SidK is introduced as a highly specific probe to visualize and quantify the proton-pumping vacuolar H+ATPases (V-ATPases) of mammalian cells and yeast.
Abstract
Acidification of secretory and endocytic organelles is required for proper receptor recycling, membrane traffic, protein degradation, and solute transport. Proton-pumping vacuolar H+ ATPases (V-ATPases) are responsible for this luminal acidification, which increases progressively as secretory and endocytic vesicles mature. An increasing density of V-ATPase complexes is thought to account for the gradual decrease in pH, but available reagents have not been sufficiently sensitive or specific to test this hypothesis. We introduce a new probe to localize and quantify V-ATPases. The probe is derived from SidK, a Legionella pneumophila effector protein that binds to the V-ATPase A subunit. We generated plasmids encoding fluorescent chimeras of SidK1-278, and labeled recombinant SidK1-278 with Alexa Fluor 568 to visualize and quantify V-ATPases with high specificity in live and fixed cells, respectively. We show that V-ATPases are acquired progressively during phagosome maturation, that they distribute in discrete membrane subdomains, and that their density in lysosomes depends on their subcellular localization.
Introduction
The steady-state pH of individual cellular compartments is a key determinant of their function and must be regulated stringently. The luminal pH directs receptor recycling and membrane traffic, regulates protein degradation, and contributes to the transmembrane protonmotive force that drives the transport of a variety of organic and inorganic solutes (Maxfield and McGraw, 2004; Saftig and Klumperman, 2009; Huotari and Helenius, 2011). It is therefore not surprising that dysregulation of organellar pH has been implicated in various human diseases such as cancer, neurological disorders, osteoporosis, and autoimmunity (Marshansky et al., 2014; Capecci and Forgac, 2013; Sun-Wada et al., 2006; Eaton et al., 2021).
Most endocytic and secretory organelles maintain an acidic lumen, with acidification increasing progressively as these organelles approach their terminal stages (Mellman et al., 1986). Thus, the endocytic pathway progresses from slightly acidic early endosomes (pH 6.5) to highly acidic lysosomes (pH 4–5). The main driver of organellar acidification is the ATP-dependent H+ pump known as the vacuolar H+ATPase (V-ATPase). The V-ATPase is present in membranes of the endocytic and secretory pathways, and is also found in the plasma membrane (PM) of specialized cell types involved in the active extrusion of cytosolic H+, such as osteoclasts and renal intercalated cells (Toei et al., 2010; Futai et al., 2019). The V-ATPase is a large complex with 16 different subunits in mammals. ATP hydrolysis occurs in the soluble catalytic V1 region (subunits A–H), driving rotation of the enzyme’s rotor subcomplex and H+ translocation through the VO regions (subunits a, d, e, f, c, cʺ, ATP6AP1/Ac45, and ATP6AP2/PRR; Abbas et al., 2020). V-ATPase activity is regulated by several mechanisms, including reversible dissociation of the V1 and VO regions (Tabke et al., 2014; Kawasaki-Nishi et al., 2001; Parra and Kane, 1998; Poëa-Guyon et al., 2013), phosphorylation (Voss et al., 2007; Alzamora et al., 2010), and changes in membrane lipid composition (Banerjee et al., 2019; Vasanthakumar et al., 2019; Uchida et al., 1985).
Several parameters determine the steady-state pH of the lumen of an organelle. Because H+ pumping by V-ATPases is electrogenic, the rate of pumping can be limited by the permeability of the membrane to neutralizing counterions. In addition, the accumulation of H+ is opposed by ongoing H+ backflux or “leak” via a collection of incompletely characterized channels and transporters. A final fundamental parameter is the density of V-ATPase complexes in the membrane of any particular organelle. It has been tacitly assumed that V-ATPase density increases progressively as the components of endocytic and secretory pathways mature and become more acidic. In support of this assumption, increased assembly of V-ATPases has been reported on endomembranes as their luminal acidity increases (Trombetta et al., 2003). However, the assumption has not been widely validated for two main reasons. First, it is difficult to isolate individual stages of these pathways with sufficient purity for reliable biochemical analysis. Second, reagents with sufficient resolution and accuracy are lacking for localization and quantification of V-ATPase in cells. Several antibodies to different V-ATPase subunits are available commercially, and some have yielded satisfactory results in analyses of tissues like the kidney, where specialized cell types are uniquely enriched in V-ATPases. However, these same antibodies show low signal-to-noise ratio when used to stain nonspecialized single cells, which have a lower abundance of V-ATPases (Yajima et al., 2007; McGuire et al., 2019; Michel et al., 2013; Ramirez et al., 2019; Chung et al., 2011; Asare and Abu Kwaik, 2007; Santic et al., 2008; Hinton et al., 2009). The interpretation of the resulting immunostaining can be ambiguous.
The paucity of reagents currently available to study the V-ATPase motivated us to develop a novel tool for its specific intracellular labeling. To this end, we took advantage of SidK, an effector protein deployed by Legionella pneumophila to inhibit the V-ATPase (Xu et al., 2010). SidK was recently shown to bind directly to yeast and mammalian V-ATPases (Zhao et al., 2017; Abbas et al., 2020), allowing use of the effector to purify H+ pumps from tissue extracts (Abbas et al., 2020). In this study, we describe the generation and labeling of a recombinant fragment of SidK and its use to localize and quantify V-ATPases in eukaryotic cells with high sensitivity and specificity. We used this reagent to study the distribution of the V-ATPase in various cell types, and to monitor its acquisition by membrane-bound compartments as they mature along the endocytic pathway. Last, we used SidK to estimate the number of V-ATPase complexes in individual compartments as a function of their position within the cell.
Results
Generation of fluorescent SidK chimeras
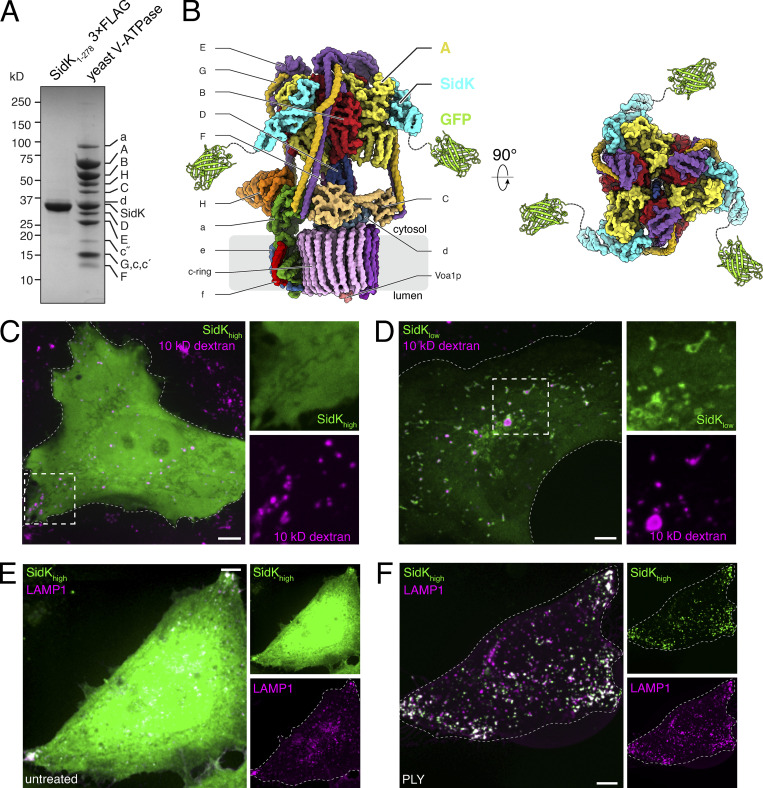
Because SidK binds with high affinity to the A subunit (Xu et al., 2010; Sharma and Wilkens, 2017; Abbas et al., 2020; Zhao et al., 2017), we reasoned that it would be an effective probe to visualize V-ATPases by fluorescence microscopy. To this end, we generated chimeric constructs consisting of amino acids 1–278 of SidK (SidK1-278) attached to a fluorescent protein. SidK1-278 suffices to interact with the A subunit with high specificity; indeed, this fragment was used for the affinity purification of V-ATPases from cell and tissue extracts (Fig. 1 A; see also Abbas et al., 2020). GFP or mCherry were linked to SidK1-278 via its C terminus because the structure of the V-ATPase:SidK1-278 complex suggested that attachment at this position was unlikely to affect association of the chimera with the V-ATPase (Fig. 1 B; Abbas et al., 2020). For brevity, the resulting construct is referred to simply as SidK.
Figure 1.
SidK interacts with the V-ATPase in vitro and when expressed in mammalian cells. (A) SDS-PAGE of 3×FLAG SidK1-278 illustrates purity of the probe and its ability to allow isolation of yeast V-ATPases by affinity chromatography. Bands corresponding to SidK1-278 and individual subunits of V-ATPase are labeled. (B) Model of yeast V-ATPase with SidK1-278-GFP bound, generated from PDB accession nos. 5VOX, 1GFL, and 6O7T. (C) Visualization of SidKhigh cells. HeLa cells were transfected with SidK-GFP (green) and lysosomes loaded with Alexa Fluor 647–labeled dextran (magenta). Side panels show individual SidK and dextran channels in dotted square, 2.2× magnification. (D) Visualization of SidKlow cells (green) labeled as in C. Side panels show individual SidK and dextran channels, 2.0× magnification. (E and F) Lysosomes identified by LAMP1 immunostaining (magenta) after expression of high levels of SidK-GFP (SidKhigh; green). Prior to fixation and immunostaining, cells were left untreated (E) or permeabilized with PLY to remove excess cytosolic SidK-GFP (F). Side panels show individual SidK and LAMP1 channels. Here and elsewhere, cell outlines are indicated by dotted lines. Images in C-F are compressions of confocal stacks representative of ≥30 fields from three or more experiments of each type. Scale bars: 5 µm.
Varying levels of expression were observed following transient transfection of fluorescent SidK in HeLa cells. The fluorescence intensity of the soluble (excess) SidK seen in high expressers (SidKhigh) precluded resolution of the construct bound to V-ATPase–containing organelles, such as lysosomes (Fig. 1 C). In contrast, SidK showed clear association with vesicular and tubular structures in low-expressing cells (SidKlow; Fig. 1 D), some of which were identifiable as lysosomes by loading with fluorescent dextran. Because the excess fluorescence of SidKhigh cells appeared to be cytosolic, we predicted that selective removal of soluble material would reveal the more tightly bound, organelle-associated probe. This prediction was tested by comparing SidKhigh cells before (Fig. 1 E) and after (Fig. 1 F) selective leaching of cytosolic components following permeabilization of the PM with the pore-forming toxin, pneumolysin (PLY). As anticipated, the cytosolic fluorescence was largely depleted in PLY-treated cells, revealing membrane-associated SidK, a fraction of which was clearly colocalized with LAMP1, a marker of late endosomes/lysosomes. We concluded that the fluorescent chimeras of SidK can be used for detection of membrane-associated ligands, presumably V-ATPases, particularly in cells with a low expression level or permeabilized with PLY or similar reagents.
Expression of SidK constructs affects lysosomal positioning and pH
In untreated HeLa cells, late endosomes/lysosomes are largely located near the nucleus, as revealed by LAMP1 immunostaining (Fig. 2 A). However, we noted that endo/lysosomes tended to accumulate at the cell periphery of SidK-transfected cells, particularly those where the construct was highly expressed (Fig. 1 F and Fig. 2 B). Because SidK has been shown to inhibit the V-ATPase partially in vitro and in mammalian cells (Zhao et al., 2017; Xu et al., 2010), we considered the possibility that inhibition of H+ pumping was responsible for the altered distribution of LAMP1-positive compartments. This was validated by treating cells with concanamycin A, a specific V-ATPase inhibitor, which resulted in a similar margination of a fraction of the LAMP1-positive vesicles (Fig. 2 C). These observations are consistent with previous reports that the positioning of lysosomes correlates with their luminal pH (Johnson et al., 2016).
Figure 2.
Overexpression of SidK affects lysosome positioning and pH. (A) Localization of endogenous LAMP1 (magenta) in untreated cells. (B) Localization of LAMP1 (magenta) in cells expressing SidK-mCherry (green). Side panels show individual SidK and LAMP1 channels in area denoted by dotted square, 2.0× magnification. Arrowheads mark sites of peripheral LAMP1 accumulation. (C) Localization of LAMP1 (magenta) in cells treated with 250 nM concanamycin A for 1 h. Arrowheads mark sites of peripheral LAMP1 accumulation. (D) Retention of cresyl violet (magenta) in cells expressing SidK-GFP (green) with lysosomes preloaded with Alexa Fluor 647–conjugated dextran (dex; blue). Side panels show individual channels of area denoted by dotted square, 2.7× magnification. (E) Cells expressing SidK-GFP (green) and labeled for lysosomes (blue) were treated with 250 nM concanamycin A and 10 mM NH4Cl for 30 min to neutralize luminal pH. After treatment, cells were incubated with cresyl violet as in D. Side panels: 2.7× magnification. (A–E) Extended focus images representative of ≥30 fields from three or more experiments of each type. All scale bars: 5 µm. (F) Lysosomal pH determined in SidK– and SidK+ HeLa cells. Lysosomes of control or SidK-mCherry–transfected HeLa cells were loaded overnight with FITC-dextran. pH was measured by ratiometric fluorescence microscopy. For each condition, three independent experiments were quantified, with ≥20 cells per replicate. Data are mean ± SEM. P calculated using unpaired, two-tailed Student’s t test. (G) Lysosomal V-ATPase activity was measured in control and SidK-mCherry–expressing HeLa cells acutely treated with 500 nM concanamycin A. V-ATPase activity was determined from the inverse rate of alkalization upon addition of concanamycin A. For each condition, three independent experiments were quantified, with ≥10 cells per replicate. Data are mean ± SEM. P calculated using unpaired, two-tailed Student’s t test.
To verify that SidK caused lysosomal alkalinization, we used cresyl violet (Ostrowski et al., 2016), an acidotropic fluorescent dye. Unexpectedly, and in apparent disagreement with the findings of Xu et al. (2010), the lysosomes remained acidic (retained cresyl violet) despite the presence of SidK in the cytosol (Fig. 2 D). Additionally, we did not detect any differences in lysosomal degradative capacity (measured using DQ-BSA) in SidK-overexpressing cells, another, indirect, indicator of lysosome acidification (Fig. S1, A and B). The reliability of cresyl violet as an indicator of lysosomal acidification was confirmed using concanamycin A, which prevented the accumulation of the dye (Fig. 2 E). It is noteworthy, however, that like other acidotropic dyes, cresyl violet is a coarse indicator, unable to sense moderate changes in pH, and lysosomal hydrolases have functional optima between pH 4 and 6 (Brockman and Murphy, 1993). In this regard, in vitro determinations using purified yeast V-ATPase showed that saturating concentrations of SidK caused only a ∼30% inhibition of ATPase activity (Zhao et al., 2017). To more precisely assess the effect of SidK on HeLa cells, their lysosomes were loaded with FITC-dextran, and pH was measured ratiometrically. Using this approach, the pH of lysosomes in SidK-expressing cells was found to be moderately, yet significantly, more alkaline (pH 5.22 ± 0.12 [mean ± SEM]) than that of control cells (pH 4.97 ± 0.07 [mean ± SEM]; Fig. 2 F). To more directly assess the effect of SidK on H+ pumping by the V-ATPase, we quantified the effect of concanamycin A on proton flux across the lysosomal membrane. These measurements are based on the assumption that at steady-state pH, V-ATPase activity is precisely offset by an equivalent but opposite backflux of H+. The initial rate of H+ leakage unmasked by addition of concanamycin A can be considered as a measure of pumping by the V-ATPase at steady-state. This analysis (Fig. 2 G) indicated that SidK reduced H+ pumping by 29%, in good agreement with Zhao et al. (2017).
Figure S1.
Overexpression of SidK-fluorescent protein does not affect lysosome degradation or mTORC1 signaling. (A) Dequenching of DQ-BSA (magenta) in control HeLa (top) and RAW264.7 (bottom) cells transfected with GFP (green), where lysosomes had been coloaded with Alexa Fluor 647–conjugated 10 kD dextran (blue). See Materials and methods. Side panels show the individual DQ-BSA and dextran channels of the area denoted by the dotted square, at 1.4× magnification. Images are extended focus compressions of confocal images representative of ≥10 fields from three or more separate experiments of each type. Scale bars: 5 µm. (B) Quantification of DQ-BSA dequenching in SidK– and SidK+ HeLa or RAW264.7 cells, normalized to the 10 kD dextran coloaded into lysosomes. For each condition, three independent experiments were quantified, with ≥10 cells per replicate. Data (magenta circles) are replicate mean ± SEM. P value was calculated using unpaired, two-tailed Student’s t test. n = 14,010, 11,371, 10,955, and 9,864 lysosomes analyzed, respectively. (C) Effects of SidK overexpression on mTORC1 activity. HeLa cells transiently expressing mCherry or SidK-mCherry for 16–24 h were analyzed by immunoblotting for the phosphorylation of mTORC1 targets S6K and ULK1. pS6K-Thr389 (70 kD) is shown in comparison to endogenous S6K (70 kD), and pULK1-Ser757 (140 kD) is shown in comparison to tubulin (55 kD). Blots shown are representative of three independent experiments. Source data are available for this figure: SourceData FS1.
The function of mTORC1 depends in part on the V-ATPase (Zoncu et al., 2011), but the modest inhibition exerted by SidK was insufficient to affect the activity of the kinase, assessed by quantifying pS6K and pULK1 by immunoblotting (Fig. S1 C).
Localization of the V-ATPase using SidK-AL568
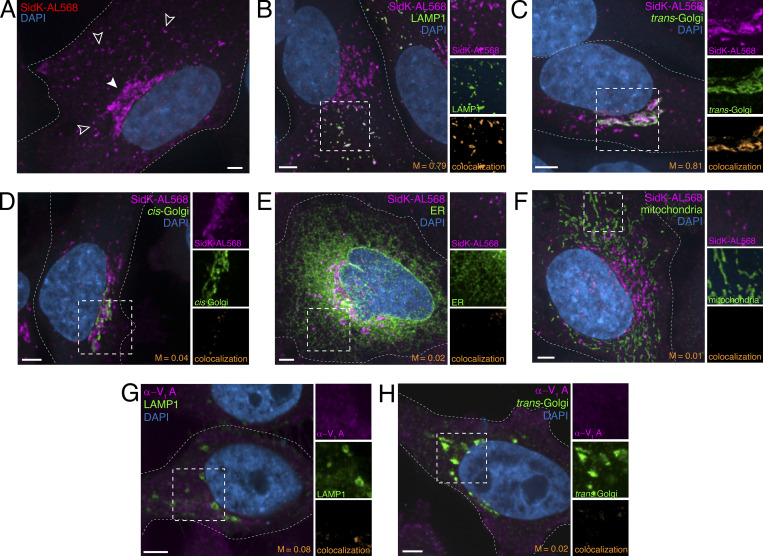
While capable of detecting the V-ATPase under appropriate conditions, expression of a genetically encoded form of fluorescent SidK had obvious limitations, most notably that chronic inhibition of the pump, even if only partial, alters the distribution of the organelles. To avoid inhibition of the pump before its detection, we developed an alternative probe based on SidK that could be used to label the V-ATPase in fixed and permeabilized cells. Recombinant SidK1-278 was expressed in bacteria and, after purification (Fig. 1 A), was covalently labeled with Alexa Fluor 568 (referred to as SidK-AL568) and used to stain cells. In HeLa cells, SidK-AL568 labeled vesicular and cisternal compartments reminiscent of endosomes and the Golgi complex with remarkably little background (Fig. 3 A, left). This staining was blocked by pretreatment with unlabeled SidK (Fig. 3 A, right), implying that binding is saturable.
Figure 3.
SidK-AL568 specifically labels the V-ATPase of mammalian and yeast cells. (A) HeLa cells were stained with SidK-AL568 (magenta) either directly (left) or after incubation with a fivefold excess of unlabeled SidK1-278 (right). F-actin labeled with fluorescent phalloidin (blue). (B) Spheroplasts of wild-type or vma1Δ S. cerevisiae were stained with SidK-AL568 (magenta) as detailed in Materials and methods. Nuclei labeled with DAPI (cyan). Outlines of spheroplasts, visualized by differential interference contrast microscopy, are indicated by dotted lines. (C and D) HeLa cells transfected with VOa2- (C) or VOa3-GFP (D) constructs (green) were stained using SidK-AL568 (magenta). Nuclei labeled with DAPI (blue). Cell outlines indicated by dotted lines. Side panels show individual SidK-AL568 and VOa channels. Colocalization of SidK-AL568 and the corresponding VOa subunit is shown in orange, and the Mander’s coefficient (M) between the subunit and SidK-AL568 is indicated. (A–D) Compressions of confocal stacks representative of ≥30 fields from three or more experiments. Scale bars: 5 µm.
The specificity of SidK-AL568 for the A subunit of the V-ATPase was validated by comparing staining of wild-type Saccharomyces cerevisiae to that of a mutant strain lacking subunit A (vma1Δ). SidK-AL568 clearly labeled the vacuole and prevacuolar compartment of wild-type spheroplasts, but staining was absent in the vma1Δ strain (Fig. 3 B). Additional evidence that SidK-AL568 associates with V-ATPase complexes in mammalian cells was obtained by ectopic expression in HeLa cells of fluorescently tagged Voa2 or Voa3 subunits that localize predominantly to the Golgi complex or endosomes, respectively (Saw et al., 2011). SidK-AL568 colocalized with both Voa2- and Voa3-GFP, with highly significant Mander’s coefficients (M = 0.74 and 0.79, respectively; Fig. 3, C and D).
We next used SidK-AL568 to assess the presence and density of V-ATPase complexes in defined intracellular compartments. As above and illustrated in more detail in Fig. 4 A, SidK-AL568 stained peripheral vesicles (Fig. 4 A, open arrowheads) as well as juxtanuclear vesicles and cisternae (closed arrowheads) likely corresponding to endocytic and Golgi components, respectively. These assumptions were confirmed by simultaneously visualizing LAMP1 and the trans-Golgi (Fig. 4, B and C), demonstrating that SidK-AL568 colocalizes with these compartments, which are known to have a markedly acidic lumen (Casey et al., 2010). In contrast, SidK-AL568 showed minimal colocalization with markers of the ER and mitochondria (Fig. 4, E and F), which have a near neutral or slightly alkaline lumen (Casey et al., 2010). Of note, SidK-AL568 labeled poorly the cis-Golgi (Fig. 4 D), which is thought to be less acidic than the mid- and trans-cisternae.
Figure 4.
SidK-AL568 detects the V-ATPase in acidic intracellular organelles. (A) HeLa cells were stained with SidK-AL568 (magenta). Nuclei labeled with DAPI (blue). Arrowheads mark peripheral, likely endocytic, vesicles (open) and juxtanuclear vesicles and cisternae, likely including the Golgi compartment (closed). (B–F) V-ATPase in HeLa cells labeled with SidK-AL568 (magenta) and costained for (B) LAMP1, (C) trans-Golgi, (D) cis-Golgi, (E) ER, and (F) mitochondria (green). Nuclei labeled with DAPI (blue). Side panels show individual SidK-AL568 and organelle channels in area denoted by dashed square, at 1.1, 0.9, 1.1, 1.7, and 1.5× magnification, respectively. (G and H) HeLa cells were immunostained using the V1A antibody, α-ATP6V1A (Abnova; catalog no. H00000523-M02; magenta) in cells costained for LAMP1 (G) or trans-Golgi (H), in green. Nuclei labeled with DAPI (blue). Side panels show individual α-V1A and organelle channels in dotted square, at 1.1 and 1.4× magnification, respectively. (A–H) are extended focus images representative of ≥30 fields from three or more experiments. Colocalization and Mander’s coefficients (M) between the organellar marker and V-ATPase probe shown in orange. All scale bars: 5 µm.
Based on these observations, we believe that SidK-AL568 is an excellent probe to detect V-ATPases by fluorescence microscopy. Indeed, when compared with a commercially available antibody raised against the same subunit to which SidK-AL568 binds (subunit A), our probe yielded better results. The antibody chosen for comparison (α-ATP6V1A from Abnova) is widely used in the literature, often as the basis for important functional conclusions (e.g., Ramirez et al., 2019). In our hands, the α-ATP6V1A antibody recognized the V1A subunit in mammalian cell membranes and lysates by immunoblotting (Fig. S2 C), although binding to other proteins was obvious, particularly when longer incubations or higher lysate concentrations were used (Fig. S2 D). The yeast V1A subunit was not recognized by α-ATP6V1A, while SidK-AL568 effectively labeled the yeast protein (Fig. 3 B). When tested for immunostaining of HeLa cells (Fig. 4, G and H), the α-ATP6V1A antibody yielded a diffuse punctate pattern reminiscent of that reported by Ramirez et al. (2019) for tumor cells and elsewhere for a variety of cells stained with other A subunit antibodies (Yajima et al., 2007; McGuire et al., 2019; Michel et al., 2013). However, its colocalization with acidic compartments was poor: the Mander’s coefficient of the antibody with LAMP1 was M = 0.08 (compared with an M = 0.79 with SidK-AL568), while that with a trans-Golgi marker was M = 0.05 (compared with an M = 0.81 with SidK-AL568). Most of the α-ATP6V1A staining was background, which was removed by applying the unbiased Costes thresholding method before colocalization analysis (Costes et al., 2004; see Materials and methods); this was not the case for SidK-AL568 staining (Fig. S2). We therefore suggest that SidK-AL568 is a more specific, preferable probe.
Figure S2.
Comparison of SidK-AL568 and V1A antibody specificity. (A and B) Demonstration of the Costes thresholding method used before colocalization analyses. HeLa cells were stained with SidK-AL568 (A; magenta) or α-V1A (B; magenta), and the trans-Golgi was immunostained as described in Materials and methods. For both A and B, the left panel shows staining of the magenta channel before Costes thresholding, while the right panel shows the same channel after thresholding. Outlines of cells are indicated by dotted lines. Scale bars: 5 µm. Images in A and B correspond to Fig. 4, C and H, respectively. (C) Immunoblotting of the indicated protein amounts of yeast membranes, rat membranes, and HeLa and RAW264.7 lysates for the V-ATPase using the V1A antibody, α-ATP6V1A. The V1A band is marked with an open arrowhead. V1A blot is shown in comparison to its GAPDH loading control. (D) Immunoblotting of 20 µg protein samples of HeLa and RAW264.7 lysates with the V1A antibody. The V1A band is marked with an open arrowhead. V1A blot is shown in comparison to its GAPDH loading control. Blots in C and D are representative of three similar experiments. Source data are available for this figure: SourceData FS2.
Assessment of V-ATPase acquisition by maturing phagosomes
The phagosomes formed by macrophages are specialized compartments with microbicidal and degradative functions. The lumen of nascent phagosomes is near neutral pH, but becomes gradually acidic as the compartment matures, reaching a pH ≤5 (e.g., Fig. 5 A). This acidification depends on the activity of V-ATPases (Lukacs et al., 1990), which are undetectable on the macrophage PM that forms the initial phagosomal enclosure. The graded acidification is thought to result from accumulation of V-ATPases owing to fusion of the nascent phagosome with early and late endosomes and, ultimately, with lysosomes. Remarkably, to our knowledge, this purported mechanism has not been documented experimentally. We therefore used the SidK-AL568 probe to detect V-ATPases during the phagosomal maturation process in murine macrophages that had been transfected with various membrane markers of maturation state (Fig. 5 B).
Figure 5.
Acidification of macrophage phagosomes parallels recruitment of the V-ATPase to the phagosomal membrane. (A) RAW264.7 cells were incubated with IgG-opsonized FITC-zymosan to initiate phagocytosis. Phagosomal pH changes were determined by ratiometric fluorescence microscopy. Time 0 marks the point where acidification became evident. Data points (open circles) represent the average of two independent experiments, with ≥10 cells per replicate. (B) Schematic showing the stages of phagosome engulfment and maturation, with associated membrane markers. (C) V-ATPase localization in resting RAW264.7 cells stained with SidK-AL568 (magenta). (D–H) RAW264.7 cells transfected with the indicated phagosomal maturation markers (green) were allowed to internalize IgG-opsonized SRBCs for various times before fixation. Cells were then fixed, permeabilized, and stained with SidK-AL568 (magenta). (D) Cells expressing a PM marker, fixed 2 min after initiation of phagocytosis. (E) Cells expressing Rab5A, fixed 5 min after initiation of phagocytosis. (F) Cells expressing a PtdIns(3)P-specific probe, fixed 5 min after initiation of phagocytosis. (G) Cells expressing a Rab7 marker, fixed 30 min after initiation of phagocytosis. (H) Cells immunostained for LAMP1, fixed 30 min after initiation of phagocytosis. (C–H) XY optical slices acquired near the middle of the cell or phagosome, representative of ≥30 fields from three or more experiments of each type. Scale bars: 5 µm.
Like HeLa cells, resting RAW264.7 macrophages showed distinct vesicular staining with SidK-AL568, as well as larger vacuoles that likely form by constitutive macropinocytosis (Fig. 5 C). Nascent phagosomes, enriched by arresting phagocytosis shortly (2 min) after exposure to target particles, were identified by the persistence of plasmalemmal markers (e.g., PM-GFP). As expected, nascent phagosomes were essentially devoid of V-ATPases, although V-ATPase–rich organelles seemed to accumulate in their immediate vicinity (Fig. 5 D). In contrast, early phagosomes, which are measurably acidic and were identified by the acquisition of Rab5 and PtdIns(3)P, showed distinct acquisition of SidK-AL568 into discrete areas (Fig. 5, E and F). Late phagosomes/phagolysosomes, identified by possessing Rab7 or LAMP1, were even more enriched with SidK-AL568 (Fig. 5, G and H), consistent with their highly acidic pH. Interestingly, a patchy localization of SidK-AL568 was noted at all stages of phagosome maturation, suggesting that the recruitment of V-ATPases may occur at restricted sites, which may have important implications for traffic and pH regulation.
The V-ATPase is heterogeneously distributed on the membrane of acidic organelles
The discontinuous pattern of SidK-AL568 on the late-phagosome membrane is reminiscent of the spatial segregation of other membrane components that was observed previously and associated with the formation of ER–phagosome contacts (Levin-Konigsberg et al., 2019). These contacts are established, at least partly, by interaction of phagosomal ORP1L with the ER proteins VapA and VapB (Rocha et al., 2009; Loewen and Levine, 2005). The SidK-AL568 patches may represent similar regions of V-ATPase exclusion from ER contact sites. To assess this possibility, phagosomes of cells transfected with ORP1L-GFP were stained with SidK-AL568. Following transfection (Fig. 6 A), ORP1L and the V-ATPase showed an inverse distribution, the latter accumulating in regions where ORP1L was depleted. This segregation could be quantified and is represented as a ratio of SidK-568:ORP1L fluorescence in Fig. 6 A. In contrast, SidK-AL568 codistributed with Arl8b (Fig. 6 B), which is excluded from ER contact sites (Levin-Konigsberg et al., 2019). It was imperative to ensure that the apparent depletion of V-ATPases from contact areas was not caused by limited access of SidK-AL568. Accessibility was verified by immunostaining VapB alongside SidK-AL568 (Fig. 6 C). As was the case for ORP1L, the regions that were rich in VapB were comparatively depleted of SidK-AL568. Because the antibody used to immunostain VapB is much larger (∼150 kD) than SidK-AL568 (∼35 kD), exclusion of the V-ATPase probe cannot account for the observed segregation, indicating the genuine existence of V-ATPase–enriched microdomains.
Figure 6.
V-ATPase accumulates in subdomains along the phagosomal membrane and is excluded from ER contact sites. (A) RAW264.7 cells transfected with ORP1L-GFP were allowed to internalize IgG-opsonized SRBCs and fixed after 60 min. Cells were stained with SidK-AL568 (magenta) and visualized along with ORP1L (cyan). Individual SidK-AL568 and ORP1L channels and the SidK-AL568:ORP1L fluorescence ratio (RatioSidK-AL568:ORP1L) are shown. Fluorescence ratio is pseudocolored in a rainbow look up table, corresponding to ratio values from 0 to 2. (B) RAW264.7 cells transfected with Arl8b-GFP allowed to internalize IgG-opsonized SRBCs, fixed after 30 min, then stained with SidK-AL568 (magenta) and visualized along with Arl8b-GFP (cyan). Individual channels and the pseudocolored fluorescence ratio are shown. (C) Human macrophages were allowed to internalize IgG-opsonized SRBCs and fixed after 30 min. Cells were fixed, permeabilized, and stained with SidK-AL568 (magenta) and immunostained for endogenous VapB (cyan). Smaller panels show (top to bottom) individual SidK-AL568, VapB, and SidK-AL568:VapB fluorescence ratio channels (RatioSidK-AL568:VapB) for three individual phagosomes (left to right) identified by dotted squares, all at 1.8× magnification. (A–C) Central xy optical slices acquired near the middle of the cell or phagosome representative of ≥30 fields from three or more experiments of each type. Scale bars: 5 µm.
We next asked whether similar subdomains exist in other organelles. To facilitate visualization, we generated enlarged lysosomes using sucrose (Cohn and Ehrenreich, 1969; Swanson et al., 1986; Ferris et al., 1987) in cells that had been transfected with ORPL1 (Fig. 7 A) or Arl8b (Fig. 7 B). As with late phagosomes, V-ATPase (SidK-AL568) was depleted from membrane domains where ORPL1 was found, while coinciding with Arl8b.
Figure 7.
Lysosomes and lysosomal tubules show V-ATPase subdomains. (A) RAW264.7 cells transfected with ORP1L-GFP were subjected to lysosome enlargement by overnight treatment with 30 mM sucrose. Cells were stained with SidK-AL568 (magenta) and the distribution of V-ATPase in sucrosomes labeled with ORP1L (cyan) visualized. Side panels show individual SidK-AL568, ORP1L, and colocalization channels (left to right). Colocalization and Mander’s coefficient between ORP1L and SidK-AL568 (M) shown in orange. (B) RAW264.7 cells transfected with Arl8b-GFP were subjected to lysosome enlargement by treatment with 30 mM sucrose. Cells were stained with SidK-AL568 (magenta) and the distribution of V-ATPase in sucrosomes labeled with Arl8b (cyan) visualized. Side panels: Individual SidK-AL568, Arl8b, and colocalization channels (left to right). Colocalization and Mander’s coefficients between Arl8b and SidK-AL568 (M) shown in orange. (C–E) Localization of V-ATPases in tubular lysosomal structures in RAW264.7 cells (C), human macrophages (D), and HeLa cells (E). Cells were stained using SidK-AL568 (magenta) and immunostained for endogenous LAMP1 (cyan). Solid white arrowheads mark lysosomal regions that are SidK+ LAMP1–; open arrowheads mark regions that are SidK– LAMP1+. Side panels show individual SidK-AL568, LAMP1, and colocalization channels (top to bottom). Colocalization and Mander’s coefficients between LAMP1 and SidK-AL568 (M) shown in orange. (A–E) xy optical slices representative of ≥30 fields from three or more experiments of each type. Scale bars: 5 µm.
The existence of comparatively long tubular lysosomes in macrophages (Knapp and Swanson, 1990; Swanson et al., 1987) also enabled us to assess segregation in resting cells. In RAW264.7 macrophages (Fig. 7 C) and human monocyte–derived macrophages (Fig. 7 D), regions of late endosome/lysosomes, identified by LAMP1, were preferentially enriched with V-ATPase. This pattern could also be observed in some HeLa cells where the LAMP1-positive structures were sufficiently large (Fig. 7 E). In all cases, regions of V-ATPase exclusion from LAMP1-stained areas were observed. The observed segregation may be an indication of selective delivery or removal of V-ATPases at varying stages of organellar maturation.
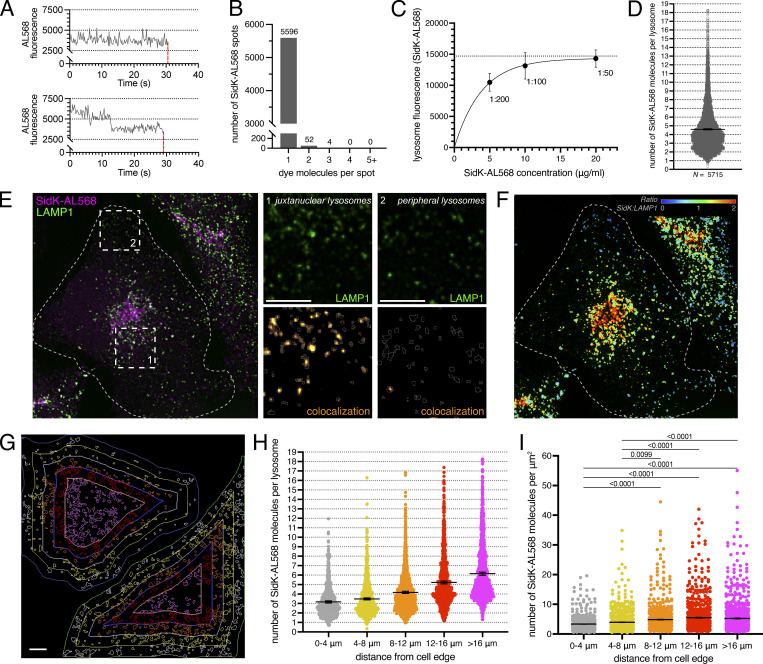
Quantification of the number of V-ATPases per lysosome
To date, estimates of the number of V-ATPase complexes in vesicular compartments have been global approximations based on measurements in pooled whole-cell preparations (de Araujo et al., 2020; Takamori et al., 2006). In an effort to refine estimates and provide topological information, we attempted to quantify the number of SidK-AL568 molecules bound per organelle. This required determination of the fraction of SidK molecules labeled by the Alexa dye and the number of fluorophores attached per SidK-AL568 molecule, followed by comparison of the single molecule fluorescence to the total fluorescence associated with the organelle of interest. We determined that 96.7% of SidK molecules were labeled, and analysis of the photobleaching pattern of monodisperse SidK (Fig. 8 A) indicated that 99% of these molecules had reacted with a single fluorophore (Fig. 8 B). We next determined the concentration of SidK-AL568 needed to saturate all the available binding sites on lysosomes (Fig. 8 C). With these parameters and conditions, the total fluorescence associated with individual lysosomes was converted to a corresponding number of V-ATPases, assuming that all three A subunits of every V-ATPase are accessible for SidK-AL568 binding. In HeLa cells, where individual lysosomes can be delineated more readily, the number of SidK-AL568 molecules associated per lysosome varied (Fig. 8 D), with an average of 4.42 ± 0.03 (mean ± SEM) SidK molecules per lysosome, equivalent to 1.47 V-ATPase complexes per lysosome.
Figure 8.
Estimation of the number of V-ATPases per lysosome. (A) Photobleaching of monodisperse SidK-AL568. Representative intensity-time traces for a single SidK-AL568 labeled with one (top) or two (bottom) Alexa Fluor 568 moieties. Loss of fluorescence after complete photobleaching shown by the dotted red lines. (B) Histogram of the measured number of dye molecules per SidK-AL568 monomer determined analyzing >5,000 single-molecule photobleaching traces. Number of SidK-AL568 molecules in each category is indicated above the bar. (C) HeLa cells stained with SidK-AL568 at varying concentrations. Lysosomal fluorescence was plotted to determine the concentration of SidK-AL568 required to reach maximal lysosome fluorescence, indicative of saturation of binding sites. (D) HeLa cells stained with SidK-AL568 using the near-saturation conditions determined in C (1:50). Total fluorescence per lysosome is compared with the average fluorescence of a single molecule of SidK-AL568; the average number of SidK-AL568 molecules bound per lysosome is indicated by the horizontal line, as is the SE (whiskers). A total of 5,715 lysosomes from two independent experiments were quantified. (E) HeLa cells stained with SidK-AL568 (magenta) using the near-saturating conditions determined in C (1:50), and costained for LAMP1 (green). Side panels show individual LAMP1 (top) and SidK-AL568:LAMP1 colocalization channels (bottom) for juxtanuclear (left) or peripheral (right) lysosomes in the areas denoted by dotted squares. Cell and lysosome outlines indicated by dotted and solid lines, respectively. (F) SidK-AL568:LAMP1 fluorescence ratio (RatioSidK-AL568:LAMP1) corresponding to image in E. The fluorescence ratio was pseudocolored, representing ratio values from 0 to 2. (G) Shell analysis of SidK distribution as a function of distance from the cell outer edge. HeLa cells were stained using SidK-AL568 and costained for LAMP1. Cell outlines were drawn and degraded iteratively inward by 4 µm to create concentric shells (colored differentially) within each cell that were used to subgroup LAMP1+ lysosomes for subsequent analysis. Images in E-G are compressions of confocal stacks representative of ≥10 fields from two or more experiments of each type. (H) For each shell, total fluorescence per lysosome was compared with the average fluorescence of a single molecule of SidK-AL568, and the average number of SidK-AL568 molecules bound per lysosome calculated as a function of distance from the cell outer edge. Data are mean ± SEM of two independent experiments with five or more cells per replicate. n = 791, 1,112, 1,203, 1,275, and 1,334 lysosomes analyzed for shells 0–4, 4–8, 8–12, 12–16, and >16 µm from the cell edge, respectively. (I) For the data in H, the number of SidK-AL568 molecules bound per lysosome was divided by the lysosome surface area (μm2), and the ratio graphed as a function of distance from the cell outer edge. Data are mean ± SEM of two experiments with five or more cells per replicate. Scale bars: 5 µm.
Previous research had shown that lysosomes are heterogeneous within a cell (Bright et al., 1997, 2016; Butor et al., 1995; Cheng et al., 2018). Additionally, lysosomal pH can vary with vesicle positioning, with peripheral lysosomes generally being more alkaline, and lysosomes near the center of the cell more acidic (Johnson et al., 2016; Webb et al., 2021). A simple explanation for this finding could be that peripheral lysosomes have fewer V-ATPases than central lysosomes, although other mechanisms could also cause this effect (see Introduction). To explore this experimentally, lysosomes of HeLa cells were identified using LAMP1 and their V-ATPase density quantified with SidK-AL568. The resultant labeling with SidK-AL568 was more intense in juxtanuclear lysosomes than in more peripheral ones (Fig. 8 E). This differential distribution was quantified and is presented as a ratio of SidK-AL568:LAMP1 fluorescence (Fig. 8 F). To more precisely assess whether the number of V-ATPases per lysosome varies as a function of their subcellular localization, the SidK-AL568 fluorescence of individual LAMP1-positive structures was analyzed relative to their distance from the edge of the cell. Cell outlines were drawn and degraded inward by 4 µm iteratively, to create concentric shells within each cell (Fig. 8 G). Comparison of these subgrouped LAMP1-positive vesicles confirmed the existence of a gradient of SidK-AL568 that correlates with the distance of the lysosomes from the cell center. The most juxtanuclear lysosomes bind approximately twice as many SidK-AL568 molecules as peripheral ones (Fig. 8 H). This change in V-ATPase density corresponded to 2.1 V-ATPase complexes per lysosome at >16 µm from the cell edge, compared with 1.1 lysosomal V-ATPases per lysosome for lysosomes 0–4 µm from the cell edge. Interestingly, ∼18% of the lysosomes closest to the nucleus showed more than the average number of V-ATPase complexes (two to six per lysosome). This change could not be explained merely by increased lysosomal size, as the number of SidK-AL568 per lysosome surface area was nevertheless lower in peripheral than in juxtanuclear lysosomes (Fig. 8 I). Note that the size of lysosomes that are smaller than the limit of resolution of the confocal microscope would have been overestimated. We concluded that the pH heterogeneity of lysosomes within individual cells can be explained, at least in part, by differences in their V-ATPase density.
Discussion
Despite its clear importance in the maintenance of organellar pH and tissue homeostasis, the study of V-ATPases has been hampered by the lack of reagents to accurately localize and quantify these complexes. To address this problem, we developed a detection tool based on SidK, an L. pneumophila protein. SidK was an attractive choice for several reasons. First, it was reported to bind to the V-ATPase A subunit with high specificity (Abbas et al., 2020; Zhao et al., 2017; Xu et al., 2010) and affinity (dissociation constant ∼3.5 nM; Sharma and Wilkens, 2017), which is comparable to the affinity of many immunoglobulins for their cognate antigens. Second, because the primary sequence of the V-ATPase A subunit is highly conserved across Eukarya, a SidK-based reagent should be applicable to a variety of model organisms. Indeed, we found SidK to interact equally with yeast and mammalian cells, while a commercially available V1A antibody was mammalian-specific. Finally, unlike other V-ATPase subunits, the A subunit exists as a single isoform, so that the SidK reagent would be expected to bind V-ATPase wherever present in organelles, cells, and tissues.
We first expressed the V-ATPase–binding region of SidK as a fusion with GFP or mCherry. At modest expression levels, this construct enabled the detection of V-ATPase–rich membranes. However, at higher expression levels, the unbound (cytosolic) probe obscured the binding to organelles. This problem could be mitigated by leaching the cytosolic excess out the cells by permeabilizing the plasmalemma with PLY, revealing the specifically bound constructs. However, when expressing SidK, a process that requires hours, we observed a change in lysosomal positioning that we attributed to its inhibitory effect, as it mimicked the effects of concanamycin A. Accordingly, we measured a ∼30% inhibition of the H+-pumping rate in cells expressing SidK-mCherry. While our findings were not unexpected considering the observations made earlier in vitro and in cells (Abbas et al., 2020; Zhao et al., 2017; Xu et al., 2010; Johnson et al., 2016), they nonetheless underscored a limitation inherent to the use of ectopically expressed SidK. Indeed, inhibition of V-ATPases and the resultant changes in luminal pH can affect not only lysosome localization but also the ability of the V-ATPase to interact with other molecules (Maranda et al., 2001; Hurtado-Lorenzo et al., 2006; Hosokawa et al., 2013), altering intracellular signaling (Balgi et al., 2011; Hu et al., 2016). However, we were unable to detect any effects of SidK expression on lysosomal degradative ability or mTORC1 signaling (Fig. S1). It is also noteworthy that SidK-GFP/mCherry did not properly label acidic portions of the Golgi complex. This lack of labeling could have resulted from obstruction caused by other molecules that associate with V-ATPases in the Golgi, but not with those of the endocytic compartment.
To circumvent the problems associated with SidK expression, we used fluorescently conjugated recombinant SidK as an overlay staining reagent, followed by washing to remove unbound probe. SidK-AL568 localized to punctate and cisternal structures in mammalian cells, with little nonspecific background. The high signal-to-noise ratio, along with specific organellar markers, allowed us to discern V-ATPase–positive organelles clearly, primarily endo/lysosomes and the trans-Golgi. This labeling was in sharp contrast to the staining we obtained using a commercial antibody to the A subunit and the conditions for its use described in the literature (Ramirez et al., 2019), which yielded results similar to those reported by others using other available antibodies (Yajima et al., 2007; McGuire et al., 2019; Michel et al., 2013). SidK did not detect much diffuse (cytosolic) staining, which would be expected if the V1 fully dissociates from VO (Poëa-Guyon et al., 2013; Bodzęta et al., 2017; Jaskolka et al., 2021; McGuire et al., 2019; Benlekbir et al., 2012; Parra and Kane, 1998; Tabke et al., 2014). However, some authors have suggested that the dissociated V1 remains in the immediate vicinity of the membrane-embedded VO (Tabke et al., 2014), in which case the dissociated subunits could not be resolved by optical microscopy.
V-ATPase complexes have been shown to interact with several accessory proteins involved in endocytic sorting and signaling (Merkulova et al., 2015; Maxson and Grinstein, 2014; Marshansky et al., 2014). These interacting proteins include sorting nexins SNX27, SNX10, and SNX11 (Merkulova et al., 2015; Chen et al., 2012; Xu et al., 2020), and the Ragulator complex, which controls mTOR activation during nutrient sensing (Zoncu et al., 2011). Additionally, the V-ATPase is hypothesized to function as a sensor of luminal pH that regulates endocytosis through a pH-dependent interaction with ARF nucleotide-binding site opener (Hosokawa et al., 2013; Maranda et al., 2001; Hurtado-Lorenzo et al., 2006). Recently, some members of the Tre2/Bub2/Cdc16, lysin motif, domain catalytic protein family have been found to interact with V-ATPase domains and modulate V-ATPase activity or stability (Merkulova et al., 2015; Castroflorio et al., 2021). These reports have relied on immunoprecipitation with available V-ATPase antibodies to study interactions. However, use of SidK could provide more sensitive and/or specific measurements of interactions, as well as the ability to visualize novel interactions by high-resolution microscopy (Tan et al., 2021 Preprint). Of interest, the 34.6 kD SidK1-278 fragment we used is considerably smaller than conventional primary antibodies (IgG ∼150 kD), which are often used in combination with an equally large secondary antibody. Our smaller probe is better suited to access V-ATPases, and could therefore be advantageous for immunogold-labeling for electron microscopy, where the penetrance of bulky antibodies can limit detection. The smaller size of SidK-AL568 compared with SidK-GFP/mCherry may also explain why the former is better able to detect V-ATPase in the Golgi complex, although fixation and permeabilization may have also facilitated its access.
The successful labeling of the acidic intracellular compartments emboldened us to use the probe to monitor the dynamics of recruitment of the V-ATPase to a specialized vesicular compartment, the phagosome. The progressive development of phagosomal acidification is well established and was assumed, yet not proven, to be associated with increasing V-ATPase density. We found that while the nascent phagosome is virtually devoid of V-ATPases, the presence of the complex becomes evident in early (Rab5- and PtdIns[3]P-positive) phagosomes, and their number increases further as they attain the phagolysosomal stage. Therefore, the increasing acidity during maturation can be attributed, at least in part, to an increase in V-ATPase density. This increase correlates well with the recent finding that acidification of phagosomes to a pH <6 during the transition between early and late phagosome results in the dissociation of Vps34 class III phosphatidylinositol-3-kinase from these organelles (Naufer et al., 2018). V-ATPase recruitment to the phagosome could be expected to control the cessation of PtdIns(3)P synthesis on these compartments, which is necessary for transition to the late phagosome/phagolysosome stage (Vieira et al., 2001; Naufer et al., 2018).
We also observed that V-ATPases were not homogeneously localized throughout the membrane of the phagosome, and similar observations were made in enlarged and normal lysosomes. The existence of specialized subdomains was recently reported in phagosomes, where it was associated with the formation of contacts with the ER and the extension of tubular structures (Levin-Konigsberg et al., 2019). In the endocytic system, similar microdomains have been postulated to play roles in cargo sorting and receptor recycling through tubulation (Wijdeven et al., 2016; Rocha et al., 2009), and are likely also involved in phagosome resolution. Once internalized material has been degraded by acidic hydrolases, essential phagosomal and lysosomal membrane proteins must be parsed and redistributed within the cell for reutilization. The reformation of the lysosomal compartment by membrane retrieval following content condensation was predicted long ago (Bright et al., 1997, 2016; Mullock et al., 1998), but the fate of specific components has not been well documented. In the case of the V-ATPase, retrieval from maturing phagosomes was demonstrated in Dictyostelium discoideum (Clarke et al., 2010), but to our knowledge has not been studied in mammalian cells. The observed V-ATPase–rich domains may be a precursor to the formation of recycling vesicles or tubules. In this regard, pH gradients have been observed recently in tubules formed by macrophages (Suresh et al., 2020 Preprint; Naufer et al., 2018). Together, these findings suggest that H+-pumping microdomains may form on acidic organelles during the tubulation and fission that accompany resolution.
The high signal-to-noise ratio provided by the SidK-AL568 probe and its defined labeling stoichiometry enabled us to quantify the number of V-ATPases per organelle. Quantification relied on the use of saturating concentrations of the probe, a requirement we fulfilled by analyzing the concentration dependence of SidK-AL568 binding. We calculated the number of V-ATPases per lysosome to range between 1 and 6, averaging ∼1.5. This number is similar to estimates for synaptic vesicles, with one V-ATPase per vesicle (Takamori et al., 2006). The paucity of pumps per organelle likely accounts for the difficulty encountered in their detection and highlights the need for highly specific probes. It is important to emphasize that the calculated numbers of V-ATPases are likely underestimates, for several reasons. First, our estimates rely on the assumption that all three A subunits of every pump are accessible to the probe. Because an increasing number of proteins are appreciated to bind to the V-ATPase, steric hindrance may curtail the number of accessible subunits, which would result in an underestimate of the number of pumps. Additionally, while Alexa dyes are not particularly prone to self-quenching (Panchuk-Voloshina et al., 1999; Szabó et al., 2018), molecular crowding could have resulted in quenched emission (Chen et al., 2010; Ogawa et al., 2009).
Through these experiments, we noted that the distribution of V-ATPases in association with LAMP1-positive structures was not homogeneous throughout the cell: more SidK-AL568 bound to juxtanuclear lysosomes when compared with peripheral lysosomes. The density of SidK-AL568 increased with distance from the cell edge, with the most central lysosomes containing up to six V-ATPases. These data are in line with recent reports showing that juxtanuclear lysosomes are, on average, more acidic than the more peripheral ones (Johnson et al., 2016; Webb et al., 2021).
While bearing in mind the caveats raised above, our estimates can be used to calculate the rate of pumping of individual lysosomal V-ATPases in situ. Experiments like those in Fig. 2 H indicated that concanamycin A alkalinized the lysosomes at an initial rate of 0.198 ± 0.1 pH · min−1. Because at steady-state the activity of the pumps must have matched the H+ leak unmasked by concanamycin A, the measured rate can be used to estimate pump activity. Considering the buffering power of the lysosomes, estimated to be 23.5 mM · pH−1 by pulsing with ammonium, we calculate a H+ flux per lysosome volume of 9.5 × 10−2 mmol · min−1 · liter−1. Assuming an approximate lysosomal volume of 2 × 10−17 liter (de Araujo et al., 2020; Yordanov et al., 2019), this measurement is equivalent to a flux of 953 H+ · s−1 per lysosome. As 10 H+ are pumped by the V-ATPase for every 3 ATP hydrolyzed (Zhao et al., 2017; Abbas et al., 2020), the flux corresponds to ∼191 molecules of ATP hydrolyzed per lysosome per second. Considering that there are one to two pumps on average per lysosome, the calculated flux is within the same order as the V-ATPase activity measured previously in cell-free systems in mammals (29.2 ATP per V-ATPase · s−1; Abbas et al., 2020) and yeast (from 3.7 to 300 ATP per V-ATPase · s−1; Vasanthakumar et al., 2019; Uchida et al., 1985; Sharma and Wilkens, 2017; Kawasaki-Nishi et al., 2001).
In summary, we have introduced a new, powerful probe to visualize and quantify V-ATPases in eukaryotic cells. This probe enabled us to assess the subcellular distribution of V-ATPases and, in combination with ratiometric pH determinations, estimate the rate of flux for individual lysosome-associated V-ATPases in intact cells. These results reveal heterogeneity in the lysosomal compartment that is a result of vesicle location–dependent differences in V-ATPase density. We expect this probe to be a useful label for super-resolution imaging and electron microscopy, and anticipate that SidK-AL568 and similar probes will contribute not only to studies of V-ATPases in cells but also to our understanding of endocytic and secretory processes more generally.
Materials and methods
Cell culture
HeLa and RAW264.7 cells were obtained from and authenticated by the American Type Culture Collection. Both cell lines tested negative for mycoplasma contamination by DAPI staining. HeLa cells were grown in DMEM containing L-glutamine and 10% heat-inactivated FCS (MultiCell; Wisent) at 37°C under 5% CO2. RAW264.7 cells were grown in RPMI-1640 medium containing L-glutamine and 10% heat-inactivated FCS at 37°C under 5% CO2.
To obtain nonpolarized human monocyte-derived macrophages, peripheral blood mononuclear cells were isolated from the blood of healthy donors by density-gradient separation with Lympholyte-H (Cedarlane). Human monocytes were then separated by adherence and incubated in RPMI-1640 containing L-glutamine, 10% heat-inactivated FCS, 100 U · ml−1 penicillin, 100 µg · ml−1 streptomycin, 250 ng · ml−1 amphotericin B, and 25 ng · ml−1 hM-CSF (PeproTech) for 5–7 d, at 37°C under 5% CO2, before experimentation.
Reagents
Mammalian expression vectors were obtained from the following sources: pmCherry-N1 (Clontech) and pEGFP-N1 (Clontech), pSAB35 (Addgene; plasmid no. 175787), sec61b-GFP (Addgene; plasmid no. 121159), PM-GFP (Teruel et al., 1999), Rab5A-GFP (Roberts et al., 2000), PX-GFP (Kanai et al., 2001), Rab7-GFP (Bucci et al., 2000), LAMP1-GFP (Martinez et al., 2000), ORP1L-GFP (Rocha et al., 2009), and Arl8b-GFP (Johnson et al., 2016). VOa1-, VOa2-, and VOa3-GFP were the kind gift from Dr. Shuzo Sugita (University Health Network, Toronto, Canada).
Primary antibodies were purchased from the following vendors: human LAMP1 (Developmental Studies Hybridoma Bank; catalog no. H4A3-s), VapB (Sigma-Aldrich; catalog no. HPA013144), TGN46 (Abcam; catalog no. ab50595), GM130 (BD Transduction Laboratories; catalog no. 610822), ATP6V1A (Abnova; catalog no. H00000523-M02), pS6K-Thr389 (Cell Signaling; catalog no. 9205), S6K (Cell Signaling; catalog no. 2708), pULK1-Ser757 (Cell Signaling; catalog no. 6888), α-tubulin (Sigma-Aldrich; catalog no. T5168), and GAPDH (EMD Millipore; catalog no. MAB374). Secondary antibodies conjugated with Alexa Fluor 405, 488, 555, 647, or HRP were purchased from Jackson ImmunoResearch.
Fluorescently conjugated 10 kD dextran, Alexa Fluor 405 and Alexa Fluor 568 N-HydroxySuccinimide (NHS) esters, and fluorescently conjugated phalloidin and streptavidin were purchased from Invitrogen. Concanamycin A, nigericin, DAPI, propidium iodide, and cresyl violet were from Sigma-Aldrich. Sheep RBCs (SRBCs; 10% suspension) were from MP Biomedicals. Anti-SRBC antibodies were from Cedarlane Laboratories. PFA (16% wt/vol) was from Electron Microscopy Sciences.
SidK-GFP/mCherry plasmid construction
SidK1-278 was amplified by PCR using plasmid pSAB35 (Abbas et al., 2020) as a template, with the following forward and reverse primers, respectively: 5′-GAGGAGGAATTCATGTCTTTTATCAAGGTAGGTATAAAAATG-3′ and 5′-GAGGAGGGATCCCCTTTGCTTAAAGCATTTAATTTTTCG-3′. The PCR product was digested with EcoRI and BamHI (New England Biolabs) and ligated into pmCherry-N1 and pEGFP-N1 plasmids that had been digested with the same restriction enzymes.
For illustrative purposes, a model of yeast V-ATPase with SidK-GFP bound (Fig. 1 B) was generated from PDB accession nos. 5VOX, 1GFL, and 6O7T, using UCSF ChimeraX (Goddard et al., 2018).
Immunoblotting
HeLa cells or RAW264.7 cells were grown in 6-well plates at a concentration of 2 × 105 cells · ml−1 or 8 × 105 cells · ml−1, respectively. In some cases, wells were transfected with the indicated constructs for 16 h. Wells were lysed in 150 µl RIPA buffer containing protease and phosphatase inhibitors. Protein concentrations quantified by BCA protein assay (Pierce). Lysates were diluted in Laemmli buffer (Bio-Rad) and analyzed by 10% SDS-PAGE. Gels were transferred to a polyvinylidene difluoride membrane and blocked in TBS containing 5% BSA and 0.05% Tween-20 for 1 h at room temperature. Primary antibody staining was done in blocking buffer for 1 h at room temperature. Primary antibody dilutions were as follows: ATP6V1A (1:1,000), pS6K (1:1,000), S6K (1:1,000), pULK1 (1:1,000), tubulin (1:5,000; loading control), and GAPDH (1:5,000; loading control). After washing the membrane in TBS containing 0.05% Tween-20, samples were incubated 1 h at room temperature with an HRP-conjugated secondary antibody at 1:3,000 dilution. Blots were developed using the ECL Prime Western Blot detection reagent and visualized with a Bio-Rad ChemiDoc MP Imaging System and Image Lab software 5.2.1.
Purification of SidK1-278 protein
To purify SidK1-278-3×FLAG, Escherichia coli strain BL21 was transformed with pSAB35 (Abbas et al., 2020) and grown at 30°C with shaking in 1 liter Luria Broth medium (BioShop) supplemented with 50 mg · liter−1 kanamycin. At an OD600 of 0.6–0.8, protein expression was induced with 1 mM IPTG, and cells were grown overnight at 16°C. All subsequent steps were performed at 4°C. Cells were harvested by centrifugation at 5,250 × g, resuspended in 25 ml HisTrap Buffer (50 mM Tris-HCl, pH 7.4, 25 mM imidazole, and 300 mM NaCl), and lysed by sonication. The cell lysate was centrifuged at 38,000 × g, and the supernatant was loaded onto a 5-ml HisTrap Ni-NTA column (GE Healthcare). The column was washed with HisTrap Buffer and protein eluted with a linear gradient of imidazole from 25 to 300 mM in HisTrap Buffer over 10 column volumes.
Fractions containing 6×his-SidK1-278-3×FLAG were pooled, mixed with Tobacco Etch Virus protease, and dialyzed against 2 liter Dialysis Buffer (50 mM Tris-HCl, pH 7.4, and 300 mM NaCl) with 1 mM DTT overnight. Cleaved protein was dialyzed against 2 × 1 liter Dialysis Buffer to remove imidazole and DTT and passed through a 5-ml HisTrap column. The column was washed with HisTrap Buffer, and the flowthrough and wash were collected, pooled, and concentrated in a centrifugal concentrating device (EMD Millipore). To remove aggregated protein, SidK1-278-3×FLAG was further purified with a Superdex 200 10/300 Increase gel filtration column (GE Healthcare) equilibrated with 50 mM Tris-HCl, pH 7.4, and 150 mM NaCl. Fractions containing protein were pooled, concentrated, flash-frozen in liquid N2, and stored at −80°C.
Purification of S. cerevisiae V-ATPase using SidK
S. cerevisiae strain BJ2168 was grown in 11 liter yeast extract peptone dextrose medium (YPD; BioShop) in a Microferm fermenter (New Brunswick Scientific) at 30°C, with aeration of 34 cubic feet per hour, and stirring at 300 rpm. Yeast were harvested after 18 h (OD660 = 4.5) by centrifugation at 4,000 × g for 15 min at 4°C. All subsequent steps were performed at 4°C. Cell walls were broken by bead beating in lysis buffer (phosphate-buffered saline, pH 7.4, 8% [wt/vol] sucrose, 2% [wt/vol] sorbitol, 2% [wt/vol] glucose, 5 mM ɛ-amino-n-caproic acid, 5 mM benzamidine, 5 mM EDTA, and 0.001% [wt/vol] PMSF). Cellular debris was removed by centrifugation at 3,000 × g for 10 min, and cell membranes were collected by ultracentrifugation at 152,957 × g for 40 min. The membrane pellet was resuspended in 36 ml lysis buffer, divided into four aliquots, flash-frozen in liquid N2, and stored at −80°C. Two membrane pellets (corresponding to half a fermenter growth) were thawed and solubilized with addition of n-dodecyl-β-D-maltopyranoside (Anatrace) to 1% (wt/vol) final concentration, and n-dodecyl-β-D-maltopyranoside–solubilized S. cerevisiae V-ATPase was isolated with M2 Affinity agarose gel (Sigma-Aldrich) preloaded with SidK1-278-3×FLAG as described previously (Abbas et al., 2020). Protein purity was confirmed by SDS-PAGE using 4–20% Mini-PROTEAN TGX protein Gels (Bio-Rad).
Preparation of yeast and rat brain membranes
Yeast membranes were prepared from the strain SABY31 as described previously (Benlekbir et al., 2012) and resuspended in lysis buffer (phosphate-buffered saline, pH 7.4, 8% [wt/vol] sucrose, 2% [wt/vol] sorbitol, 2% [wt/vol] glucose, 5 mM ɛ-amino-n-caproic acid, 5 mM benzamidine, 5 mM EDTA, and 3.5 mM PMSF). Rat brain membranes were prepared as described previously (Abbas et al., 2020) and resuspended in solubilization buffer (50 mM Tris-HCl, pH 7, 300 mM NaCl, 10% [wt/vol] sucrose, 10% [vol/vol] glycerol, 5 mM ɛ-amino-n-caproic acid, 5 mM benzamidine, 5 mM EDTA, and 0.2 mM PMSF).
Transient DNA transfection
HeLa cells were plated on 18-mm glass coverslips at a concentration of ∼5 × 104 cells · ml−1, 16–24 h before transfection. FuGENE 6 (Promega) transfection reagent was used according to the manufacturer’s instructions to transfect HeLa cells at a 3:1 ratio (using 1.5 µl FuGENE 6 and 0.5 µg DNA per well). RAW264.7 cells were plated on 18-mm glass coverslips at ∼2 × 105 cells · ml−1, 16–24 h before transfection. FuGENE HD (Promega) transfection reagent was used to transfect RAW264.7 cells at a 3.5:1 ratio (using 1.75 µl FuGENE HD and 0.5 µg DNA per well). In all cases, monolayers were used for experiments 16 h after transfection.
Ratiometric fluorescence microscopy for the measurement of lysosomal pH, V-ATPase activity, and buffering power
HeLa cells were plated on 18-mm glass coverslips and incubated overnight with 250 µg · ml−1 FITC-conjugated 10 kD dextran, which was then chased for 1 h in complete medium before imaging to visualize lysosomes. Coverslips were then mounted in a Chamlide magnetic chamber and incubated in HBSS medium for fluorescence-based pH determinations.
Steady-state lysosomal pH was determined by exciting FITC-dextran labeled lysosomes sequentially at 481 ± 15 nm and 436 ± 20 nm, collecting emitted light at 520 ± 35 nm. The fluorescence intensity of FITC when excited at ∼490 nm is highly pH-dependent and was used to determine the pH of lysosomes. The fluorescence when excited at ∼440 nm is much less pH-dependent and was used to correct for potential photobleaching or focal changes during image acquisition. The 490-nm/440-nm fluorescence ratio of FITC is used to determine pH values. Multiple fields of cells were imaged, and the data were processed with Volocity.
The 490-nm/440-nm fluorescence ratios were converted to pH by sequentially incubating the cells for 5 min in isotonic K+ solutions (143 mM KCl, 5 mM glucose, 1 mM MgCl2, and 1 mM CaCl2) of different pH (pH 4.5, buffered with 20 mM acetic acid; pH 5.5, 6.5, and 7.5 solutions buffered with 20 mM MES), containing 10 µM nigericin and 5 µM monensin. Background-subtracted 490-nm/440-nm fluorescence ratios were then plotted against pH, and the data fitted by least squares were used to interpolate the lysosomal pH.
For V-ATPase activity determinations, HeLa cells were acutely treated with 500 nM concanamycin A, and images were acquired at 30-s intervals for 10 min. The time dependence of the resulting pH changes was then plotted in GraphPad Prism, and the initial rates of alkalinization (per cell) were determined within 2 min of addition of concanamycin A.
For the determination of buffering power, HeLa cells containing FITC-dextran–loaded lysosomes were challenged with 0.5 mM NH4Cl, and the resulting change in fluorescence intensity (490 nm/440 nm) was measured immediately. The corresponding pH change, calculated as above, was used to calculate the concentration of ammonium (NH4+) formed inside the lysosome (lys), using the Henderson–Hasselbalch equation, and the buffering capacity estimated as Δ[NH4+]lys/ΔpHlys.
Lysosome labeling, cresyl violet staining, and dissipation of organellar pH
For identification of lysosomes, HeLa cells were incubated overnight with 100 µg · ml−1 fluorescently conjugated 10 kD dextran at the time of transient transfection, 16–24 h before experiments. The next day, monolayers were washed 3× with PBS and placed in HBSS medium at 37°C for 1 h to chase the dextran to lysosomes. Monolayers were then imaged live by confocal microscopy.
For cresyl violet staining, HeLa cells, seeded on 18-mm glass coverslips at a concentration of 1 × 105 cells · ml−1, were labeled with fluorescent dextran as described above. Monolayers were then incubated at 37°C with 1 µM cresyl violet in HBSS (MultiCell; Wisent) for 5 min, washed three times, and imaged live by confocal microscopy. In some cases, organellar pH was dissipated before cresyl violet staining. To this end, the cells were treated for 30 min with 250 nM concanamycin A and 10 mM NH4Cl in HBSS. After this treatment, monolayers were stained with 1 µM cresyl violet and imaged live, as above.
DQ-BSA assay
HeLa or RAW264.7 cells were seeded on 18-mm glass coverslips at a concentration of 1 × 105 cells · ml−1 and 5 × 105 cells · ml−1, respectively, and transiently transfected with GFP or SidK-GFP as described above. 16–24 h later, cells were loaded 5 h with 10 µg · ml−1 DQ-BSA Red (Molecular Probes) and 100 µg · ml−1 fluorescently conjugated 10 kD dextran. After this incubation, monolayers were washed three times, fixed in 3% PFA, and analyzed by confocal microscopy. The fluorescence intensity of DQ-BSA per lysosome was normalized to 10 kD dextran fluorescence, to control for differences in lysosomal content.
Fluorescent labeling of SidK1-278
Prior to labeling, purified SidK1-278-3×FLAG (see above; referred to in the text as SidK) was buffer-exchanged into PBS using a centrifugal concentrator (EMD Millipore). Following this, SidK was directly labeled by conjugation with Alexa Fluor 568 NHS ester (Invitrogen). A 10:1 dye:SidK molar ratio was prepared in 0.05 M borate buffer, vortexed, and incubated shaking at 500 rpm for 1 h at room temperature. Labeled SidK (referred to as SidK-AL568) was then dialyzed in 4× 1 liter PBS to remove unincorporated dye. One volume of 100% glycerol was added to the SidK-AL568 for stability, and the SidK-AL568 stored at 4°C.
The fraction of SidK labeled with Alexa Fluor 568 was determined using absorbance spectroscopy on a Nanodrop 2000 instrument (Thermo Fisher Scientific). SidK and SidK-AL568 were diluted to 0.1 mg · ml−1 in PBS. The A280 of SidK and its molecular weight (34,646 g · mol−1) were used to calculate its extinction coefficient (ε SidK = 41,690 cm−1 · M−1). Then the A280 and A577 (λmax of Alexa Fluor 568) of SidK-AL568 were measured, and the degree of labeling (moles of dye per mole of protein) for SidK-AL568 was calculated, using the following two equations, along with the ε dye value for Alexa Fluor 568 (91,300 cm−1 · M−1) and the correction factor for the A280 contribution of Alexa Fluor 568 (CF280 = 0.46):
| (1) |
and
| (2) |
The number of dye particles per SidK-AL568 molecule was determined by single-molecule photobleaching imaging (Liesche et al., 2015). Briefly, a 106 dilution of SidK-AL568 was prepared and added to clean coverslips, creating a monodispersed sample. Monodispersed SidK-AL568 was then imaged continuously by confocal microscopy at ∼5 frames · s−1, until spot disappearance occurred through photobleaching. Time series images were analyzed using Volocity v6.3 software (Quorum Technologies), and SidK-AL568 molecule fluorescence over time was plotted to generate traces where the number of steps required for SidK-AL568 spot disappearance was counted. Each step corresponded to a single Alexa Fluor 568 dye molecule conjugated to SidK. The average fluorescence value for bona fide single-bleach-step SidK-AL568 was used to calculate the percentage of SidK-AL568 in the monodispersed population with 1, 2, 3, 4, or 5+ dye molecules per SidK.
Culturing of yeast and immunofluorescence of yeast spheroplasts
Wild-type (MATα ura3–52 leu2–3,112 his4–519 ade6) and vma1Δ (MATα ura3–52 leu2–3,112 his4–519 ade6 vma1Δ::LEU2) S. cerevisiae strains were kindly gifted by Dr. Morris Manolson (University of Toronto, Toronto, Canada). Yeast cultures were routinely grown at 30°C, with shaking, in YPD medium (BD Biosciences).
For spheroplast preparation, overnight cultures were diluted 1:100 in YPD broth and grown at 30°C for 2 h. 1 ml of yeast culture was then fixed with 4% PFA for 30 min while shaking at room temperature. After washing cells 2× with 0.1 M KHPO4, pH 6.5, yeast were washed 1× in K-Sorb (1.2 M sorbitol in 0.1 M KPHO4) and resuspended in 0.5 ml K-Sorb containing 5 µl β-mercaptoethanol and 10 mg · ml−1 zymolyase 20T (MP Biomedicals). Cells were shaken at room temperature for 30 min and gently washed 2× with K-Sorb. After final resuspension in 0.5 ml K-Sorb, 20 µl of spheroplast suspension was spread on a concanavalin A–coated slide and allowed to adhere for 15 min at room temperature. Following this, spheroplasts were permeabilized/blocked for 30 min in PBS 0.1% Tween-20 containing 5% BSA and 5% skim milk. Samples were then incubated in the same buffer containing 1:200 SidK-AL568 and 1 µg · ml−1 DAPI for 30 min at room temperature. After staining, slides were washed 3× with PBS and visualized by confocal microscopy.
Immunofluorescence of mammalian cells
After fixing in 3% PFA for 10 min at room temperature, cells were washed in PBS and permeabilized/blocked in PBS containing 5% BSA and 0.1% Triton X-100 for 30 min at room temperature. Samples were then incubated with primary staining reagents for 30 min at room temperature. Primary antibody dilutions were as follows: LAMP1 (1:50), VapB (1:100), TGN46 (1:100), GM130 (1:100), and ATP6V1A (1:100). For mitochondrial staining, fluorescently conjugated streptavidin was used at 1:100 dilution. SidK-AL568 was used at 1:100, 1:50, or 1:25 dilution, as indicated in the text. Where indicated, a fivefold excess of unlabeled SidK1-278 was used in the blocking/permeabilization step to block SidK-AL568 binding sites before the addition of SidK-AL568. After primary staining, monolayers were washed 3× with PBS, and samples were incubated 30 min at room temperature with Alexa Fluor–conjugated secondary antibodies at a 1:1,000 dilution. Where indicated, 1:1,000 fluorescent phalloidin or 1 µg · ml−1 DAPI was added together with the secondary antibodies. Samples were washed 3× with PBS and viewed by confocal microscopy in PBS.
Phagosome and sucrosome analyses
For phagocytosis assays, RAW264.7 cells were plated on 18-mm glass coverslips at a concentration of 2 × 105 cells · ml−1 and grown for 16–24 h. Cells were transfected as described above with the constructs indicated in the text. The day of experiments, SRBCs were prepared for use in phagocytosis. Briefly, 100 µl of SRBC suspension was washed with 3× with PBS and labeled with Alexa Fluor 405 NHS ester for 20 min, shaking at room temperature. SRBC were then opsonized with 2 µl of rabbit anti-SRBC IgG at 37°C for 1 h. Prepared SRBCs were washed 3× with PBS and resuspended to a final volume of 1 ml in PBS. After 1:10 dilution in PBS, 25 µl of this suspension was added to the RAW264.7 cells. Alternatively, FITC-labeled zymosan was diluted to 10 mg · ml−1 in PBS and opsonized by incubation with human IgG (final IgG concentration, 5 mg · ml−1) for 30 min at room temperature. Prepared zymosan was then washed 3× with PBS and resuspended to a final volume of 20 µl in PBS. 1 µl of this suspension was added to RAW264.7 seeded onto coverslips. In all cases, phagocytosis was synchronized by sedimenting particles onto the cells using centrifugation at 300 × g for 1 min. After phagocytosis, monolayers were fixed in 3% PFA for 10 min at room temperature and stored in PBS until used. FITC-zymosan phagocytosis, which was used to determine the rate of acidification of the nascent phagosome, was imaged immediately after sedimentation of the particles.
To generate sucrosomes, transfected RAW264.7 cells were incubated for 16–24 h in RPMI-1640 medium containing L-glutamine, 10% heat-inactivated FCS, and 30 mM sucrose at 37°C under 5% CO2. The next day, cells were washed 3× with PBS, placed in RPMI-1640 medium containing L-glutamine and 10% heat-inactivated FCS, and used for experiments as described in the text. After experiments, monolayers were fixed in 3% PFA for 10 min at room temperature and stored in PBS.
Quantitation of the number of SidK-AL568 per lysosome
The average fluorescence value of monodispersed SidK-AL568 molecules was used to estimate the number of SidK-AL568 monomers per lysosome in HeLa cells that had been costained with SidK-AL568 and LAMP1. 1:25, 1:50, and 1:100 SidK-AL568 dilutions were assessed to determine the conditions required for saturation of SidK-AL568 staining, needed to estimate the maximum number of SidK-AL568 molecules bound per LAMP1+ lysosome.
Microscopy
Confocal images were acquired using a spinning disk system (Quorum Technologies Inc.). The instrument consists of a microscope (Axiovert 200M; Zeiss), scanning unit (CSU10; Yokogawa Electric Corp.), electron-multiplied charge-coupled device camera (C9100-13; Hamamatsu Photonics), five-line (405-, 443-, 491-, 561-, and 655-nm) laser module (Spectral Applied Research), and filter wheel (MAC5000; Ludl) and is operated by Volocity v6.3. Images were acquired using a 63×/1.4 NA oil objective (Zeiss), with an additional 1.5× magnifying lens and the appropriate emission filter. For live experiments, cells were maintained at 37°C using an environmental chamber (Live Cell Instruments).
Ratiometric fluorescence pH measurements were acquired on an epifluorescence microscope (Axiovert 200M; Zeiss) running on Volocity v6.3, equipped with a camera (Flash 4.0v2; Hamamatsu), excitation lamp (X-Cite 120; EXFO Life Sciences Group), a 63×/1.4 NA oil objective (Zeiss), the appropriate dichroic mirror (CFP/YFP; Chroma Technology), and filter wheels containing the necessary filters for FITC ratiometric fluorescence determinations (excitation at 481 ± 15 nm or 436 ± 20 nm, emission 520 ± 35 nm). Cells were imaged in in PBS (after fixation) or in culture medium (live cells), as indicated in the text. Experiments were performed at room temperature (fixed cells) or while maintaining the temperature at 37°C (live cells) with an environmental controller (Medical Systems Corp.).
Image analysis
Image processing and analyses were performed using Volocity v6.3. Image deconvolution was done on acquired Z-stacks within the Volocity Restoration module, using the iterative restoration function. Calculated fluorochrome point spread functions were used to deconvolve individual channels for five to eight iterations, until a confidence limit of >90% was achieved. For colocalization analyses, the Volocity Colocalization module was used to calculate the positive product of the differences of the mean channels (Li et al., 2004), which was then overlaid on merged images for visualization. Alternatively, for some colocalization analyses, Mander’s overlap coefficients were calculated in Volocity, which describe the percent of various organelle markers that colocalize with SidK-AL568 (referred to as M). Costes thresholding, integrated in the Volocity software, was used to automatically identify colocalization threshold values. The Costes algorithm for estimating thresholds is a robust, reproducible method that speeds processing and eliminates user bias (Costes et al., 2004). Briefly, it measures correlation coefficients for all pixels, starting at the highest intensities. This is repeated down a regression line until the correlation coefficient is ≤0. The channel intensity values at this point become background threshold values, and only pixels above the background intensity values are considered in colocalization analyses. Mander’s coefficients are then calculated as the fraction of total fluorescence in the region of interest that occurs in these pixels. The Costes method provides a quick and effective method for distinguishing labeled structures from background, thus supporting accurate measurement of Mander’s coefficients.
The Volocity Ratio function was used for SidK-AL568 ratio calculations. This divides background-subtracted intensities of SidK-AL568 and lysosomal, sucrosomal, or phagosomal markers (as indicated in the text) to calculate the ratio of SidK-AL568 fluorescence to that of the chosen organellar marker. The Ratio function generates a rainbow look up table ratio channel that is applied as an overlay on merged images, with a scale representing ratio values from 0 to 2.
General methodology and statistics
Data calculations and normalizations were done using Microsoft Excel 2011 (Microsoft Corp.) or GraphPad Prism v9 software (GraphPad Software, Inc.). Because experiments were, for the most part, in vitro imaging determinations of individual cells, samples were assigned to groups according to specific experimental treatments (control versus experimental group). The number of individual experiments and the number of determinations per experiment were selected to attain an estimate of the variance compatible with the statistical tests used, primarily Student’s t test. Each type of experiment was performed a minimum of 3 separate times (biological replicates) with a minimum of 10 individual event determinations (equivalent, but not identical to technical replicates). Data were tested for normality using the D’Agostino–Pearson test, and appropriate testing was applied. Throughout the text and figures, data are reported as mean ± SEM. No data were excluded as outliers. All statistics were calculated using GraphPad Prism v9.
Online supplemental material
Fig. S1 shows the effect of SidK expression on DQ-BSA dequenching (Fig. S1, A and B) or mTORC1 signaling (Fig. S1 C) in HeLa cells transiently transfected with control or SidK plasmids. Fig. S2 shows a comparison of the specificity of SidK-AL568 (Fig. S2 A) versus immunostaining with α-V1A antibody, α-ATP6V1A (Fig. S2 B). Images of the purple channel from Fig. 4, C and H, respectively, are shown before and after thresholding by the Costes method (Costes et al., 2004). Fig. S2, C and D, shows immunoblotting of membrane preparations and cell lysates using the α-ATP6V1A antibody.
Supplementary Material
contains original blots for Fig. S1.
contains original blots for Fig. S2.
Acknowledgments
Y.M. Abbas was supported by a Canadian Institutes of Health Research Postdoctoral Fellowship. J.L. Rubinstein was supported by the Canada Research Chairs program and Canadian Institutes of Health Research Project Grant PJT166152. S. Grinstein was supported by Canadian Institutes of Health Research grant FDN-143202.
The authors declare no competing financial interests.
Author contributions: Conceptualization: M.E. Maxson, Y.M. Abbas, J.L. Rubinstein, and S. Grinstein; investigation: M.E. Maxson, Y.M. Abbas, J.Z. Wu, J.D. Plumb, and S. Grinstein; formal analysis: M.E. Maxson and J.Z. Wu; resources: Y.M. Abbas, J.L. Rubinstein, and S. Grinstein; writing–original draft: M.E. Maxson, Y.M. Abbas, J.L. Rubinstein, and S. Grinstein; writing–review and editing: M.E. Maxson, Y.M. Abbas, J.L. Rubinstein, and S. Grinstein; and supervision: J.L. Rubinstein and S. Grinstein.
Data availability
Experimental datasets that support the findings of this study are available from the corresponding authors upon reasonable request.
References
- Abbas, Y.M., Wu D., Bueler S.A., Robinson C.V., and Rubinstein J.L.. 2020. Structure of V-ATPase from the mammalian brain. Science. 367:1240–1246. 10.1126/science.aaz2924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alzamora, R., Thali R.F., Gong F., Smolak C., Li H., Baty C.J., Bertrand C.A., Auchli Y., Brunisholz R.A., Neumann D., et al. 2010. PKA regulates vacuolar H+-ATPase localization and activity via direct phosphorylation of the a subunit in kidney cells. J. Biol. Chem. 285:24676–24685. 10.1074/jbc.M110.106278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asare, R., and Abu Kwaik Y.. 2007. Early trafficking and intracellular replication of Legionella longbeachaea within an ER-derived late endosome-like phagosome. Cell. Microbiol. 9:1571–1587. 10.1111/j.1462-5822.2007.00894.x [DOI] [PubMed] [Google Scholar]
- Balgi, A.D., Diering G.H., Donohue E., Lam K.K.Y., Fonseca B.D., Zimmerman C., Numata M., and Roberge M.. 2011. Regulation of mTORC1 signaling by pH. PLoS One. 6:e21549. 10.1371/journal.pone.0021549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee, S., Clapp K., Tarsio M., and Kane P.M.. 2019. Interaction of the late endo-lysosomal lipid PI(3,5)P2 with the Vph1 isoform of yeast V-ATPase increases its activity and cellular stress tolerance. J. Biol. Chem. 294:9161–9171. 10.1074/jbc.RA119.008552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benlekbir, S., Bueler S.A., and Rubinstein J.L.. 2012. Structure of the vacuolar-type ATPase from Saccharomyces cerevisiae at 11-Å resolution. Nat. Struct. Mol. Biol. 19:1356–1362. 10.1038/nsmb.2422 [DOI] [PubMed] [Google Scholar]
- Bodzęta, A., Kahms M., and Klingauf J.. 2017. The Presynaptic v-ATPase Reversibly Disassembles and Thereby Modulates Exocytosis but Is Not Part of the Fusion Machinery. Cell Rep. 20:1348–1359. 10.1016/j.celrep.2017.07.040 [DOI] [PubMed] [Google Scholar]
- Bright, N.A., Reaves B.J., Mullock B.M., and Luzio J.P.. 1997. Dense core lysosomes can fuse with late endosomes and are re-formed from the resultant hybrid organelles. J. Cell Sci. 110:2027–2040. 10.1242/jcs.110.17.2027 [DOI] [PubMed] [Google Scholar]
- Bright, N.A., Davis L.J., and Luzio J.P.. 2016. Endolysosomes Are the Principal Intracellular Sites of Acid Hydrolase Activity. Curr. Biol. 26:2233–2245. 10.1016/j.cub.2016.06.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockman, S.A., and Murphy R.F.. 1993. Endosomal and Lysosomal Hydrolases. In Biological Barriers to Protein Delivery. Audus K.L., and Raub T.J., editors. Springer US, Boston, MA. 51–70. 10.1007/978-1-4615-2898-2_3 [DOI] [Google Scholar]
- Bucci, C., Thomsen P., Nicoziani P., McCarthy J., and van Deurs B.. 2000. Rab7: a key to lysosome biogenesis. Mol. Biol. Cell. 11:467–480. 10.1091/mbc.11.2.467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butor, C., Griffiths G., Aronson N.N. Jr., and Varki A.. 1995. Co-localization of hydrolytic enzymes with widely disparate pH optima: implications for the regulation of lysosomal pH. J. Cell Sci. 108:2213–2219. 10.1242/jcs.108.6.2213 [DOI] [PubMed] [Google Scholar]
- Capecci, J., and Forgac M.. 2013. The function of vacuolar ATPase (V-ATPase) a subunit isoforms in invasiveness of MCF10a and MCF10CA1a human breast cancer cells. J. Biol. Chem. 288:32731–32741. 10.1074/jbc.M113.503771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casey, J.R., Grinstein S., and Orlowski J.. 2010. Sensors and regulators of intracellular pH. Nat. Rev. Mol. Cell Biol. 11:50–61. 10.1038/nrm2820 [DOI] [PubMed] [Google Scholar]
- Castroflorio, E., den Hoed J., Svistunova D., Finelli M.J., Cebrian-Serrano A., Corrochano S., Bassett A.R., Davies B., and Oliver P.L.. 2021. The Ncoa7 locus regulates V-ATPase formation and function, neurodevelopment and behaviour. Cell. Mol. Life Sci. 78:3503–3524. 10.1007/s00018-020-03721-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, H., Ahsan S.S., Santiago-Berrios M.B., Abruña H.D., and Webb W.W.. 2010. Mechanisms of quenching of Alexa fluorophores by natural amino acids. J. Am. Chem. Soc. 132:7244–7245. 10.1021/ja100500k [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Y., Wu B., Xu L., Li H., Xia J., Yin W., Li Z., Shi D., Li S., Lin S., et al. 2012. A SNX10/V-ATPase pathway regulates ciliogenesis in vitro and in vivo. Cell Res. 22:333–345. 10.1038/cr.2011.134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, X.-T., Xie Y.-X., Zhou B., Huang N., Farfel-Becker T., and Sheng Z.-H.. 2018. Characterization of LAMP1-labeled nondegradative lysosomal and endocytic compartments in neurons. J. Cell Biol. 217:3127–3139. 10.1083/jcb.201711083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung, C., Mader C.C., Schmitz J.C., Atladottir J., Fitchev P., Cornwell M.L., Koleske A.J., Crawford S.E., and Gorelick F.. 2011. The vacuolar-ATPase modulates matrix metalloproteinase isoforms in human pancreatic cancer. Lab. Invest. 91:732–743. 10.1038/labinvest.2011.8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke, M., Maddera L., Engel U., and Gerisch G.. 2010. Retrieval of the vacuolar H-ATPase from phagosomes revealed by live cell imaging. PLoS One. 5:e8585. 10.1371/journal.pone.0008585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohn, Z.A., and Ehrenreich B.A.. 1969. The uptake, storage, and intracellular hydrolysis of carbohydrates by macrophages. J. Exp. Med. 129:201–225. 10.1084/jem.129.1.201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costes, S.V., Daelemans D., Cho E.H., Dobbin Z., Pavlakis G., and Lockett S.. 2004. Automatic and quantitative measurement of protein-protein colocalization in live cells. Biophys. J. 86:3993–4003. 10.1529/biophysj.103.038422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Araujo, M.E.G., Liebscher G., Hess M.W., and Huber L.A.. 2020. Lysosomal size matters. Traffic. 21:60–75. 10.1111/tra.12714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton, A.F., Merkulova M., and Brown D.. 2021. The H+-ATPase (V-ATPase): from proton pump to signaling complex in health and disease. Am. J. Physiol. Cell Physiol. 320:C392–C414. 10.1152/ajpcell.00442.2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferris, A.L., Brown J.C., Park R.D., and Storrie B.. 1987. Chinese hamster ovary cell lysosomes rapidly exchange contents. J. Cell Biol. 105:2703–2712. 10.1083/jcb.105.6.2703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futai, M., Sun-Wada G.-H., Wada Y., Matsumoto N., and Nakanishi-Matsui M.. 2019. Vacuolar-type ATPase: A proton pump to lysosomal trafficking. Proc. Jpn. Acad., Ser. B, Phys. Biol. Sci. 95:261–277. 10.2183/pjab.95.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddard, T.D., Huang C.C., Meng E.C., Pettersen E.F., Couch G.S., Morris J.H., and Ferrin T.E.. 2018. UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci. 27:14–25. 10.1002/pro.3235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinton, A., Sennoune S.R., Bond S., Fang M., Reuveni M., Sahagian G.G., Jay D., Martinez-Zaguilan R., and Forgac M.. 2009. Function of a subunit isoforms of the V-ATPase in pH homeostasis and in vitro invasion of MDA-MB231 human breast cancer cells. J. Biol. Chem. 284:16400–16408. 10.1074/jbc.M901201200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosokawa, H., Dip P.V., Merkulova M., Bakulina A., Zhuang Z., Khatri A., Jian X., Keating S.M., Bueler S.A., Rubinstein J.L., et al. 2013. The N termini of a-subunit isoforms are involved in signaling between vacuolar H+-ATPase (V-ATPase) and cytohesin-2. J. Biol. Chem. 288:5896–5913. 10.1074/jbc.M112.409169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, Y., Carraro-Lacroix L.R., Wang A., Owen C., Bajenova E., Corey P.N., Brumell J.H., and Voronov I.. 2016. Lysosomal pH Plays a Key Role in Regulation of mTOR Activity in Osteoclasts. J. Cell. Biochem. 117:413–425. 10.1002/jcb.25287 [DOI] [PubMed] [Google Scholar]
- Huotari, J., and Helenius A.. 2011. Endosome maturation. EMBO J. 30:3481–3500. 10.1038/emboj.2011.286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurtado-Lorenzo, A., Skinner M., El Annan J., Futai M., Sun-Wada G.-H., Bourgoin S., Casanova J., Wildeman A., Bechoua S., Ausiello D.A., et al. 2006. V-ATPase interacts with ARNO and Arf6 in early endosomes and regulates the protein degradative pathway. Nat. Cell Biol. 8:124–136. 10.1038/ncb1348 [DOI] [PubMed] [Google Scholar]
- Jaskolka, M.C., Winkley S.R., and Kane P.M.. 2021. RAVE and Rabconnectin-3 Complexes as Signal Dependent Regulators of Organelle Acidification. Front. Cell Dev. Biol. 9:698190. 10.3389/fcell.2021.698190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, D.E., Ostrowski P., Jaumouillé V., and Grinstein S.. 2016. The position of lysosomes within the cell determines their luminal pH. J. Cell Biol. 212:677–692. 10.1083/jcb.201507112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanai, F., Liu H., Field S.J., Akbary H., Matsuo T., Brown G.E., Cantley L.C., and Yaffe M.B.. 2001. The PX domains of p47phox and p40phox bind to lipid products of PI(3)K. Nat. Cell Biol. 3:675–678. 10.1038/35083070 [DOI] [PubMed] [Google Scholar]
- Kawasaki-Nishi, S., Bowers K., Nishi T., Forgac M., and Stevens T.H.. 2001. The amino-terminal domain of the vacuolar proton-translocating ATPase a subunit controls targeting and in vivo dissociation, and the carboxyl-terminal domain affects coupling of proton transport and ATP hydrolysis. J. Biol. Chem. 276:47411–47420. 10.1074/jbc.M108310200 [DOI] [PubMed] [Google Scholar]
- Knapp, P.E., and Swanson J.A.. 1990. Plasticity of the tubular lysosomal compartment in macrophages. J. Cell Sci. 95:433–439. 10.1242/jcs.95.3.433 [DOI] [PubMed] [Google Scholar]
- Levin-Konigsberg, R., Montaño-Rendón F., Keren-Kaplan T., Li R., Ego B., Mylvaganam S., DiCiccio J.E., Trimble W.S., Bassik M.C., Bonifacino J.S., et al. 2019. Phagolysosome resolution requires contacts with the endoplasmic reticulum and phosphatidylinositol-4-phosphate signalling. Nat. Cell Biol. 21:1234–1247. 10.1038/s41556-019-0394-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Q., Lau A., Morris T.J., Guo L., Fordyce C.B., and Stanley E.F.. 2004. A syntaxin 1, Galpha(o), and N-type calcium channel complex at a presynaptic nerve terminal: analysis by quantitative immunocolocalization. J. Neurosci. 24:4070–4081. 10.1523/JNEUROSCI.0346-04.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liesche, C., Grussmayer K.S., Ludwig M., Wörz S., Rohr K., Herten D.-P., Beaudouin J., and Eils R.. 2015. Automated Analysis of Single-Molecule Photobleaching Data by Statistical Modeling of Spot Populations. Biophys. J. 109:2352–2362. 10.1016/j.bpj.2015.10.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewen, C.J.R., and Levine T.P.. 2005. A highly conserved binding site in vesicle-associated membrane protein-associated protein (VAP) for the FFAT motif of lipid-binding proteins. J. Biol. Chem. 280:14097–14104. 10.1074/jbc.M500147200 [DOI] [PubMed] [Google Scholar]
- Lukacs, G.L., Rotstein O.D., and Grinstein S.. 1990. Phagosomal acidification is mediated by a vacuolar-type H(+)-ATPase in murine macrophages. J. Biol. Chem. 265:21099–21107. 10.1016/S0021-9258(17)45332-4 [DOI] [PubMed] [Google Scholar]
- Maranda, B., Brown D., Bourgoin S., Casanova J.E., Vinay P., Ausiello D.A., and Marshansky V.. 2001. Intra-endosomal pH-sensitive recruitment of the Arf-nucleotide exchange factor ARNO and Arf6 from cytoplasm to proximal tubule endosomes. J. Biol. Chem. 276:18540–18550. 10.1074/jbc.M011577200 [DOI] [PubMed] [Google Scholar]
- Marshansky, V., Rubinstein J.L., and Grüber G.. 2014. Eukaryotic V-ATPase: novel structural findings and functional insights. Biochim. Biophys. Acta. 1837:857–879. 10.1016/j.bbabio.2014.01.018 [DOI] [PubMed] [Google Scholar]
- Martinez, I., Chakrabarti S., Hellevik T., Morehead J., Fowler K., and Andrews N.W.. 2000. Synaptotagmin VII regulates Ca(2+)-dependent exocytosis of lysosomes in fibroblasts. J. Cell Biol. 148:1141–1149. 10.1083/jcb.148.6.1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxfield, F.R., and McGraw T.E.. 2004. Endocytic recycling. Nat. Rev. Mol. Cell Biol. 5:121–132. 10.1038/nrm1315 [DOI] [PubMed] [Google Scholar]
- Maxson, M.E., and Grinstein S.. 2014. The vacuolar-type H+-ATPase at a glance - more than a proton pump. J. Cell Sci. 127:4987–4993. 10.1242/jcs.158550 [DOI] [PubMed] [Google Scholar]
- McGuire, C.M., Collins M.P., Sun-Wada G., Wada Y., and Forgac M.. 2019. Isoform-specific gene disruptions reveal a role for the V-ATPase subunit a4 isoform in the invasiveness of 4T1-12B breast cancer cells. J. Biol. Chem. 294:11248–11258. 10.1074/jbc.RA119.007713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellman, I., Fuchs R., and Helenius A.. 1986. Acidification of the endocytic and exocytic pathways. Annu. Rev. Biochem. 55:663–700. 10.1146/annurev.bi.55.070186.003311 [DOI] [PubMed] [Google Scholar]
- Merkulova, M., Păunescu T.G., Azroyan A., Marshansky V., Breton S., and Brown D.. 2015. Mapping the H(+) (V)-ATPase interactome: identification of proteins involved in trafficking, folding, assembly and phosphorylation. Sci. Rep. 5:14827. 10.1038/srep14827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel, V., Licon-Munoz Y., Trujillo K., Bisoffi M., and Parra K.J.. 2013. Inhibitors of vacuolar ATPase proton pumps inhibit human prostate cancer cell invasion and prostate-specific antigen expression and secretion. Int. J. Cancer. 132:E1–E10. 10.1002/ijc.27811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullock, B.M., Bright N.A., Fearon C.W., Gray S.R., and Luzio J.P.. 1998. Fusion of lysosomes with late endosomes produces a hybrid organelle of intermediate density and is NSF dependent. J. Cell Biol. 140:591–601. 10.1083/jcb.140.3.591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naufer, A., Hipolito V.E.B., Ganesan S., Prashar A., Zaremberg V., Botelho R.J., and Terebiznik M.R.. 2018. pH of endophagosomes controls association of their membranes with Vps34 and PtdIns(3)P levels. J. Cell Biol. 217:329–346. 10.1083/jcb.201702179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa, M., Kosaka N., Choyke P.L., and Kobayashi H.. 2009. H-type dimer formation of fluorophores: a mechanism for activatable, in vivo optical molecular imaging. ACS Chem. Biol. 4:535–546. 10.1021/cb900089j [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrowski, P.P., Fairn G.D., Grinstein S., and Johnson D.E.. 2016. Cresyl violet: a superior fluorescent lysosomal marker. Traffic. 17:1313–1321. 10.1111/tra.12447 [DOI] [PubMed] [Google Scholar]
- Panchuk-Voloshina, N., Haugland R.P., Bishop-Stewart J., Bhalgat M.K., Millard P.J., Mao F., Leung W.-Y., and Haugland R.P.. 1999. Alexa dyes, a series of new fluorescent dyes that yield exceptionally bright, photostable conjugates. J. Histochem. Cytochem. 47:1179–1188. 10.1177/002215549904700910 [DOI] [PubMed] [Google Scholar]
- Parra, K.J., and Kane P.M.. 1998. Reversible association between the V1 and V0 domains of yeast vacuolar H+-ATPase is an unconventional glucose-induced effect. Mol. Cell. Biol. 18:7064–7074. 10.1128/MCB.18.12.7064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poëa-Guyon, S., Ammar M.R., Erard M., Amar M., Moreau A.W., Fossier P., Gleize V., Vitale N., and Morel N.. 2013. The V-ATPase membrane domain is a sensor of granular pH that controls the exocytotic machinery. J. Cell Biol. 203:283–298. 10.1083/jcb.201303104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez, C., Hauser A.D., Vucic E.A., and Bar-Sagi D.. 2019. Plasma membrane V-ATPase controls oncogenic RAS-induced macropinocytosis. Nature. 576:477–481. 10.1038/s41586-019-1831-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts, R.L., Barbieri M.A., Ullrich J., and Stahl P.D.. 2000. Dynamics of rab5 activation in endocytosis and phagocytosis. J. Leukoc. Biol. 68:627–632. [PubMed] [Google Scholar]
- Rocha, N., Kuijl C., van der Kant R., Janssen L., Houben D., Janssen H., Zwart W., and Neefjes J.. 2009. Cholesterol sensor ORP1L contacts the ER protein VAP to control Rab7-RILP-p150 Glued and late endosome positioning. J. Cell Biol. 185:1209–1225. 10.1083/jcb.200811005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saftig, P., and Klumperman J.. 2009. Lysosome biogenesis and lysosomal membrane proteins: trafficking meets function. Nat. Rev. Mol. Cell Biol. 10:623–635. 10.1038/nrm2745 [DOI] [PubMed] [Google Scholar]
- Santic, M., Asare R., Skrobonja I., Jones S., and Abu Kwaik Y.. 2008. Acquisition of the vacuolar ATPase proton pump and phagosome acidification are essential for escape of Francisella tularensis into the macrophage cytosol. Infect. Immun. 76:2671–2677. 10.1128/IAI.00185-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saw, N.M.N., Kang S.-Y.A., Parsaud L., Han G.A., Jiang T., Grzegorczyk K., Surkont M., Sun-Wada G.-H., Wada Y., Li L., and Sugita S.. 2011. Vacuolar H(+)-ATPase subunits Voa1 and Voa2 cooperatively regulate secretory vesicle acidification, transmitter uptake, and storage. Mol. Biol. Cell. 22:3394–3409. 10.1091/mbc.e11-02-0155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma, S., and Wilkens S.. 2017. Biolayer interferometry of lipid nanodisc-reconstituted yeast vacuolar H+ -ATPase. Protein Sci. 26:1070–1079. 10.1002/pro.3143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun-Wada, G.H., Toyomura T., Murata Y., Yamamoto A., Futai M., and Wada Y.. 2006. The a3 isoform of V-ATPase regulates insulin secretion from pancreatic beta-cells. J. Cell Sci. 119:4531–4540. 10.1242/jcs.03234 [DOI] [PubMed] [Google Scholar]
- Suresh, B., Saminathan A., Chakraborty K., Cui C., Becker L., and Krishnan Y.. 2020. Tubular lysosomes harbor active ion gradients and poise macrophages for phagocytosis. bioRxiv. (Preprint posted December 6, 2020). 10.1101/2020.12.05.413229 [DOI] [PMC free article] [PubMed]
- Swanson, J., Yirinec B., Burke E., Bushnell A., and Silverstein S.C.. 1986. Effect of alterations in the size of the vacuolar compartment on pinocytosis in J774.2 macrophages. J. Cell. Physiol. 128:195–201. 10.1002/jcp.1041280209 [DOI] [PubMed] [Google Scholar]
- Swanson, J., Burke E., and Silverstein S.C.. 1987. Tubular lysosomes accompany stimulated pinocytosis in macrophages. J. Cell Biol. 104:1217–1222. 10.1083/jcb.104.5.1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabó, Á., Szendi-Szatmári T., Ujlaky-Nagy L., Rádi I., Vereb G., Szöllősi J., and Nagy P.. 2018. The Effect of Fluorophore Conjugation on Antibody Affinity and the Photophysical Properties of Dyes. Biophys. J. 114:688–700. 10.1016/j.bpj.2017.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabke, K., Albertmelcher A., Vitavska O., Huss M., Schmitz H.-P., and Wieczorek H.. 2014. Reversible disassembly of the yeast V-ATPase revisited under in vivo conditions. Biochem. J. 462:185–197. 10.1042/BJ20131293 [DOI] [PubMed] [Google Scholar]
- Takamori, S., Holt M., Stenius K., Lemke E.A., Grønborg M., Riedel D., Urlaub H., Schenck S., Brügger B., Ringler P., et al. 2006. Molecular anatomy of a trafficking organelle. Cell. 127:831–846. 10.1016/j.cell.2006.10.030 [DOI] [PubMed] [Google Scholar]
- Tan, Y.Z., Abbas Y.M., Wu J.Z., Wu D., Hesketh G.G., Bueler S.A., Gingras A.-C., Robinson C.V., Grinstein S., and Rubinstein J.L.. 2021. Structure of mammalian V-ATPase with the TLDc domain protein mEAK7 bound. bioRxiv. (Preprint posted November 3, 2021). 10.1101/2021.11.03.466369 [DOI]
- Teruel, M.N., Blanpied T.A., Shen K., Augustine G.J., and Meyer T.. 1999. A versatile microporation technique for the transfection of cultured CNS neurons. J. Neurosci. Methods. 93:37–48. 10.1016/S0165-0270(99)00112-0 [DOI] [PubMed] [Google Scholar]
- Toei, M., Saum R., and Forgac M.. 2010. Regulation and isoform function of the V-ATPases. Biochemistry. 49:4715–4723. 10.1021/bi100397s [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trombetta, E.S., Ebersold M., Garrett W., Pypaert M., and Mellman I.. 2003. Activation of lysosomal function during dendritic cell maturation. Science. 299:1400–1403. 10.1126/science.1080106 [DOI] [PubMed] [Google Scholar]
- Uchida, E., Ohsumi Y., and Anraku Y.. 1985. Purification and properties of H+-translocating, Mg2+-adenosine triphosphatase from vacuolar membranes of Saccharomyces cerevisiae. J. Biol. Chem. 260:1090–1095. 10.1016/S0021-9258(20)71211-1 [DOI] [PubMed] [Google Scholar]
- Vasanthakumar, T., Bueler S.A., Wu D., Beilsten-Edmands V., Robinson C.V., and Rubinstein J.L.. 2019. Structural comparison of the vacuolar and Golgi V-ATPases from Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA. 116:7272–7277. 10.1073/pnas.1814818116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira, O.V., Botelho R.J., Rameh L., Brachmann S.M., Matsuo T., Davidson H.W., Schreiber A., Backer J.M., Cantley L.C., and Grinstein S.. 2001. Distinct roles of class I and class III phosphatidylinositol 3-kinases in phagosome formation and maturation. J. Cell Biol. 155:19–25. 10.1083/jcb.200107069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voss, M., Vitavska O., Walz B., Wieczorek H., and Baumann O.. 2007. Stimulus-induced phosphorylation of vacuolar H(+)-ATPase by protein kinase A. J. Biol. Chem. 282:33735–33742. 10.1074/jbc.M703368200 [DOI] [PubMed] [Google Scholar]
- Webb, B.A., Aloisio F.M., Charafeddine R.A., Cook J., Wittmann T., and Barber D.L.. 2021. pHLARE: a new biosensor reveals decreased lysosome pH in cancer cells. Mol. Biol. Cell. 32:131–142. 10.1091/mbc.E20-06-0383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijdeven, R.H., Janssen H., Nahidiazar L., Janssen L., Jalink K., Berlin I., and Neefjes J.. 2016. Cholesterol and ORP1L-mediated ER contact sites control autophagosome transport and fusion with the endocytic pathway. Nat. Commun. 7:11808. 10.1038/ncomms11808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, L., Shen X., Bryan A., Banga S., Swanson M.S., and Luo Z.-Q.. 2010. Inhibition of host vacuolar H+-ATPase activity by a Legionella pneumophila effector. PLoS Pathog. 6:e1000822. 10.1371/journal.ppat.1000822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, T., Gan Q., Wu B., Yin M., Xu J., Shu X., and Liu J.. 2020. Molecular Basis for PI(3,5)P2 Recognition by SNX11, a Protein Involved in Lysosomal Degradation and Endosome Homeostasis Regulation. J. Mol. Biol. 432:4750–4761. 10.1016/j.jmb.2020.06.010 [DOI] [PubMed] [Google Scholar]
- Yajima, S., Kubota M., Nakakura T., Hasegawa T., Katagiri N., Tomura H., Sasayama Y., Suzuki M., and Tanaka S.. 2007. Cloning and expression of vacuolar proton-pumping ATPase subunits in the follicular epithelium of the bullfrog endolymphatic sac. Zool. Sci. 24:147–157. 10.2108/zsj.24.147 [DOI] [PubMed] [Google Scholar]
- Yordanov, T.E., Hipolito V.E.B., Liebscher G., Vogel G.F., Stasyk T., Herrmann C., Geley S., Teis D., Botelho R.J., Hess M.W., and Huber L.A.. 2019. Biogenesis of lysosome-related organelles complex-1 (BORC) regulates late endosomal/lysosomal size through PIKfyve-dependent phosphatidylinositol-3,5-bisphosphate. Traffic. 20:674–696. 10.1111/tra.12679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, J., Beyrakhova K., Liu Y., Alvarez C.P., Bueler S.A., Xu L., Xu C., Boniecki M.T., Kanelis V., Luo Z.-Q., et al. 2017. Molecular basis for the binding and modulation of V-ATPase by a bacterial effector protein. PLoS Pathog. 13:e1006394. 10.1371/journal.ppat.1006394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoncu, R., Bar-Peled L., Efeyan A., Wang S., Sancak Y., and Sabatini D.M.. 2011. mTORC1 senses lysosomal amino acids through an inside-out mechanism that requires the vacuolar H(+)-ATPase. Science. 334:678–683. 10.1126/science.1207056 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
contains original blots for Fig. S1.
contains original blots for Fig. S2.
Data Availability Statement
Experimental datasets that support the findings of this study are available from the corresponding authors upon reasonable request.