Abstract
Pseudomonas (formerly Flavimonas) oryzihabitans is an uncommon pathogen that may cause catheter-associated infections. Although it has occasionally been isolated from the environment, the source of human infection has not previously been documented. We describe an AIDS patient who developed Pseudomonas oryzihabitans bacteremia due to colonization of a Hickman catheter. The patient reported having strictly followed the recommendations for catheter hygiene. The only flaw detected was the use of a synthetic bath sponge in the shower. The sponge was cultured and yielded P. oryzihabitans among other nonfermentative, gram-negative bacilli. To determine the prevalence of P. oryzihabitans in sponges, we cultured 15 samples from unrelated households. The microorganism was isolated from 3 of the 15 samples. Molecular typing by arbitrarily primed PCR (AP-PCR) was performed with the environmental and clinical isolates. Three different profiles were obtained for the six isolates analyzed from the patient's sponge. The strain from the AIDS patient was identical to one of those from his sponge and was different from all the remaining strains. The AP-PCR typing results were subsequently confirmed by pulsed-field gel electrophoresis. It can be concluded that sponges are occasionally colonized by P. oryzihabitans. For the first time a probable source of an indwelling catheter contamination with this bacterium has been found. Patients carrying these devices should avoid using sponge-like materials, as these are suitable environments for nonfermentative, gram-negative bacilli.
Pseudomonas (formerly Flavimonas) oryzihabitans (1) is a yellow-pigmented, gram-negative, oxidase-negative, nonfermenting bacillus which has been isolated from damp environments, such as rice paddies and sink drains (6).
P. oryzihabitans has only rarely been associated with human infections. However, since 1977 at least 80 cases of infection with this microorganism, including bloodstream infections and peritonitis in patients undergoing peritoneal dialysis, have been reported. Most of them were infections related to foreign materials or indwelling catheters (5, 10, 13).
Diverse epidemiological studies have found P. oryzihabitans in the hospital environment, although the potential sources of human infection have not been precisely demonstrated (3, 16).
To the best of our knowledge, the case we report here is the first case of infection caused by P. oryzihabitans directly tracked to an environmental source.
MATERIALS AND METHODS
A 30-year-old man with AIDS (CDC stage C3) was admitted to our hospital with fever, chills, and low blood pressure. The patient had a Hickman catheter for administration of drugs related to treatment of Kaposi's sarcoma. On admission, physical examination revealed inflammatory signs at the catheter insertion site.
Blood samples for culture obtained through the catheter connections and from a peripheral vein were processed by the Isolator method (Wampole Laboratory, Cranbury, N.J.).
Parenteral ciprofloxacin was started, with an initial good clinical outcome, but 5 days later the patient developed a high temperature, chills, and shock. The catheter was removed and the tip was cultured on blood agar by the method of Maki et al. (11).
A yellow-pigmented, gram-negative, catalase-positive, oxidase-negative, motile, and nonfermentative rod was isolated from lysis-centrifugation blood samples taken through the catheter hubs (white port, 30 CFU/ml; red port, countless CFU per milliliter) and from the culture of the catheter tip after its removal (more than 15 CFU). The culture of peripheral blood was negative. Cultures of skin from the catheter insertion site yielded only coagulase-negative staphylococci.
The microorganism was identified as P. oryzihabitans, and it was resistant to ampicillin, amoxicillin-clavulanic acid, and cefazolin and was susceptible to broad-spectrum cephalosporins, aztreonam, imipenem, aminoglycosides, ciprofloxacin, and trimethoprim-sulfamethoxazole.
Identification and susceptibility testing of isolates were performed with MicroScan Negative Combo 6I panels (Microscan, Baxter Diagnostics, Inc., West Sacramento, Calif.), and the results were confirmed by standard microbiological methods (17), with the API 20 NE system (bioMérieux, Marcy l'Etoile, France), and by disk diffusion testing, performed according to the guidelines of the National Committee for Clinical Laboratory Standards (12). Microbiological cultures of heparinized flasks, gauze, antiseptic solution, and sponges were done on blood and MacConkey agar plates at 30 and 37°C. The patient mentioned the use of a sponge for personal hygiene, which was also cultured. Several pieces of each sponge were blotted onto the surface of the agar. The sponges from different households included as controls were produced by different manufacturers.
Molecular biology-based methods. (i) AP-PCR.
Single colonies were picked and grown in Luria-Bertani medium. The cultures were stopped when the exponential stage was reached. Growth from the cultures were plated onto blood agar to rule out the presence of other contaminant bacteria. Gram staining, oxidase determination, and the API 20 NE system were used to identify P. oryzihabitans in the cultures. The cells were harvested, and genomic DNA was extracted by the Genome DNA kit system (Bio 101, Inc., Vista, Calif.). The chromosomal DNA was checked by electrophoresis in agarose gels, and the concentrations of the different extracts were standardized by spectrophotometric measurements.
Arbitrarily primed PCR (AP-PCR) was performed with Ready-to-go RAPD analysis beads (Pharmacia-Biotech). The primers selected were primers 1 (5′-GGTGCGGGAA-3′), 4 (5′-AAGAGCCCGT-3′), 5 (5′-AACGCGCAAC-3′), and 6 (5′-CCCGTCAGCA-3′) from Bio-Rad and OPA-1 (5′-CAGGCCCTTC-3′) and OPA-2 (5′-TGCCGAGCTG-3′) from Operon Technologies. The reaction mixtures included 20 ng of DNA and 25 pmol of each primer in a final volume of 25 μl.
Parallel reactions with 1:5 DNA dilutions were performed to rule out potential differences in the profiles due to concentration effects. The amplification profile was 95°C for 5 min and 45 amplification cycles of 95°C for 1 min, 36°C for 1 min, and 72°C for 2 min, with a tail end of 5 min at 72°C. Ten microliters of each reaction mixture was loaded onto 2% agarose gels for fingerprinting analysis.
(ii) PFGE.
Bacterial cultures were performed and checked as indicated for AP-PCR. Agarose plugs containing the amount of bacterial cells included in 1.5 ml of the exponential-phase culture were obtained with a contour-clamped homogeneous electric field (CHEF) genomic DNA plug kit (Bio-Rad). The cells were lysed, and the genomic DNA was extracted from the plugs, according to the supplier's instructions. The low-frequency-of-cleavage restriction endonucleases XbaI and SpeI (Promega) were selected for use for DNA digestion. Restricted DNA fragments were separated by pulsed-field gel electrophoresis (PFGE) performed in a CHEF system (Bio-Rad) with 1% agarose gels and 0.5× Tris-borate-EDTA buffer. The gels were run for 17 h at 195 V and 14°C with pulses ramping from 5 to 50 s.
Typing data analysis.
All the typing profiles obtained by the AP-PCR assays with the six selected primers were processed to represent the similarity dendrogram by the unweighted pair group method matched with averages by applying the Dice coefficient (specific software, Lane Manager and A.D.A. programs [Technología para diagnóstico e investigación]).
RESULTS
The patient's routine for catheter care was reviewed to find a potential source of contamination of the catheter with P. oryzihabitans. The only flaw detected was the repeated use of heparinized flasks and the use of a synthetic bath sponge in the shower. Microbiological cultures of the heparinized flasks and other materials used for assessment of sterility of materials used in catheter care were all negative. However, several P. oryzihabitans colonies were isolated from the patient's bath sponge, among other gram-negative bacilli (Ochrobactrum sp. and Pseudomonas sp.).
In order to determine the prevalence of P. oryzihabitans in bath sponges, we cultured samples of sponges from 15 unrelated households. P. oryzihabitans was isolated from three sponges.
To analyze the potential relationships between the isolates obtained from the patient's catheter and those obtained from his sponge, a molecular biology-based typing analysis was performed. Eleven P. oryzihabitans isolates were analyzed. Two were clinical isolates: one from our patient and another from a different unrelated patient. The remaining isolates corresponded to nine environmental sources: six from our patient's sponge and three from sponges belonging to a further three households included as controls.
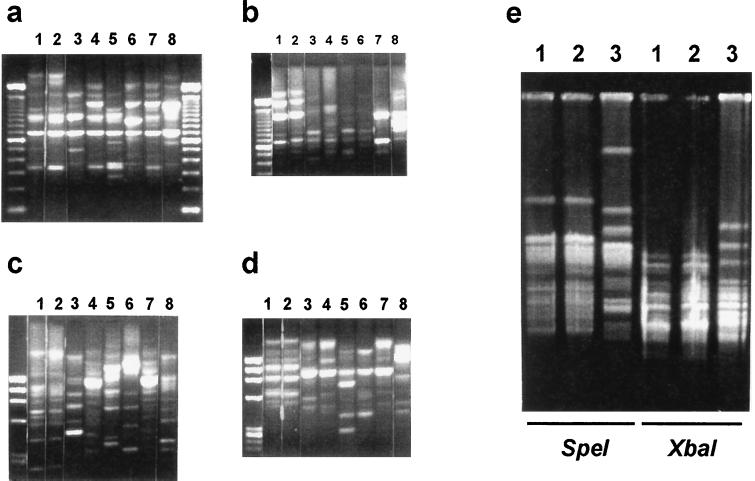
Genomic DNA fingerprints of all the strains were obtained by AP-PCR. The fingerprint obtained for our patient's clinical isolate was identical to the one for an isolate from his sponge (Fig. 1a to d, lanes 1 and 2) with all primers selected. Among the other five isolates from the patient's sponge, two different patterns were obtained, and both patterns were different from that for the clinical isolate (Fig. 1a to d, lanes 3 and 4). The fingerprints for the control isolates of expected unrelatedness, the environmental isolates from unrelated households (lanes 5 to 7) and the clinical strain corresponding to the control patient (lane 8), were clearly different from those obtained for the isolates from our patient.
FIG. 1.
(a to d) DNA fingerprints obtained by AP-PCR. (a) Primer 1; (b) primer 4; (c) primer 5; (d) primer 6 (data for the OPA primers are not shown). Lanes: 1, clinical isolate from our patient; 2 to 4, isolates from our patient's sponge; 5 to 7, isolates from three different sponges from other households; 8, clinical strain from control patient. In all cases, the first lane on the left corresponds to DNA ladders used as weight markers. (e) PFGE analysis with isolates iñ lanes 1, 2, and 3 from panels a to d. The three lanes on the left correspond to DNA digestion with SpeI, and the three lanes on the right correspond to DNA digestion with XbaI.
PFGE analysis was subsequently performed with the two related isolates (isolates 1 and 2) and one of those from the patient's sponge with a different fingerprint, included as an unrelated control. The macrorestriction DNA patterns obtained with two different restriction enzymes confirmed the similarity previously found for clones 1 and 2 (Fig. 1e) and showed a fingerprint clearly different from that for the unrelated strain (Fig. 1e, lane 3).
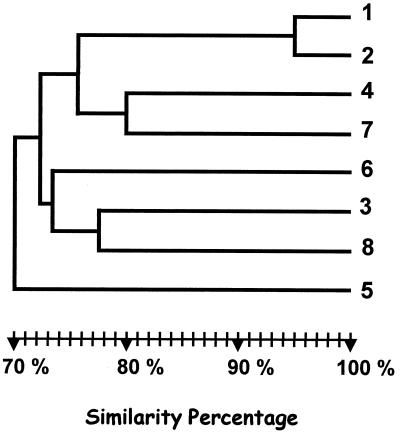
Similarity dendrograms were obtained from an analysis of all the AP-PCR typing profiles obtained with the six different primers selected (Fig. 2). Our patient's clinical isolate (isolate 1) clustered with a high degree of similarity (95%) with isolate 2, confirming their clonal relationship. The other strains were unclustered.
FIG. 2.
Similarity dendrogram obtained by analysis by the unweighted pair group method matched with arithmetic averages of all AP-PCR profiles with the six primers selected. Isolates are positioned according to the Dice similarity index.
DISCUSSION
Infections due to P. oryzihabitans have increasingly been associated with episodes of catheter-related bacteremia, peritonitis in patients undergoing continuous ambulatory peritoneal dialysis, wound infections, and meningitis following neurosurgery (10, 13), especially in patients with underlying debilitating diseases. Most cases are nosocomially acquired, and only six cases of community-acquired infections with this microorganism have been described (4, 7). For our patient, the infection was considered to be community acquired, as it was detected on admission.
In previous studies, molecular epidemiological analysis has been performed with P. oryzihabitans strains involved in nosocomial or community-acquired infections by different molecular typing methods. No homologies were found among clinical and environmental isolates, and therefore, the potential sources of the infections caused by this bacterium remain unknown (7, 8, 9, 16). In our case, the molecular biology-based typing analysis clearly indicated a high degree of similarity between the fingerprint of the clinical strain isolated from our patient and that of a strain from the sponge used for hygienic purposes. The homologies found between their respective fingerprints could not be due to a poor resolution by our technical approach as, at the time, differences from other isolates within the same patient's sponge could clearly be detected. Additionally, the homologies observed by AP-PCR were confirmed by PFGE analysis, a molecular typing method considered to have a higher discriminatory potential than AP-PCR (2, 15). All these data strongly suggest for the first time an epidemiological link between a P. oryzihabitans clinical isolate and an environmental source.
The clonal variety of the P. oryzihabitans population involved in the colonization of the patient's sponge (three different typing profiles found for the isolates assayed) is worth mentioning. A potential preferential role of some of the environmental clones in catheter-associated infections in terms of a greater fitness for colonization or invasion of these devices has not been analyzed.
P. oryzihabitans should be considered an increasingly relevant nosocomial or community-acquired pathogen in patients with intravascular devices. We have found that P. oryzihabitans is an occasional inhabitant of bath sponges, and our results describe the first tracking of an environmental source for these infections. Sponges may be the primary source, but it seems more probable that the sponge initially became colonized from water, the patient's skin, or some other unidentified environmental source. It should be recommended that catheter-bearing patients avoid the use of bath sponges and other permanently wet items for skin care.
ACKNOWLEDGMENT
We thank Thomas O'Boyle for help with preparation of the manuscript.
REFERENCES
- 1.Anzai Y, Kudo Y, Oyaizu H. The phylogeny of the genera Chryseomonas, Flavimonas, and Pseudomonas supports synonymy of these three genera. Int J Syst Bacteriol. 1997;47:249–251. doi: 10.1099/00207713-47-2-249. [DOI] [PubMed] [Google Scholar]
- 2.Bingen E H, Weber M, Derelle J, Brahimi N, Lambert-Zechovsky N Y, Vidailhet M, Navarro J, Elion J. Arbitrarily primed polymerase chain reaction as a rapid method to differentiate crossed from independent Pseudomonas cepacia infections in cystic fibrosis patients. J Clin Microbiol. 1993;31:2589–2593. doi: 10.1128/jcm.31.10.2589-2593.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Conlu A, Rothman J, Staszewski H, Schoch P E, Domenico P, Quadri S M H, Cunha B A. Flavimonas oryzihabitans (CDC group Ve-2) bacteraemia associated with Hickman catheters. J Hosp Infect. 1992;20:293–299. doi: 10.1016/0195-6701(92)90007-9. [DOI] [PubMed] [Google Scholar]
- 4.Giacometti A, Cironi O, Quarta M, Schimizzi A M, del Prete M S, Scalise G. Unusual clinical presentation of infection due to Flavimonas oryzihabitans. Eur J Clin Microbiol Infect Dis. 1998;17:645–648. doi: 10.1007/BF01708348. [DOI] [PubMed] [Google Scholar]
- 5.Hawkins R E, Moriarty R A, Lewis D E, Oldfield E C. Serious infections involving the CDC group Ve bacteria Chryseomonas luteola and Flavimonas oryzihabitans. Rev Infect Dis. 1991;13:257–260. doi: 10.1093/clinids/13.2.257. [DOI] [PubMed] [Google Scholar]
- 6.Kodama K, Kimura K, Komagata K. Two new species of Pseudomonas: P. oryzihabitans isolated from a rice paddy and P. luteola isolated from clinical specimens. Int J Syst Bacteriol. 1985;35:467–474. [Google Scholar]
- 7.Lam S, Isenberg H D, Edwards B, Hilton E. Community-acquired soft-tissue infections caused by Flavimonas oryzihabitans. Clin Infect Dis. 1994;18:808–809. doi: 10.1093/clinids/18.5.808. [DOI] [PubMed] [Google Scholar]
- 8.Lin R D, Hsueh P R, Chang J C, Teng L J, Chang S C, Ho S W, Hsieh W C, Luh K T. Flavimonas oryzihabitans bacteremia: clinical features and microbiological characteristics of isolates. Clin Infect Dis. 1997;24:867–873. doi: 10.1093/clinids/24.5.867. [DOI] [PubMed] [Google Scholar]
- 9.Liu P Y, Shi Z Y, Lau Y J, Hu B S, Shyr J M, Tsai W S, Lin Y H, Tseng C Y. Epidemiological typing of Flavimonas oryzihabitans by PCR and pulsed-field gel electrophoresis. J Clin Microbiol. 1996;34:68–70. doi: 10.1128/jcm.34.1.68-70.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lucas K G, Kiehn T E, Sobeck K A, Armstrong D, Brown D E. Sepsis caused by Flavimonas oryzihabitans. Medicine (Baltimore) 1994;73:209–214. doi: 10.1097/00005792-199407000-00003. [DOI] [PubMed] [Google Scholar]
- 11.Maki D G, Weise C E, Serafin H W. A semiquantitative culture method for identifying intravenous catheter-related infections. N Engl J Med. 1977;296:1305–1309. doi: 10.1056/NEJM197706092962301. [DOI] [PubMed] [Google Scholar]
- 12.National Committee for Clinical Laboratory Standards. Performance standards for antimicrobial susceptibility testing: eighth informational supplement. Document MS100–S8, vol. 18, no. 1. Wayne, Pa: National Committee for Clinical Laboratory Standards; 1998. [Google Scholar]
- 13.Rahav G, Simhon A, Mattan Y, Moses A E, Sacks T. Infections with Chryseomonas luteola (CDC group Ve-1) and Flavimonas oryzihabitans (CDC group Ve-2) Medicine (Baltimore) 1995;74:83–88. doi: 10.1097/00005792-199503000-00003. [DOI] [PubMed] [Google Scholar]
- 14.Saulnier P, Bourneix C, Prevost G, Andremont A. Random amplified polymorphic DNA assay is less discriminant than pulsed-field electrophoresis for typing strains of methicillin-resistant Staphylococcus aureus. J Clin Microbiol. 1993;31:982–985. doi: 10.1128/jcm.31.4.982-985.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.VanCouwenberghe C J, Cohen S H, Tang Y J, Gumerlock P H, Silva J., Jr Genomic fingerprinting of epidemic and endemic strains of Stenotrophomonas maltophilia (formerly Xanthomonas maltophilia) by arbitrarily primed PCR. J Clin Microbiol. 1995;33:1289–1291. doi: 10.1128/jcm.33.5.1289-1291.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Verhasselt B, Claeys G, Elaichouni A, Verschraegen G, Laureysa G, Vaneechoutte M. Case of recurrent Flavimonas oryzihabitans bacteremia associated with an implanted central venous catheter (Port-A-Cath): assessment of clonality by arbitrarily primed PCR. J Clin Microbiol. 1995;33:3047–3048. doi: 10.1128/jcm.33.11.3047-3048.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.von Graevenitz A. Acinetobacter, Alcaligenes, Moraxella, and other nonfermentative gram-negative bacteria. In: Murray P R, Baron E J, Pfaller M A, Tenover F C, Yolken R H, editors. Manual of clinical microbiology. 6th ed. Washington, D.C.: American Society for Microbiology; 1995. pp. 520–532. [Google Scholar]