi.2016.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Worldometer. Coronavirus update (live): cases and deaths from COVID-19 virus pandemic. Worldometers (2021). https://www.worldometers.info/coronavirus/?. Accessed 4 Mar 2021
- 15.Tahamtan A, Ardebili A. Real-time RT-PCR in COVID-19 detection: issues affecting the results. Expert Rev Mol Diagn. 2020;20(5):453–454. doi: 10.1080/14737159.2020.1757437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Corman VM, et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020 doi: 10.2807/1560-7917.ES.2020.25.32.2001483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lauer SA, et al. The incubation period of coronavirus disease 2019 (CoVID-19) from publicly reported confirmed cases: estimation and application. Ann Intern Med. 2020;172(9):577–582. doi: 10.7326/M20-0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Islam MM, Karray F, Alhajj R, Zeng J. A review on deep learning techniques for the diagnosis of novel coronavirus (COVID-19) IEEE Access. 2021;9:30551–30572. doi: 10.1109/ACCESS.2021.3058537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.COVID-19 Database | SIRM [Online] (2020). https://www.sirm.org/en/category/articles/covid-19-database/. Accessed 10 Feb 2021
- 20.Gaillard, F.: Radiopaedia.org, the wiki-based collaborative Radiology resource. Radiopaedia.org [Online] (2014). http://radiopaedia.org/. Accessed 22 Feb 2021
- 21.COVID 19 chest X-ray [Online] (2020). https://github.com/agchung. Accessed 10 Feb 2021
- 22.Frederick Nat Lab: The cancer imaging archive (TCIA). The Cancer Imaging Archive: 1 [Online] (2018). https://www.cancerimagingarchive.net. Accessed 23 Feb 2021
- 23.Mooney P. Chest X-ray images (pneumonia) | Kaggle. Kaggle.com; 2018. https://www.kaggle.com/paultimothymooney/chest-xray-pneumonia%0A. https://data.mendeley.com/datasets/rscbjbr9sj/2. Accessed 30 Aug 2020
- 24.NIH chest X-rays (2018) Kaggle [Online]. Available: https://www.kaggle.com/nih-chest-xrays/data. Accessed 10 Feb 2021
- 25.Alqudah AM, Qazan S (2020) Augmented COVID-19 X-ray images dataset: 4. 10.17632/2FXZ4PX6D8.4.
- 26.Malik HA, Tayyaba NA (2020) Chest radiographs of COVID-19 infected. Mendeley Data. 10.17632/67dmnmx33v.1
- 27.Kermany DS, Goldbaum M, Cai W, Valentim C, Liang H, Baxter SL, et al. Identifying medical diagnoses and treatable diseases by image-based deep learning. Cell. 2018;172:1122–1131. doi: 10.1016/j.cell.2018.02.010. [DOI] [PubMed] [Google Scholar]
- 28.Shiraishi J, Katsuragawa S, Ikezoe J, Matsumoto T, Kobayashi T, Komatsu KI, et al. Development of a digital image database for chest radiographs with and without a lung nodule: receiver operating characteristic analysis of radiologists' detection of pulmonary nodules. Am J Roentgenol. 2000;174(1):71–74. doi: 10.2214/ajr.174.1.1740071. [DOI] [PubMed] [Google Scholar]
- 29.Oh Y, Park S, Ye JC. Deep learning COVID-19 features on CXR using limited training data sets. IEEE Trans Med Imag. 2020;39(8):2688–2700. doi: 10.1109/TMI.2020.2993291. [DOI] [PubMed] [Google Scholar]
- 30.Zhang J, Xie Y, Li Y, Shen C, Xia Y (2020) COVID-19 screening on chest X-ray images using deep learning based anomaly detection 1. aeXiv.2002.12338
- 31.Apostolopoulos ID, Mpesiana TA. COVID-19: automatic detection from X-ray images utilizing transfer learning with convolutional neural networks. Phys Eng Sci Med. 2020;43(2):635–640. doi: 10.1007/s13246-020-00865-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tsiknakis N, et al. Interpretable artificial intelligence framework for COVID-19 screening on chest X-rays. Exp Ther Med. 2020 doi: 10.3892/etm.2020.8797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sethy PK, Behera SK, Ratha PK, Biswas P. Detection of coronavirus disease (COVID-19) based on deep features and support vector machine. Int J Math Eng Manag Sci. 2020;5(4):643–651. doi: 10.33889/IJMEMS.2020.5.4.052. [DOI] [Google Scholar]
- 34.Saha P, Sadi MS, Islam MM. EMCNet: Automated COVID-19 diagnosis from X-ray images using convolutional neural network and ensemble of machine learning classifiers. Inform Med unlocked. 2021;22:100505. doi: 10.1016/j.imu.2020.100505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Singh H, Bawa S. Predicting COVID-19 statistics using machine learning regression model: Li-MuLi-Poly. Multimedia Syst. 2021 doi: 10.1007/s00530-021-00798-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mahmud T, Rahman MA, Fattah SA. CovXNet: a multi-dilation convolutional neural network for automatic COVID-19 and other pneumonia detection from chest X-ray images with transferable multi-receptive feature optimization. Comput Biol Med. 2020 doi: 10.1016/j.compbiomed.2020.103869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Horry, M., et al. (2020) X-ray image based COVID-19 detection using pre-trained deep learning models. engrXiv. 10.31224/osf.io/wx89s
- 38.Cohen JP, Morrison P, Dao L, Roth K, Duong TQ, Ghassemi M (2020) COVID-19 image data collection: prospective predictions are the future [Online]. Available: http://arxiv.org/abs/2006.11988
- 39.Frederick Nat Lab. The cancer imaging archive (TCIA). The Cancer Imaging Archive; 2018. p. 1 [Online]. Available: https://www.cancerimagingarchive.net/about-the-cancer-imaging-archive-tcia/%0A. https://www.cancerimagingarchive.net/%0A. https://www.cancerimagingarchive.net/%0A. http://www.cancerimagingarchive.net/.
- 40.Mooney P (2018) Chest X-ray images (pneumonia) | Kaggle. Kaggle.com. https://www.kaggle.com/paultimothymooney/chest-xray-pneumonia%0A. https://data.mendeley.com/datasets/rscbjbr9sj/2. Accessed 3 Mar 2021
- 41.Rastgarpour M, Shanbehzadeh J (2011) Application of AI techniques in medical image segmentation and novel categorization of available methods and tools. In: IMECS 2011—international Multi Conference of engineers and computer scientists 1: 519–23.
- 42.Hwang EJ, et al. Development and validation of a deep learning-based automated detection algorithm for major thoracic diseases on chest radiographs. JAMA Netw Open. 2019;2(3):e191095. doi: 10.1001/jamanetworkopen.2019.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cireşan DC, Giusti A, Gambardella LM, Schmidhuber J. Mitosis detection in breast cancer histology images with deep neural networks. Lect Notes Comput Sci. 2013;8150:411–418. doi: 10.1007/978-3-642-40763-5_51. [DOI] [PubMed] [Google Scholar]
- 44.Mohsen H, El-Dahshan ESA, El-Horbaty ESM, Salem ABM. Classification using deep learning neural networks for brain tumors. Futur Comput Informatics J. 2018;3(1):68–71. doi: 10.1016/j.fcij.2017.12.001. [DOI] [Google Scholar]
- 45.Quang D, Xie X. DanQ: a hybrid convolutional and recurrent deep neural network for quantifying the function of DNA sequences. Nucleic Acids Res. 2016;44(11):e107. doi: 10.1093/nar/gkw226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Simonyan K, Zisserman A (2015) Very deep convolutional networks for large-scale image recognition. In International Conference on Learning Representations, ICLR
- 47.He K, Zhang X, Ren S, Sun J (2016) Deep residual learning for image recognition. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, IEEE: 770–778
- 48.Wang Q, Yang D, Li Z, Zhang X, Liu C. Deep regression via multi-channel multi-modal learning for pneumonia screening. IEEE Access. 2020;8:78530–78541. doi: 10.1109/ACCESS.2020.2990423. [DOI] [Google Scholar]
- 49.Huang G, Liu Z, Van Der Maaten L, Weinberger, Q. (2017) Densely connected convolutional networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, IEEE: 4700–4708
- 50.Janizek JD, Erion G, DeGrave AJ, Lee SI (2020) An adversarial approach for the robust classification of pneumonia from chest radiographs. In: Proceedings of the ACM Conference on Health, Inference, and Learning: 69–79
- 51.Malik H, Farooq MS, Khelifi A, Abid A, Qureshi JN, Hussain M. A comparison of transfer learning performance versus health experts in disease diagnosis from medical imaging. IEEE Access. 2020 doi: 10.1109/ACCESS.2020.3004766. [DOI] [Google Scholar]
- 52.He K, Zhang X, Ren S, Sun J (2016)Deep residual learning for image recognition. Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition 2016: 770–778
- 53.Szegedy C, Ioffe S, Vanhoucke V, Alemi AA (2017) Inception-v4, inception-ResNet and the impact of residual connections on learning. Presented at the Proceedings of the Thirty-First AAAI Conference on Artificial Intelligence, San Francisco, California, USA
- 54.Russakovsky O, Deng J, Su H, et al. ImageNet large scale visual recognition challenge. Int J Comput Vision. 2015;115:211–252. doi: 10.1007/s11263-015-0816-y. [DOI] [Google Scholar]
- 55.Loey M, Smarandache F, Khalifa NEM. Within the lack of chest COVID-19 X-ray dataset: a novel detection model based on GAN and deep transfer learning. Symmetry. 2020;12(4):651. doi: 10.3390/SYM12040651. [DOI] [Google Scholar]
- 56.Rahimzadeh M, Attar A. A modified deep convolutional neural network for detecting COVID-19 and pneumonia from chest X-ray images based on the concatenation of Xception and ResNet50V2. Inform Med Unlocked. 2020;19:100360. doi: 10.1016/j.imu.2020.100360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jiang X, Coffee M, Bari A, Wang J, Jiang X, Huang J, et al. Towards an artificial intelligence framework for data-driven prediction of coronavirus clinical severity. Comput Mater Continua. 2020;63(1):537–551. doi: 10.32604/cmc.2020.010691. [DOI] [Google Scholar]
- 58.Wang X, Peng Y, Lu L, Lu Z, Bagheri M, Summers RM (2017) ChestX-ray8: hospital-scale chest X- ray database and benchmarks on weakly-supervised classification and localization of common thorax diseases. In: Proceedings of the 30th IEEE conference on computer vision and pattern recognition (CVPR). (Honolulu, Hawaii, USA, 21–26 July 2017): 3462–3471. 10.1109/CVPR.2017.369.
- 59.Naeem A, Farooq MS, Khelifi A, Abid A. Malignant melanoma classification using deep learning: datasets, performance measurements, challenges and opportunities. IEEE Access. 2020;8:110575–110597. doi: 10.1109/ACCESS.2020.3001507. [DOI] [Google Scholar]
- 60.Hassantabar S, Ahmadi M, Sharifi A. Diagnosis and detection of infected tissue of COVID-19 patients based on lung X-ray image using convolutional neural network approaches. Chaos Solitons Fractals. 2020;140:110170. doi: 10.1016/j.chaos.2020.110170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jain R, Gupta M, Taneja S, Hemanth DJ. Deep learning-based detection and analysis of COVID-19 on chest X-ray images. Appl Intell. 2021;51(3):1690–1700. doi: 10.1007/s10489-020-01902-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xiao B, Yang Z, Qiu X, Xiao J, Wang G, Zeng W, et al. PAM-DenseNet: a deep convolutional neural network for computer-aided COVID-19 diagnosis. IEEE Trans Cybern. 2021 doi: 10.1109/TCYB.2020.3042837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chowdhury NK, Kabir MA, Rahman MM, Rezoana N. ECOVNet: a highly effective ensemble based deep learning model for detecting COVID-19. PeerJ Comput Sci. 2021;7:e551. doi: 10.7717/peerj-cs.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Marques G, Agarwal D, de la Torre Díez I. Automated medical diagnosis of COVID-19 through EfficientNet convolutional neural network. Appl Soft Comput. 2020;96:106691. doi: 10.1016/j.asoc.2020.106691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Canayaz M. C+ EffxNet: a novel hybrid approach for COVID-19 diagnosis on CT images based on CBAM and EfficientNet. Chaos Solitons Fractals. 2021;151:111310. doi: 10.1016/j.chaos.2021.111310. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.
Not applicable.