Abstract
Typing of hepatitis C virus (HCV) isolates from Argentine patients was performed by using different methodologies in a population of 243 patients. HCV subtype was assigned based upon restriction fragment length polymorphism (RFLP). HCV RNA genomes obtained from serum samples were classified as belonging to clade 1 (53.5%), 2 (23.0%), or 3 (8.6%); 14.8% of samples showed HCV mixed infections, more frequently implying different subtypes within the same clade. In addition to RFLP typing, phylogenetic relatedness among sequences from both 5′ untranslated region (n = 50) and nonstructural 5B coding region (n = 15) was established.
Hepatitis C virus (HCV), the causative agent of most cases of non-A, non-B hepatitis, exists as a heterogeneous group of viruses sharing at least 65% homology among different strains. This virus has a positive-sense, single-stranded RNA genome of approximately 9.5 kb. There are three functional regions of the genome, the 5′ untranslated region (5′UTR), the coding region encoding the structural and nonstructural viral proteins, and the 3′UTR. When different strains of HCV are compared, nucleotide sequence variation is unevenly distributed throughout the genome, ranging from segments within the coding regions with high variability (such as for envelope proteins) to the highly conserved 5′UTR region. Sequence analysis performed on isolates from different geographic areas around the world has revealed the presence of six genetic clusters which have been recently classified as clades 1 to 6 (42).
Several methodologies have been developed for identification of genetic groups of HCV. Among them, two noncommercial ones are widely used when a large number of samples are being studied: restriction fragment length polymorphism (RFLP) analysis of PCR amplicons from the 5′UTR (9) and core-based PCR typing (28, 30, 31). However, the subtype assignment is strongly recommended to be based upon sequence analysis on HCV coding regions such as core, E1 or NS5B (42).
Since data regarding HCV genomic characteristics from South America are still scarce, the aim of the present study was to characterize the clade and subtype distribution of different isolates from chronically HCV-infected Argentine patients. To reach this goal, three different methodologies were used, including cDNA sequencing and phylogenetic analysis of selected samples, supported by bootstrap resampling.
MATERIALS AND METHODS
Patients.
From 1993 to 1999 we characterized 243 HCV isolates from three different groups of viremic patients (nonhemophiliac and naive of antiviral therapy). The first group comprised 129 patients with parenteral risk of viral infection, including 56 blood transfusion patients, 40 intravenous drug users (IVDU), 15 hemodialysis patients, and an additional 18 individuals who had a history of potential parenteral routes for viral exposure (undergoing surgery, having a tattoo or acupuncture, being a laboratory professional, or practicing homosexual behavior). The second group (n = 8) included patients with nonparenteral risk (either having nonmarital household contact with an infected individual or being a drug addict who uses oral or nasal routes of drug entry). The third group (n = 106) was composed of sporadic or community-acquired cases. The mean age ± standard deviation (SD) in the whole study group was 43.3 ± 24.8 years (range, 2 to 73 years), with a gender distribution showing male predominance (60.5%). Age ranges were not equally represented among different groups of patients. The mean ± SD, median, and range (1st to 3rd quartile) for each group were, respectively, as follows: for transfused patients, 24.6 ± 21.2, 16, and 8 to 36; for IVDU, 33.3 ± 10.2, 30, and 26.5 to 35.5; for dialyzed patients, 28.4 ± 14.4, 31, and 21 to 35; for sporadic cases, 50.0 ± 17.4, 55.0, and 39.5 to 64; for health workers, 47.9 ± 9.5, 45, and 41 to 56; for other parentally infected patients, 41.6 ± 20.0, 44.5, and 30.7 to 58.7; and, lastly, for other nonparentally infected patients, 43.0 ± 10.1, 39, and 38.2 to 43.7.
Informed consent to participate in the study was provided by all patients or their parents. Two hundred twenty-five patients were adults, and 18 were pediatric patients. All serum samples were analyzed by 5′UTR RFLP. In addition, selected cases were studied using other methodologies, as shown in Table 1.
TABLE 1.
HCV genomic characterization studies performed with 243 samples
| Group | No. of samples positive by:
|
|||
|---|---|---|---|---|
| 5′UTR RFLP analysis | Core PCR | 5′UTR cDNA sequencing, phylogenetic analysis, and predicted RFLP | NS5B cDNA sequencing and phylogenetic analysisa | |
| 5′UTR RFLP only (n = 148) | 148 | |||
| Concordant 5′UTR RFLP analysis and 5′UTR cDNA sequencingb(n = 37) | 37 | 37 | ||
| Concordant 5′UTR RFLP analysis and 5′UTR cDNA sequencing but discrepant versus core PCR (n = 1) | 1c | 1 | 1 | 1 |
| Discrepant 5′UTR RFLP analysis versus core PCR (n = 6) | 6 | 6 | ||
| Concordant 5′UTR RFLP analysis and 5′UTR cDNA sequencing but nontypeable by core PCR (n = 12) | 12 | 12 | 12 | 12 |
| Typed by 5′UTR RFLP analysis but nontypeable by core PCR (n = 14) | 14 | 14 | ||
| Concordant 5′UTR RFLP analysis and core PCR (n = 23) | 23 | 23 | ||
| Unrecognized subtype by 5′UTR RFLP analysis (n = 2) | 2 | 2 | ||
| Total | 243 | 56 | 50 | 15 |
Only 15 out of 34 studied sera were NS5B PCR positive.
Four samples exhibited mixed infections by RFLP, showing the predominant population also documented by cDNA sequencing, in addition to minor HCV population(s).
Minor population of type 1 (plus 2a/c) was also detected in 3 out of 10 RT-nested 5′UTR PCR amplifications followed by RFLP analysis of isolate 760.
Serum alanine aminotransferase (ALT) levels were determined by using a commercial kit (BioSystems) according to the manufacturer's instruction. Normal ALT levels were ≤35 U/liter when testing was done at 37°C.
RNA extraction, RT-nested PCR of the 5′UTR, and RFLP analysis.
RNA was extracted from 200 μl of serum using guanidinium isothiocyanate and acidic phenol followed by reverse transcription (RT)-nested PCR amplification of the 5′UTR as previously described (33). The HCV 5′UTR amplicons obtained (210 bp) were characterized by RFLP (9) as modified by the authors (33, 39, 41). This procedure was applied to the whole population analyzed (Table 1).
PCR amplification of other genome regions.
Core-based genotyping was performed as previously described (33) and was initially used to analyze HCV strains from 56 consecutive patients included within this study (core PCR [Table 1]). This methodology was subsequently discontinued, since a significant percentage of the isolates could not be typed. For selected samples an improved version of this method was employed in order to detect the HCV 2c subtype (26).
RT-nested PCR amplification of an NS5B fragment (nucleotides 7975 to 8196) as previously described (6, 11) was performed on 34 samples selected because they (i) had discrepant results between core-based genotyping and 5′UTR RFLP (n = 6), (ii) were nontypeable by the core-based methodology (n = 26), or (iii) had unrecognized RFLP patterns within the 5′UTR, resulting in inconclusive subtype assignment (n = 2). Since only 56 specimens had been previously tested by the core-based RT-nested PCR, we cannot rule out the possibility that more specimens would have yielded discrepant results between such a method and 5′UTR RFLP.
5′UTR-core RFLP.
In order to confirm mixed infections involving subtypes 1a and 1b detected by 5′UTR RFLP, amplicons of 647 bp partially encompassing both the 5′UTR and the core were obtained by RT-heminested PCR using primers HCV2 (outer sense), HCV4 (inner sense), and 186 (antisense) as described previously (33, 41), using the same cycling conditions reported for core-based PCR (33). Bands of the expected size were purified and digested for 6 h with AccI. As reported elsewhere (2, 3), this endonuclease recognizes two restriction sites within 1a amplicons (fragments of 225, 192, and 230 bp) and one restriction site within 1b (or 1c) amplicons (fragments of 225 and 417 bp) (Fig. 1).
FIG. 1.

5′UTR-core RFLP. Amplicons (647 bp) were digested with AccI and run on a 4% agarose gel which was stained with ethidium bromide. Lane a, mixed HCV infection involving subtypes 1a (predominant) and 1b; lane b, 25-bp ladder; lane c, subtype 1b restriction pattern.
Avoidance of PCR contamination.
PCR amplification of all three genome regions (5′ UTR, core, and NS5B) strictly followed the recommendations of Kwok and Higuchi (21). In addition, different sets of micropipettes and special aerosol-resistant tips (Molecular Bio-Products, Inc.) were used for each procedure from serum collection to agarose gel analysis of the PCR products. To validate results, a negative control was included from the extraction step for every four samples, and another negative control was also added from RT. A positive control was included from RNA extraction. Separate rooms were used for PCR preamplification, amplification, and gel loading. Mixed infections were confirmed by duplicate analysis from two different serum aliquots.
Sequencing of 5′UTR and NS5B amplicons.
The 5′UTR (n = 50) and NS5B (n = 15) PCR products were purified as previously described (40), and the fragments were sequenced (43) using both sense and antisense primers with 5′ -end-fluorescent-labeled dideoxynucleotides in an automatic sequencer (ABI 373A; Applied Biosystems, Foster City, Calif.). To avoid misinterpretations, each template was obtained from at least two different aliquots of RNA and sequenced bidirectionally.
Selected samples for 5′UTR cDNA sequencing were (i) 13 out of the 33 samples which had exhibited either nontypeable (n=26) or discrepant results compared with 5′UTR RFLP (n = 7), according to the core-based typing method; and (ii) 37 others representative of the different groups studied, including parenteral risk of infection (n = 25), nonparenteral risk (n = 7), and sporadic cases (n = 5). The above-mentioned set of 13 samples was randomly selected in order to test the accuracy of data already obtained by 5′UTR RFLP (Table 1).
Phylogenetic methods.
The clade and subtype for each sample was determined by sequence comparison with prototypic strains followed by further phylogenetic analysis. The DNA alignments were generated with the Clustal X program (48). Evolutionary distances between sequences were determined with the DNADIST program (Kimura two-parameter method) of the PHYLIP package, version 3.5c (13). The phylogenetic tree was constructed with the TREEVIEW program, as previously described (40). Bootstrap analysis was performed by using the programs SEQBOOT (to generate 1,000 reshuffled sequences), CONSENSE, and RETREE at the midpoint of the longest branch for comparative purposes.
HCV serologic genotyping.
Selected samples (n = 3) showing mixed infections ascribed to different genotypes were studied by enzyme-linked immunosorbent assay by means of a commercial test to detect antibodies against type-specific HCV NS4 peptides (MUREX HCV, Serotyping 1–6 Assay; Murex, Buenos Aires, Argentina).
Statistical analysis.
Statistical differences were calculated by means of parametric tests (comparison of two sample proportions or Student's t test) or nonparametric tests (by the chi-square test with Yates' correction, and by Fisher's exact test when 8 ≤ N ≤ 75, where N is the sum of the values in a given table). Tadpole III program (Biosoft, Cambridge, United Kingdom) was used throughout this study. The median age and the 1st and 3rd quartile range from each population group were calculated by using the Excel program from the Office 97 Microsoft package.
Nucleotide sequence accession numbers.
Nucleotide sequence data reported in this paper were deposited in the GenBank database with the following accession numbers: AF041264 to AF041313 (5′UTR sequences) and AF041314 to AF041328 (NS5B sequences).
RESULTS
RFLP analysis of 5′UTR amplicons.
Out of 243 samples, clade 1 was detected in 130 (53.5%), clade 2 was detected in 56 (23.0%), and clade 3 was detected in 21 (8.6%) samples. Thirty-six samples (14.8%) showed mixed infections, involving different clade and subtype combinations (Table 2). Infections ascribed by RFLP to a unique subtype within clades 1 or 2 were classified as follows: within clade 1, subtype 1a/c was found in 57 samples (23.4%), while subtype 1b accounted for the remaining 73 (30.0%); clade 2 included 47 samples regarded as subtype 2a/c (19.3%) and 9 samples regarded as subtype 2b/c (3.7%); within clade 3, all samples (n = 21) were characterized as subtype 3a/c/d/e (8.6%). No samples belonging to subtype 3b were detected.
TABLE 2.
HCV genotype distribution among Argentine subjects as determined by 5′UTR RFLP analysis
| Group (n) | No. of isolates (% relative prevalence) ascribed to subtype(s)
|
|||||
|---|---|---|---|---|---|---|
| 1a/c | 1b | 2a/c | 2b/c | 3a/c/d/e | Mixed infectionsa | |
| Sporadic case (106) | 26 (24.5) | 25 (23.6) | 26 (24.5) | 4 (3.8) | 5 (4.7) | 1a + 1b = 17; 2a + 2b = 2; 1a + 2a = 1; (18.9) |
| Transfusion patient (56) | 10 (17.9) | 24c (42.8) | 11 (19.6) | 3 (5.4) | 2 (3.6) | 1a + 1b = 5; 1a + 2a = 1; (10.7) |
| Hemodialysis patient (15) | 4 (26.7) | 3 (20.0) | 4 (26.7) | 0 (0.0) | 2 (13.3) | 1a + 1b = 2 (13.3) |
| IVDU (40) | 10 (25.0) | 12 (30.0) | 3c (7.5) | 0 (0.0) | 10d (25.0) | 1a + 1b = 1; 1a + 1b + 3a = 1; 1b + 2a = 1; 1b + 3a = 1; 2b + 3a = 1; (12.5) |
| Health worker (9) | 3 (33.3) | 2 (22.2) | 2 (22.2) | 0 (0.0) | 0 (0.0) | 2a + 1b = 1; 2a + 3a = 1; (22.2) |
| Other parenteral (9) | 1 (11.1) | 2 (22.2) | 1 (11.1) | 2d (22.2) | 2 (22.2) | 1a + 1b = 1 (11.1) |
| Other nonparenteral (8) | 3 (37.5) | 5 (62.5) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.00) |
| Total (243) | 57 (23.4) | 73 (30.0) | 47 (19.3) | 9 (3.7) | 21 (8.6) | 36 (14.8) |
1a = 1a/c; 2a = 2a/c; 2b = 2b/c; 3a = 3a/c/d/e.
5′UTR RFLP analysis was performed without subtyping since the previous version of the method employed for the remaining samples (8) was used.
P < 0.05 compared with the proportion of the same subtype within the whole population studied (two unpaired proportions, two-sided test).
P < 0.01 compared with the proportion of the same subtype within the whole population studied (two unpaired proportions, two-sided test).
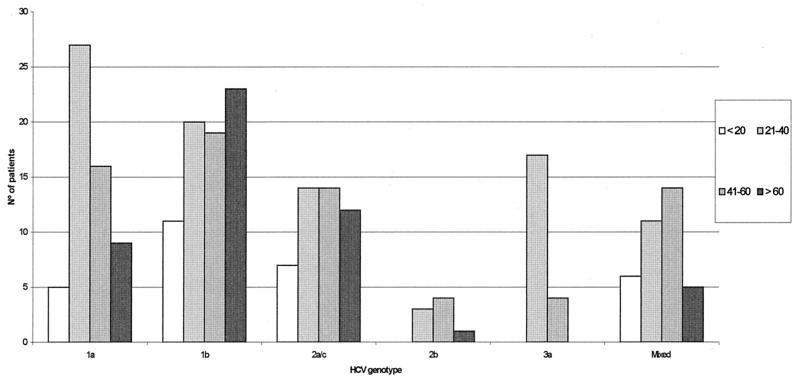
The relative prevalence of each subtype within the different groups studied showed interesting features since significant or highly significant differences were observed among transfused patients and IVDU, respectively, comparing the specific subtypes with their prevalence within the whole population (two unpaired proportions, two-sided test). Although two other groups of patients showed statistically significant differences as well, the scarce number of patients included within each of them precludes any further conclusions (Table 2). Among transfusion patients a higher prevalence of 1b subtype (P < 0.05) was observed. In contrast, among IVDU, the contribution of subtype 3a/c/d/e was significantly higher compared to its own incidence within the (non-IVDU) whole population (P < 0.01), while there was less subtype 2a/c compared to the overall rates (P < 0.05). Interestingly, when HCV genotype 3a/c/d/e was plotted against age, a striking contribution was ascribed to young ([21- to 40-year old] mainly IVDU) patients (Fig. 2). A statistically significant association was observed between subtype 3a with both the above-mentioned age range group and IVDU (chi-square test: P = 0.0026 versus the remaining subjects [n = 222] and P = 0.0088 versus other parentally infected patients [n = 83]). Likewise, subtype 1a was significantly more prevalent among the 21- to 40-year-old group compared with the remaining age ranges (chi-square test: P = 0.0001 versus those <20 years old; P = 0.0317 versus those 41 to 60 years old; and P = 0.001 versus those >60 years old).
FIG. 2.
HCV genotype distribution according to age range (years).
Out of the 243 HCV infected sera, 36 mixed infections (14.8%) were detected which were predominantly 1a/c and 1b (n = 26; 72.2% of mixed infections). The majority of these 1a/c and 1b mixed infections were mainly detected among sporadic cases, which was a significant contribution compared to the whole incidence of these subtypes among other groups (P < 0.01). Randomly selected mixed infections were entirely confirmed either by HCV serologic genotyping (for mixed types, see below) or by documentation of a unique AccI restriction site only present among 1a isolates at the core region (Fig. 1) (2, 3).
Primer-specific amplification of the core (core PCR).
PCR amplification using subtype-specific primers within the core genomic region was performed as described by Okamoto et al. (30, 31) on 56 samples (Table 1). We observed no amplification products from 26 isolates (46.4%). Six samples exhibited discordant typing results compared to 5′UTR RFLP analysis. The subtype assignment in the remaining 24 samples was consistent with the data obtained by RFLP. Subsequent use of an improved version of this methodology (26) allowed us to detect initially nontypeable 2c isolates (i.e., serum 874).
Sequence analysis.
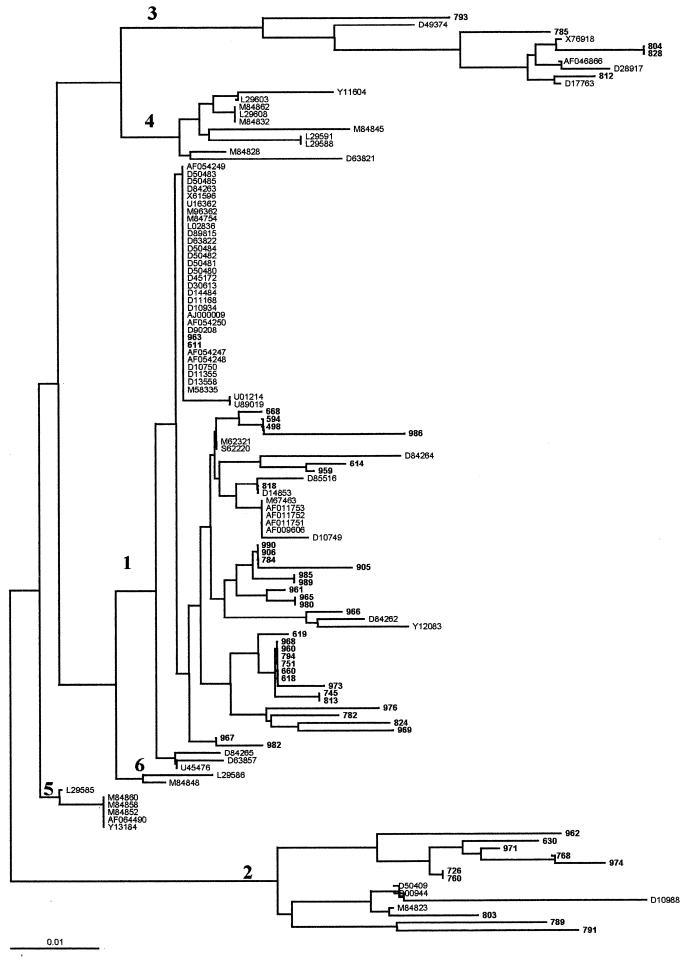
Fifty samples were selected for 5′UTR sequence comparison by phylogenetic analysis of clades and types and are identified in Fig. 3. These studies demonstrated that 35 isolates belonged to clade 1 (70%), 10 belonged to clade 2 (20%), and 5 belonged to clade 3 (10%). However, when results from these 50 sequenced isolates were compared with the experimental results from 5′UTR RFLP analysis it was shown (by the latter methodology) that four samples contained mixed infections (i.e., 1a plus 1b, n = 3; 1a plus 1b plus 3a, n = 1) composed of predominant genomes (detected by both cDNA sequencing and RFLP analysis), in addition to minor HCV populations detected only by RFLP analysis (Table 1).
FIG. 3.
The phylogenetic tree shown is based on 124 sequences derived from the 5′UTR of HCV. Argentine HCV sequences (n = 50) are indicated by their isolate numbers (in boldface type): samples 498, 594, 611, 614, 618, 619, 630, 660, 668, 726, 745, 751, 760, 768, 782, 784, 785, 789, 791, 793, 794, 803, 804, 812, 813, 818, 824, 828, 905, 906, 959, 963, 965, 969, 971, 973, 974, 976, 980, 982, 985, 986, 989, and 990 correspond to accession numbers from GenBank AF041264 to AF041313, respectively. Sequences used as references for the phylogenetic tree (n = 74) are identified by their accession number from GenBank. Note that major HCV clades from 1 to 6 are indicated. The DNA alignments were generated with the Clustal X program. The phylogenetic tree was constructed with the TREEVIEW program. Bootstrap analysis was performed by using the programs SEQBOOT (to generate 1,000 reshuffled sequences), CONSENSE, and RETREE at the midpoint of the longest branch for comparative purposes (PHYLIP package, version 3.5c). Bar, number of nucleotide substitutions per site.
With regard to the NS5B isolates studied, only 15 out of 35 (42.9%) could be detected by RT-nested PCR amplification (Table 1), reflecting the known different sensitivity to detect HCV RNA based upon RT-PCR amplification on this region with regard to the most conserved 5′UTR (5, 49). NS5B sequencing and the corresponding phylogenetic analysis showed that five samples belonged to subtype la, five samples belonged to subtype 1b and the remaining five samples belonged to 2c (Fig. 4). Ten out of 15 samples (66.6%) showed results concordant with 5′UTR RFLP analysis (see below).
FIG. 4.
The phylogenetic tree shown is based on sequences derived from the NS5B region of HCV. Argentine HCV sequences (n = 15) are indicated by their isolate number (in boldface type): samples 619, 760, 794, 803, 812, 874, 959, 967, 968, 969, 974, 980, 986, 1274, and 1470 correspond to accession numbers from GenBank AF041314 to AF041328, respectively. Sequences used as references for the phylogenetic tree (n = 99) are identified by their accession number from GenBank. Note that major HCV clades from 1 to 6 are indicated. Shaded boxes indicate bootstrap values. Bar, number of nucleotide substitutions per site.
Both phylogenetic trees were supported by bootstrap values higher than 70% for clade and HCV subtype assignment, respectively.
The chance of PCR contamination, and therefore potential mistyping, within samples inter se from both 5′UTR and NS5B regions was exhaustively investigated and ruled out since different controls confirmed the accuracy of data, with (i) RNA from two different aliquots, (ii) cDNA bidirectionally sequenced, and (iii) absence of 100% homology with any other product from the same regions.
HCV serologic genotyping.
Three selected samples were confirmed as showing mixed infections ascribed to different genotypes (i.e., 1 plus 2, n = 1; 1 plus 3, n = 2).
Discrepant HCV typing results.
Table 3 summarizes the discrepancies we observed using the three different approaches for subtype assignment where NS5B amplicons could be obtained. Subtype assignment based upon 5′UTR RFLP and NS5B sequence analysis was concordant in 10 of the 15 samples. One of these discrepant samples was the initial serum from a health care worker (sample 760) which appeared to have a mixed population detected in the 5′UTR, the minor one of which was selectively amplified within the NS5B region (subtype 1a). Later during infection (sample 874), the major population (subtype 2a/c) was the only one detected in both 5′UTR RFLP analysis and within the NS5B sequence analysis. The four remaining discrepant samples were subtype la and 2c based upon NS5B sequence analysis, but subtypes la/c, lb, 3a/c/d/e, and 2a/c based upon 5′UTR RFLP analysis. Six other samples failed to show concordant results between 5′UTR RFLP analysis and core PCR (i.e., 2a/c versus 1a; 2a/c versus 3a; la versus 3a (n = 2); 1b and 3a versus 1a, 1b, and 3a; and 3a versus 1b and 2b, respectively), but NS5B PCR amplification consistently rendered negative results.
TABLE 3.
Discrepancies among different methodologies for HCV genotyping of Argentine isolates
| Isolate no. (populationa) | Type(s) according to:
|
||
|---|---|---|---|
| Core PCR | 5′UTR RFLP analysis | NS5B sequencing | |
| 619 (SC) | NTb | 1a | 1a |
| 760 (HW)c | 1b | 2a/c + 1d | 1a |
| 794 (SC) | NT | 1b | 1b |
| 803 (T) | NT | 2a/c | 1a |
| 812 (IVDU) | NT | 3a | 1a |
| 874 (HW)c | NTe | 2a/c | 2c |
| 959 (NONPR) | NT | 1a | 1a |
| 967 (IVDU) | NT | 1b | 1b |
| 968 (T) | NT | 1b | 2c |
| 969 (HW) | NT | 1a | 1a |
| 974 (SC) | NTe | 2a/c | 2c |
| 980 (NOPR) | NT | 1a | 2c |
| 986 (SC) | NT | 1b | 1b |
| 1274 (SC) | NTe | 2a/c | 2c |
| 1470 (T) | NT | 1b | 1b |
SC, sporadic cases; HW, health workers; T, transfusion patients; NONPR, nonspecified other nonparenteral risks; NOPR, nonspecified other parenteral risks.
NT, not typeable.
Nucleotide sequences obtained from two different samples from the same patient after 6 months of elapsed time.
Minor population: type 1 (plus 2 a/c) was also detected in 3 out of 10 RT-nested 5′UTR PCR amplifications followed by RFLP analysis.
When 2c-specific primers were used, this sample became characterized. Another group of six sera showed discrepant results between core PCR and 5′UTR RFLP, but NS5B amplification rendered negative results.
5′UTR-core RFLP.
Mixed infections involving 1a plus 1b detected by 5′UTR RFLP were confirmed by AccI digestion (2, 3) of amplicons from 10 randomly selected samples (Fig. 1).
Biochemical evaluation of liver function.
To determine the clinical significance of infection with different HCV genotypes, we analyzed the relationship of serum ALT levels from 97 samples, approximately representing the same genotype distribution observed in the whole population studied. Data are shown in Table 4. No statistical difference was observed when ALT levels from serum samples exhibiting different HCV types or subtypes were compared. However, ALT values from patients infected with more than one type or subtype were significantly higher than those observed in patients infected with genotype 1 (P = 0.0108) or 3 (P = 0.02) but not with genotype 2; remarkably, when a specific subtype was considered, lb, ALT values differed with high statistical significance compared to values for mixed infections (P = 0.0032).
TABLE 4.
Serum ALT values according to HCV genotype distribution
| Genotype (no. of samples) | ALT ± SD (IU/liter) |
|---|---|
| Type 1 (51) | 63 ± 63 |
| Subtype 1a/c (24) | 79 ± 86 |
| Subtype 1b (27) | 48 ± 27 |
| Type 2 (17) | 62 ± 60 |
| Subtype 2a/c (14) | 61 ± 64 |
| Subtype 2b/c (3) | 67 ± 41 |
| Type 3 (12)a | 62 ± 54 |
| Mixed infection (17) | 122 ± 119b |
All 3a/c/d/e.
P = 0.0032 versus 1b; P = 0.0108 versus 1; P = 0.02 versus 3.
DISCUSSION
These studies provide the most comprehensive data to date on HCV subtypes present in chronically infected patients referred to medical treatment in Argentina between 1993 and 1999.
Out of 547 viremic patients, 243 sera were included within these investigations, and the HCV genotype was determined by RFLP analysis of the 5′UTR region (9) while selected subsets were also evaluated by primer-specific core region amplification (30, 31) and amplification and sequencing of a fragment derived from the NS5B genome region (6, 11).
At least four major points deserve to be highlighted: (i) the overall HCV subtype distribution; (ii) the relatively high prevalence of mixed infections; (iii) the inferred high prevalence of subtype 2c within 2a/c isolates; and (iv) the observation of discrepant results when different HCV genomic regions were analyzed.
Firstly, only genotypes 1, 2, and 3 were detected with decreasing prevalence, as shown in Table 2. Genetic analysis using 5′UTR RFLP indicates that 23.5% of infections are subtype 1a/c, 30.0% are subtype 1b, 23.0% are subtype 2a/c or 2b/c, 8.6% are subtype 3a/c/d/e, and 14.8% are mixed infections. These results confirm and strengthen our initial data (33, 41). Thus, HCV genotype distribution in Argentina closely resembles the reported data from other Western countries. However, it is worth mentioning that other Argentine researchers have recently observed the circulation of a small percentage of strains ascribed to either genotype 4 or 5 among hemophiliacs (36). The higher prevalence of clade 3 observed among IVDU with regard to other groups studied (P < 0.01) is in agreement with previous reports from Europe (35). Together with a recent report from Argentina (14), these are the first studies from another part of the world which also suggest that IVDU may carry selective subtypes.
Secondly, the presence of HCV mixed infections is an interesting field of current research. Two previous reports from the authors showed dissimilar results at this specific point. We had initially detected a high proportion of mixed infections, reaching up to 45.4% (33), while in a later study we did not detect mixed infections (41). These discrepancies might be explained by considering the typing methodologies that we employed each time. In the former, a core-based specific PCR method was applied (30, 31), and it is known that this procedure may readily detect different genotypes but produces a certain degree of mispriming (23). In the later study, a majority of typing results was obtained by predicted RFLP patterns from 5′UTR nucleotide sequencing. It seems plausible that only predominant genomes would have been detected, since purified PCR products were then directly sequenced. Therefore, the existence of minor HCV populations could not be ruled out. Besides, distinct populations studied exhibited differential risk of multiple exposure to HCV sources (e. g., hemophiliacs, not included in our second survey [41]).
In the present study, we have analyzed all samples by using RFLP, which is reportedly known to need roughly equimolar concentrations for detecting mixed infections implying at least two different HCV subtypes (9). Among 243 HCV-infected patients, 36 exhibited mixed infections (14.8%). This percentage is higher than the observed value for samples from other geographic areas in previous epidemiological studies using the same methodology (46) although it is approximately similar to the rate reported in Venezuela (38). The possibility of RT-nested PCR contaminations was entirely ruled out since RNA was extracted from two separate serum aliquots and subsequently amplified. All RFLP results regarding the predominant viral population were in agreement with data obtained by cDNA sequencing of the same 5′UTR (n = 50). Subtypes 1a/c and 1b accounted for most of the mixed infections (72.2%), although 5′UTR RFLP methodology might have mistyped a very low number of the samples due to the unusual presence of A or G at position −99 within subtypes 1b or 1a (9). This contribution was also observed among sporadic cases, reflecting the high prevalence of these subtypes in the general population of Buenos Aires, Argentina.
Data regarding mixed infections were documented using different approaches, including (i) 5′UTR RFLP analysis as mentioned above; (ii) analysis of a second genomic region looking for a specific AccI restriction site (cleavage at nucleotide 184 from the putative AUG start codon) at the core region (n = 10), which was reported to be indicative of subtype la (Fig. 1, lane a, corresponding to a mixed infection involving 1a plus 1b) but was absent among 1b isolates (Fig. 1, lane c) (2, 3); and (iii) detecting the immune response against type-specific NS4 peptides. Concordant results obtained were in agreement with data already reported (27) when comparing 5′UTR RFLP analysis, NS4 serologic genotyping, and 5′UTR sequencing, which also showed 100% concordance among them.
Several hypotheses might be considered for mixed HCV infections: (i) simultaneous coinfection with different types and/or subtypes; (ii) superinfection of HCV-infected but nonprotected individuals, as documented in experimentally inoculated primates (29) or naturally infected patients (22); (iii) HCV genotype turnover as observed among dialyzed (37) and hemophilic (12, 39) patients; (iv) HCV genotype overtake phenomena (which may last up to 1 to 8 months) (22); and (v) selective replicative advantage of genotype 1 (22). Moreover, it should be taken into account that the daily produced heterogeneous quasispecies belonging to a given genotype could temporarily fluctuate (10, 15). The simultaneous coinfection with different genotypes might imply a large cohort of individuals that already have mixed infections and the efficient transmission of all strains. Conversely, if superinfection were the main cause of mixed infections, it should be envisaged to occur more frequently among IVDU. Since this was not the case (only 5 out of 40 patients [12.5%]) potential explanations should consider both their younger age (Fig. 2) and (short) time of intravenous drug addiction. HCV genotype overtake phenomena and subsequent replacement have been observed in longitudinal studies of hemophiliacs (12, 39) where multiple exposures were documented. Likewise, this fact might account for a certain percentage of mixed infections within the population studied. For unknown reasons, type 1 (and, if present, subtype 1b) is frequently predominant in donor-recipient pairs of genotypes when orthotopic liver transplantation patients are monitored, thus suggesting a selective replicative advantage (22). Coincidently, 29 out of 36 mixed infections (80.6%) involved subtype 1b, a much higher rate than the overall prevalence of such subtype within the whole population studied (P < 0.01). Interestingly, ALT values from patients exhibiting HCV mixed infections showed higher values than those infected with genotype 1 or 3 (P = 0.0108 and 0.02, respectively). However, a cautious interpretation of these data is needed, considering both the limited number of mixed infections analyzed in this study and the usual fluctuation of ALT levels in serum during the natural course of the infection.
Thirdly, regarding HCV type 2 assignment by NS5B sequence analysis, three out of three 2a/c isolates (according to 5′UTR RFLP analysis) were finally ascribed to subtype 2c (samples 874, 974, and 1274), in agreement with results obtained by an improved version of the core-based typing assay (Table 3) (26). It is concluded that this subtype might potentially represent an important contribution to local HCV epidemiology, taking into account that (i) 23.0% of the population studied exhibited type 2 infections (n = 56) and (ii) both subtypes 2a and 2b are indistinguishable from subtype 2c at the 5′UTR. A high prevalence of subtype 2c has been found among individuals in Italy (4, 16, 25, 44, 47), from which a substantial proportion of the Buenos Aires population is descended, due to immigration within this century.
Fourthly, type assignment by 5′UTR RFLP and NS5B phylogenetic analyses exhibited concordant results in 10 of 15 cases studied (66.7%), while five samples were discrepant with regard to the previously observed 5′UTR RFLP pattern (Table 3). Discrepant results according to the analyzed HCV genomic region have been previously reported as well (23, 27, 34). In this regard, at least three potential explanations should be considered: (i) the coexistence of HCV genotypes for which primers used during RT-PCR displayed different sensitivity and/or specificity (Table 3, see sample 760: 1b at core 2a/c plus 1 [minor population] at 5′UTR, and 1a at NS5B genomic regions); (ii) in vitro template shuffling during RT-PCR amplification; or (iii) hypothetical possibility of recombination between different HCV variants showing nucleotide sequences belonging to different genotypes throughout viral genomes, as inferred from early reports (17, 19). In this regard, thorough studies could not subsequently demonstrate an eventual phenomenon of recombination between HCV genotypes (44, 46), but this hypothesis remains to be further explored.
The above-mentioned five discrepant samples were assayed by the core-based specific PCR technique (30, 31), but unfortunately only one of them proved positive; therefore, the remaining four samples were classified as nontypeable by this methodology. Factors such as low viral load (50) and lack of exact complementarity between 2a primers and 2c sequences accounted for most nontypeable samples (data not shown). This hypothesis was confirmed when samples 874, 974, and 1274 were assayed with a 2c-specific core primer (26).
Considering some limitations of 5′UTR RFLP analysis for HCV subtyping (9, 46), despite recently proposed improvements (3), it is currently agreed that it is mandatory to perform simultaneous sequencing of coding genomic regions, which exhibit a greater degree of nucleotide heterogeneity within a given type (42, 45). Nevertheless, RFLP is still a widely used tool for epidemiological studies when massive genomic analysis regarding other hepatitis viruses is undertaken (24, 32, 33).
By means of different methodologies including phylogenetic analysis, this study depicts a general view of circulating HCV strains during 1993 to 1999 in Buenos Aires, Argentina, and adds information on HCV molecular epidemiology in South America (14, 20, 33, 36–39, 41). After cross-protection studies are performed with different genotypes in primates—as previously investigated with wild-type strains (29)—potential immunogens for humans are expected to be developed (1, 7, 8, 18, 51); M. Houghton, Q.-L. Choo, G. Kuo, D. Chien, A. Weiner, M. Selby, L. Consens, S. Coates, R. Ralston, H. Davis, J. Kansopon, K. Berger, S. Wong, M. Wininger, C. Dong, K. Crawford, M. Chin, E. Glazer, M. Jennings, E. Muchmore, D. Rosa, and S. Abrignani, Abstr. IX Triennial Int. Symp. Viral Hepat. Liver Dis., abstr. 150, 1996). Our data should be taken into account when future HCV vaccines are constructed based upon geographical and epidemiological criteria.
ACKNOWLEDGMENTS
This study was supported partly by grants HDP/HDR (DRC/RG/ARG/92-804) from the Pan-American Health Organization, PIP 6554/97 and PIP 842/98 from CONICET, BID 802/OC-AR- PICT 04977/99 from FONCYT, from the Fundación Florencio Fiorini, and from Universidad del Salvador.
REFERENCES
- 1.Abrignani S, Rosa D. Perspectives for a hepatitis C virus vaccine. Clin Diagn Virol. 1998;15:181–185. doi: 10.1016/s0928-0197(98)00028-2. [DOI] [PubMed] [Google Scholar]
- 2.Andonov A, Chaudhary R K. Genotyping of Canadian hepatitis C virus isolates by PCR. J Clin Microbiol. 1994;32:2031–2034. doi: 10.1128/jcm.32.8.2031-2034.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buoro S, Pizzighella S, Boschetto R, Pellizzari L, Cusan M, Bonaguro R, Mengoli C, Caudai C, Padula M, Valensin P E, Palù G. Typing of hepatitis C virus by a new method based on restriction fragment length polymorphism. Intervirology. 1999;42:1–8. doi: 10.1159/000024953. [DOI] [PubMed] [Google Scholar]
- 4.Cammarota G, Maggi F, Vatteroni M L, Da Prato L, Barsanti L, Bendinelli M, Pistello M. Partial nucleotide sequencing of six subtype 2c hepatitis C viruses detected in Italy. J Clin Microbiol. 1995;33:2781–2784. doi: 10.1128/jcm.33.10.2781-2784.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Castillo I, Bartolome J, Quiroga J A, Carreño V. Comparison of several PCR procedures for detection of serum HCV-RNA using different regions of the HCV genome. J Virol Methods. 1992;38:71–79. doi: 10.1016/0166-0934(92)90170-i. [DOI] [PubMed] [Google Scholar]
- 6.Chan S W, McOmish F, Holmes E C, Dow B, Peutherer J F, Follett E, Yap P L, Simmonds P. Analysis of a new hepatitis C virus type and its phylogenetic relationship to existing variants. J Gen Virol. 1992;73:1131–1141. doi: 10.1099/0022-1317-73-5-1131. [DOI] [PubMed] [Google Scholar]
- 7.Choo Q-L, Kuo G, Ralston R, Weiner A, Chien D, Van Nest G, Han J, Berger K, Thudium K, Kuo C, Kansopon J, McFarland J, Tabrizi A, Ching K, Moss B, Cummins L B, Houghton M, Muchmore E. Vaccination of chimpanzees against infection by the hepatitis C virus. Proc Natl Acad Sci USA. 1994;91:1294–1298. doi: 10.1073/pnas.91.4.1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cooreman M P, Schoondermark-Van de Ven E M. Hepatitis C virus: biological and clinical consequences of genetic heterogeneity. Scand J Gastroenterol Suppl. 1996;218:106–115. doi: 10.3109/00365529609094740. [DOI] [PubMed] [Google Scholar]
- 9.Davidson F, Simmonds P, Ferguson J C, Jarvis L M, Dow B C, Follet E A C, Keller A J, Kruisius T, Lin C, Medgyesu G A, Kiyokawa H, Olim G, Duraisamy G, Cuypers T, Seed A A, Ten D, Conradie J, Kew M C, Lin M, Nuchaprayoon C, Ndimbe O K, Yap P L. Survey of major genotypes and subtypes of hepatitis C virus using RFLP of sequences amplified from the 5′ non-coding region. J Gen Virol. 1995;76:1197–1204. doi: 10.1099/0022-1317-76-5-1197. [DOI] [PubMed] [Google Scholar]
- 10.Domingo E, Baranowski E, Ruiz-Jarabo C M, Martin-Hernández A M, Sáiz J C, Escarmís C. Quasispecies structure and persistence of RNA viruses. Emerg Infect Dis. 1998;4:521–527. doi: 10.3201/eid0404.980402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Enomoto N, Takada A, Nakao T, Date T. There are two major types of hepatitis C virus in Japan. Biochem Biophys Res Commun. 1990;170:1021–1025. doi: 10.1016/0006-291x(90)90494-8. [DOI] [PubMed] [Google Scholar]
- 12.Eyster M E, Sherman K E, Goedert J J, Katsoulidou A, Hatzakis A for the Multicenter Hemophilia Cohort Study. Prevalence and changes in hepatitis C virus genotypes among multitransfused persons with hemophilia. J Infect Dis. 1999;179:1062–1069. doi: 10.1086/314708. [DOI] [PubMed] [Google Scholar]
- 13.Felsenstein J. PHYLIP inference package, version 3.5c. Seattle: Department of Genetics, University of Washington; 1993. [Google Scholar]
- 14.Findor J A, Sorda J A, Daruich J, Bruch Igartua E, Manero E, Avagnina A, Benbassat D, Rey J, Nakatsuno M. Distribution of the genotypes of hepatitis C virus in intravenous drug addicts in Argentina. Medicina (Buenos Aires) 1999;59:49–54. [PubMed] [Google Scholar]
- 15.Forns X, Purcell R H, Bukh J. Quasispecies in viral persistence and pathogenesis of hepatitis C virus. Trends Microbiol. 1999;7:402–410. doi: 10.1016/s0966-842x(99)01590-5. [DOI] [PubMed] [Google Scholar]
- 16.Guadagnino V, Stroffolini T, Rapicetta M, Costantino A, Kondili L A, Menniti-Ippolito F, Caroleo B, Costa C, Griffo G, Loiacono L, Pisani V, Foca A, Piazza M. Prevalence, risk factors, and genotype distribution of hepatitis C virus infection in the general population: a community-based survey in southern Italy. Hepatology. 1997;26:1006–1011. doi: 10.1002/hep.510260431. [DOI] [PubMed] [Google Scholar]
- 17.Honda M, Kaneko S, Unoura M, Kobayashi K, Murakami S. Sequence analysis of putative structural regions of hepatitis C virus isolated from 5 Japanese patients with hepatocellular carcinoma. Arch Virol. 1993;128:163–169. doi: 10.1007/BF01309797. [DOI] [PubMed] [Google Scholar]
- 18.Inchauspé G. DNA vaccine strategies for hepatitis C. J Hepatol. 1999;30:339–346. doi: 10.1016/s0168-8278(99)80084-1. [DOI] [PubMed] [Google Scholar]
- 19.Kato N, Ootsuyama Y, Tanaka T, Nakagawa M, Nakazawa T, Muraiso K, Ohkoshi S, Hijikata M, Shimotohno K. Marked sequence diversity in the putative envelope proteins of hepatitis C viruses. Virus Res. 1992;22:107–123. doi: 10.1016/0168-1702(92)90038-b. [DOI] [PubMed] [Google Scholar]
- 20.Krug L P, Lunge V R, Ikuta N, Fonseca A S K, Cheinquer H, Ozaki L S, Barros S G. Hepatitis C virus genotypes in Southern Brazil. Braz J Med Biol Res. 1996;29:1229–1232. [PubMed] [Google Scholar]
- 21.Kwok S, Higuchi R. Avoiding false positives with PCR. Nature (London) 1989;339:237–238. doi: 10.1038/339237a0. [DOI] [PubMed] [Google Scholar]
- 22.Laskus T, Wang L-F, Rakela J, Vargas H, Pinna A D, Tsamandas A C, Demetris A J, Fung J. Dynamic behavior of hepatitis C virus in chronically infected patients receiving liver graft from infected donors. Virology. 1996;220:171–176. doi: 10.1006/viro.1996.0297. [DOI] [PubMed] [Google Scholar]
- 23.Lau J Y N, Mizokami M, Kolberg J A, Davis G L, Prescott L E, Ohno T, Perrillo R P, et al. Application of six hepatitis C virus genotyping systems to sera from chronic hepatitis C patients in the United States. J Infect Dis. 1995;171:281–289. doi: 10.1093/infdis/171.2.281. [DOI] [PubMed] [Google Scholar]
- 24.Lindh M, Anderson A-S, Gusdal A. Genotypes, nt 1858 variants, and geographic origin of hepatitis B virus—large-scale analysis using a new genotyping method. J Infect Dis. 1997;175:1285–1293. doi: 10.1086/516458. [DOI] [PubMed] [Google Scholar]
- 25.Maggi F, Vatteroni M L, Fornai C, Morraica A, Giorgi M, Bendinelli M, Pistello M. Subtype 2c of hepatitis C virus is highly prevalent in Italy and is heterogeneous in the NS5a region. J Clin Microbiol. 1997;35:161–164. doi: 10.1128/jcm.35.1.161-164.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mondelli M, Cerino A, Bono F, Cividini A, Maccabruni A, Arico M, Malfitano A, Barbarini G, Piazza V, Minoli L, Silini E. Hepatitis C virus (HCV) core serotypes in chronic HCV infection. J Clin Microbiol. 1994;32:2523–2527. doi: 10.1128/jcm.32.10.2523-2527.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Navas S, Castillo I, Martín J, Quiroga J A, Bartolomé J, Carreño V. Concordance of hepatitis C virus typing methods based on restriction fragment length polymorphism analysis in 5′ noncoding region and NS4 serotyping but not in core PCR or a line probe assay. J Clin Microbiol. 1997;35:317–321. doi: 10.1128/jcm.35.1.317-321.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Okamoto H, Kobata S, Tokita H, Inoue T, Woodfield G D, Holland P V, Al-Knawy B A, Uzunalimoglu O, Miyakawa Y U, Mayumi M. A second-generation method of genotyping hepatitis C virus by the polymerase chain reaction with sense and antisense primers deduced from the core gene. J Virol Methods. 1996;57:31–45. doi: 10.1016/0166-0934(95)01960-x. [DOI] [PubMed] [Google Scholar]
- 29.Okamoto H, Mishiro S, Tokita H, Tsuda F, Miyakawa Y, Mayumi M. Superinfection of chimpanzees carrying hepatitis C virus genotype II/1b with that of genotype III/2a or I/1a. Hepatology. 1994;20:11431–11436. [PubMed] [Google Scholar]
- 30.Okamoto H, Sujiyama Y, Okada S, Kurai K, Akahane Y, Sugai Y, Tanaka T, Sato K, Tsuda F, Miyakawa Y, Mayumi M. Typing hepatitis C virus by polymerase chain reaction with type-specific primers: application to clinical surveys and tracing infection sources. J Gen Virol. 1992;73:673–679. doi: 10.1099/0022-1317-73-3-673. [DOI] [PubMed] [Google Scholar]
- 31.Okamoto H, Tokita H, Sakamoto M, Horikita M, Kojima M, Iizuka H, Mishiro S. Characterization of the genomic sequence of type V (or 3a) hepatitis C virus isolates and PCR primers for specific detection. J Gen Virol. 1993;74:2385–2390. doi: 10.1099/0022-1317-74-11-2385. [DOI] [PubMed] [Google Scholar]
- 32.Oubiña J R, Mathet V, Feld M, Della Latta M P, Ferrario D, Verdun R, Libonatti O, Fernández J, Carballal G, Sánchez D O, Quarleri J F. Genetic diversity of GBV-C/HGV strains among HIV infected-IVDU and blood donors from Buenos Aires, Argentina. Virus Res. 1999;65:121–129. doi: 10.1016/s0168-1702(99)00109-4. [DOI] [PubMed] [Google Scholar]
- 33.Oubiña J R, Quarleri J F, Rudzinski M, Parks C, Badía I, González Cappa S M. Genomic characterization of hepatitis C virus from Argentina. J Med Virol. 1995;47:97–104. doi: 10.1002/jmv.1890470118. [DOI] [PubMed] [Google Scholar]
- 34.Panigrahi A K, Roca J, Acharya S K, Jameel S, Panda S K. Genotype determination of hepatitis C virus from northern India: identification of a new subtype. J Med Virol. 1996;48:191–198. doi: 10.1002/(SICI)1096-9071(199602)48:2<191::AID-JMV12>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 35.Pawlotsky J-M, Tsakiris L, Roudot-Thoraval F, Pellet C, Stuyver L, Duval J, Dhumeaux D. Relationship between hepatitis C virus genotypes and sources of infection in patients with chronic hepatitis C. J Infect Dis. 1995;171:1607–1610. doi: 10.1093/infdis/171.6.1607. [DOI] [PubMed] [Google Scholar]
- 36.Picchio G R, Nakatsuno M, Boggiano C, Sabbe R, Corti M, Daruich J, Perez-Bianco R, Tezanos-Pinto M, Kokka R, Wilber J, Mosier D. Hepatitis C (HCV) genotype and viral titer distribution among Argentinean hemophilic patients in the presence or absence of human immunodeficiency virus (HIV) co-infection. J Med Virol. 1997;52:219–225. [PubMed] [Google Scholar]
- 37.Pujol F H, Devesa M, Loureiro C L, Capriles F, Liprandi F. Turnover of hepatitis C virus genotypes in hemodialysis patients. Arch Virol. 1998;143:823–827. doi: 10.1007/s007050050334. [DOI] [PubMed] [Google Scholar]
- 38.Pujol F H, Loureiro C L, Devesa M, Blitz L, Parra K, Beker S, Liprandi F. Determination of genotypes of hepatitis C virus in Venezuela by restriction fragment length polymorphism. J Clin Microbiol. 1997;35:1870–1872. doi: 10.1128/jcm.35.7.1870-1872.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Quarleri J F, Badía I, Mathet V, Oubiña J R. Epidemiología molecular del virus de la hepatitis C (HCV) en pacientes hemofílicos de edad pediátrica. Pren Med Argent. 1999;86:181–185. [Google Scholar]
- 40.Quarleri J F, Mathet V L, Feld M, Ferrario D, Della Latta M P, Verdun R, Sánchez D O, Oubiña J R. GBV-C/HGV groups and subgroups: classification by a restriction fragment length polymorphism method based on the phylogenetic analysis of the 5′ untranslated region. J Clin Microbiol. 1999;37:1340–1347. doi: 10.1128/jcm.37.5.1340-1347.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Quarleri J F, Robertson B H, Mathet V, Sinha S D, Badía I, Frider B, Ferro A, Galoppo C, Sookoián S, Castaño G, Oubiña J R. Genomic and phylogenetic analysis of hepatitis C virus strains from Argentina. Medicina (Buenos Aires) 1998;58:153–159. [PubMed] [Google Scholar]
- 42.Robertson B, Myers G, Howard C, Brettin T, Bukh J, Gaschen B, Gojobori T, Maaertens G, Mizokami M, Nainan O, Netesov S, Nishioka K, Simmonds P, Smith D, Stuyver L, Weiner A. Classification, nomenclature, and database development for hepatitis C virus (HCV) and related viruses: proposals for standardization. Arch Virol. 1998;143:2493–2503. doi: 10.1007/s007050050479. [DOI] [PubMed] [Google Scholar]
- 43.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain- terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Silini E, Bono F, Cerino A, Piazza V, Solcia E, Mondelli H U. Virological features of hepatitis C virus infections in hemodialysis patients. J Clin Microbiol. 1993;31:2913–2917. doi: 10.1128/jcm.31.11.2913-2917.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Simmonds P, Smith D B, McOmish F, Yap P L, Kolberg J, Urdea M S, Holmes E C. Identification of genotypes of hepatitis C virus by sequence comparisons in the core, E1 and NS-5 regions. J Gen Virol. 1994;75:1053–1061. doi: 10.1099/0022-1317-75-5-1053. [DOI] [PubMed] [Google Scholar]
- 46.Smith B D, Mellor J, Jarvis L M, Davidson F, Kolberg J, Urdea M, Yap P-L, Simmonds P International HCV Collaborative Study Group. Variation of the hepatitis C virus 5' noncoding region: implications for secondary structure virus detection and typing. J Gen Virol. 1995;76:1749–1761. doi: 10.1099/0022-1317-76-7-1749. [DOI] [PubMed] [Google Scholar]
- 47.Spada E, Ciccaglione A R, Dettori S, Chionne P, Kondili L A, Amoroso P, Guadagnino V, Greco M, Rapicetta M. Genotyping HCV isolates from Italy by type-specific PCR assay in the core region. Res Virol. 1998;149:209–218. doi: 10.1016/s0923-2516(98)80002-2. [DOI] [PubMed] [Google Scholar]
- 48.Thompson J D, Gibson T J, Plewniak F, Jeanmougin F, Higgins D G. The CLUSTAL X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xu L Z, Martinot-Peignoux M, Marcellin P, Benhamou J P, Larzul D. Comparison of the sensitivity of nested PCR in the 5′ non-coding and the NS5 regions of the HCV genome. J Hepatol. 1994;20:598–602. doi: 10.1016/s0168-8278(05)80346-0. [DOI] [PubMed] [Google Scholar]
- 50.Yuki N, Hayashi N, Kasahara A, Hagiwara H, Ohkawa K, Fusamoto H, Kamada T. Hepatitis C virus replication and antibody responses toward specific hepatitis C virus proteins. Hepatology. 1994;19:1360–1365. [PubMed] [Google Scholar]
- 51.Zuckerman A J, Zuckerman J N. Prospects for hepatitis C vaccine. J Hepatol. 1995;22:97–100. [PubMed] [Google Scholar]